Significance
The prokaryotic CRISPR/Cas9 system has recently been applied in genome editing in mammalian cells with the potential to bring curative therapies to patients with genetic diseases. However, efficient in vivo delivery of this machinery remains challenging for most tissue types. We now developed a method to locally deliver Cas9/sgRNA ribonucleoproteins into the skin of postnatal mice, which was used to correct genetic defects in skin stem cells of postnatal recessive dystrophic epidermolysis bullosa (RDEB) mice. Our study provides proof-of-principle evidence that Cas9/sgRNA ribonucleoprotein-based gene therapies can be applied to restore collagen VII protein function in postnatal RDEB mice, suggesting that the Cas9/sgRNA ribonucleoprotein-based gene therapy may offer curative treatment for RDEB and other genetic disorders.
Keywords: in vivo gene editing, Cas9/sgRNA ribonucleoproteins, skin stem cell, electroporation, RDEB
Abstract
The prokaryotic CRISPR/Cas9 system has recently emerged as a powerful tool for genome editing in mammalian cells with the potential to bring curative therapies to patients with genetic diseases. However, efficient in vivo delivery of this genome editing machinery and indeed the very feasibility of using these techniques in vivo remain challenging for most tissue types. Here, we show that nonreplicable Cas9/sgRNA ribonucleoproteins can be used to correct genetic defects in skin stem cells of postnatal recessive dystrophic epidermolysis bullosa (RDEB) mice. We developed a method to locally deliver Cas9/sgRNA ribonucleoproteins into the skin of postnatal mice. This method results in rapid gene editing in epidermal stem cells. Using this method, we show that Cas9/sgRNA ribonucleoproteins efficiently excise exon80, which covers the point mutation in our RDEB mouse model, and thus restores the correct localization of the collagen VII protein in vivo. The skin blistering phenotype is also significantly ameliorated after treatment. This study provides an in vivo gene correction strategy using ribonucleoproteins as curative treatment for genetic diseases in skin and potentially in other somatic tissues.
CRISPR/Cas9-mediated genome editing has recently emerged as a powerful tool for genome engineering and for use as a potential therapeutic method to treat patients with genetic diseases (1–3). CRISPR/Cas9-mediated somatic genome editing of multiple mice organs, including lung, liver, brain, pancreas, and muscle, have been reported (4–10). To date, most approaches have relied on virus-based delivery systems, which have drawbacks such as potential integration of viral DNA into a host genome, off-target effects due to prolonged expression of genome editing machinery, and possible activation of virus-triggered host immune responses. Additionally, most therapeutic applications will require the tissue-specific delivery of the genome editing machinery in vivo. So far, this has remained challenging for most tissues, such as skin.
In healthy skin, epidermal keratinocytes and dermal fibroblasts secrete collagen VII protein and it in turn forms stable homotrimers to assemble into networks of anchoring fibrils (11, 12). The anchoring fibrils are located within the basement membrane zone (BMZ) between the epidermis and dermis, where they participate in stabilizing the association of the epidermis to the underlying dermis (13). In patients with recessive dystrophic epidermolysis bullosa (RDEB), mutations in the Col7a1 gene cause absent or dysfunctional collagen VII protein production, which leads to defective epidermal–dermal adhesion (14). The main clinical manifestations of RDEB include: chronic and severe cutaneous blistering, especially on hands and feet; damage to internal epithelia, such as oral, esophageal, and anal structures; an increased risk for developing aggressive forms of squamous cell carcinoma; and overall reduced life expectancy (15).
Treatments currently under development for patients with RDEB mainly include protein therapy (16, 17) or combined gene and cell therapy. In particular, the cellular therapies of RDEB rely on introducing donor cells that are capable of producing normal collagen VII protein, such as allogeneic dermal fibroblasts, gene-corrected RDEB fibroblasts (18–20), gene-corrected keratinocytes in autografts (21–23), and gene-corrected keratinocytes derived from reverted-induced pluripotent stem cells (24–27). Based on what is known from these studies, clinical trials for RDEB that combine gene and cell therapies have been initiated. Examples include the intradermal injection of allogeneic fibroblasts (20) and the grafting of epidermal tissues generated ex vivo that express virally introduced normal collagen VII (28). Bone marrow transplantation has also been reported to ameliorate the skin blistering phenotype (29, 30). Despite all of these advances, there are still no curative treatments for RDEB.
Here, we demonstrate the in vivo use of Cas9/sgRNA ribonucleoproteins to mediate gene correction in skin stem cells of postnatal RDEB mice. First we generate a RDEB mouse model based on a patient-specific point mutation. Then we establish a mouse model missing the exon containing the point mutation in Col7a1 gene to prove the efficiency and safety of exon skipping as a gene correction method. To apply the gene correction system in intact postnatal skin, we developed a method to deliver nonreplicable protein/RNA complexes in vivo; it induces one-time, permanent modification of genomic DNA in skin stem cells. Finally, we demonstrate the feasibility and efficiency of our curative gene editing system to permanently restore the function of the collagen VII protein in vivo.
Results
Col7a1 c.6485G > A mut/mut Mice Show RDEB Hallmarks.
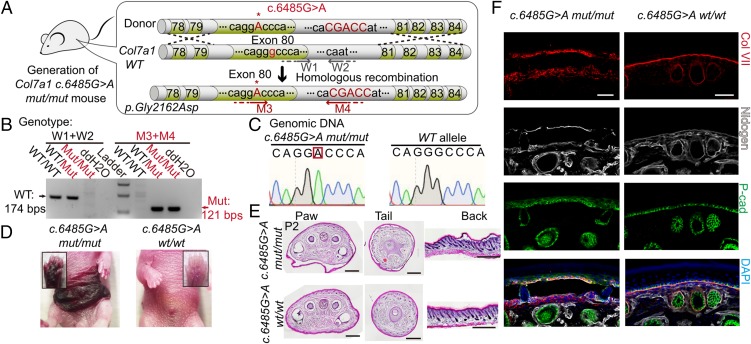
First, we established a RDEB mouse model based on the genome sequencing data of a patient with RDEB in Beijing. CRISPR/Cas9 system-facilitated homologous recombination was used to generate the c.6485G > A point mutation within exon80 of the mouse Col7a1 gene (Fig. 1A). Using specific primer pairs targeting wild-type (WT) and mutant alleles in PCR-based genotyping assays, we can distinguish among WT, heterozygous, and homozygous mutant mice (Fig. 1B). Sequencing of genomic DNA confirmed the existence of the expected specific c.6485G > A mutation within the Col7a1 locus (Fig. 1C). After birth, Col7a1 c.6485G > A mut/mut mice exhibited striking skin blistering phenotypes similar to the patient with RDEB whose point mutation was used to create this mouse model (Fig. 1D). The Col7a1 c.6485G > A wt/mut mice appeared normal, consistent with the patient’s carrier parent. Histological examinations demonstrated separation of the epidermis from the dermis at all of the locations examined, including the mouse tail, paw, and back skin (Fig. 1E). Collagen VII immunofluorescence staining revealed striking changes in protein-localization patterns: in WT skin, collagen VII proteins were localized at the BMZ in a linear pattern; in the Col7a1 c.6485G > A mut/mut skin, collagen VII proteins were scattered in a dotted pattern in the basal epidermis layer and the dermis near the BMZ zone (Fig. 1F and Fig. S1A). Most of the Col7a1 c.6485G > A mut/mut mice died within a week after birth, due to complications from the disease (Fig. S1B). Overall, our newly generated Col7a1 c.6485G > A mut/mut mouse line, based on a patient-specific point mutation, exhibits typical pathologic features encountered in patients with RDEB. And it is a disease-relevant animal model that can enable us to develop effective gene correction therapy.
Fig. 1.
Establishment of a RDEB mouse model based on patient-specific mutations. (A) A portion of the murine Col7a1 gene from exon78 to exon 84 (about 1.5 kb) is used as the donor DNA to generate a knockin mouse line. Col7a1 c.6485G > A mutation in exon80 is highlighted with uppercase letters. A 5-bp (CGACC) insertion in intron80 was also introduced to facilitate genotyping identification of the mutant allele. The W1/W2 and M3/M4 primer pairs are used to identify the WT Col7a1 locus and the recombined Col7a1 locus, respectively. (B) Genotyping results of Col7a1 c.6485G > A WT (wt/wt), heterozygous (wt/mut), and homozygous (mut/mut) mice using the W1/W2 and M3/M4 primers. (C) Partial Col7a1 locus genomic DNA sequencing results for WT and Col7a1 c.6485G > A mut/mut mice. (D) Spontaneous blisters form on the paws and ventral skin of Col7a1 c.6485G > A mut/mut mice. (E) H&E staining of postnatal day 2 (P2) Col7a1 c.6485G > A mut/mut and WT tissue sections from paw, tail, and back skin. Note the separation of epidermis from dermis in Col7a1 c.6485G > A mut/mut skins. (Scale bar, 500 μm.) (F) Immunofluorescence stainings of collagen VII (Col VII), P-cad, and Nidogen in Col7a1 c.6485G > A mut/mut and WT tail skins. (Scale bar, 50 μm.)
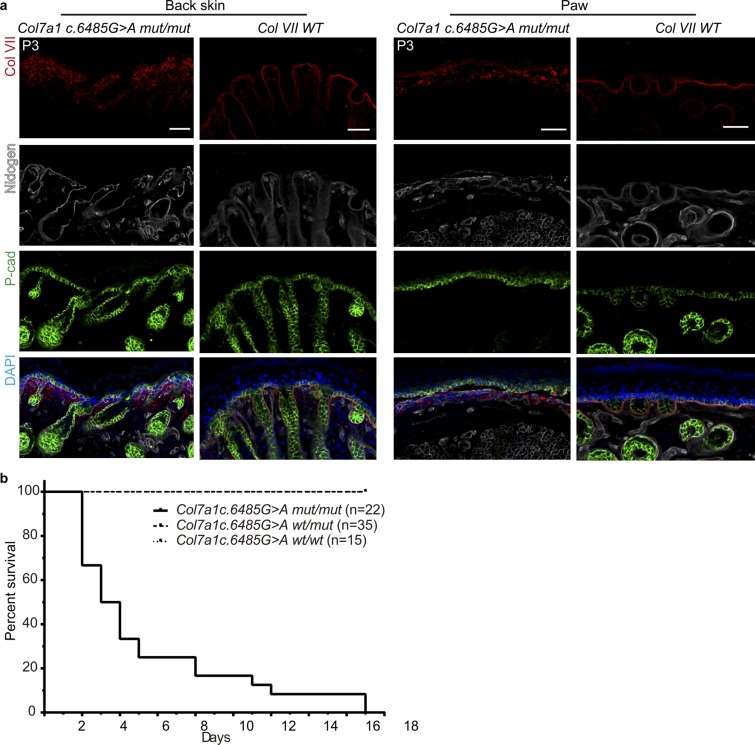
Fig. S1.
Phenotypic analysis of Col7a1 c.6485G > A mut/mut compared with WT mice. (A) Immunofluorescence staining for Col VII, P-cad, and Nidogen in the back skin and paw skin of Col7a1 c.6485G > A mut/mut and WT mice. (Scale bar, 40 μm.) (B) Survival curve of Col7a1 c.6485G > A wt/wt, wt/mut, and mut/mut mice.
Evidence from both an in Vivo Animal Model and an in Vitro Cell Line Demonstrates the Feasibility and Safety of Exon Skipping as a Gene Correction Method.
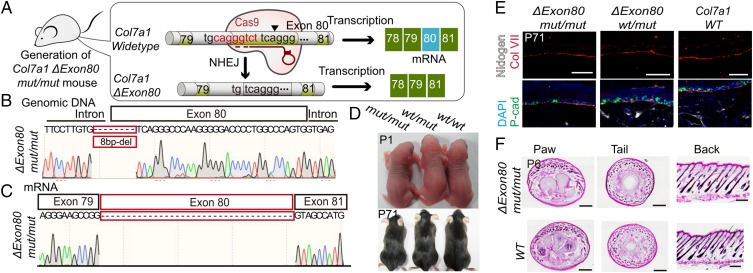
Col7a1 exon80 encodes part of the collagenous domain that contains stretches of bases coding for G-X-Y (G: glycine, X and Y: two other random amino acids) repeat sequences; it is in-frame with neighboring exons. In theory, deleting the point mutation containing exon80 should result in a collagen VII protein with a slightly shortened collagenous domain that would ostensibly retain its normal function (Fig. S2A). However, the feasibility and safety of this gene correction strategy must of course be tested rigorously in vivo, with particular attention paid to whether or not the shortened form of the collagen VII protein may act in a dominant negative manner to disrupt homotrimer formation. To this end, we generated Col7a1 exon80-skipped mice (Col7a1 Δexon80) using the CRISPR/Cas9 system (Fig. 2A). In this mutant strain, there is an 8-bp deletion that spans the junction between intron79 and exon80 (3 bp in intron79 and 5 bp in exon80). This deletion abrogates the acceptor site for intron splicing and causes exon80 to be skipped during splicing. Genomic DNA and mRNA sequencing results confirmed, respectively, the occurrence of the 8-bp deletion at the Col7a1 locus and the skipping of exon80 (Fig. 2 B and C). The Col7a1 Δexon80 mut/mut and importantly wt/mut mice are indistinguishable from their WT littermates; no skin blistering phenotypes were observed in newborns or in adults (Fig. 2D). The localization of the collagen VII protein was identical in Col7a1 Δexon80 mut/mut and wt/mut, and WT mice, exhibiting a linear pattern at the BMZ (Fig. 2E). Thorough histological examination revealed an intact epidermis and dermis adhesion at all of the locations examined, including the mouse tail, paw, and back skin (Fig. 2F). These results demonstrate that exon80 deletion in vivo is an efficient and safe strategy to restore the function of the collagen VII protein.
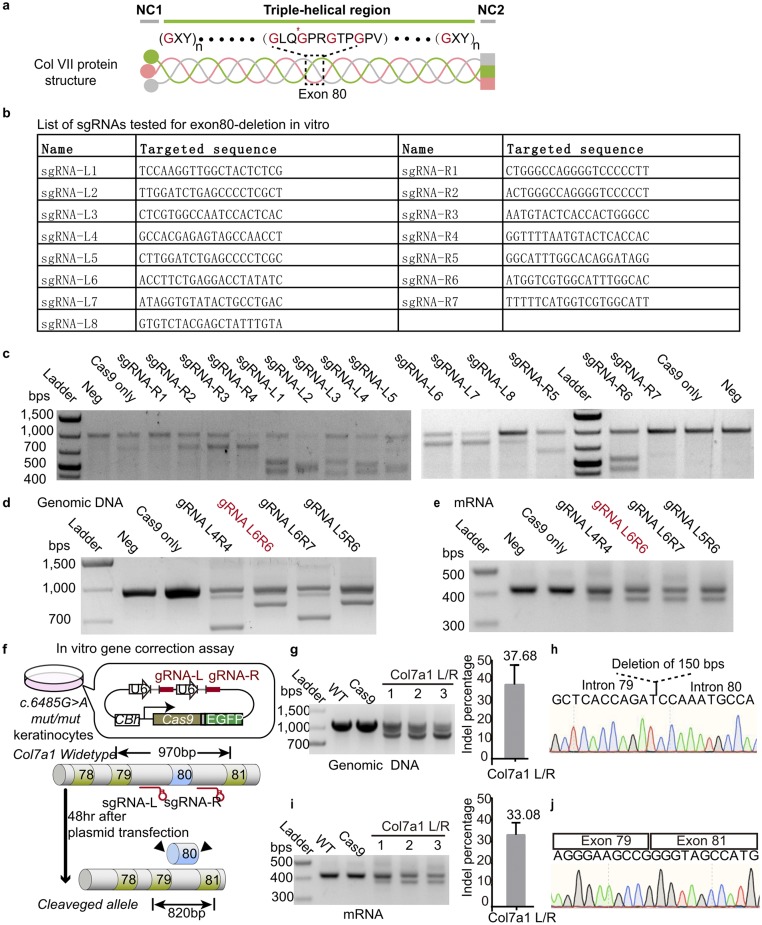
Fig. S2.
In vitro cell culture sgRNA functional test. (A) Schematic diagram of the structure of collagen VII. (B) List of sgRNA molecules tested for Col7a1 exon80 deletion. (C) T7 endonuclease I assay for each sgRNA functional test. (D) PCR amplifications of the Col7a1 genomic DNA demonstrate truncation at exon80 after transfection with different sgRNA pairs. The sgRNA pair used for in vivo Col7a1 gene editing is marked in red. (E) RT-PCR of Col7a1 mRNA indicates deletion at exon80 after transfection with different sgRNA pair combinations. (F) Schematic diagram of the in vitro gene correction assay. Modified pSpCas9-2A-GFP plasmid expresses Cas9 and two different sgRNAs (gRNA-L/R). (G) PCR amplifications of the Col7a1 genomic DNA demonstrate truncation at exon80 after transfection. Percentage of truncated genomic DNA is shown on the Right. (H) Partial Col7a1 gene sequencing result confirms the deletion of exon80. (I) RT-PCR of Col7a1 mRNA indicates deletion of exon80 after transfection. Percentage of truncated mRNA product is shown on the Right. (J) Partial Col7a1 mRNA sequencing result confirms deletion of exon80.
Fig. 2.
Col7a1-ΔExon80 mut/mut and wt/mut mice have normal skin with correct collagen VII localization. (A) Schematic diagram of the generation of Col7a1-ΔExon80 mut/mut mice. (B) Partial Col7a1 genomic DNA sequencing result shows the 8-bp deletion spanning the intron79 and exon80 junction in Col7a1-ΔExon80 mut/mut mice. (C) Partial Col7a1 mRNA sequencing result shows skipping of exon80 in Col7a1-ΔExon80 mut/mut mice. (D) Images of Col7a1-ΔExon80 mut/mut, wt/mut, and WT mice at P1 and P71. (E) Immunofluorescence staining for Col VII, P-cad, and Nidogen in the paws of Col7a1-ΔExon80 mut/mut, wt/mut, and WT mice. (Scale bar, 50 μm.) (F) H&E staining of P6 Col7a1-ΔExon80 mut/mut and WT tissue sections in paw, tail, and back skin. (Scale bar, 500 μm.)
To screen for sgRNA pairs that can efficiently and specifically mediate DNA excision in introns flanking exon80 and thus lead to direct exon80 deletion, we evaluated several sgRNAs in Col7a1 c.6485G > A mut/mut keratinocytes in vitro (Fig. S2B). Heteroduplex DNA resulting from Cas9/sgRNA-mediated indel formation was detected within most of the tested sgRNAs (Fig. S2C). One pair of sgRNAs that exhibited both high cutting efficiency and high specificity was selected for further use (Fig. S2 D and E). A modified pSpCas9(BB)-2A-EGFP (PX458) plasmid was adopted to express the Cas9 nuclease and both of the sgRNAs (sgRNA-L and sgRNA-R that target, respectively, the 5′ and 3′ introns of exon80) (Fig. S2F). Sequencing confirmed the deletion of exon80 at genomic DNA level and mRNA level with high efficiency in vitro (Figs. S2 G–J). These results demonstrate that we can use exon deletion to restore the normal function of the collagen VII protein in vivo and demonstrate the suitability of our selected sgRNA pairs to efficiently and specifically delete exon80 of the Col7a1 locus.
In Vivo Delivered Cas9/sgRNA Ribonucleoproteins via Electroporation Mediates Efficient and Precise DNA Editing in Skin Stem Cells.
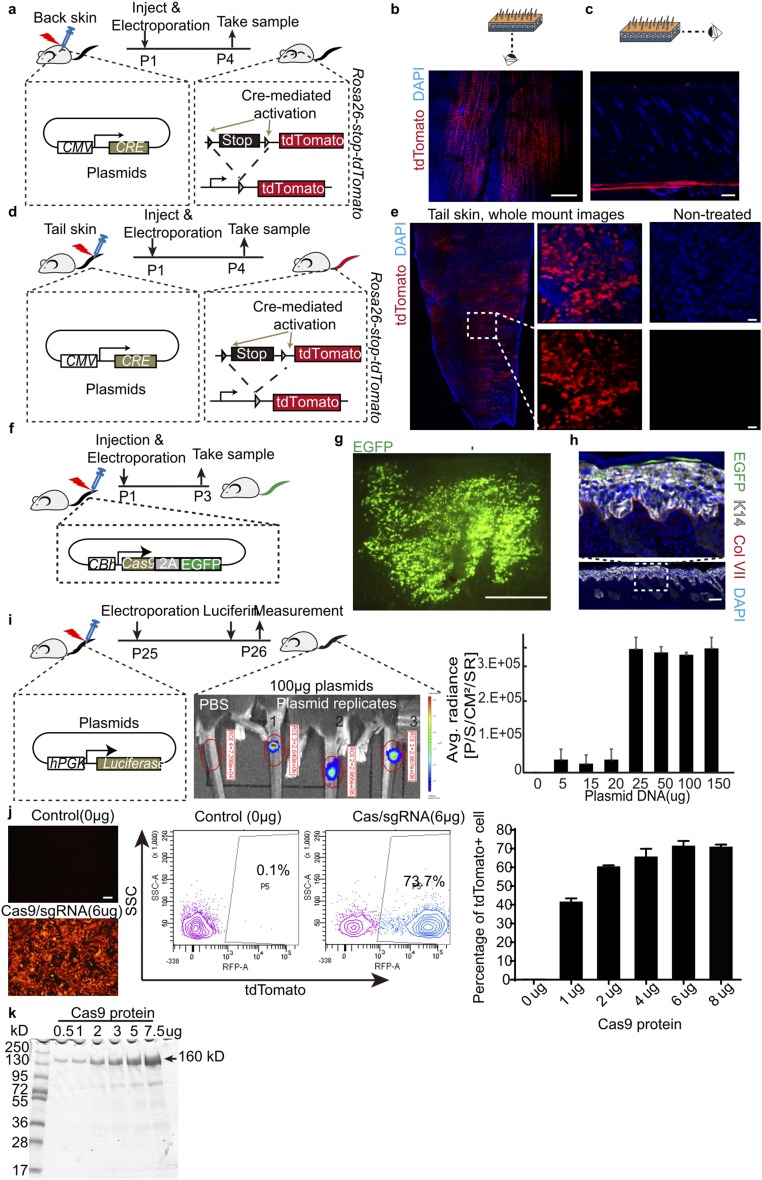
To have practical and long-term therapeutic effect for our gene correction strategy, it would be essential to adapt the CRISPR/Cas9-mediated gene editing system in postnatal skin stem cells in vivo. We initially tried using electroporation-based delivery of plasmid DNA to the skin of postnatal mice. To unambiguously detect functional DNA delivery in vivo, we used Cre-expressing plasmid in the skin of Rosa26-stop-tdTomato reporter mice. Successfully delivering DNA into the skin cells in vivo would lead to Cre-recombinase–mediated excision of the floxed-stop-floxed (lsl) codon and would thus allow tdTomato expression. Cre-expressing plasmids were first intradermally injected into mouse back skin; this step was followed by electroporation. We observed that most of the tdTomato+ cells were present only in the panniculus carnosus of the mouse back skin (Fig. S3 A–C). The panniculus carnosus is a sheet of muscle cells underneath the mouse back skin dermis; it is vestigial or absent in human skin (31). To circumvent this problem, we switched to newborn mouse tail skin, which has a thick epidermis and no panniculus carnosus, similar to human skin. Using the same optimized electroporation conditions, whole mount staining clearly revealed tdTomato+ cells in the epidermis (Fig. S3 D and E). The same in vivo electroporation method also works efficiently to deliver plasmid into adult mouse tail skin (Fig. S3I).
Fig. S3.
Electroporation condition test. (A) Schematic diagram for testing electroporation in back skin with Cre-expressing plasmids. Representative whole mount (B) and section (C) immunofluorescence stainings of Rosa26-stop-tdTomato reporter mouse back skin posttreatment. (Scale bar, 500 μm for whole mount, 100 μm for sections.) (D) Schematic diagram for testing electroporation in tail skin with Cre-expressing plasmids. (E) Representative whole mount immunofluorescence images of Rosa26-stop-tdTomato reporter mouse tail skin with or without treatment. (Scale bar, 500 μm for Left image, 100 μm for Right images.) (F) Schematic diagram of electroporation experiments with the pSpCas9-2A-GFP plasmid at the tail of newborn mice. Representative whole mount (G) and section (H) immunofluorescence staining of tail skin at 2 d after pSpCas9-2A-GFP plasmid electroporation. (Scale bar, 1 mm for whole mount images, 100 μm for section staining images.) (I) Schematic diagram of electroporation with plasmid in adult mice tail skin. Luciferase activity is measured at 24 h posttreatment using an in vivo luciferase imaging system. Quantification data are shown at Right. (J) Electroporation delivered Cas9/sgRNA complexes in cultured Rosa26-stop-tdTomato reporter keratinocytes in vitro. Representative fluorescence images of cultured cells posttreatment are shown at Left. FACS quantification of tdTomato+ cells are used to optimize the dosage of Cas9/gRNA ribonucleoproteins for maximum efficiency. (K) Evaluation of the purity of recombinant Cas9 via Coomassie Blue staining.
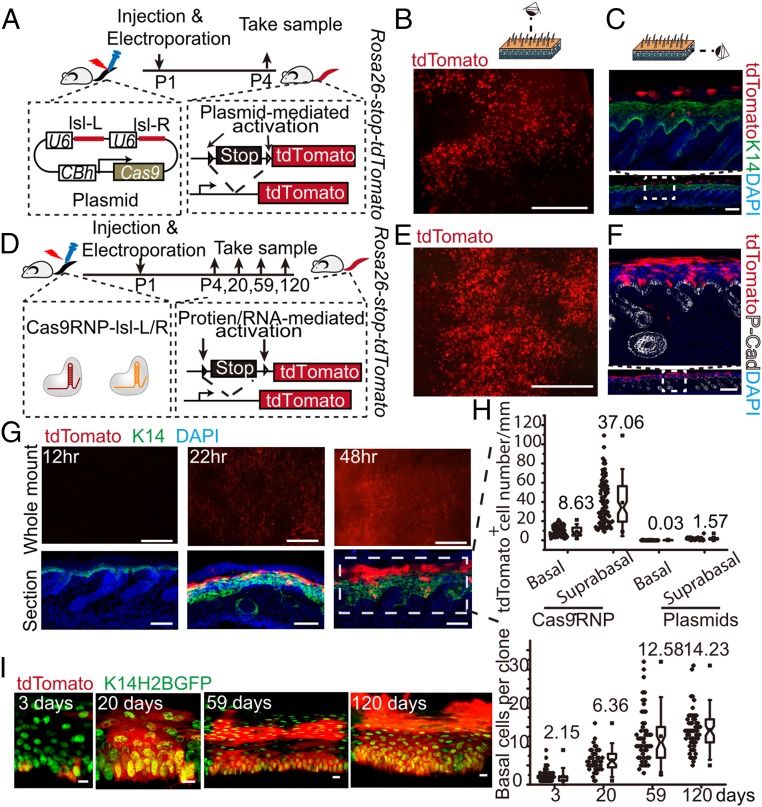
Next, we used a plasmid expressing Cas9-2A-EGFP and could easily detect abundant GFP+ cells in whole mount tail skin epidermis at 2 d posttreatment (Fig. S3 F–H). However, upon closer inspection, most if not all of the GFP+ cells were located in the stratum corneum; none were present in the basal epidermal layer where stem cells locate (Fig. S3H). Similar results were observed using a plasmid expressing the Cas9 nuclease and two sgRNAs targeting the floxed stop codon in Rosa26-stop-tdTomato reporter mice, which are termed sgRNA lsl-L and lsl-R, respectively (Fig. 3 A–C). So, even though the plasmid delivery method worked in vivo, its failure to target either epidermal stem cells or dermal fibroblasts renders this method ineffective for use in gene therapy applications.
Fig. 3.
Efficient in vivo deliveries of Cas9/sgRNA ribonucleoproteins into skin stem cells of postnatal mice using electroporation. (A) Schematic diagram of electroporation with pSpCas9-lsl-L/R plasmid at the tail of Rosa26-stop-tdTomato reporter newborn mice. Representative whole mount (B) and section (C) immunofluorescence staining of Rosa26-stop-tdTomato mouse tail skin after pSpCas9-lsl-L/R plasmid electroporation. (Scale bar, 1 mm for whole mount images, 100 μm for section staining images.) (D) Schematic diagram of electroporation with Cas9/sgRNA ribonucleoproteins at the tails of Rosa26-stop-tdTomato reporter newborn mice. Representative whole mount (E) and section (F) immunofluorescence staining of Rosa26-stop-tdTomato reporter tail skin at 3 d after treatment. (Scale bar, 1 mm for whole mount images, 500 μm for section staining images.) (G) Representative whole mount and section immunofluorescence stainings of Rosa26-stop-tdTomato reporter tail skin at 12 h, 22 h, and 48 h post-Cas9/sgRNA ribonucleoprotein electroporation. (Scale bar, 500 μm for whole mount images, 70 μm for section images.) (H) Quantifications of the number of tdTomato+ cells in the basal and suprabasal layers of epidermis at 3 d postelectroporation treatments with either ribonucleoproteins or plasmid DNA. (I) Expansion of basal cell per colony after one electroporation treatment with Cas9/sgRNA ribonucleoproteins. Representative images at 3, 20, 59, and 120 d posttreatment are shown at Left; quantifications are shown at Right. (Scale bar, 10 μm.)
Cas9/sgRNA ribonucleoproteins have been reported to have a higher rate of cleavage and clearance compared with plasmid DNA in in vitro cell culture systems (32). When high-purity recombinant Cas9 and in vitro-transcribed sgRNAs lsl-L and lsl-R were electroporated into Rosa26-stop-tdTomato keratinocytes, as high as 70% of cells expressed tdTomato (Fig. S3 J and K). Previous reports used lipid-mediated delivery of Cas9/sgRNA complexes into the mouse inner ear in vivo, resulting in genome modification in ear hair cells (33). Here we used electroporation to deliver Cas9/sgRNA ribonucleoproteins into intact mouse tail skin. Unlike the results observed in our DNA plasmid electroporation experiments, this combination resulted in efficient targeting of cells in all layers of the epidermis, including in the basal epidermis (Fig. 3 D–F). The earliest detectable tdTomato expression occurred 22 h after electroporation (Fig. 3G). At 48 h after one electroporation treatment, dramatically higher numbers of basal and suprabasal epidermal cells expressed tdTomato, compared with the very low efficiency using plasmid DNA (Fig. 3 G and H). Because epidermis is a tissue with fast cellular turnover rate during normal homeostasis, to have long-term therapeutic effect following gene correction in vivo, it is essential that skin stem cells with long-term self-renewing ability are targeted for gene editing so that they can continuously produce functional collagen VII. To determine whether epidermal stem cells were targeted successfully, we followed the long-term fate of the labeled basal epidermis cells after one electroporation and found that up to 120 d later the basal cell colonies laterally expanded and gave continuous progenies in all of the upper epidermis layers (Fig. 3I). These results strongly indicate that we are able to efficiently deliver Cas9/sgRNA ribonucleoproteins into postnatal skin stem cells in live animals, which is a prerequisite for carrying out effective gene correction therapies in vivo.
Cas9/sgRNA Ribonucleoproteins Mediated in Vivo Exon80 Deletion in Col7a1 c.6485G > A mut/mut Mice Rescues Skin Defects.
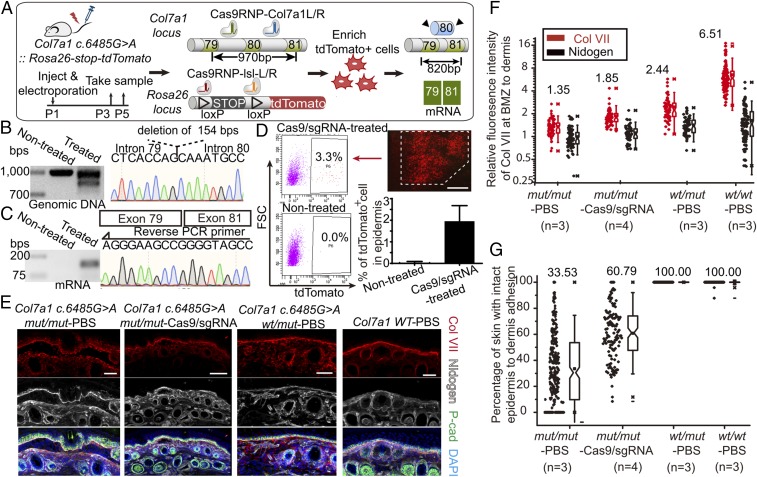
Combining the exon skipping gene correction strategy and the in vivo Cas9/sgRNA ribonucleoproteins delivery system, we carried out the curative gene editing treatment in our postnatal RDEB mutant mice. Cas9/sgRNA ribonucleoproteins targeting exon80 of the Col7a1 locus were injected intradermally into Col7a1 c.6485G > A mut/mut mouse tail skin at P1 or P2, followed by electroporation. To isolate the target cells for sequence analysis, Rosa26-stop-tdTomato allele was bred into the mutant mouse line and Cas9/sgRNA ribonucleoproteins targeting the floxed stop codon of tdTomato were injected simultaneously during treatment. Tail skin samples were analyzed 3–5 d posttreatment (Fig. 4A). Fluorescence-activated cell sorting (FACS) isolated tdTomato+ cells were used for genomic DNA and mRNA sequence analysis. Truncated genomic DNA resulting from Cas9/sgRNA-mediated excision was detected in gene-corrected skin samples, and sequencing results revealed the successful deletion of Col7a1 exon80 (Fig. 4B). RT-PCR of transcripts from gene-corrected skin samples also revealed deletion of exon80 at the mRNA level (Fig. 4C). To estimate the efficiency of in vivo gene editing in our mutant mice, percentage of tdTomato+ cells in whole epidermis after a single treatment was quantified by FACS, and ∼2% epidermal cells were targeted (Fig. 4D). We next examined the collagen VII protein distribution patterns in vivo using immunofluorescence staining (Fig. 4 E and F). In WT skin treated with PBS injection and electroporation, collagen VII proteins localized at the BMZ in a linear pattern. Although Col7a1 c.6485G > A wt/mut mice have functionally normal skin, the collagen VII proteins of these mice were distributed at the BMZ in a linear pattern as well as scattered throughout the rest of the skin in a dotted pattern. This phenomenon suggests as long as there are functional collagen VII protein forming anchoring fibrils at the BMZ, the intact adhesion of epidermis to dermis will be maintained, despite the presence of defective collagen VII protein elsewhere in the skin. When Cas9/sgRNA ribonucleoproteins targeting Col7a1 exon80 were injected intradermally into the Col7a1 c.6485G > A mut/mut mouse tail skin followed by electroporation, there was clear enrichment of collagen VII localization at the BMZ 3–5 d later, a phenomenon not observed when Col7a1 c.6485G > A mut/mut mice skin was treated with the control PBS injection and electroporation. We quantified these changes in the distribution of collagen VII using Nidogen as a marker for where the BMZ is (Fig. 4F). The ratio of fluorescence intensities of collagen VII at the BMZ and neighboring lower dermis showed significant increase after one treatment with Cas9/sgRNA ribonucleoproteins in the Col7a1 c.6485G > A mut/mut mice skin. Under the same measurement condition, the ratio of fluorescence intensity for Nidogen did not show difference, thus excluding the possibility of measurement bias. As the ultimate functional test for the in vivo gene editing efficiency in our RDEB mice, we measured the degree of the epidermis-to-dermis adhesion. Both WT and the Col7a1 c.6485G > A wt/mut skin had 100% adhesion between the epidermis and the dermis. After one treatment with Cas9/sgRNA ribonucleoproteins, the average adhesion area of Col7a1 c.6485G > A mut/mut skin increased from ∼30% to ∼60% (Fig. 4G). All these findings provide direct evidence that Cas9/sgRNA ribonucleoproteins can efficiently mediate gene editing for RDEB at the genomic DNA, mRNA, protein, and phenotypic levels.
Fig. 4.
Cas9/sgRNA ribonucleoprotein-mediated in vivo gene editing in postnatal RDEB mice. (A) Schematic diagram of in vivo gene editing in Col7a1 c.6485G > A::Rosa26-stop-tdTomato mice. (B) PCR amplifications of the Col7a1 genomic DNA from FACS isolated in vivo tdTomato+ cells demonstrate truncation at exon80 after treatment. Partial Col7a1 gene sequencing result shows the deletion of exon80. (C) RT-PCR of Col7a1 mRNA from FACS isolated in vivo tdTomato+ cells indicates deletion of exon80, confirmed by sequencing result of the Col7a1 mRNA. (D) FACS analysis of the percentage of tdTomato+ cells in epidermis of Col7a1 c.6485G > A::Rosa26-stop-tdTomato mice at day 2 posttreatment. (Scale bar, 1 mm.) (E) Immunofluorescence stainings of Col VII, P-cad, and Nidogen in Col7a1 c.6485G > A mut/mut, wt/mut and WT tail skins with indicated electroporation treatments with either PBS or Cas9/sgRNA ribonucleoproteins targeting Col7a1 exon80. (Scale bar, 50 μm.) (F) Relative fluorescence intensity of Col VII at BMZ compared with lower dermis. Nidogen marks the location of BMZ and its quantification serves as a quantification control. (G) Percentage of skin with intact epidermis-to-dermis adhesion in different samples.
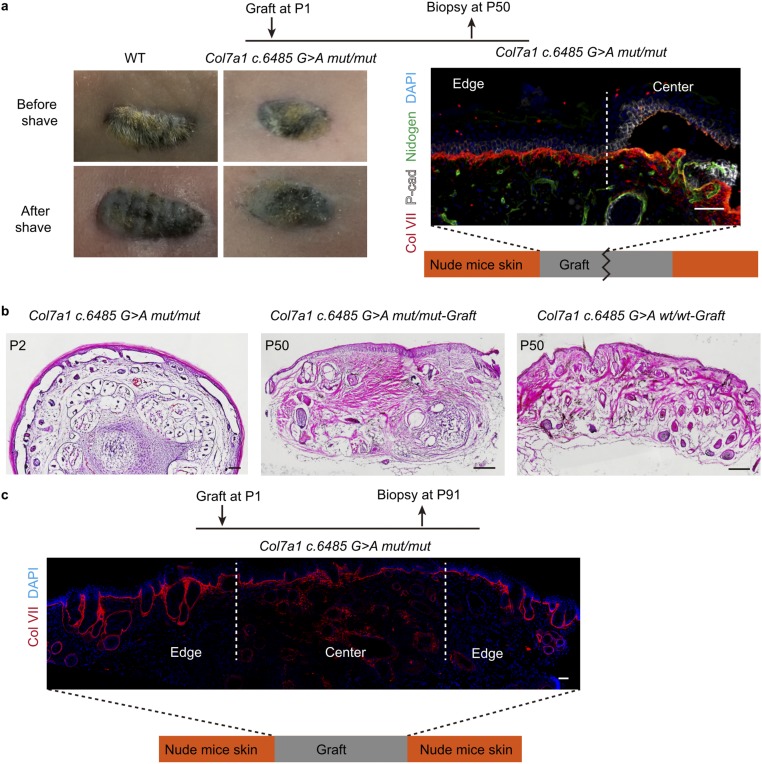
It is worth noting that the effects described above were observed only 3–5 d following a single treatment. Whether or not the corrective effect lasts over the long term is a key remaining question. And given the fact that we observed the long-term expansion of basal cell colonies following treatment, it will be interesting to see whether the correction efficiency may actually increase over time. To circumvent the problem of early postnatal lethality of the Col7a1 c.6485G > A mut/mut mice, we tried grafting the tail skin onto nude mice to pursue long-term analysis. Both WT and mutant skin grafts survived on the backs of nude mice. However, it became obvious that the epidermis-to-dermis adhesion defect in mutant skin was rescued on the backs of nude mice over time without treatment (Fig. S4 A–C). Immunofluorescence staining revealed the presence of mouse collagen VII protein at the BMZ close to the edge of the graft, and this pattern expanded even further into the center of the graft at later time points (Fig. S4 A and C). This phenomenon made the long-term characterization of the gene correction effect impossible in these grafted tissues. However, it offers additional evidence supporting that not all of the mutant cells need to be genetically corrected to functionally rescue RDEB, because the secreted functional collagen VII protein from gene-corrected cells can spread to neighboring tissues to rescue the skin blistering defect.
Fig. S4.
Col7a1 c.6485G > A mut/mut tail skin graft on nude mice back skin. (A) Representative images and section immunofluorescence staining of Col7a1 c.6485G > A mut/mut mice tail skin 50 d postgraft. (Scale bar, 50 μm.) (B) Representative H&E staining of Col7a1 c.6485G > A mut/mut mice tail skin at P2 before graft, as well as Col7a1 c.6485G > A mut/mut mice tail skin graft and WT tail skin at 50 d postgraft. (Scale bar, 200 μm.) (C) Representative immunofluorescence staining of WT and Col7a1 c.6485G > A mut/mut mice grafts after 91 d. (Scale bar, 50 μm.)
Discussion
Although several gene and cell therapies have been developed to deliver functional collagen VII protein to the RDEB tissue, no curative treatment exists for this disease as yet. In this study, we developed a therapeutic strategy based on the Cas9/sgRNA ribonucleoprotein-mediated gene editing system to restore collagen VII protein function in postnatal RDEB mice. The strategy creates an in-frame deletion of the genomic DNA covering exon80 that commonly harbors the recessive point mutation known to be responsible for the disease. Our results demonstrate that Cas9/sgRNA ribonucleoproteins mediate gene editing efficiently excising targeted genomic DNA in skin stem cells in postnatal animals. This restores the function of the collagen VII protein, correcting the epidermis–dermis adhesion defect.
The Cas9/sgRNA ribonucleoprotein-based gene therapy developed here has several advantages over traditional gene/cell therapies and virus-based CRISPR gene-editing systems. First, our Cas9/sgRNA ribonucleoprotein-based gene therapy works at the genomic level in stem cells in vivo. This implies long-term effectiveness in restoring the defective gene and should obviate the need for repeated treatment over time. Also, because this therapy works in intact somatic tissue in vivo, the complexity and full function of the tissue is maintained. These advantages would significantly reduce the cost and complications associated with traditional gene or cell therapies for RDEB. Second, using the nonreplicable protein/RNA complexes as gene editing system in vivo circumvents multiple drawbacks associated with virus-based delivery systems described earlier.
At the same time, several issues still remain for this in vivo gene editing system. First of all, our current electroporation system can only be applied to local skin areas, and the percentage of targeted cells after a single treatment still needs to be improved. Even though we do see significant effect in terms of restoration of correct collagen VII protein localization and improvement of skin adhesion in the treated area, further optimization of the delivery condition that can increase the target area as well as editing efficiency will have profound clinical impact. Secondly, whether or not this gene editing system can result in effective treatment over the long term needs to be addressed. Other skin disease mouse models that do not result in early postnatal lethality or do not involve secreted protein can circumvent our current technical problem. Additionally the human RDEB skin model generated ex vivo and grafted onto nude mice can be used to address the long-term effect question, and it will also provide a necessary system to study whether the condition developed here can also be applied to human skin tissue. Last but not least, even though the electroporation is performed immediately after ribonucleoprotein complex injection, the potential in vivo immunogenicity against the injected ribonucleoprotein complexes needs to be addressed when multiple injections might be needed to achieve sufficient genome editing in vivo.
In sum, our study provides proof-of-principle evidence that Cas9/sgRNA ribonucleoprotein-based gene therapies can be applied to restore collagen VII protein function in postnatal RDEB mice, suggesting that the Cas9/sgRNA ribonucleoprotein-based gene editing system may offer curative treatment for RDEB and other genetic disorders.
Materials and Methods
Detailed materials and methods are provided in SI Materials and Methods.
All mice were maintained in an specific pathogen-free (SPF) facility, and procedures were conducted in a manner consistent with the National Institute of Biological Sciences Guide for the Care and Use of Laboratory Animals. For the offspring identification, genotyping was performed using specific primers against mutant allele and WT allele, respectively. The 1.5-kb donor harboring the c.6485G > A mutation as well as a “CGACC” insertion within an intron was generated by PCR. The pSpCas9(BB)-2A-GFP (PX458, plasmid 48138) plasmid was purchased from Addgene. The CMV promoter-driven pLJM1-Cre plasmid was modified from the pLJM1-EGFP (Addgene, plasmid 19319) plasmid. The sgRNAs were designed using the on-line CRISPR design tool of Feng Zhang’s laboratory (crispr.mit.edu/). Primary keratinocytes were isolated from newborn back skin. Lipofectamine 2000 (Thermo Fisher Scientific) was used to transfect keratinocyte cells with plasmids. Within in vitro keratinocytes, Cas9/sgRNA complexes were electroporated with neon electroporation system (Thermo Fisher Scientific). T7 endonuclease I assay was used to detect sgRNA function in vitro. In vitro transfected keratinocytes and in vivo treated skin were analyzed and enriched by fluorescence-activated cell sorting with standard procedures. Genomic DNA was extracted with a TIAMamp genomic DNA kit (TIANGEN). Total RNA was isolated using a direct-Zol RNA mini prep kit (Zymo Research) and reverse transcripted into cDNA with reverse-transcriptase reaction mix (Takara). Single colony sequencing was performed to detect indels within genome and mRNA. Recombinant Cas9 was expressed using pET28-b(+)-Cas9 transformed into BL21(DE3). The Cas9 protein was purified using Ni-NTA agarose resin (Qiagen). SgRNA was transcripted using a MEGAscript T7 Transcription kit (Thermo Fisher Scientific) and purified with a Gel DNA Recovery kit (Zymo Research). In vivo mice electroporation experiments were performed with a square wave pulse generator (ECM 830). Standard cryosection fluorescence staining and hematoxylin and eosin (H&E) staining procedures were performed to detect protein expression in indicated tissue. Digital images were acquired by confocal microscope and processed by Imaris software. Each newborn Col7a1 c.6485G > a mut/mut mouse tail skin together with a WT littermate tail skin was grafted onto the same CD1 nude mouse back skin. Box-and-whisker plots were prepared with Origin 9 software (OriginLab), which was also used in the statistical analysis of data. GraphPad Prism 6 software was used to make the survival curves and histograms.
SI Materials and Methods
Mice.
All mice were maintained in an SPF facility, and procedures were conducted in a manner consistent with the National Institute of Biological Sciences Guide for the Care and Use of Laboratory Animals.
Donor Template Preparation for Knockin Mice Generation.
The original 1.5-kb template was amplified from the C57BL/6J mouse genome with primers: forward primer (Fw): GGGTGAGGCTTCCTACCCTA; reverse primer (Rv): CGACAAAGGACACGGGAGAA. QuikChange PCR was performed to generate the c.6485G > A mutation as well as a “CGACC” insertion within an intron with the primers: c.6485G > A-Fw: CCTTGTGCAGGGTCTTCAGGACCCAAGGGGGACCCCTGG; c.6485G > A-Rv: CCAGGGGTCCCCCTTGGGTCCTGAAGACCCTGCACAAGG; insertion-Fw: ATGTTTTTCATGGTCGTGGCATTTGGCACAGGATAGG, insertion-Rv: ATGTTTTTCATGGTCGTGGCATTTGGCACAGGATAGG.
PCR assays were performed with PrimeSTAR GXL DNA Polymerase kit (Takara) according to the manufacturer’s instructions. The PCR thermocycling program was as follows: 98 °C for 2 min; 35× (98 °C for 10 s, 60 °C for 15 s, and 68 °C for 1 min/1 kb); 68 °C for 7 min; followed by 4 °C. PCR products were analyzed by agarose gel electrophoresis and purified from the gel using a Gel DNA Recovery kit (Zymo Research). The original donor template PCR products were subcloned into the pMD-19T vector (Takara) according to the manufacturer’s instructions. Individual clones were picked and the inserted DNA was sequenced. QuikChange PCR was performed using the pMD-19T vector with the insertion as the template. Final donor templates were cut from the pMD-19T vector and purified for microinjection.
sgRNA Design.
The sgRNAs were designed by searching for “GG” or “CC” sequences near the sites of interest using the on-line CRISPR design tool of Feng Zhang’s laboratory (crispr.mit.edu/).
The sequence of sgRNAs for Col7a1 c.6485G > A and Col7a1 exon80 deletion mouse line generation was TTCCTTGTGCAGGGTCTTCA (GGG). The sequences of sgRNA for floxed transcription blocker deletion were (i) lsl-L: AAAGAATTGATTTGATACCG (CGG); (ii) lsl-R: GTATGCTATACGAAGTTATT (AGG).
Genotyping.
For founder mutant mice (F0), genomic DNA was extracted from the digits of pups, and the genomic sequences around the sgRNA target sites were PCR amplified using the primers: Fw: GGGTGAGGCTTCCTACCCTA; Rv: ATGACTGTCCCCTCCCTCTC. PCR products were subcloned into the pMD-19T vector (Takara) according to the manufacturer’s instructions. Individual clones were picked and the DNA was sequenced. Mutants were mated with C57BL/6J mice to generate F1 mutant mice. The genomic sequences around the sgRNA target sites of each F1 mouse were PCR amplified and sequenced, as descried above. Mutants with the intended mutations were then mated to generate the desired genotypes for the experimental analyses. For the offspring, genotyping was performed using premix Taq (Takara). For Col7a1 c.6485G > A mouse line genotyping, the primers c.6485G > A_WT-Fw: CCTGTGCCAAATGCCAATG and c.6485G > A_WT-Rv: GATGACTGTCCCCTCCCTCT were used to identify the wild-type allele. The primers c.6485G > A_mut-Fw: GCAGGTAATGTTTTTCATGGTCG and c.6485G > A_mut-Rv: GCAGGTAATGTTTTTCATGGTCG were used to identify the mutated allele. For Col7a1 ΔExon80 mouse line genotyping, the primers ΔExon80_WT-Fw: CCTTCCTTGTGCAGGGTCT and ΔExon80_WT-Rv: GATGACTGTCCCCTCCCTCT were used to identify the wild-type allele. The primers ΔExon80_mut-Fw: CCCTTCCTTGTGTCAGGGCC and ΔExon80_mut-Rv: GATGACTGTCCCCTCCCTCT were used to identify the mutated allele of col7a1 c.6485G > A mouse.
Primary Keratinocytes Cell Culture Experiment.
Primary keratinocytes were isolated from newborn back skin. Briefly, newborn back skin was removed and placed, dermis side down, in dispase (Life Technologies, 0.4 mg/mL in PBS) for 1 h at 37 °C. The epidermis was separated and digested by 0.25% trypsin solution (Gibco) at 37 °C until most of the keratinocytes had dissociated from the skin. To transfect keratinocyte cells with pSpCas9(BB)-2A-GFP (PX458), Lipofectamine 2000 Transfection Reagent (Thermo Fisher Scientific) was used according to the manufacturer’s instructions. The cells were incubated for 48–72 h before harvesting for further analysis. To obtain Cas9/gRNA complexes for electroporation, purified Cas9 protein was added to 10 μL of Resuspension Buffer R (Invitrogen), followed by addition of indicated gRNA to maintain a 1:1 ratio. The mixture was incubated at room temperature for 10 min, before being mixed with cells for electroporation using neon electroporation system (Thermo Fisher Scientific) according to the manufacturer’s instructions. After electroporation, cells were transferred immediately to a 24-well plate containing 0.5 mL of the keratinocyte growth medium and then incubated for 40 h in a 5% (vol/vol) CO2 incubator. The cells were harvested after incubation and subjected to fluorescence-activated cell sorting (FACS) analysis.
T7 Endonuclease I Assay.
Targeted genomic loci were amplified from genomic DNA using primers: Fw: GGGTGAGGCTTCCTACCCTA and Rv: ATGACTGTCCCCTCCCTCTC. The PCR was performed with a PrimeSTAR GXL DNA Polymerase kit (Takara). Hybridization PCR condition was as follows: 5 min, 95 °C; ramp down to 85 °C at −2 °C/s; ramp down to 25 °C at −0.1 °C/s; hold at 4 °C. T7 endo I (NEB) was added to the PCR and incubated at 37 °C for 15 min. A total of 0.25 M EDTA was added to stop the reaction, and products were loaded immediately on a 1.5% (wt/vol) agarose gel for analysis.
FACS.
For FACS isolation of transfected keratinocyte cells, cells were dissociated using 0.25% trypsin solution (Gibco). The mixture was incubated for 5 min at 37 °C, and warm keratinocyte growth medium supplemented with 10% FBS was added. The cells were centrifuged at 300 × g for 5 min at room temperature. The medium was removed and the cells were resuspended in 1× PBS supplemented with 5% (vol/vol) FBS. Cells were filtered into a cell strainer tube through its mesh cap. Sorted single cells were separated into microfuge tubes for further analysis. For isolation of genome-edited cells from the treated mice specified in the text, tail skin was removed, cut into two to three small pieces, and placed in collagenase for 1 h with shaking at 37 °C. Cell suspensions were then harvested and centrifuged at 300 × g for 5 min. Cell pellets were resuspended and further digested in a 0.25% trypsin solution (Gibco) at 37 °C for 30 min. Single-cell suspensions were obtained by triturating the skin gently. After neutralization with 5% FBS and spinning down, cells were resuspended with 5% FBS in PBS. After filtering with strainers (70 μm followed by 40 μm), sorted single cells were separated into microfuge tubes for further analysis. For quantifying percentage of tdTomato+ cells in the epidermis after electroporation treatment, the tdTomato+ area within tail skin was removed and treated with 20 mM EDTA at 37 °C for 10–15 min. Then epidermis was separated from dermis using tweezers under a stereoscopic microscope and then digested with 0.25% trypsin for 15 min. After neutralization, the digested cells were centrifuged for FACS analysis.
Isolation of Genomic DNA and mRNA from Sorted Cells.
Genomic DNA was extracted with a TIAMamp genomic DNA kit (TIANGEN) according to the manufacturer’s instructions. Total RNA was isolated from FACS-purified cells lysed with TRIzol (Life Technologies) followed by extraction using a direct-Zol RNA mini prep kit (Zymo Research). Equal amounts of RNA were added to reverse-transcriptase reaction mix (Takara) with Oligo(dT) to obtain cDNA for subsequence sequence analysis. Both genomic DNA, as well as cDNA, was measured with a NanoDrop instrument (Thermo Fisher Scientific).
Corrected Col7a1 Genomic DNA and mRNA Sequence Analysis.
Genomic DNA and cDNA PCR were performed with a PrimeSTAR GXL DNA Polymerase kit (Takara), according to manufacturer’s instructions. Primers were as follows: for genomic DNA PCR: Fw (GGGTGAGGCTTCCTACCCTA) and Rev (ATGACTGTCCCCTCCCTCTC); primers for cDNA used for in vitro gene-edited cells PCR: Fw (GGGGTGTACCAGGCATCAAA) and Rev (ATCTCCGTCCTTTCCACTGC); and primers for cDNA used for in vivo gene-edited cells PCR: Fw (AGGAGACAGGGGTGTACCAG); Rev (TCTCCATGGCTACCCGGCT, note this primer spans exon79 and exon81, omitting exon80). PCR products were purified via agarose gel electrophoresis and then subcloned into the pMD-19T vector for sequencing.
In Vitro Transcription of sgRNA.
The T7 promoter sequence was added to the sgRNA template by PCR. The gel-purified PCR products were used as templates for in vitro transcription using a MEGAscript T7 Transcription kit (Thermo Fisher Scientific). sgRNA templates were purified with a Gel DNA Recovery kit (Zymo Research) and eluted with nuclease-free water. The concentration of guide RNA was measured by a NanoDrop instrument (Thermo Fisher Scientific).
Recombinant Cas9 Protein Purification.
Recombinant Cas9 was purified using pET28-b(+)-Cas9 transformed into BL21(DE3), and protein expression was induced with 0.2 mM IPTG at 18 °C for 17 h. The Cas9 protein was purified using Ni-NTA agarose resin (Qiagen) and was dialyzed against stock solution (20 mM Hepes pH 7.5, 150 mM KCl, 1 mM DTT, and 50% glycerol).
Intradermal Injection and in Vivo Electroporation.
For adult mice experiments, mice were anesthetized using tribromoethanol. Before injection, plasmids were mixed with dynabeads (beads dilution ratio: 1/30; Invitrogen) to mark the electroporation area for later analysis. Plasmids in 1× DPBS (50 µL per site for back skin, 30 µL per site for tail skin, at concentrations as indicated for specific experiments) were intradermally injected using insulin syringes with a 29-gauge needle (BD) into the middle of the back skin or tail root. To keep variability to a minimum, the same skilled operator performed all injections. For Cas9-ribonucleoprotein electroporation, the molar ratio of gRNA-to-Cas9 protein was kept at approximately 1:1, and the molar ratio among the different sgRNAs was kept at 1. For each newborn, around 50 µg of purified recombination Cas9 protein was mixed with 5 µg total sgRNAs (2.5 µg/sgRNA) in a 20 µL volume with 1× DPBS. The Cas9/sgRNAs mixture was incubated at room temperature for 15 min. Before injection, the dynabeads (beads dilution ratio: 1/30; Invitrogen) were added into the Cas9/sgRNAs mixture to mark the electroporation area for later analysis. The in vivo electroporation was performed with a square wave pulse generator (ECM 830) and 5 mm-diameter tweezertrodes (45-0489, BTX). The distance between the two electrodes depends on the thickness of the skin fold or tail diameter. After intradermal injection, electroporation was performed with the following condition: 10 pulses of 50 ms each, 18 V/mm, 100-ms interval time. For the experiments with back skin, the electroporation was performed once. For tail skin, two electroporations were performed, with orientations perpendicular to each other.
Histology, Immunofluorescence, Confocal Microscopy, and Image Processing.
For section staining, tissues were embedded in O.C.T. compound (Tissue-Tek) and frozen. After cryosectioning (20–30 mm) and fixing for 10 min in 4% (vol/vol) paraformaldehyde in PBS, sections were permeabilized for 10 min in 0.5% Triton (PBST) and blocked for 1 h in a solution of 2% (vol/vol) normal donkey serum, 1% BSA, and 0.3% Triton in PBS. The following antibodies were used: anti-Collagen VII (Acris, AP23438PU-N; 1:100), anti–P-cad (R&D, BAF761; 1:500), anti-Nidogen (Santa Cruz, ELM1; 1:500), and anti-K14 (Chen Ting laboratory; 1:1,000). The primary antibodies were incubated overnight at 4 °C. The following secondary antibodies were used in the study: 488-anti-goat (Jackson Immuno Research; 1:1,000), 546-anti-rabbit (Jackson Immuno Research; 1:1,000), 647-anti-rat (Jackson Immuno Research; 1:1,000). The secondary antibodies were incubated at room temperature for 1 h. For H&E staining, after cryosectioning (10 μm) and fixation for 10 min in 4% (wt/vol) paraformaldehyde in PBS, sections were stained in hematoxylin (Sigma) for 20 s and then rinsed in running tap water and 0.3% acid alcohol. Eosin (Sigma) was used to treat the sections for 30 s. For whole mount tail skins, images were acquired using a fluorescence dissecting stereomicroscope (Leica, M165 FC). Section samples were imaged on a Nikon A1-R confocal microscope. H&E staining sections were imaged with a VS120 microscope (Olympus Life Science). Microscopy data were analyzed using Imaris (3D software) with the 3D visualization module. Red/blue/green (RBG) images were assembled and labeled with Adobe Illustrator CS6.
Collagen VII Protein Fluorescence Quantification Analysis.
At time points indicated in the text, tails were embedded into O.C.T. compound, with the anterior–posterior axis perpendicular to the bottom side of the embedding chamber. Collagen VII localization was analyzed using Fiji software. A rectangular box was drawn at the BMZ, according to the expression pattern of Nidogen. The total fluorescence intensity of collagen VII and the total fluorescence intensity of Nidogen was then measured. The same box was then moved to the lower dermis at a consistent distance, and measurements were repeated. The ratio between the fluorescence intensities at the two different locations was calculated.
Newborn Tail Skin Transplantation onto Nude Mice.
CD1 nude mice were anesthetized using tribromoethanol. For each newborn Col7a1 c.6485G > A mut/mut mouse tail skin, a WT littermate tail skin was also grafted onto the same nude mouse back skin as control. For each pair of grafts, two full thickness patches of skin from a nude mouse back skin (about 5 × 15 mm) were removed. Grafts were secured by sterile gauze and cloth bandages, which were removed after healing (10 d).
Software for Data Analysis.
Box-and-whisker plots were prepared with Origin 9 software (Origin Lab), which was also used in the statistical analysis of data. GraphPad Prism 6 software was used to make the survival curves and histograms.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1614775114/-/DCSupplemental.
References
- 1.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 3.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maddalo D, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516(7531):423–427. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yin H, et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32(6):551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swiech L, et al. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol. 2015;33(1):102–106. doi: 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiou SH, et al. Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing. Genes Dev. 2015;29(14):1576–1585. doi: 10.1101/gad.264861.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tabebordbar M, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351(6271):407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long C, et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351(6271):400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson CE, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351(6271):403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lunstrum GP, Sakai LY, Keene DR, Morris NP, Burgeson RE. Large complex globular domains of type VII procollagen contribute to the structure of anchoring fibrils. J Biol Chem. 1986;261(19):9042–9048. [PubMed] [Google Scholar]
- 12.Sakai LY, Keene DR, Morris NP, Burgeson RE. Type VII collagen is a major structural component of anchoring fibrils. J Cell Biol. 1986;103(4):1577–1586. doi: 10.1083/jcb.103.4.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uitto J, Pulkkinen L. Molecular complexity of the cutaneous basement membrane zone. Mol Biol Rep. 1996;23(1):35–46. doi: 10.1007/BF00357071. [DOI] [PubMed] [Google Scholar]
- 14.Burgeson RE. Type VII collagen, anchoring fibrils, and epidermolysis bullosa. J Invest Dermatol. 1993;101(3):252–255. doi: 10.1111/1523-1747.ep12365129. [DOI] [PubMed] [Google Scholar]
- 15.Bruckner-Tuderman L. Dystrophic epidermolysis bullosa: Pathogenesis and clinical features. Dermatol Clin. 2010;28(1):107–114. doi: 10.1016/j.det.2009.10.020. [DOI] [PubMed] [Google Scholar]
- 16.Remington J, et al. Injection of recombinant human type VII collagen corrects the disease phenotype in a murine model of dystrophic epidermolysis bullosa. Mol Ther. 2009;17(1):26–33. doi: 10.1038/mt.2008.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woodley DT, et al. Injection of recombinant human type VII collagen restores collagen function in dystrophic epidermolysis bullosa. Nat Med. 2004;10(7):693–695. doi: 10.1038/nm1063. [DOI] [PubMed] [Google Scholar]
- 18.Ortiz-Urda S, et al. Injection of genetically engineered fibroblasts corrects regenerated human epidermolysis bullosa skin tissue. J Clin Invest. 2003;111(2):251–255. doi: 10.1172/JCI17193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woodley DT, et al. Normal and gene-corrected dystrophic epidermolysis bullosa fibroblasts alone can produce type VII collagen at the basement membrane zone. J Invest Dermatol. 2003;121(5):1021–1028. doi: 10.1046/j.1523-1747.2003.12571.x. [DOI] [PubMed] [Google Scholar]
- 20.Wong T, et al. Potential of fibroblast cell therapy for recessive dystrophic epidermolysis bullosa. J Invest Dermatol. 2008;128(9):2179–2189. doi: 10.1038/jid.2008.78. [DOI] [PubMed] [Google Scholar]
- 21.Gache Y, et al. Construction of skin equivalents for gene therapy of recessive dystrophic epidermolysis bullosa. Hum Gene Ther. 2004;15(10):921–933. doi: 10.1089/hum.2004.15.921. [DOI] [PubMed] [Google Scholar]
- 22.Chen M, et al. Restoration of type VII collagen expression and function in dystrophic epidermolysis bullosa. Nat Genet. 2002;32(4):670–675. doi: 10.1038/ng1041. [DOI] [PubMed] [Google Scholar]
- 23.Ortiz-Urda S, et al. Stable nonviral genetic correction of inherited human skin disease. Nat Med. 2002;8(10):1166–1170. doi: 10.1038/nm766. [DOI] [PubMed] [Google Scholar]
- 24.Sebastiano V, et al. Human COL7A1-corrected induced pluripotent stem cells for the treatment of recessive dystrophic epidermolysis bullosa. Sci Transl Med. 2014;6(264):264ra163. doi: 10.1126/scitranslmed.3009540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wenzel D, et al. Genetically corrected iPSCs as cell therapy for recessive dystrophic epidermolysis bullosa. Sci Transl Med. 2014;6(264):264ra165. doi: 10.1126/scitranslmed.3010083. [DOI] [PubMed] [Google Scholar]
- 26.Umegaki-Arao N, et al. Induced pluripotent stem cells from human revertant keratinocytes for the treatment of epidermolysis bullosa. Sci Transl Med. 2014;6(264):264ra164. doi: 10.1126/scitranslmed.3009342. [DOI] [PubMed] [Google Scholar]
- 27.Bilousova G, Roop DR. Induced pluripotent stem cells in dermatology: Potentials, advances, and limitations. Cold Spring Harb Perspect Med. 2014;4(11):a015164. doi: 10.1101/cshperspect.a015164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferrari S, Pellegrini G, Matsui T, Mavilio F, De Luca M. Towards a gene therapy clinical trial for epidermolysis bullosa. Rev Recent Clin Trials. 2006;1(2):155–162. doi: 10.2174/157488706776876472. [DOI] [PubMed] [Google Scholar]
- 29.Kiuru M, Itoh M, Cairo MS, Christiano AM. Bone marrow stem cell therapy for recessive dystrophic epidermolysis bullosa. Dermatol Clin. 2010;28(2):371–382, xii–xiii. doi: 10.1016/j.det.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 30.Wagner JE, et al. Bone marrow transplantation for recessive dystrophic epidermolysis bullosa. N Engl J Med. 2010;363(7):629–639. doi: 10.1056/NEJMoa0910501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naldaiz-Gastesi N, et al. Identification and characterization of the dermal panniculus carnosus muscle stem cells. Stem Cell Reports. 2016;7(3):411–424. doi: 10.1016/j.stemcr.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang X, et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J Biotechnol. 2015;208:44–53. doi: 10.1016/j.jbiotec.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 33.Zuris JA, et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol. 2015;33(1):73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]