Significance
Circadian rhythms depend upon the precise coordination of protein interactions within the transcription–translation feedback loop of the molecular clock. Period (PER) and cryptochrome (CRY) rhythmically repress activity of the circadian transcription factor, CLOCK:BMAL1 (brain and muscle Arnt-like protein 1), to establish daily patterns of gene expression. CRY1 binds to CLOCK:BMAL1 with and without PER to inhibit CLOCK:BMAL1 activity. Here we show that CRY1 interacts with the CLOCK PAS-B domain to dock the transcription factor into the secondary pocket of CRY1. Studies of a CRY1:CLOCK:BMAL1 complex highlight critical interfaces for the direct regulation of CLOCK:BMAL1 by cryptochromes. A molecular understanding of the clock mechanism is fundamental for the development and application of therapies for circadian-related disorders.
Keywords: circadian rhythms, cryptochrome, PAS domains, integrative modeling
Abstract
The basic helix–loop–helix PAS domain (bHLH-PAS) transcription factor CLOCK:BMAL1 (brain and muscle Arnt-like protein 1) sits at the core of the mammalian circadian transcription/translation feedback loop. Precise control of CLOCK:BMAL1 activity by coactivators and repressors establishes the ∼24-h periodicity of gene expression. Formation of a repressive complex, defined by the core clock proteins cryptochrome 1 (CRY1):CLOCK:BMAL1, plays an important role controlling the switch from repression to activation each day. Here we show that CRY1 binds directly to the PAS domain core of CLOCK:BMAL1, driven primarily by interaction with the CLOCK PAS-B domain. Integrative modeling and solution X-ray scattering studies unambiguously position a key loop of the CLOCK PAS-B domain in the secondary pocket of CRY1, analogous to the antenna chromophore-binding pocket of photolyase. CRY1 docks onto the transcription factor alongside the PAS domains, extending above the DNA-binding bHLH domain. Single point mutations at the interface on either CRY1 or CLOCK disrupt formation of the ternary complex, highlighting the importance of this interface for direct regulation of CLOCK:BMAL1 activity by CRY1.
Circadian rhythms allow animals to coordinate behavior and physiology with the environmental light/dark cycle (1). Although a host of cellular processes contribute to the generation of ∼24-h timing at the molecular level (i.e., transcriptional, posttranscriptional, translational, posttranslational), the mammalian transcription factor CLOCK:BMAL1 (brain and muscle Arnt-like protein 1) sits at the core of integrated transcription–translation feedback loops that regulate the rhythmic expression of over 40% of the genome throughout the body (2). In support of its central role, the loss of Bmal1 renders mice arrhythmic in the absence of external time cues, the only single clock gene deletion to do so in mice (3). Disruption of circadian rhythms has been linked to altered cellular homeostasis and disease, yet we still lack fundamental insight into basic mechanisms of clock function, including how core clock proteins interact with each other to control the ∼24-h periodicity of gene expression (4).
Recent studies have suggested the presence of several regulatory complexes of core clock proteins that form throughout the day to establish a dynamic balance of CLOCK:BMAL1 activation and repression. In the morning, CLOCK:BMAL1 is bound at E-box sites on DNA with its coactivator CBP/p300, driving expression of the core clock repressors Period (Per) and cryptochrome (Cry) along with other clock-controlled output genes. Repression begins early in the evening, defined by large hetero-multimeric PER:CRY complexes bound to CLOCK:BMAL1 (5–7). The structural basis for formation of these complexes, and whether they occur primarily on or off DNA, is still not well understood (8). Based on ChIP-Seq studies, these complexes appear to remodel or reform over time, evolving to a late repressive complex where CRY1 is bound to CLOCK:BMAL1 on DNA, apparently independently of PER (7).These findings suggest that cryptochromes can work both together and separately from PER to repress CLOCK:BMAL1 activity (8–10). We showed that tuning affinity of CRY1 for the transactivation domain (TAD) of BMAL1 controls circadian period by competing with the coactivator CBP/p300 (11). CRY1 also binds to CLOCK, although it is not yet understood how multivalent interactions with CLOCK:BMAL1 contribute to CRY1 function. Therefore, understanding the molecular basis for recruitment of regulators to CLOCK:BMAL1 will shed light on mechanisms that are crucial for establishing the ∼24-h periodicity of the circadian clock.
Here, we set out to identify how CRY1 interacts with CLOCK:BMAL1 to form a stable ternary complex. We found that the photolyase homology region (PHR) of CRY1 binds directly to the second of two tandem PAS domains (PAS-B) of CLOCK and identified single point mutations on CRY1 and CLOCK PAS-B that eliminate complex formation. Using these data to guide HADDOCK (High Ambiguity Driven protein–protein DOCKing) modeling, we found that CLOCK PAS-B docks directly into the secondary pocket of the CRY1 PHR. This pocket is evolutionarily conserved with photolyase, where it serves as the binding site for an antenna chromophore that is important for repair of UV-induced DNA damage (12). Small angle X-ray scattering (SAXS) studies of CRY1, CLOCK:BMAL1, and the CRY1:CLOCK:BMAL1 ternary complex highlight structural dynamics of these complexes and validate our low-resolution model of the ternary complex. Together, these data illustrate how CRY1 exploits a conserved binding pocket to form a ternary complex with CLOCK:BMAL1 that maintains the transcription factor in a repressed state to close the circadian feedback loop.
Results
CRY1 Interacts Directly with the CLOCK:BMAL1 PAS Domain Core.
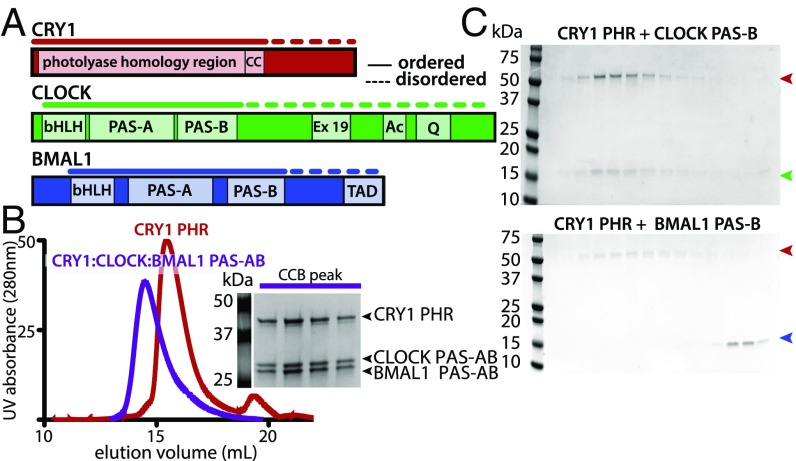
The repressive activity of CRY1 is essential to generate circadian rhythms (13–15); one way that CRY1 does this is by binding the BMAL1 TAD to sequester it from coactivators (11, 16). However, CRY1 has only moderate affinity (Kd ∼1 µM) for the isolated TAD (11, 17), suggesting that it makes at least one other interaction with CLOCK:BMAL1 that allows it to serve as a potent repressor when expressed to near stoichiometric levels (18). Previous studies suggest the CLOCK PAS-B domain is important for repression by CRY1 (11, 19, 20), but evidence for a direct interaction is lacking. To further explore the biochemical basis for interactions between CRY1 and CLOCK:BMAL1, we purified the core PHR of mouse CRY1 and a tandem PAS domain heterodimer (comprising PAS-A and PAS-B domains, PAS-AB) of mouse CLOCK:BMAL1 (Fig. 1A). Using size-exclusion chromatography (SEC) to follow complex formation, we found that the CRY1 PHR directly bound the PAS-AB core of CLOCK:BMAL1 to form a ternary complex (Fig. 1B). Further dissection of this interaction revealed that the CLOCK PAS-B domain alone was sufficient to bind CRY1 PHR. Moreover, although BMAL1 PAS-B shares the same protein fold as CLOCK PAS-B, it did not interact with CRY1, highlighting the specificity of this interaction (Fig. 1C).
Fig. 1.
CRY interacts directly with CLOCK:BMAL1 PAS domain core. (A) Domain schematic of CRY1, CLOCK and BMAL1. Solid lines indicate regions used in this current study. Dashed lines indicate regions with a high degree of disorder. (B) SEC analysis of complex formation with CRY1 PHR alone or mixed with the CLOCK:BMAL1 tandem PAS-AB domain dimer. Proteins were mixed and incubated at 4 °C overnight and then injected on a S200 10/300 GL column. The peak fractions of CRY1 PHR with CLOCK:BMAL1 PAS-AB was analyzed by SDS/PAGE [CRY1:CLOCK:BMAL1 ternary complex (CCB) peak] and stained by Coomassie. (C) SEC analysis of CRY1 PHR with CLOCK PAS-B (Upper) or BMAL1 PAS-B (Lower) in isolation. Identical peak fractions were analyzed by SDS/PAGE and stained by Coomassie. Red arrow indicates CRY1 PHR, green arrow indicates CLOCK PAS-B, and blue arrow indicates BMAL1 PAS-B.
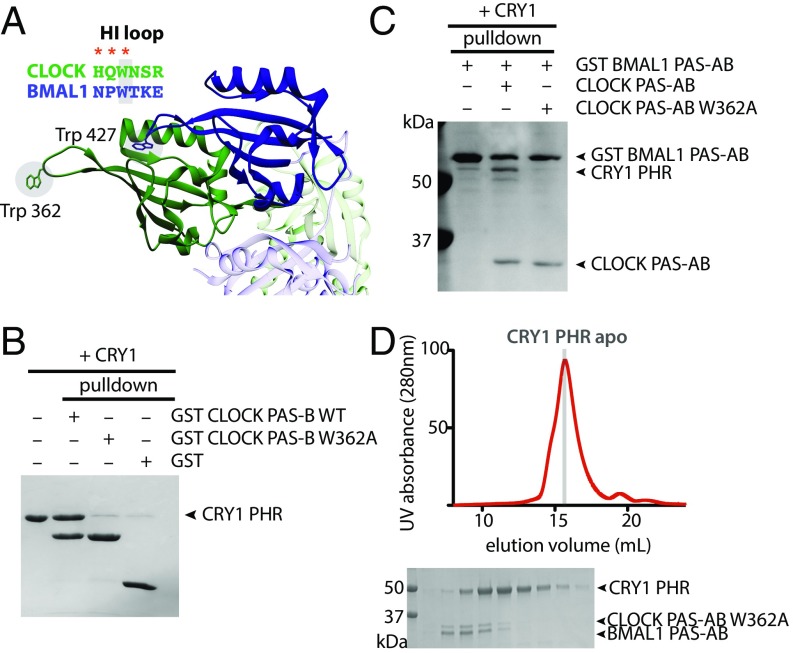
Several residues in the HI loop (connecting the Hβ and Iβ strands) of CLOCK PAS-B are important for CRY1-mediated repression of CLOCK:BMAL1 (11, 19, 20). The entire HI loop is freely accessible in the crystal structure of the CLOCK:BMAL1 basic helix–loop–helix (bHLH)-PAS dimer, protruding out from the PAS-B dimer interface (Fig. 2A) (21). To test the role of the HI loop in binding CRY1, we made a W362A substitution in CLOCK PAS-B and tested its ability to bind CRY1 using a GST pull-down experiment. This single point mutation disrupted formation of the stoichiometric CRY1:CLOCK PAS-B complex (Fig. 2B). We then explored the importance of W362 for the CRY1:CLOCK interaction in the context of a larger, tandem PAS domain dimer. Although GST-BMAL1 PAS-AB was able to pull down similar amounts of wild-type and W362A CLOCK PAS-AB, CRY1 was only present in a ternary complex with wild-type CLOCK PAS-AB (Fig. 2C). Furthermore, a CLOCK:BMAL1 PAS-AB dimer possessing the W362A mutation no longer comigrated with CRY1 on SEC (Fig. 2D). Collectively, these data demonstrate that stable association of CRY1 with the CLOCK:BMAL1 PAS domain core is predicated on a single, solvent-accessible tryptophan on CLOCK PAS-B.
Fig. 2.
A single point mutation disrupts CRY1:CLOCK:BMAL1 complex formation. (A) PAS-B domains of CLOCK:BMAL1 (PDB ID code 4F3L; CLOCK, green; BMAL1, blue) with conserved tryptophan in HI loop shown in sticks. Red asterisks indicate mutations in CLOCK that disrupt CRY1 repression of CLOCK:BMAL1. Adjacent PAS-A domains are shown in light blue (BMAL1) and light green (CLOCK). (B) GST pull-down assay of GST-CLOCK PAS-B and GST-CLOCK PAS-B W362A with CRY1 PHR. (C) GST pull-down assay of GST-BMAL1 PAS-AB alone, in the presence of CLOCK PAS-AB or CLOCK PAS-AB W362A with CRY1 PHR. (D) S200 10/300 GL SEC analysis of complex formation with CRY1 PHR and the PAS-AB dimer with W362A CLOCK mutation.
The CLOCK PAS-B Domain Docks into the CRY1 Secondary Pocket.
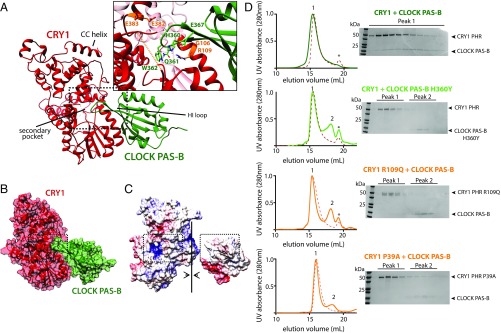
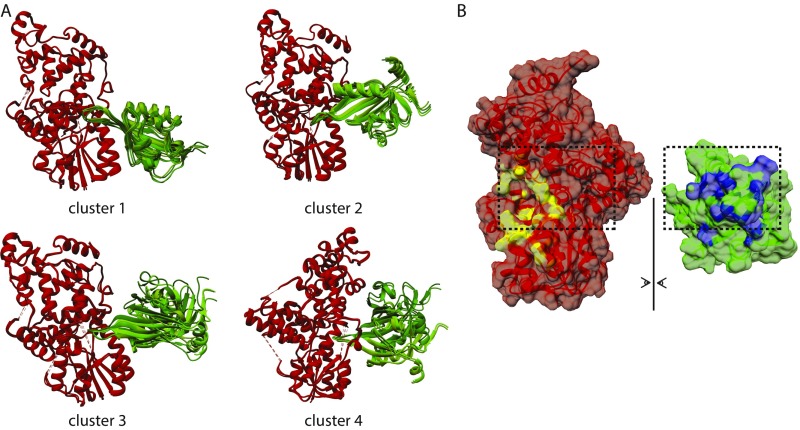
To better understand the nature of the CRY1:CLOCK PAS-B interface, we generated a computational model of the complex using HADDOCK (22, 23). HADDOCK uses residues identified from experimental studies to guide selection of probable protein–protein interfaces and then performs rigid body docking and simulated annealing protocols to provide clusters of hits that are ranked by energetic considerations and their similarity to one another. Based on previous mutagenesis data and our own studies herein, we used the following residues as active restraints, defined by their importance for binding and solvent accessibility: CRY1: G106, R109, E383, E382 (17, 24); and CLOCK PAS-B: G332, H360, Q361, W362, E367 (Fig. 3A) (11, 19, 20). The CRY1 restraints cluster around the secondary pocket in the PHR, which is structurally conserved with photolyase where it serves as a chromophore binding pocket (12). The existing crystal structure of mouse CRY1 lacks a short, flexible loop adjacent to this pocket (17), so we solved a structure of the mouse CRY1 PHR (1.8-Å resolution) in a new space group with the goal of visualizing this loop (PDB ID code 5T5X) (Table S1). Although our new structure also lacked density for this loop, it was of higher resolution so we used it along with the CLOCK PAS-B domain (isolated from PDB ID code 4F3L) for HADDOCK modeling. Clusters were ranked using electrostatic, van der Waals and ambiguous interaction restraint energy terms. All four clusters docked the HI loop of CLOCK PAS-B into secondary pocket of CRY1 in similar orientations (Fig. S1A), with the top cluster populated by the greatest number of models (118 models) and the best overall HADDOCK score (Table S2).
Fig. 3.
CLOCK PAS-B docks into secondary pocket of CRY1. (A) Representative PDB from top HADDOCK cluster (cluster 1). Active residues used to guide the docking are shown in orange (CRY1) and light green (CLOCK). CRY1 PHR unstructured secondary pocket loop is shown in an orange dashed line. Table S2 for details on HADDOCK cluster statistics. (B) Surface representation of CRY1:CLOCK PAS-B HADDOCK model. (C) Electrostatic representation of CRY1:CLOCK PAS-B HADDOCK model. Surface potential maps were generated using the Adaptive Poisson-Boltzmann Solver in University of California, San Francisco Chimera (43). The secondary pocket of CRY1 and HI loop of CLOCK PAS-B are highlighted in the dashed box analogous to A. (D) SEC analysis of the CRY1:CLOCK PAS-B interaction with mutants. Proteins were mixed and incubated at ∼30 min at 4 °C and then injected on a S200 10/300 GL column. Asterisk, slight UV-absorbing contaminant. Numbers above peaks correspond to fractions analyzed by SDS/PAGE and Coomassie stain at right. Residue P39 is located in the disordered loop shown in orange dashed line in A. Elution profiles of CRY1 PHR WT or mutant alone are shown in each respective panel in the red dashed line.
Table S1.
Statistics of data collection and refinement
| Data collection and refinement | Data |
| Data collection | |
| Space group | P1 |
| Unit cell dimensions | 46.63, 51.03, 54.17 |
| a, b, c (Å), α, β, γ (°) | 71.62, 84.12, 88.89 |
| Resolution range (Å) (highest shell) | 51.15–1.80 (1.83–1.80) |
| Wavelength (Å) | 0.999 |
| Total observations | 138,880 (7431) |
| Unique reflections | 38,528 (2114) |
| Completeness (%) | 93.5 (85.9) |
| Rmerge | 11.0 (47.2) |
| CC1/2 | 0.98(0.74) |
| <I/σ> | 45.4 (4.6) |
| Redundancy | 3.6 (3.5) |
| Refinement | |
| No. of reflections | 35,812 |
| Rwork (%)/Rfree (%) | 16.8/23.1 |
| No. of atoms | |
| Protein | 3,829 |
| Ligand/ion | 1 |
| Water | 455 |
| Average B-factor (Wilson) | 20.5 |
| Rmsd bond length (Å) | 0.013 |
| Rmsd bond angle | 1.35 |
Details of structure for apo mouse cryptochrome 1 PDB ID code 5T5X. Values reported in parentheses are for higher resolution shell.
Fig. S1.
CRY1:CLOCK PAS-B HADDOCK modeling. (A) All docking solutions from HADDOCK 2.2 analysis of CRY1:CLOCK PAS-B interaction. Clusters are ranked numerically according to lowest HADDOCK scoring. See Table S2 for details of cluster scoring. (B) Buried surface area representation of CRY1 and CLOCK PAS-B for top HADDOCK cluster 1 PDB using built-in chimera tools. Buried surface of CRY1 shown in yellow and CLOCK PAS-B in blue. Buried surface area calculated by HADDOCK: 1944.5 ± 83.2 Å2.
Table S2.
HADDOCK cluster statistics
| Variable | Cluster 1 | Cluster 2 | Cluster 3 | Cluster 4 |
| HADDOCK score | −147.3 | −131.6 | −109.5 | −132.1 |
| Cluster size | 118 | 43 | 15 | 13 |
| Rmsd lowest-energy structure | 12.7 ± 0.2 | 13.3 ± 0.2 | 11.0 ± 0.8 | 2.8 ± 2.0 |
| Van der Walls energy | −47.4 ± 3.8 | −47.4 ± 4.1 | −40.1 ± 7.3 | −51.9 ± 9.9 |
| Electrostatic energy | −289.5 ± 42.7 | −243.0 ± 31.0 | −213.8 ± 52.9 | −206.9 ± 18.3 |
| Desolvation energy | −44.5 ± 7.6 | −36.7 ± 5.9 | −30.9 ± 7.7 | −41.0 ± 7.2 |
| Restraints violation energy | 24.1 ± 25.04 | 11.5 ± 14.27 | 43.7 ± 11.91 | 22.2 ± 17.36 |
| Buried surface area | 1944.5 ± 83.2 | 1869.0 ± 46.2 | 1446.6 ± 114.9 | 1839.6 ± 105.4 |
| Z-score | −1.3 | −0.1 | 1.5 | −0.1 |
Output statistics for CRY1 PHR (PDB: 5T5X) docking with CLOCK PAS-B (PDB: 4F3L, CLOCK PAS-B) using HADDOCK 2.2.
A representative model from the top cluster is characterized by a large buried surface area (1994.5 ± 83.2 Å2) mediated by burial of the HI loop and additional sites of contact between the β-sheet of CLOCK PAS-B and CRY1 (Fig. 3B and Fig. S1B). We also noted complementary electrostatic contacts at the interface (Fig. 3C). To test this model experimentally, we made additional point mutations at the observed interface. CLOCK PAS-B H360Y and two mutations in CRY1 (P39A and R109Q) each disrupted formation of a CRY1:CLOCK PAS-B complex as shown by loss of CLOCK PAS-B comigration with CRY1 (peak 1) and the presence of a new peak for the isolated CLOCK PAS-B domain (peak 2) by SEC (Fig. 3D). This finding is consistent with the inability of CRY1 R109Q to coimmunoprecipitate with CLOCK:BMAL1 and reconstitute circadian rhythms in cell-based cycling assays (24). Additionally, mutations that eliminate CRY1:CLOCK PAS-B complex formation in vitro also significantly reduce repressive activity of full-length mCRY1 in steady-state luciferase reporter assays (Fig. S2), demonstrating that these phenotypes are mediated by a direct interaction between CRY1 and the CLOCK:BMAL1 complex at the secondary pocket.
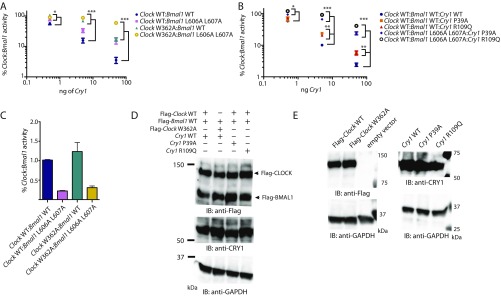
Fig. S2.
Point mutations in CLOCK PAS-B and CRY1 secondary pocket reduce repression of Clock:Bmal1 by Cry1. (A) Per1-Luc assay with increasing amounts of plasmids encoding mCry1 (0.5, 5, and 50 ng) with Clock wild-type or Clock W362A and Bmal1 WT or L606A L607A (100 ng each). Relative activity normalized to Clock:Bmal1 (wild-type or mut) without mCry1 cotransfected set to 100. (B) Per1-Luc assay with increasing amounts of mCry1 wild-type or point mutants (P39A and R109Q) with Clock wild-type and Bmal1 wild-type or L606A L607A. L606A and L607A mutations in the transactivation domain of Bmal1 reduce repression by mCry1 and synergistically reduce repression when paired with mutations in CLOCK PAS-B and the CRY1 secondary pocket. *P < 0.05, **P < 0.01, ***P < 0.001 compared by two-tailed t test. (C) Comparison of relative activity of Clock:Bmal1 constructs used in Per1-luciferase assay. The basal activity of the mutants are shown relative to wild-type Clock:Bmal1 activity set to 1.0. (D) HEK293T cells were transfected with plasmid ratios used in Per1-luciferase assay (100 ng each Flag-Clock and Flag-Bmal1, and Cry1 plasmid as indicated, scaled 4× for increase in culture dish area). Relative protein expression levels are shown by Western blotting using indicated antibodies. GAPDH is shown as a loading control. (E) HEK293T cells were transfected with 1 μg of the indicated plasmid. Relative protein expression levels are shown by Western blotting using indicated antibodies.
Solution Scattering Studies Highlight Flexibility of Clock Protein Complexes.
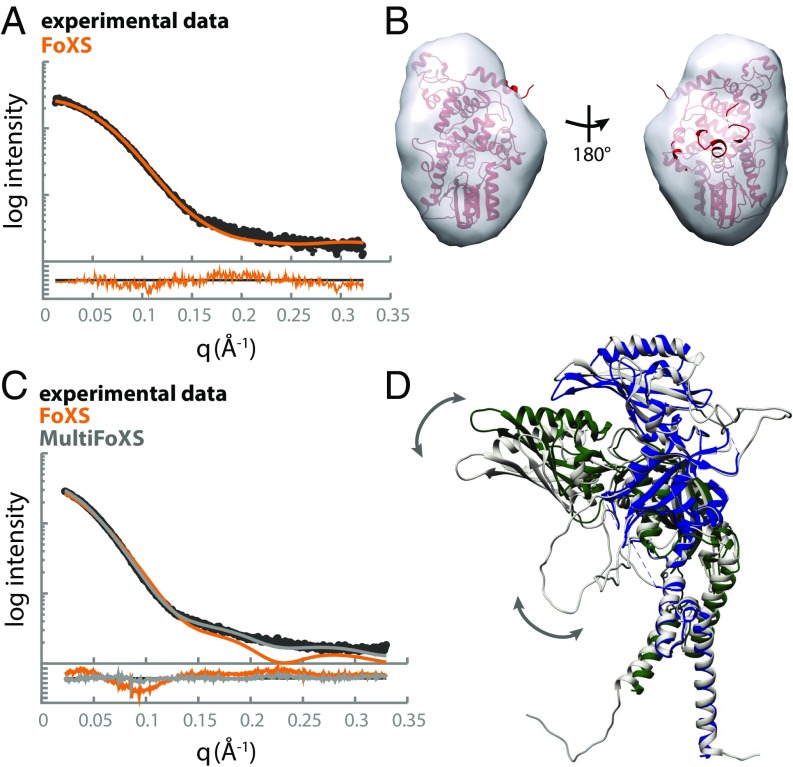
To examine the behavior of the late circadian repressive complex in more detail, we used the solution-based technique of SAXS. We first performed SAXS analysis on the isolated CRY1 PHR and CLOCK:BMAL1 bHLH PAS-AB heterodimer individually to provide insight into their behavior before assembling the ternary complex. Scattering data were collected at several concentrations; both CRY1 PHR and CLOCK:BMAL1 bHLH PAS-AB samples were well-behaved, showing no radiation damage or aggregation as demonstrated by Guinier analysis (Fig. S3). The mass and radius of gyration determined from our analysis of the SAXS data agreed with values calculated from the crystal structures of CRY1 and CLOCK:BMAL1 bHLH PAS-AB. We then used the SAXS profile calculation server FoXS to generate a theoretical scattering profile of CRY1 PHR based on our crystal structure (Fig. 4A) (25). Comparison of the theoretical scattering profile to the experimental data provided a fit within the noise (χ = 1.13), indicating that CRY1 PHR maintains a compact structure in solution that is similar to its crystal structure. Moreover, our crystal structure of CRY1 PHR fit well into a corresponding solution envelope consistent with the pairwise distribution function (Figs. 4B and 5A).
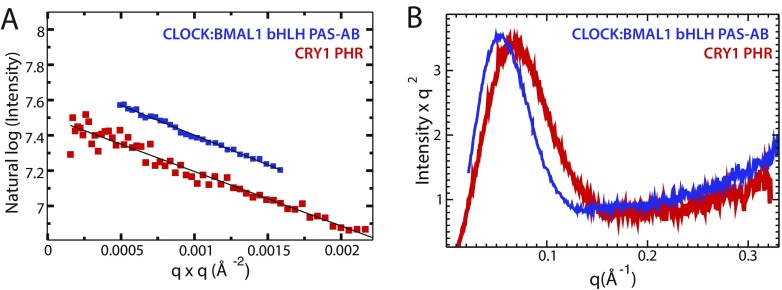
Fig. S3.
SAXS profile of CRY1 and CLOCK:BMAL1 bHLH PAS-AB. (A) Guinier analysis of both CLOCK:BMAL1 bHLH PAS-AB and CRY1 PHR show no aggregation indicated by the linear dependence of log(l(q)) vs. q2. SAXS calculated molecular weight for CRY1 PHR: 45 kDa, CLOCK:BMAL1 bHLH PAS-AB: 78 kDa. (B) Kratky plot of CLOCK:BMAL1 dimer and CRY PHR show mostly folded character and an elongated shape.
Fig. 4.
CRY1 PHR is compact and CLOCK:BMAL1 bHLH-PAS-AB dimer is highly flexible in solution. (A) SAXS profile for CRY1 PHR (black) compared with the theoretical scattering profile for CRY1 PHR (PDB ID code 5T5X) calculated by the FoXS server (orange). Residuals for the fit are shown below with an overall χ = 1.13. (B) The crystal structure of CRY1 PHR fit into the solution envelope generated from the SAXS data. (C) SAXS profile for the CLOCK:BMAL1 PAS-AB dimer (black) compared with the theoretical scattering profile calculated from PDB ID code 4F3L by the FoXS server (orange). Multistate modeling of flexible regions within the dimer was performed with HingeProt paired with MultiFoXS. (D) A representative PDB from the top MultiFoXS hit that includes flexible loops not visible in the crystal structure (gray) aligned with PDB ID code 4F3L using the PAS-A domain of BMAL1; CLOCK, green; BMAL1, blue. Arrows indicate regions of predicted flexibility.
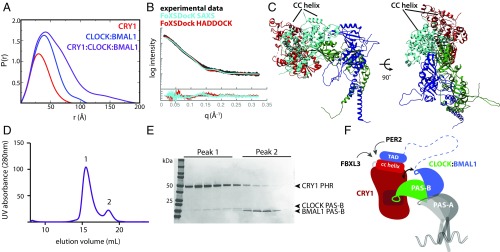
Fig. 5.
A model for the CRY1:CLOCK:BMAL1 repressive complex. (A) Pairwise distribution function of complexes in the present study. CRY1 PHR, Dmax = 86 Å (red); CLOCK:BMAL1 bHLH-PAS-AB dimer, Dmax, 115 Å (blue); CRY1:CLOCK:BMAL1 ternary complex, Dmax, 195 Å (purple). (B) SAXS curve for the ternary complex (black). Docking of CRY1 onto CLOCK:BMAL1 was restrained by the SAXS profile using FoXSDock. The model with the best combined SAXS and energy score is shown in light blue (χ = 2.22). The FoXSDock HADDOCK structure (red) is among these top scoring models that most closely represent the CRY1 PHR:CLOCK PAS-B HADDOCK model. Fig. S4D for representative PDB of the FoXSDock HADDOCK scattering profile shown in the red scattering trace. (C) Top FoXSDock model (light blue) aligned with the HADDOCK model from Fig. 2 using the CLOCK PAS-B domain. (D) SEC analysis of CRY1 PHR mixed with CLOCK:BMAL1 PAS-B heterodimer. Proteins were mixed and incubated at ∼30 min. at 4 °C and then injected on a S200 10/300 GL column. (E) Peak fractions were analyzed by SDS/PAGE and stained with Coomassie. (F) Cartoon model of the late repressive CRY1:CLOCK:BMAL1 repressive complex.
In contrast, the experimental scattering profile of the CLOCK:BMAL1 bHLH PAS-AB heterodimer was not well fit by the theoretical scattering profile calculated from its crystal structure (FoXS, χ = 5.93) (Fig. 4C). The PAS-A domains of CLOCK and BMAL1 both possess long, flexible loops that are not observed in the crystal structure (12% and 26% of the sequence, respectively) (21). To better describe the motions of these dynamic regions, we used MODELER v9.15 to build in the missing fragments (26) and MultiFoXS to sample a range of possible conformations constrained by the SAXS data. As a result, we found conformations that fit the experimental scattering profile within the noise (χ = 1.43) (Fig. 4D) (27, 28). The top structural ensemble resulting from this analysis highlighted two main findings: (i) the loops absent from the crystal structure are highly flexible in solution and contribute significantly to the scattering profile of the PAS domain core, and (ii) the interface between CLOCK and BMAL1 PAS-B domains may be dynamic. Our best fits were obtained using a model where the PAS-B domains were able to sample an undocked state, suggesting that the PAS-B domains may exist in more than one state in solution. Given that multiple regions within the PAS domain core of CLOCK:BMAL1 are known to be important for its function (21, 29), characterization of their dynamic behavior in solution could begin to shed light on their role in regulation of DNA binding and CLOCK:BMAL1 transcriptional activity.
Low-Resolution Model of the CRY1:CLOCK:BMAL1 Ternary Complex.
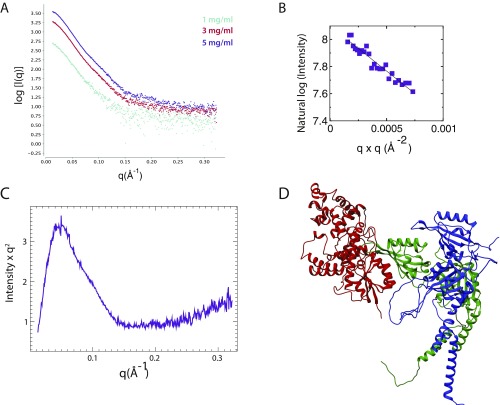
The use of SAXS to guide and validate computational models of protein complexes can be a powerful tool with high-resolution structures in hand for individual components (30). To generate a low-resolution model for the ternary complex, we purified the CRY1 PHR together with the CLOCK:BMAL1 bHLH PAS-AB dimer as a stable ternary complex by SEC and collected SAXS data (Fig. 5). Analysis of the scattering profiles confirmed the presence of all three molecules, consistent with the molecular weight of the ternary complex (Fig. S4 A–C). Furthermore, the ternary complex showed a maximum particle size (Dmax) of 195 Å, much longer than either CRY1 or CLOCK:BMAL1 alone (86 Å and 115 Å, respectively) (Fig. 5A). The elongated Dmax of the ternary complex suggests that CRY1 extends out from the CLOCK:BMAL1 bHLH PAS-AB dimer.
Fig. S4.
SAXS profile of the CRY1:CLOCK:BMAL1 repressive complex. (A) Scattering traces of CRY1:CLOCK:BMAL1 ternary complex (CCB) at different concentrations are shown. These scattering plots were merged to generate the dataset as the input for FoXSDock. (B) Guinier analysis of CCB shows little or no aggregation of sample. SAXS-calculated molecular weight of the ternary complex is 113 kDa. (C) Kratky plot shows the CCB complex indicates a folded mass with an elongated shape. (D) PDB of FoXSDock HADDOCK driven model that is among the top 20 nearly degenerate docking structures, χ = 2.74.
We assessed models for the ternary complex using two methods. First, we used FoXSDock, which combines experimental data and analysis of calculated energies at predicted interfaces to best fit the SAXS profile of a complex from two known structures. In agreement with the long Dmax, the top FoXSDock model of the ternary complex (χ = 2.22) placed CRY1 alongside the PAS-AB core, docked at the CLOCK PAS-B interface (Fig. 5 B and C). Importantly, each of the statistically degenerate top ensembles independently placed CRY1 at the CLOCK PAS-B interface. However, there was some ambiguity in the positioning of CRY1 using the SAXS data alone, as the experimental scattering profile was equally fit by several orientations of CRY1 bound to the HI loop protrusion in CLOCK PAS-B. We then examined how well our HADDOCK model fit the data when aligned onto the bHLH PAS-AB dimer via the CLOCK PAS-B domain. As shown in Fig. 5B, both methods provided reasonable fits to the experimental data, as shown by the overlay of a representative model of HADDOCK (FoXSDock HADDOCK χ = 2.74) (Fig. S4D), with the best-scored SAXS-driven model (FoXSDock SAXS). Importantly, both of these models orient CRY1 such that its coiled-coil (CC) helix sits on the top of the ternary complex, available to make interactions with the BMAL1 TAD and other clock proteins that target this critical interface (Fig. 5C) (11, 24, 31, 32). Therefore, the integration of biochemistry, SAXS, and computational modeling provide low-resolution models of the CRY1:CLOCK:BMAL1 ternary complex.
As with our SAXS studies of the CLOCK:BMAL1 heterodimer, scattering data for the ternary complex were best fit by a model where the PAS-B domains of CLOCK and BMAL1 were no longer tightly bound to each other, with the heterodimer maintained by interactions between the N-terminal PAS-A domains (Fig. 2C) and bHLH domains (Fig. 5B). To test whether CRY1 binding influences the association of CLOCK and BMAL1 PAS-B domains with one another, we performed binding assays using the heterodimer of isolated PAS-B domains. The PAS-B domains of CLOCK and BMAL1 form a complex that comigrates by SEC (Fig. S5). Using NMR and SEC, we confirmed that the PAS-B domains maintain a parallel, stacked orientation in isolation similar to that observed in the bHLH PAS-AB structure (21) (Fig. S5). We then asked if binding of CRY1 would influence the interaction between CLOCK and BMAL1 PAS-B domains in the dimer. SEC of CRY1 with a preformed CLOCK:BMAL1 PAS-B dimer demonstrated that binding of CLOCK PAS-B to CRY1 disrupted its interaction with BMAL1 PAS-B (Fig. 5 D and E). Taken together, these data indicate that CRY1 binding to CLOCK:BMAL1 may influence the architecture of the PAS domain core.
Fig. S5.
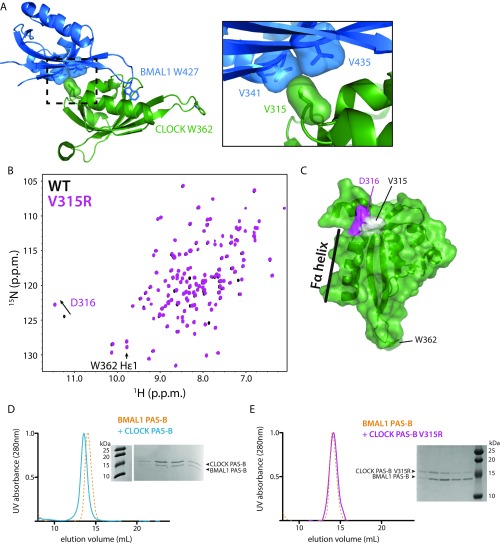
CLOCK PAS-B and BMAL1 PAS-B form a native dimer in solution. (A) Residue V315 in the Fα-helix of CLOCK PAS-B is at the native CLOCK:BMAL1 PAS-B interface in the bHLH PAS-AB dimer structure (PDB ID code 4F3L). Introduction of a charge V315R is predicted to disrupt native PAS-B dimer formation. (B) 15N HSQC of CLOCK PAS-B WT (black) and CLOCK PAS-B V315R mutant (pink). Peaks are well-dispersed indicative of a well-folded PAS domain protein. D316 shows a minor shift as it is located in close proximity to the mutation (see C). (C) CLOCK PAS-B domain highlighting residues assigned in the 5N HSQC spectra in B. (D) Superdex 75 10/300 GL analytical size-exclusion analysis of BMAL1 PAS-B and CLOCK PAS-B heterodimerization. Coelution and a shift in elution volume indicate PAS-B dimer formation. BMAL1 PAS-B alone UV trace is shown in yellow dash. CLOCK:BMAL1 PAS-B dimer trace is displayed in solid blue. Peak fractions analyzed by SDS/PAGE gel electrophoresis. (E) CLOCK PAS-B V315R mutant no longer binds BMAL1 PAS-B. SEC on Superdex 75 10/300 GL of BMAL1 PAS-B and CLOCK PAS-B V315R no longer results in a gel shift or coelution of the proteins. CLOCK PAS-B V315R incubated 1:1 with BMAL1 PAS-B is shown by the pink solid UV trace.
Discussion
Although it has been nearly two decades since the identification of cryptochromes and discovery of their essential role in circadian rhythms, it is still not clear how cryptochromes interact with CLOCK:BMAL1 to inhibit their activity and close the transcription–translation feedback loop of the clock (33). Probing the molecular details of transcription factor–regulator interactions in the clock is important, because they control 24-h timekeeping and generate a vast network of clock-controlled genes that confer circadian timing to physiology and behavior. Here we show that CRY1 interacts directly with the CLOCK:BMAL1 PAS-AB core. We previously demonstrated that multivalent interactions with CLOCK PAS-B and the BMAL1 TAD are required for repression by CRY1 (11, 19). We suggest that CRY1 binding to the PAS-B domain of CLOCK keeps the repressor stably bound to the transcription factor, facilitating its sequestration of the BMAL1 TAD from coactivators (Fig. 5F). In this way, multivalent interactions contribute to the potency of CRY1 as an essential circadian repressor even when expressed at approximately stoichiometric levels with CLOCK:BMAL1 (18).
We identify a gain-of-function interaction at the secondary pocket of mouse CRY1 and demonstrate that it is required to bind CLOCK:BMAL1. This pocket is a remnant of cryptochromes’ evolutionary relationship with the DNA damage repair enzyme, photolyase (17, 34). Photolyases use this pocket to bind an antenna chromophore that harvests photons in low-light conditions, transferring the energy to a flavin molecule buried deep within the catalytic pocket to repair UV-induced thymine dimers (35). The PHR of cryptochromes shares a high degree of structural similarity with photolyases, yet mammalian cryptochromes no longer repair DNA, and presumably have need for an antenna chromophore. We found that the bulky aromatic sidechain of CLOCK W362 is buried within the secondary pocket, exhibiting some similarity to light-harvesting chromophores that dock into the analogous pocket in photolyase. Because of the potent ability of the CLOCK W362A mutation to disrupt CRY1 binding, we propose that it could be a useful tool to specifically uncouple the direct regulation of CLOCK:BMAL1 by CRY1 in cells, allowing the functional dissection of different repressive complexes on CLOCK:BMAL1 that appear throughout the evening (7, 36).
PAS domains play crucial roles in the regulation of bHLH-PAS transcription factors by mediating interactions between bHLH-PAS partners and recruitment of regulatory proteins (37, 38). We focused here on the role that the CLOCK PAS-B domain HI loop plays in binding CRY1, but HI loops in the PAS-B domains of other clock proteins also play central roles in establishing clock protein complexes. For example, HI loop tryptophans in PER proteins mediate their PAS domain-dependent hetero- and homodimerization (39, 40), whereas the analogous tryptophan in BMAL1 PAS-B embeds itself within an internal pocket in CLOCK PAS-B to stabilize the PAS-B dimer (21). Our analysis of the SAXS data shows that the CLOCK:BMAL1 PAS-B interface likely samples open and closed states in solution. We also showed that CRY1 binding further stabilizes the open state by disrupting dimerization of PAS-B domains. Small molecules that bind to the internal pocket of the related hypoxia-inducible factor 2α PAS-B domain allosterically regulate protein interactions at the PAS-B domain (41), suggesting that CRY1 binding could act similarly to regulate docking of BMAL1 through the central pocket of CLOCK. These data highlight the potential importance of protein dynamics and allosteric regulation in controlling the architecture of clock protein complexes.
The combination of static, high-resolution structures from X-ray crystallography with solution studies of proteins by NMR and SAXS is needed to fully describe the role of flexibility in regulating formation of protein complexes. By studying the structural dynamics of the core bHLH PAS-AB dimer of CLOCK:BMAL1 in solution, we pave the way to study new, highly flexible regions of the transcription factor that control circadian rhythms. For example, our best-fit SAXS models indicate that long, flexible loops in the PAS-A domains, not observed in the crystal structure, are highly dynamic and sample a large area around the core PAS domain dimer. This flexibility could play a role in regulating CLOCK:BMAL1 function, as one of these loops, located within the BMAL1 PAS-A domain, is modified by sumoylation to control CLOCK:BMAL1 activity (29). These data also lay the foundation for future studies on the role of the disordered CRY1 C-terminal extension, which controls circadian period and amplitude (42). Understanding the structural basis for mutual exclusivity or synergy of clock protein interactions will provide a framework to elucidate the mechanistic underpinnings of the transcription-based mammalian circadian clock.
Materials and Methods
For details on protein expression and purification, SEC, HADDOCK modeling, SAXS, crystallization, structure determination, pull-down assays, and NMR, SI Materials and Methods.
SI Materials and Methods
Protein Expression and Purification.
Using the baculovirus expression system (Invitrogen), His6-tagged mouse CRY1 PHR was expressed in Sf9 suspension insect cells (Expression Systems). His6-tagged mouse CLOCK bHLH PAS-AB domains (residues 26–384) and native mouse BMAL1 bHLH PAS-AB (residues 68–453) were cloned into pFastBac HTb and pFastBac1 vectors, respectively. CLOCK PAS-B (residues 261–395), CLOCK PAS-AB (residues 93–395), and BMAL1 PAS-B (residues 329–441) were expressed in Escherichia coli Rosetta2 (DE3) cells as a fusion to the solubilizing tags His6-NusA and His6-B1 domain of Streptococcal protein G (GB1).
For His6-tagged mouse CRY1 PHR (amino acids 1–491), Sf9 suspension cells were infected with a P3 virus at 1.5 × 106 cells per milliliter and grown for 72 h. Following brief centrifugation at 3,200 × g, cells were resuspended in 50 mM Tris pH 7.5, 200 mM NaCl, 20 mM imidazole, 10% (vol/vol) glycerol, 0.2% Triton X-100, 0.1% Nonidet P-40, 0.4% Tween-20, 5 mM β-mercaptoethanol, and EDTA-free protease inhibitors (Pierce). Cells were lysed using a microfluidizer followed by brief sonication for 15-s on/30-s off for three pulses at 40% amplitude. Lysate was clarified at 140,500 × g, 4 °C for 1 h. The protein was isolated by Ni2+-nitrilotriacetic acid affinity chromatography (Qiagen). The eluted protein was further purified by ion exchange chromatography and SEC into 20 mM Hepes (pH 7.5), 125 mM NaCl, 5% (vol/vol) glycerol, and 2 mM tris(2-carboxyethyl)phosphine (TCEP).
Sf9 insect cells (Expression Systems) coexpressing His6-tagged mouse CLOCK and BMAL1 bHLH PASAB for 65 h were lysed by sonication in lysis buffer containing 50 mM Na2HPO4 pH 8.0, 300 mM NaCl, 10% (vol/vol) glycerol, 15 mM imidazole, 2.5 mM CHAPS, 5 mM β-mercaptoethanol, and EDTA-free protease inhibitors (ThermoFischer). The clarified cell lysate was applied onto a NiNTA Agarose column (Qiagen) and bound protein was eluted with a gradient of 15 mM–500 mM imidazole. Pooled fractions were buffer-exchanged into 20 mM Tris pH 8.0, 200 mM NaCl, 10% (vol/vol) glycerol, 1 mM DTT. His6 tag was removed by treatment with His6-tagged tobacco etch virus (TEV) protease overnight at 4 °C. The CLOCK:BMAL1 bHLH PAS-AB complex was further purified using a Heparin column followed by a SEC into 20 mM Hepes pH 7.5, 300 mM NaCl, 5% (vol/vol) glycerol, and 1 mM DTT.
For CLOCK PAS-B, CLOCK PAS-AB, and BMAL1 PAS-B expressed in Rosetta (DE3) cells; protein expression was induced with 0.5 mM isfopropyl-β-d-thiogalactopyranoside (IPTG) at an OD600 of 0.8, and grown for an additional 16 h at 18 °C. Soluble protein was purified by Ni2+-nitrilotriacetic acid affinity chromatography (Qiagen), followed by cleavage of the tag with His6-TEV protease overnight at 4 °C. Subsequent Ni2+-nitrilotriacetic acid affinity chromatography was performed to remove the protease and cleaved tag. The protein was further purified by SEC on a Superdex 75 16/60 prep grade column (GE Healthcare) equilibrated with 20 mM Hepes pH 7.5, 125 mM NaCl, 5% (vol/vol) glycerol, and 2 mM TCEP. Point mutations in CRY1 PHR, CLOCK PAS-B, and CLOCK PAS-AB were introduced by site-directed mutagenesis and validated by sequencing.
BMAL1 PAS-AB (residues 136–441) was expressed in E. coli SoluBL21 cells as a fusion to the solubilizing tag GST. Protein was expressed via the method described above. Soluble protein was purified by GST affinity chromatography (GE Healthcare), followed by cleavage of the tag with His6-TEV protease overnight at 4 °C. Subsequent Ni2+-nitrilotriacetic acid and GST affinity chromatography were performed to remove the protease and cleaved tag. The protein was further purified by SEC as described above.
Analytical SEC.
For analysis of complex formation by SEC, purified proteins were injected on a Superdex 200 10/300 GL or Superdex 75 10/300 GL analytical column at 10–50 μM (∼250 μL, 1:1 molar ratio) in 20 mM Hepes pH 7.5, 125 mM NaCl, 5% (vol/vol) glycerol, and 2 mM TCEP. Proteins were incubated for ∼30 min or overnight and analyzed by SEC. All size-exclusion columns were calibrated with a low molecular-weight gel filtration standards kit (GE Healthcare Life Sciences). The content of each peak was evaluated by SDS/PAGE and Coomassie staining.
Transcriptional Reporter Assays and Western Blotting.
Per1-Luc reporter gene assays investigating repression by CRY1 were performed as before (11). Briefly, the following plasmids were transfected in duplicate into HEK293T cells in a 48-well plate using LT-1 transfection reagent (Mirus): 5 ng pGL3 Per1-Luc reporter, 100 ng each pSG5 mouse Flag-Bmal1 and pSG5 mouse His6 Flag-Clock, and pcDNA3 mouse Cry1 (untagged) in increasing amounts as indicated; empty pcDNA4 vector was used to normalize total plasmid levels to 800 ng DNA per well. Cells were harvested 30 h after transfection using Passive Lysis Buffer (New England Biolabs) and luciferase activity assayed using Bright-Glo luciferin reagent (Promega). Each reporter assay was repeated three independent times. To compare expression of Flag-tagged Clock, Bmal1, and untagged Cry1 genes, cells were lysed in 50 mM Hepes pH 7.5, 150 mM NaCl, 1 mM EDTA, 5% (vol/vol) glycerol, and 1% Triton X-100. Immunoblotting was done with the HRP-conjugated rabbit polyclonal OctA-Probe antibody (D-8; Santa Cruz Biotechnology cat. no. sc-807), rabbit polyclonal anti-CRY1 (H-84; Santa Cruz Biotechnology cat. no. sc-33177) or mouse monoclonal GAPDH (G-9; Santa Cruz Biotechnology, sc-365-062). HRP-conjugated secondary antibodies were used at 1:10,000 (Santa Cruz Biotechnology) in TBST. Western signal was detected using Clarity ECL reagent (Bio-Rad) and visualized on a ChemiDoc XRS+ imager (Bio-Rad).
HADDOCK Modeling.
The CLOCK PAS-B molecule (residues 261–384) used in the modeling was generated from the CLOCK:BMAL1 bHLH-PAS-AB structure (PDB ID code 4F3L). The CRY1 input molecule was our 1.8-Å resolution structure (PDB ID code 5T5X). After generating the protein components, the structures were docked as a complex using the HADDOCK 2.2 server (22). The protocol for docking and refinement was executed using the default parameter sets on the server and defining passive residues automatically around the active residues.
GST Pull-Down Assays.
GST CLOCK PAS-B and GST BMAL1 PAS-AB were expressed in E. coli as described above. Soluble proteins were purified using GS4B resin (GE Healthcare). GST-tagged proteins were eluted with 10 mM glutathione and peak elution was desalted into 20 mM Hepes pH 7.5, 125 mM NaCl, 5% (vol/vol) glycerol, and 2 mM TCEP. GST pull downs contained 1 μM GST-tagged protein (bait), 5 μM CRY1 (prey) and ∼10 μL glutathione agarose in a 200-μL reaction volume. After rotation at 4 °C overnight, reactions were washed three times with the above buffer and eluted with 6× SDS. Samples were boiled for ∼5 min and loaded on an SDS/PAGE gel for visualization by Coomassie stain.
SAXS.
SAXS data were collected on the SIBYLS beamline (12.3.1) at the Advanced Light Source (Lawrence Berkeley National Laboratory). Multiple exposures of three concentrations (1, 3, and 5 mg/mL) of freshly purified protein were taken to check for concentration dependence of scattering and radiation damage (neither was detected). Briefly, data were merged using PRIMUS, and the radius of gyration was determined using the Guinier approximation. The pair-distance distribution function [P(r)] and maximal particle size (Dmax) were generated in GNOM, and the output data were used by GASBOR to calculate 10 independent solution envelopes that were averaged together using DAMAVER. The improved model of the solution structure was used in University of California, San Francisco Chimera to fit into the averaged solution envelope.
SAXS modeling was performed using a combination of programs: FoXS (43), MultiFoXS (28), and FoXSDock (25, 44). Conformational sampling of the CLOCK:BMAL1 bHLH PAS-AB was done by MultiFoXS starting from the crystal structure (PDB ID code 4F3L) with missing fragments built by MODELER (26). HingeProt (27) was used to determine the hinge regions within the CLOCK:BMAL1 heterodimer with PDB ID code 4F3L as the input. The input to FoXSDock was the HADDOCK active residues as the binding site and (Fig. 3) the SAXS profile of the CRY1:CLOCK:BMAL1 bHLH PAS-AB complex. The entire sequences of CRY1 and CLOCK:BMAL1 bHLH PAS-AB subject to crystallography and SAXS, including vector artifacts after TEV cleavage, were used in the modeling (21).
X-Ray Crystallography.
CRY1 protein was purified as described above. The protein was concentrated to 5 mg/mL and crystallized by hanging-drop vapor diffusion at 22 °C. Crystals formed in a 1:1 ratio of protein to precipitant in 0.1 M Mes pH 6.8, 10 mM EDTA, 15% (vol/vol) PEG3350. Crystals were frozen in the proper well buffer with 20% (vol/vol) PEG400 as a cryoprotectant. Data were collected at the Advanced Light Source, Lawrence Berkeley National Laboratory at Beamline (BL5.0.1). Diffraction spots were integrated using MOSFLM (45), and data were merged and scaled using Scala (46). Phases were first solved for by molecular replacement with a previous apo CRY1 structure (PDB ID code 4K0R) using Phaser (47). The structure was built with Coot and refined with PHENIX (48, 49). Coordinates and structure factors have been deposited in PDB (PDB ID code 5T5X).
NMR.
NMR experiments were conducted at 25 °C on a Varian INOVA 600-MHz spectrometer equipped with 1H, 13C, 15N triple resonance, z axis pulsed field gradient probes. All NMR data were processed using NMRPipe/NMRDraw (50). Chemical shift assignments were made by mutation and by analogy to a far downfield-shifted peak found in all PAS domain heteronuclear single quantum coherence (HSQC) spectra (51–53) for the residue that sits at the top of the helical dipole for the Fα helix (D316 in CLOCK PAS-B). 1H-15N HSQC spectra were collected on 100 µM 15N CLOCK PAS-B WT or V315R mutant in 50 mM Tris, pH 7.5, 50 mM NaCl, 2 mM TCEP, 10% (vol/vol) D2O. HSQC data were visualized with NMRViewJ (54).
Acknowledgments
We thank the staff at Advanced Light Source X-ray diffraction beamline (BL5.0.1) and the SIBYLS SAXS beamline (BL12.3.1) for assistance with data collection. The SIBYLS SAXS beamline is supported by United States Department of Energy Integrated Diffraction Analysis Technologies Program Grant DE-AC02-05CH11231. This work was supported from NIH Grant GM107069 (to C.L.P.). A.K.M. was supported by NIH Fellowship F31 CA189660. J.S.T. is an Investigator in the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5T5X).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1615310114/-/DCSupplemental.
References
- 1.Partch CL, Green CB, Takahashi JS. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2014;24(2):90–99. doi: 10.1016/j.tcb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc Natl Acad Sci USA. 2014;111(45):16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bunger MK, et al. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103(7):1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi JS, Hong H-K, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: Implications for physiology and disease. Nat Rev Genet. 2008;9(10):764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown SA, et al. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science. 2005;308(5722):693–696. doi: 10.1126/science.1107373. [DOI] [PubMed] [Google Scholar]
- 6.Kim JY, Kwak PB, Weitz CJ. Specificity in circadian clock feedback from targeted reconstitution of the NuRD corepressor. Mol Cell. 2014;56(6):738–748. doi: 10.1016/j.molcel.2014.10.017. [DOI] [PubMed] [Google Scholar]
- 7.Koike N, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338(6105):349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye R, et al. Dual modes of CLOCK:BMAL1 inhibition mediated by Cryptochrome and Period proteins in the mammalian circadian clock. Genes Dev. 2014;28(18):1989–1998. doi: 10.1101/gad.249417.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen R, et al. Rhythmic PER abundance defines a critical nodal point for negative feedback within the circadian clock mechanism. Mol Cell. 2009;36(3):417–430. doi: 10.1016/j.molcel.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ye R, Selby CP, Ozturk N, Annayev Y, Sancar A. Biochemical analysis of the canonical model for the mammalian circadian clock. J Biol Chem. 2011;286(29):25891–25902. doi: 10.1074/jbc.M111.254680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu H, et al. Cryptochrome 1 regulates the circadian clock through dynamic interactions with the BMAL1 C terminus. Nat Struct Mol Biol. 2015;22(6):476–484. doi: 10.1038/nsmb.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sancar A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev. 2003;103(6):2203–2237. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- 13.van der Horst GT, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398(6728):627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 14.Kume K, et al. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98(2):193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 15.Vitaterna MH, et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci USA. 1999;96(21):12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Czarna A, et al. Quantitative analyses of cryptochrome-mBMAL1 interactions: Mechanistic insights into the transcriptional regulation of the mammalian circadian clock. J Biol Chem. 2011;286(25):22414–22425. doi: 10.1074/jbc.M111.244749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Czarna A, et al. Structures of Drosophila cryptochrome and mouse cryptochrome1 provide insight into circadian function. Cell. 2013;153(6):1394–1405. doi: 10.1016/j.cell.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 18.Lee Y, Chen R, Lee HM, Lee C. Stoichiometric relationship among clock proteins determines robustness of circadian rhythms. J Biol Chem. 2011;286(9):7033–7042. doi: 10.1074/jbc.M110.207217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sato TK, et al. Feedback repression is required for mammalian circadian clock function. Nat Genet. 2006;38(3):312–319. doi: 10.1038/ng1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao W-N, et al. CIPC is a mammalian circadian clock protein without invertebrate homologues. Nat Cell Biol. 2007;9(3):268–275. doi: 10.1038/ncb1539. [DOI] [PubMed] [Google Scholar]
- 21.Huang N, et al. Crystal structure of the heterodimeric CLOCK:BMAL1 transcriptional activator complex. Science. 2012;337(6091):189–194. doi: 10.1126/science.1222804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Zundert GC, et al. The HADDOCK2.2 Web Server: User-friendly integrative modeling of biomolecular complexes. J Mol Biol. 2016;428(4):720–725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 23.Dominguez C, Boelens R, Bonvin AM. HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125(7):1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 24.Nangle SN, et al. Molecular assembly of the period-cryptochrome circadian transcriptional repressor complex. eLife. 2014;3:e03674. doi: 10.7554/eLife.03674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneidman-Duhovny D, Hammel M, Sali A. FoXS: A web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res. 2010;38(Web Server issue):W540–W544. doi: 10.1093/nar/gkq461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Webb B, Sali A. Comparative protein structure modeling using MODELLER. Curr Protoc Bioinformatics. 2014;47:5.6.1–5.6.32. doi: 10.1002/0471250953.bi0506s47. [DOI] [PubMed] [Google Scholar]
- 27.Emekli U, Schneidman-Duhovny D, Wolfson HJ, Nussinov R, Haliloglu T. HingeProt: Automated prediction of hinges in protein structures. Proteins. 2008;70(4):1219–1227. doi: 10.1002/prot.21613. [DOI] [PubMed] [Google Scholar]
- 28.Schneidman-Duhovny D, Hammel M, Tainer JA, Sali A. FoXS, FoXSDock and MultiFoXS: Single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic Acids Res. 2016;44(W1):W424–W429. doi: 10.1093/nar/gkw389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cardone L, et al. Circadian clock control by SUMOylation of BMAL1. Science. 2005;309(5739):1390–1394. doi: 10.1126/science.1110689. [DOI] [PubMed] [Google Scholar]
- 30.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: Defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40(3):191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 31.Schmalen I, et al. Interaction of circadian clock proteins CRY1 and PER2 is modulated by zinc binding and disulfide bond formation. Cell. 2014;157(5):1203–1215. doi: 10.1016/j.cell.2014.03.057. [DOI] [PubMed] [Google Scholar]
- 32.Xing W, et al. SCF(FBXL3) ubiquitin ligase targets cryptochromes at their cofactor pocket. Nature. 2013;496(7443):64–68. doi: 10.1038/nature11964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michael AK, Fribourgh JL, Van Gelder RN, Partch CL. November 28, 2016. Animal cryptochromes: Divergent roles in light perception, circadian timekeeping and beyond. Photochem Photobiol, 10.1111/php.12677.
- 34.Selby CP, Sancar A. The second chromophore in Drosophila photolyase/cryptochrome family photoreceptors. Biochemistry. 2012;51(1):167–171. doi: 10.1021/bi201536w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhong D. Electron transfer mechanisms of DNA repair by photolyase. Annu Rev Phys Chem. 2015;66:691–715. doi: 10.1146/annurev-physchem-040513-103631. [DOI] [PubMed] [Google Scholar]
- 36.Gustafson CL, Partch CL. Emerging models for the molecular basis of mammalian circadian timing. Biochemistry. 2015;54(2):134–149. doi: 10.1021/bi500731f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Partch CL, Gardner KH. Coactivator recruitment: A new role for PAS domains in transcriptional regulation by the bHLH-PAS family. J Cell Physiol. 2010;223(3):553–557. doi: 10.1002/jcp.22067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Partch CL, Gardner KH. Coactivators necessary for transcriptional output of the hypoxia inducible factor, HIF, are directly recruited by ARNT PAS-B. Proc Natl Acad Sci USA. 2011;108(19):7739–7744. doi: 10.1073/pnas.1101357108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hennig S, et al. Structural and functional analyses of PAS domain interactions of the clock proteins Drosophila PERIOD and mouse PERIOD2. PLoS Biol. 2009;7(4):e94. doi: 10.1371/journal.pbio.1000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kucera N, et al. Unwinding the differences of the mammalian PERIOD clock proteins from crystal structure to cellular function. Proc Natl Acad Sci USA. 2012;109(9):3311–3316. doi: 10.1073/pnas.1113280109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheuermann TH, et al. Allosteric inhibition of hypoxia inducible factor-2 with small molecules. Nat Chem Biol. 2013;9(4):271–276. doi: 10.1038/nchembio.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khan SK, et al. Identification of a novel cryptochrome differentiating domain required for feedback repression in circadian clock function. J Biol Chem. 2012;287(31):25917–25926. doi: 10.1074/jbc.M112.368001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 44.Schneidman-Duhovny D, Hammel M, Tainer JA, Sali A. Accurate SAXS profile computation and its assessment by contrast variation experiments. Biophys J. 2013;105(4):962–974. doi: 10.1016/j.bpj.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneidman-Duhovny D, Hammel M, Sali A. Macromolecular docking restrained by a small angle X-ray scattering profile. J Struct Biol. 2011;173(3):461–471. doi: 10.1016/j.jsb.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leslie AG. The integration of macromolecular diffraction data. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 1):48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 47.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 48.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63(Pt 1):32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 50.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Delaglio F, et al. NMRPipe: A multidimensional spectral processing Asystem based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 52.Card PB, Erbel PJ, Gardner KH. Structural basis of ARNT PAS-B dimerization: Use of a common beta-sheet interface for hetero- and homodimerization. J Mol Biol. 2005;353(3):664–677. doi: 10.1016/j.jmb.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 53.Erbel PJ, Card PB, Karakuzu O, Bruick RK, Gardner KH. Structural basis for PAS domain heterodimerization in the basic helix--loop--helix-PAS transcription factor hypoxia-inducible factor. Proc Natl Acad Sci USA. 2003;100(26):15504–15509. doi: 10.1073/pnas.2533374100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]