Significance
Photoreceptor proteins play a critical role in the light regulation of physiologically essential processes, including color vision, circadian rhythms, and photomovement. Photoexcitation of a chromophore bound to the photoreceptor protein leads to local conformational changes that propagate to distal regions of the protein and thereby drive vital chemical and physical changes. Understanding the fundamental mechanisms of these proteins is important for engineering systems that use light to control biological processes with high spatiotemporal resolution. In this study, computer simulations of a photoreceptor protein provide valuable insights into the active site conformations prior to and following photoexcitation. The position of a certain tryptophan residue, as well as the hydrogen-bonding pattern, is shown to impact the activation and efficiency of this photoreceptor.
Keywords: photoreceptor, BLUF, proton transfer, electron transfer, hydrogen bonding
Abstract
Blue light using flavin adenine dinucleotide (BLUF) proteins are essential for the light regulation of a variety of physiologically important processes and serve as a prototype for photoinduced proton-coupled electron transfer (PCET). Free-energy simulations elucidate the active site conformations in the AppA (activation of photopigment and puc expression) BLUF domain before and following photoexcitation. The free-energy profile for interconversion between conformations with either Trp104 or Met106 closer to the flavin, denoted Trpin/Metout and Trpout/Metin, reveals that both conformations are sampled on the ground state, with the former thermodynamically favorable by ∼3 kcal/mol. These results are consistent with the experimental observation of both conformations. To analyze the proton relay from Tyr21 to the flavin via Gln63, the free-energy profiles for Gln63 rotation were calculated on the ground state, the locally excited state of the flavin, and the charge-transfer state associated with electron transfer from Tyr21 to the flavin. For the Trpin/Metout conformation, the hydrogen-bonding pattern conducive to the proton relay is not thermodynamically favorable on the ground state but becomes more favorable, corresponding to approximately half of the configurations sampled, on the locally excited state. The calculated energy gaps between the locally excited and charge-transfer states suggest that electron transfer from Tyr21 to the flavin is more facile for configurations conducive to proton transfer. When the active site conformation is not conducive to PCET from Tyr21, Trp104 can directly compete with Tyr21 for electron transfer to the flavin through a nonproductive pathway, impeding the signaling efficiency.
Photoreceptor proteins are essential for the light regulation of a variety of physiologically important processes in living organisms, including color vision, circadian rhythms, and photomovement (1–3). Light excitation of a chromophore moiety bound to a photoreceptor protein leads to local conformational changes that subsequently propagate to distal parts of the protein and drive other chemical and physical changes, such as transmembrane proton pumping and protein–protein binding. Photoreceptor proteins have been divided into six broad classes (4). Rhodopsins, phytochromes, and xanthopsins each bind a different chromophore that undergoes photoinduced cis/trans isomerization. Phototropins, cryptochromes, and blue light using flavin adenine dinucleotide (BLUF) proteins each bind a flavin derivative as a chromophore that undergoes covalent bond formation with a cysteine residue, electron transfer (ET), and proton-coupled electron transfer (PCET), respectively, during the photocycle. Understanding the fundamental mechanisms of naturally occurring photoreceptor proteins is important for engineering novel systems that use light as a tool to achieve noninvasive control of biological processes with high spatiotemporal resolution (5–7).
BLUF proteins are critical for processes such as the light regulation of photosynthetic gene expression, phototaxis, and photophobia in bacteria (8). These proteins are unique among the photoreceptor proteins in that they exhibit photoinduced PCET and thus are powerful model systems for the investigation of this charge-transfer process in a protein environment. Due to the modular architecture of BLUF proteins, the N-terminal flavin-binding domain, often referred to as the BLUF domain, could potentially be fused to the C-terminal effector domains of various other proteins, leading to photocontrol of a variety of processes (9). Experimental studies have shown that photoexcitation of the flavin chromophore to an excited singlet state, denoted the locally excited state, induces ET followed by proton transfer (PT) from a nearby Tyr or Trp residue to flavin. This PCET process is followed by biradical recombination, leading to the formation of the light-adapted or signaling state (10, 11). The signaling state is formed in less than 10 ns after initial photoexcitation but can persist for up to tens of minutes. Subtle conformational changes in the active site region have been proposed to distinguish the signaling state from the initial dark-adapted state, based on the small 10-nm red shift of the flavin absorption spectrum and 10-cm−1 red shift of the stretching mode assigned to the flavin C4 = O4 carbonyl group in the signaling state (12).
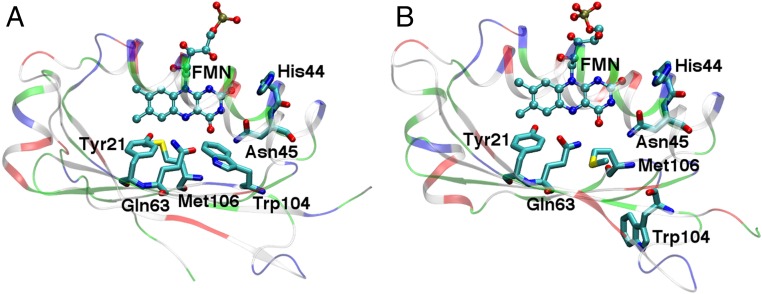
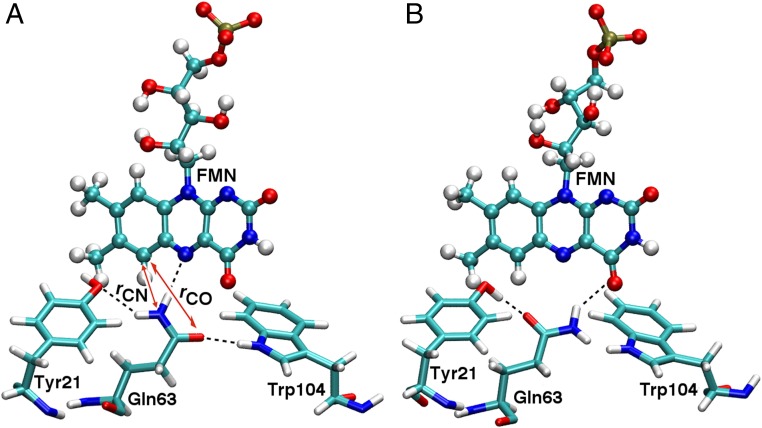
Despite numerous experimental and computational studies (8, 13–17), the exact nature of the signaling state and the mechanism of long-range signaling in BLUF proteins are not well understood. The active site conformation in even the dark-adapted state is still under debate because of discrepancies in the orientations of the key active site Trp, Met, and Gln residues among different experimental crystal and solution structures of BLUF domains (18–26). The Trp and Met residues can each be “in” or “out” of the region near the flavin mononucleotide (FMN), without simultaneously occupying the same position, resulting in the “Trpin/Metout” and “Trpout/Metin” conformations depicted in Fig. 1. Resolving these discrepancies is an important first step toward obtaining a comprehensive understanding of how long-range signaling occurs in BLUF proteins.
Fig. 1.
Protein scaffold and active site residues of the AppA BLUF domain in two X-ray crystallographic structures: (A) PDB ID code 1YRX, corresponding to the Trpin/Metout conformation and (B) PDB ID code 2IYG, corresponding to the Trpout/Metin conformation. The protein scaffold is colored by residue type: white, green, blue, and red regions represent nonpolar, neutral polar, positively charged, and negatively charged amino acid side chains, respectively. The FMN and key residues are depicted explicitly. This figure was generated using the Visual Molecular Dynamics (VMD) program (40).
Given the limited resolution of experiments in probing structural changes during the photocycle, several computational studies have been aimed at elucidating the changes in the active site hydrogen-bonding pattern during the photocycle. Quantum mechanical/molecular mechanical (QM/MM) geometry optimizations and vertical excitation energy calculations provided insight into the conformations consistent with experimental absorption data for the dark-adapted and signaling states (25). Other studies mostly used either conventional molecular dynamics trajectories or potential energy scans that were often conducted in gas-phase models of the active site (13–15, 27). The reliability of such approaches is somewhat limited by insufficient or no conformational sampling, especially given the complicated conformational landscapes of these proteins. The use of enhanced sampling techniques (16) also led to only qualitative results. Therefore, these prior calculations were not able to adequately resolve the debate regarding the active site conformation.
In this study, we use free-energy simulations to gain insight into the active site conformations of the Rhodobacter sphaeroides AppA (activation of photopigment and puc expression) (28) BLUF domain at equilibrium on the relevant electronic states. Specifically, we use the adaptively biased path optimization (ABPO) method (29) to calculate the minimum free-energy path and the associated free-energy profile for interconversion between the Trpin/Metout and Trpout/Metin conformations before photoexcitation of the AppA BLUF domain. Moreover, for each of these conformations, we calculate the free-energy profiles for the rotation and tautomerization of the active site Gln on the ground state, the locally excited state, and the charge-transfer state. Finally, we perform time-dependent density functional theory (TDDFT) (30) calculations to understand the effect of the active site hydrogen-bonding pattern on the feasibility of ET to the flavin. The results provide valuable insights into the impact of the active site hydrogen-bonding network on photocycle activation in the AppA BLUF domain.
Results and Discussion
Characterization of Excited Electronic States.
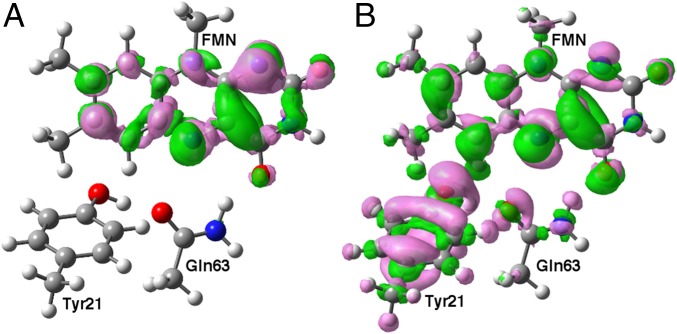
Absorption of a photon of blue light excites the flavin chromophore from the ground state (GS) to a bright locally excited (LE) singlet state with the electronic density change confined to the flavin. During the photocycle, this LE state decays at least partially to a charge-transfer (CT) state involving ET from Tyr21 to flavin. We performed TDDFT calculations with the CAM-B3LYP functional (31) on models of the BLUF domain active site to obtain insight into the nature of the LE and CT excited electronic states. The gas-phase model consisted of lumiflavin, p-cresol, and ethanamide, representing FMN, Tyr21, and Gln63, respectively. These calculations indicate that the CT state has virtually zero oscillator strength, whereas the lower-energy LE state has significant oscillator strength, consistent with photoexcitation to the LE state. Fig. 2 depicts the electronic density differences for the GS to LE state and GS to CT state vertical transitions in this model system. The LE state exhibits a shifting of electronic density within the flavin compared with the ground state, whereas the CT state clearly corresponds to the shifting of electronic density from Tyr21 to the flavin. Similar electronic density differences were observed for other gas-phase models as well as for the active site region in the presence of the protein and aqueous environment. Calculations of the energy gaps between the LE and CT excited electronic states for these systems will be discussed below.
Fig. 2.
Depiction of electronic density difference between (A) the LE and ground states and (B) the CT and ground states obtained from TDDFT calculations with the CAM-B3LYP functional and the 6-31+G** basis set for a gas-phase model consisting of lumiflavin, p-cresol, and ethanamide with initial coordinates obtained from FMN, Tyr21, and Gln63, respectively, in the AppA BLUF domain crystal structure 1YRX. Constrained geometry optimizations were performed with DFT/CAM-B3LYP/6–31+G** as described in SI Appendix. Pink and green regions represent decreased and increased electronic density upon vertical photoexcitation and correspond to isovalues of −0.0015 and 0.0015 Bohr−3, respectively.
Trp104 Conformation.
The orientation of Trp104 has been found to vary in the available experimental X-ray crystallographic and solution NMR structures of the AppA BLUF domain (Fig. 1). The crystal structure with Protein Data Bank (PDB) ID code 1YRX (18) has Trp104 positioned near FMN and Gln63, denoted the Trpin/Metout conformation, whereas the crystal structures with PDB ID codes 2IYG (21), 2IYI (21), 4HH0 (26), and 4HH1 (26) have Trp104 displaced away from the active site region with Met106 positioned near FMN, denoted the Trpout/Metin conformation. The solution NMR structure 2BUN (22) has 18 snapshots in the Trpin/Metout conformation and only 2 snapshots in the Trpout/Metin conformation.
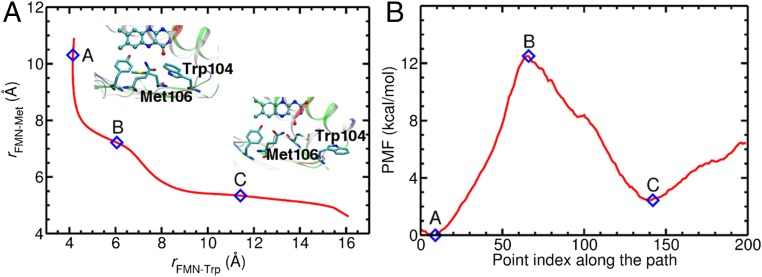
To resolve these discrepancies in the Trp104 and Met106 conformations, we calculated the minimum free-energy path (MFEP) and the associated free-energy profile for the change in Trp104 and Met106 conformations using the ABPO method. The MFEP was calculated as a function of the coordinates rFMN-Trp and rFMN-Met, which denote the O4(FMN)-NE1(Trp104) and O4(FMN)-SD(Met106) distances, respectively, where NE1 and SD represent the side-chain N and S atoms of Trp104 and Met106, respectively (Fig. 3). The MFEP associated with the interconversion between the Trpin/Metout and Trpout/Metin conformations is depicted in Fig. 4 (see SI Appendix, Figs. S1 and S2 for convergence analysis). Starting from the Trpin/Metout conformation, Met106 moves toward the active site to some extent before Trp104 starts moving “out” concertedly with Met106 moving “in.” This concerted movement continues until Met106 is almost completely in and is followed by Trp104 completing the rest of its outward motion. The free-energy profile along this path has a barrier of ∼12 kcal/mol for Trp104 to move out and Met106 to move in. The Trpin/Metout conformation is ∼3 kcal/mol lower in free energy than the Trpout/Metin conformation. Hence, at equilibrium at room temperature, the ground-state BLUF domain can undergo interconversion between the two states on a ∼100-μs timescale. This finding is consistent with the varying Trp104 conformation in different solution NMR and X-ray crystal structures of the AppA BLUF domain.
Fig. 3.
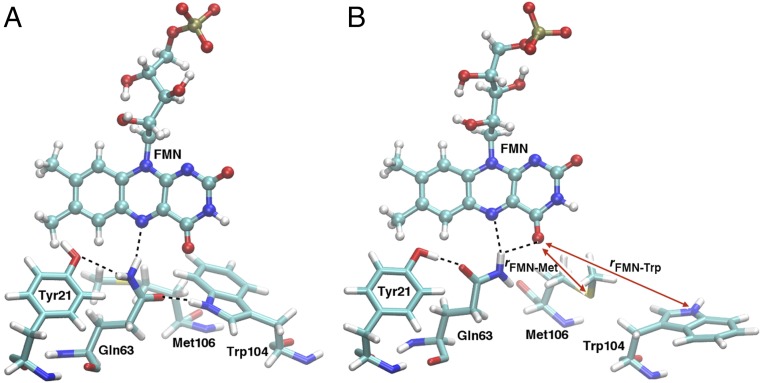
Representative snapshots depicting the (A) Trpin/Metout and (B) Trpout/Metin conformations. The red double-headed arrows labeled rFMN-Trp and rFMN-Met in B represent the O4(FMN)-NE1(Trp104) and O4(FMN)-SD(Met106) distances, respectively, that are used as reaction coordinates for the calculation of the MFEP and the free-energy profile for interconversion between the two conformations. Hydrogen-bonding interactions are depicted with dashed black lines.
Fig. 4.
Depiction of (A) the optimized MFEP and (B) the potential of mean force (PMF) associated with the interconversion between the Trpin/Metout (labeled “A”) and Trpout/Metin (labeled “C”) conformations. The MFEP is calculated as a function of the O4(FMN)-NE1(Trp104) and O4(FMN)-SD(Met106) distances, denoted as rFMN-Trp and rFMN-Met, respectively, depicted in Fig. 3B. Representative structures corresponding to the minima A and C are shown as insets and are also depicted in Fig. 3 A and B, respectively. The PMF in B is calculated along the MFEP shown in A.
Gln63 Conformation.
The conformation of the Gln63 side chain, as characterized by the positions of the side-chain O and N atoms of Gln63, is difficult to assign unambiguously based solely on X-ray diffraction data because of the similar atomic numbers of O and N. The two BLUF domain monomers in the 2IYI (21) structure differ in the assigned Gln63 side-chain conformation, despite similar Trp104 conformations (i.e., the Trpout/Metin conformation). Moreover, the solution NMR structure 2BUN (22) shows a highly flexible Gln63 side chain.
We calculated one-dimensional free-energy profiles for the rotation of the Gln63 side chain on the three electronic states of interest for both the Trpin/Metout and Trpout/Metin conformations. The partial atomic charges for the flavin and Tyr21 on the LE and CT states were obtained from TDDFT calculations on a gas-phase model system (SI Appendix, Fig. S3 and Table S1). These free-energy profiles were calculated as functions of rCN−rCO, with rCN and rCO denoting the C6(FMN)-NE2(Gln63) and C6(FMN)-OE1(Gln63) distances, respectively, where NE2(Gln63) and OE1(Gln63) represent the side-chain N and O atoms of Gln63 (Fig. 5). We found that the orientation of the hydroxyl group of Tyr21 changes in conjunction with the Gln63 rotation. Negative values of the reaction coordinate are associated with the Tyr21 hydroxyl H atom pointing away from Gln63 because the Tyr21 hydroxyl O atom accepts a hydrogen bond from NE2(Gln63). In this conformation, Tyr21 is poorly oriented for PT to either Gln63 or FMN, as depicted in Fig. 5A. In contrast, positive values of the reaction coordinate are associated with the Tyr21 hydroxyl H atom pointing toward Gln63 because OE1(Gln63) accepts a hydrogen bond from Tyr21. In this conformation, Tyr21 is well-oriented for PT to Gln63 and Gln63 is well-oriented for PT to FMN, according to the proton relay depicted in Fig. 5B.
Fig. 5.
Representative snapshots depicting two different Gln63 and Tyr21 orientations in the AppA BLUF domain in the Trpin/Metout conformation. In A, Tyr21 accepts a hydrogen bond from Gln63 and Trp104 donates a hydrogen bond to Gln63. In B, Tyr21 donates a hydrogen bond to Gln63 and Trp104 does not have a hydrogen-bonding partner for its side-chain N-H. The red double-headed arrows labeled “rCN” and “rCO” in A represent the C6(FMN)-NE2(Gln63) and C6(FMN)-OE1(Gln63) distances, respectively, that are used to define the reaction coordinate, rCN−rCO, for the calculation of the free-energy profile associated with Gln63 rotation. Note that A and B correspond to negative and positive values, respectively, of this reaction coordinate.
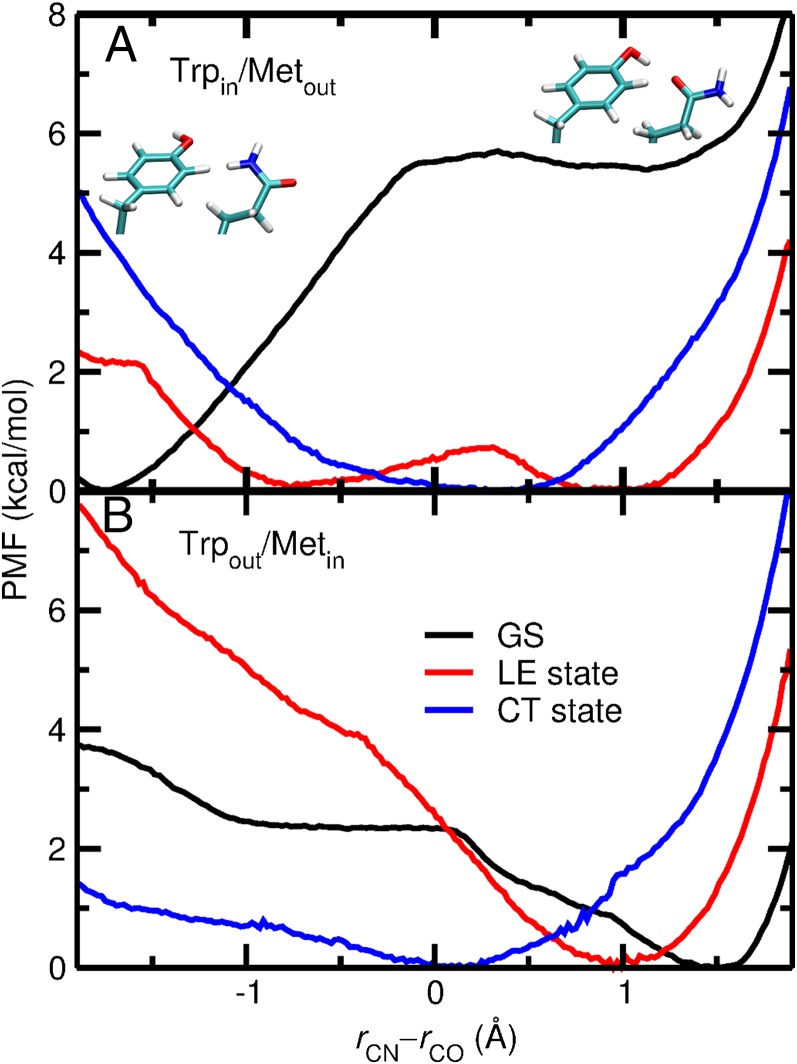
The free-energy profiles for Gln63 rotation are shown in Fig. 6. For the ground state, the conformation depicted in Fig. 5A is thermodynamically favorable by ∼5 kcal/mol in the Trpin/Metout conformation, whereas the conformation depicted in Fig. 5B is thermodynamically favorable by ∼2 kcal/mol in the Trpout/Metin conformation (black curves in Fig. 6 A and B). Thus, in the Trpin/Metout conformation, the Tyr21 side-chain conformation is less suitable for PT to Gln63 and, via the proton relay, to FMN. In the Trpout/Metin conformation, Tyr21 remains well-oriented for the proton relay. The free-energy barriers for Gln63 rotation are, however, only ∼6 kcal/mol and ∼2 kcal/mol with Trp104 in and out, respectively.
Fig. 6.
Free-energy profiles for Gln63 rotation on the ground, LE, and CT states in the (A) Trpin/Metout and (B) Trpout/Metin conformations in the AppA BLUF domain. The reaction coordinate corresponds to the difference between the C6(FMN)-NE1(Gln63) and C6(FMN)-OE2(Gln63) distances denoted as rCN and rCO, respectively, depicted in Fig. 5A. Negative and positive values of the reaction coordinate are associated with the qualitative hydrogen-bonding configurations depicted in Fig. 5 A and B, respectively, with orientations of Tyr21 and Gln63 depicted as insets in A.
The free-energy profiles are different on the two excited electronic states. In the Trpout/Metin conformation, Tyr21 remains well-oriented for PT on both excited electronic states (red and blue curves in Fig. 6B). In the Trpin/Metout conformation, two minima separated by a small barrier of ∼0.5 kcal/mol are observed for the LE state (red curve in Fig. 6A). Thus, Gln63 and Tyr21 can easily sample both conformations at room temperature on the LE state. Because the LE and ground states have similar electronic density distributions, equilibration on the LE state after photoexcitation is very fast, and the free-energy profile at equilibrium on the LE state is relevant to the dynamics following photoexcitation. For the CT state, the minimum of the free-energy profile is found to be at a value close to zero irrespective of the Trp104 conformation (blue curves in Fig. 6 A and B) because the negatively charged flavin moiety on the CT state pushes the partially negatively charged Gln63 side-chain O atom away. However, this minimum corresponds to geometries in which Tyr21 donates a hydrogen bond to the Gln63 side-chain O atom and hence is well-oriented for PT, as depicted in Fig. 5B. Thus, the CT state is consistently well-oriented for PT from Tyr21 to Gln63 for both Trp104 conformations.
The free-energy profiles in Fig. 6 are qualitatively consistent with the Gln63 and Tyr21 side-chain flexibility observed in 20-ns unrestrained MD trajectories (SI Appendix, Fig. S4). Moreover, these unrestrained MD trajectories illustrate that the conformational changes involving Gln63 and Tyr21 occur on a timescale much faster than 20 ns. In contrast, the conformational change between Trpin/Metout and Trpout/Metin was not observed during these unrestrained MD trajectories, suggesting that the conformational change involving Trp104 and Met106 occurs on a slower timescale, as also indicated by the free-energy barrier in Fig. 4. Thus, the MFEP in Fig. 4 is averaged over the accessible Gln63 and Tyr21 conformations for each Trp104 and Met106 conformation.
The tautomerization of Gln63 has been proposed to be involved in the BLUF photocycle in several studies in the literature (13, 15, 32). Our QM/MM free-energy calculations (SI Appendix, Figs. S5 and S6) indicate that the tautomerization of Gln63 to the enol form is unfavorable with a free-energy barrier of ∼40 kcal/mol at equilibrium before PT on the ground, LE, and CT states. Thus, this tautomerization does not need to be considered before PT and is not directly relevant to the present study. However, tautomerization of Gln63 may become relevant in the later stages of the photocycle (32).
Feasibility of ET from Tyr21 to FMN.
As discussed above, photoexcitation leads to the LE state, which subsequently crosses to the CT state via a nearby avoided crossing or conical intersection (27). Fig. 2 depicts electronic density differences for the LE and CT states relative to the GS for a gas-phase model consisting of p-cresol, ethanamide, and lumiflavin, representing Tyr21, Gln63, and FMN, respectively. We calculated the energy gap between the LE and CT states at the Franck–Condon geometry, defined as the partially optimized crystal structure in this case. The results for a series of gas phase models of varying sizes are provided in SI Appendix, Fig. S7 and Table S2 and are qualitatively consistent among all models containing a Gln63 analog. Additional calculations using an electrostatic embedding scheme to include the protein and solvent environment indicate that the full protein and aqueous environment does not qualitatively impact the vertical excitation energies of the LE and CT states as well as the CT-LE energy gap for different active site hydrogen-bonding patterns (SI Appendix, Table S4). The ordering of the LE and CT states predicted by the TDDFT/CAM-B3LYP calculations in this study is in agreement with previous second-order perturbation theory multiconfigurational calculations on a related model system (33) and DFT/multireference configuration interaction calculations including the protein environment at the molecular mechanical level (25).
These calculations illustrate that the energy gap between the LE and CT states depends strongly on the hydrogen-bonding pattern. For the models in which the phenolic OH group of the Tyr21 analog does not have the proper orientation for PT to the amide O of the Gln63 analog (Fig. 5A), the CT state is ∼1.2−1.6 eV higher in energy than the LE state at the Franck–Condon geometry. On the other hand, for the models in which the Tyr21 analog donates a hydrogen bond to the amide O of the Gln63 analog and has the proper conformation required for PT (Fig. 5B), the CT state is only ∼0.2−0.6 eV higher in energy than the LE state at the Franck–Condon geometry. This difference can be understood in terms of the electronic density distributions of the LE and CT states (Fig. 2). ET from the Tyr21 analog to the FMN analog is more favorable when the dipoles associated with the Gln63 amide group and the Tyr21 hydroxyl group are aligned in the opposite direction as the dipole on FMN in the CT state, thereby providing electrostatic stabilization. These calculations indicate that the hydrogen-bonding pattern in the active site of the BLUF domain affects not only the feasibility of PT from Tyr21 to FMN, but also the feasibility of ET from Tyr21 to FMN.
Comparison with Available Experimental Data.
Experimentally, the photocycle of the AppA BLUF domain has been found to have essentially the same features as that of the full AppA protein (34). It shows multiexponential decay kinetics following photoexcitation that could arise from conformational heterogeneity in the ground-state ensemble and/or multiple decay pathways (35). Our free-energy calculations indicate that the active site region of the AppA BLUF domain does indeed have significant conformational flexibility in the ground state, characterized by conformational changes of residues Tyr21, Gln63, Trp104, and Met106. This conformational flexibility is consistent with the variety of active site conformations observed in different X-ray crystallographic and solution NMR structures of the AppA BLUF domain (18, 21, 22, 26) and may be at least partially responsible for the multiexponential decay kinetics after photoexcitation.
Our data suggest that in the Trpin/Metout conformation, Tyr21 can transfer an electron to FMN more easily from conformations suitable for PT from Tyr21 to Gln63 because only this hydrogen-bonding pattern provides facile accessibility to the CT state. The Trpin/Metout conformation also samples hydrogen-bonding patterns that are not suitable for PT and therefore cannot readily access the CT state. In such conformations, Trp104 can compete with Tyr21 for ET to FMN, as indicated by our TDDFT calculations on model systems (SI Appendix, Fig. S8 and Table S3). These calculations illustrate that the CT state associated with ET from Trp104 to FMN is lower in energy than the CT state associated with ET from Tyr21 to FMN at the Franck–Condon geometry when the hydrogen-bonding pattern involving Tyr21 and Gln63 is not suitable for PT. The possible competition from Trp104 has also been deduced from experimental studies that detected formation of a Trp104 radical during the photocycle (10). As further experimental evidence, the mutation of Trp104 to Phe has been observed to lead to a greater quantum yield of the signaling state, indicating that Trp104 provides a nonproductive competing decay pathway. Trp fluorescence experiments have also indicated that Trp104 remains buried in the AppA BLUF domain active site throughout the photocycle (36). The significant population of the Trpin/Metout conformation found in our simulations is consistent with these experimental observations.
Further support for our results is provided by studies of the Slr694 BLUF domain. In this system, 9 out of the 10 subunits in the experimental crystal structure 3MZI (23) exhibit the Trpout/Metin conformation, and the photocycle is much faster than in the AppA BLUF domain (37). According to our calculations, the Trpout/Metin conformation in Slr1694 would lead to an orientation of Tyr21 that is well-suited for both ET and PT to FMN and facilitates the photocycle.
Conclusion
In this work, free-energy simulations were used to address important questions regarding the active site conformations in the AppA BLUF domain before and following photoexcitation of the flavin chromophore. We calculated the MFEP and the associated free-energy profile for interconversion between the in and out conformations of Trp104 and Met106, whose orientations have been debated for the AppA as well as other BLUF domains even in the dark-adapted state. Our calculations reveal a modest free-energy difference of ∼3 kcal/mol between the Trpin/Metout and Trpout/Metin conformations, slightly favoring the former, and a free-energy barrier of ∼12 kcal/mol for interconversion on the ground state, allowing both conformations to be sampled at equilibrium on the ground state. Our results are consistent with the observation of both conformations in the various experimental X-ray crystallographic and solution NMR structures of the AppA BLUF domain (18, 21, 22, 26) and support the hypothesis that conformational heterogeneity in the ground-state ensemble could be partially responsible for the multiexponential decay kinetics observed after photoexcitation (35).
Our calculated free-energy profiles for Gln63 rotation in both the Trpin/Metout and Trpout/Metin conformations on three different electronic states provide information about the hydrogen-bonding patterns in the active site. For the Trpin/Metout conformation, Gln63 and Tyr21 are not properly oriented for the proton relay from Tyr21 to FMN via Gln63 on the ground state. After photoexcitation to the LE state, the conformations with and without the hydrogen-bonding pattern conducive to the proton relay become approximately equally probable, separated by a free-energy barrier of only ∼0.5 kcal/mol. On the other hand, for the Trpout/Metin conformation, the hydrogen-bonding pattern conducive to the proton relay is thermodynamically favorable on both the ground state and the LE state.
The conformations of Tyr21, Gln63, and Trp104 have important implications for the photocycle. Our TDDFT calculations indicate that the energy gap between the LE and CT states is significantly greater when the hydrogen-bonding pattern is not suitable for the proton relay. This finding implies that after photoexcitation to the LE state, ET from Tyr21 to FMN will be accessible predominantly when the hydrogen-bonding pattern is conducive to the proton relay, namely for approximately half of the configurations sampled in the Trpin/Metout conformation. Furthermore, when Trp104 is sufficiently close to FMN, it can compete directly with Tyr21 for ET to FMN, especially if ET from Tyr21 is hindered by the absence of the conducive hydrogen-bonding network. Thus, occupation of the Trpin/Metout conformation is predicted to hamper the signaling efficiency, which requires ET from Tyr21.
In summary, this study has revealed several fundamental and crucial features of the AppA BLUF domain active site hydrogen-bonding network in relation to the photocycle. The knowledge gained from this work lays the foundation for future studies of proton transfer pathways and signaling state formation in the AppA BLUF domain, which in turn will pave the way for understanding the mechanism of long-range signaling by this photoreceptor.
Materials and Methods
Details of the computational methodology used in this article are provided in the SI Appendix. These methods include descriptions of the classical molecular dynamics simulations, hybrid QM/MM simulations, and TDDFT calculations. The Gaussian 09 (38) and CHARMM (39) software packages were used for the calculations.
Supplementary Material
Acknowledgments
This material is based upon work supported by the Air Force Office of Scientific Research (AFOSR) under AFOSR Award FA9550-14-1-0295. This research is part of the Blue Waters Sustained-Petascale Computing Project, which is supported by the National Science Foundation (Awards OCI-0725070 and ACI-1238993) and the state of Illinois. Blue Waters is a joint effort of the University of Illinois at Urbana–Champaign and its National Center for Supercomputing Applications.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1621393114/-/DCSupplemental.
References
- 1.Armitage JP, Hellingwerf KJ. Light-induced behavioral responses (’phototaxis’) in prokaryotes. Photosynth Res. 2003;76(1-3):145–155. doi: 10.1023/A:1024974111818. [DOI] [PubMed] [Google Scholar]
- 2.van der Horst MA, Hellingwerf KJ. Photoreceptor proteins, “star actors of modern times”: A review of the functional dynamics in the structure of representative members of six different photoreceptor families. Acc Chem Res. 2004;37(1):13–20. doi: 10.1021/ar020219d. [DOI] [PubMed] [Google Scholar]
- 3.Paul KN, Saafir TB, Tosini G. The role of retinal photoreceptors in the regulation of circadian rhythms. Rev Endocr Metab Disord. 2009;10(4):271–278. doi: 10.1007/s11154-009-9120-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conrad KS, Manahan CC, Crane BR. Photochemistry of flavoprotein light sensors. Nat Chem Biol. 2014;10(10):801–809. doi: 10.1038/nchembio.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ziegler T, Möglich A. Photoreceptor engineering. Front Mol Biosci. 2015;2:30. doi: 10.3389/fmolb.2015.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ziegler T, Schumacher CH, Möglich A. Guidelines for photoreceptor engineering. Methods Mol Biol. 2016;1408:389–403. doi: 10.1007/978-1-4939-3512-3_27. [DOI] [PubMed] [Google Scholar]
- 7.Ganguly A, et al. Changes in active site histidine hydrogen bonding trigger cryptochrome activation. Proc Natl Acad Sci USA. 2016;113(36):10073–10078. doi: 10.1073/pnas.1606610113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kennis JTM, Mathes T. Molecular eyes: Proteins that transform light into biological information. Interface Focus. 2013;3(5):20130005. doi: 10.1098/rsfs.2013.0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han Y, Braatsch S, Osterloh L, Klug G. A eukaryotic BLUF domain mediates light-dependent gene expression in the purple bacterium Rhodobacter sphaeroides 2.4.1. Proc Natl Acad Sci USA. 2004;101(33):12306–12311. doi: 10.1073/pnas.0403547101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gauden M, et al. On the role of aromatic side chains in the photoactivation of BLUF domains. Biochemistry. 2007;46(25):7405–7415. doi: 10.1021/bi7006433. [DOI] [PubMed] [Google Scholar]
- 11.Laptenok SP, et al. Electron transfer quenching in light adapted and mutant forms of the AppA BLUF domain. Faraday Discuss. 2015;177:293–311. doi: 10.1039/c4fd00189c. [DOI] [PubMed] [Google Scholar]
- 12.Masuda S, Hasegawa K, Ono TA. Light-induced structural changes of apoprotein and chromophore in the sensor of blue light using FAD (BLUF) domain of AppA for a signaling state. Biochemistry. 2005;44(4):1215–1224. doi: 10.1021/bi047876t. [DOI] [PubMed] [Google Scholar]
- 13.Sadeghian K, Bocola M, Schütz M. A conclusive mechanism of the photoinduced reaction cascade in blue light using flavin photoreceptors. J Am Chem Soc. 2008;130(37):12501–12513. doi: 10.1021/ja803726a. [DOI] [PubMed] [Google Scholar]
- 14.Sadeghian K, Bocola M, Schütz M. A QM/MM study on the fast photocycle of blue light using flavin photoreceptors in their light-adapted/active form. Phys Chem Chem Phys. 2010;12(31):8840–8846. doi: 10.1039/b925908b. [DOI] [PubMed] [Google Scholar]
- 15.Udvarhelyi A, Domratcheva T. Glutamine rotamers in BLUF photoreceptors: A mechanistic reappraisal. J Phys Chem B. 2013;117(10):2888–2897. doi: 10.1021/jp400437x. [DOI] [PubMed] [Google Scholar]
- 16.Meier K, van Gunsteren WF. On the use of advanced modelling techniques to investigate the conformational discrepancy between two X-ray structures of the AppA BLUF domain. Mol Simul. 2013;39(6):472–486. [Google Scholar]
- 17.Mathes T, Götze JP. A proposal for a dipole-generated BLUF domain mechanism. Front Mol Biosci. 2015;2:62. doi: 10.3389/fmolb.2015.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson S, et al. Structure of a novel photoreceptor, the BLUF domain of AppA from Rhodobacter sphaeroides. Biochemistry. 2005;44(22):7998–8005. doi: 10.1021/bi0502691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jung A, et al. Structure of a bacterial BLUF photoreceptor: Insights into blue light-mediated signal transduction. Proc Natl Acad Sci USA. 2005;102(35):12350–12355. doi: 10.1073/pnas.0500722102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kita A, Okajima K, Morimoto Y, Ikeuchi M, Miki K. Structure of a cyanobacterial BLUF protein, Tll0078, containing a novel FAD-binding blue light sensor domain. J Mol Biol. 2005;349(1):1–9. doi: 10.1016/j.jmb.2005.03.067. [DOI] [PubMed] [Google Scholar]
- 21.Jung A, Reinstein J, Domratcheva T, Shoeman RL, Schlichting I. Crystal structures of the AppA BLUF domain photoreceptor provide insights into blue light-mediated signal transduction. J Mol Biol. 2006;362(4):717–732. doi: 10.1016/j.jmb.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 22.Grinstead JS, et al. The solution structure of the AppA BLUF domain: Insight into the mechanism of light-induced signaling. ChemBioChem. 2006;7(1):187–193. doi: 10.1002/cbic.200500270. [DOI] [PubMed] [Google Scholar]
- 23.Yuan H, et al. Crystal structures of the Synechocystis photoreceptor Slr1694 reveal distinct structural states related to signaling. Biochemistry. 2006;45(42):12687–12694. doi: 10.1021/bi061435n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu Q, Gardner KH. Structure and insight into blue light-induced changes in the BlrP1 BLUF domain. Biochemistry. 2009;48(12):2620–2629. doi: 10.1021/bi802237r. [DOI] [PubMed] [Google Scholar]
- 25.Hsiao Y-W, Götze JP, Thiel W. The central role of Gln63 for the hydrogen bonding network and UV-visible spectrum of the AppA BLUF domain. J Phys Chem B. 2012;116(28):8064–8073. doi: 10.1021/jp3028758. [DOI] [PubMed] [Google Scholar]
- 26.Winkler A, et al. A ternary AppA-PpsR-DNA complex mediates light regulation of photosynthesis-related gene expression. Nat Struct Mol Biol. 2013;20(7):859–867. doi: 10.1038/nsmb.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Udvarhelyi A, Domratcheva T. Photoreaction in BLUF receptors: Proton-coupled electron transfer in the flavin-Gln-Tyr system. Photochem Photobiol. 2011;87(3):554–563. doi: 10.1111/j.1751-1097.2010.00884.x. [DOI] [PubMed] [Google Scholar]
- 28.Gomelsky M, Kaplan S. appA, a novel gene encoding a trans-acting factor involved in the regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J Bacteriol. 1995;177(16):4609–4618. doi: 10.1128/jb.177.16.4609-4618.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dickson BM, Huang H, Post CB. Unrestrained computation of free energy along a path. J Phys Chem B. 2012;116(36):11046–11055. doi: 10.1021/jp304720m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Casida ME, Huix-Rotllant M. Progress in time-dependent density-functional theory. Annu Rev Phys Chem. 2012;63:287–323. doi: 10.1146/annurev-physchem-032511-143803. [DOI] [PubMed] [Google Scholar]
- 31.Yanai T, Tew DP, Handy NC. A new hybrid exchange-correlation functional using the Coulomb-attenuating method (CAM-B3LYP) Chem Phys Lett. 2004;393(1-3):51–57. [Google Scholar]
- 32.Domratcheva T, Hartmann E, Schlichting I, Kottke T. Evidence for tautomerisation of glutamine in BLUF blue light receptors by vibrational spectroscopy and computational chemistry. Sci Rep. 2016;6:22669. doi: 10.1038/srep22669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Udvarhelyi A, Olivucci M, Domratcheva T. Role of the molecular environment in flavoprotein color and redox tuning: QM cluster versus QM/MM modeling. J Chem Theory Comput. 2015;11(8):3878–3894. doi: 10.1021/acs.jctc.5b00197. [DOI] [PubMed] [Google Scholar]
- 34.Kraft BJ, et al. Spectroscopic and mutational analysis of the blue-light photoreceptor AppA: A novel photocycle involving flavin stacking with an aromatic amino acid. Biochemistry. 2003;42(22):6726–6734. doi: 10.1021/bi030055o. [DOI] [PubMed] [Google Scholar]
- 35.Dragnea V, Waegele M, Balascuta S, Bauer C, Dragnea B. Time-resolved spectroscopic studies of the AppA blue-light receptor BLUF domain from Rhodobacter sphaeroides. Biochemistry. 2005;44(49):15978–15985. doi: 10.1021/bi050839x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toh KC, et al. On the signaling mechanism and the absence of photoreversibility in the AppA BLUF domain. Biophys J. 2008;95(1):312–321. doi: 10.1529/biophysj.107.117788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonetti C, et al. Hydrogen bond switching among flavin and amino acid side chains in the BLUF photoreceptor observed by ultrafast infrared spectroscopy. Biophys J. 2008;95(10):4790–4802. doi: 10.1529/biophysj.108.139246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frisch MJ, et al. Gaussian 09, Revision E.01. Gaussian, Inc.; Wallingford, CT: 2009. [Google Scholar]
- 39.Brooks BR, et al. CHARMM: The biomolecular simulation program. J Comput Chem. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14(1):33–38, 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.