Significance
We established human embryonic stem (hES) cell-inducible lines to express specific transcription factors [GS Homeobox 2 (Gsx2) and Early B-cell factor 1 (Ebf1)] to improve medium spiny neuron (MSN) differentiation and to study human striatal development in vitro. We also used a nonintegrating system, by utilizing modified mRNAs to transiently overexpress Gsx2 and Ebf1 in a different hES cell line. These data can help to improve the differentiation protocols that aim to produce high-quality cell preparation suitable for cell transplantation in animal models of Huntington’s disease. Finally, we show that hES-derived striatal progenitors can be transplanted in the striatum of animal models and can differentiate and integrate into the host, extending fibers over a long distance (including the substantia nigra, a striatal target).
Keywords: MSNs, Gsx2, Ebf1, hES cells, HD
Abstract
Medium spiny neurons (MSNs) are a key population in the basal ganglia network, and their degeneration causes a severe neurodegenerative disorder, Huntington's disease. Understanding how ventral neuroepithelial progenitors differentiate into MSNs is critical for regenerative medicine to develop specific differentiation protocols using human pluripotent stem cells. Studies performed in murine models have identified some transcriptional determinants, including GS Homeobox 2 (Gsx2) and Early B-cell factor 1 (Ebf1). Here, we have generated human embryonic stem (hES) cell lines inducible for these transcription factors, with the aims of (i) studying their biological role in human neural progenitors and (ii) incorporating TF conditional expression in a developmental-based protocol for generating MSNs from hES cells. Using this approach, we found that Gsx2 delays cell-cycle exit and reduces Pax6 expression, whereas Ebf1 promotes neuronal differentiation. Moreover, we found that Gsx2 and Ebf1 combined overexpression in hES cells achieves high yields of MSNs, expressing Darpp32 and Ctip2, in vitro as well in vivo after transplantation. We show that hES-derived striatal progenitors can be transplanted in animal models and can differentiate and integrate into the host, extending fibers over a long distance.
The striatum is the largest component of the basal ganglia, it is the hub of converging excitatory connections from the cortex and thalamus, and it originates the direct and indirect pathways, which are distinct basal ganglia circuits involved in motor control (1). In humans, the degeneration of the principal striatal neuronal population, the medium spiny neurons (MSNs), causes a severe neurodegenerative condition, Huntington’s disease (HD). A main goal in the field is the study of the mechanisms underlying neuronal specification and degeneration. A large number of studies performed in model organisms, such as the mouse model organism, have provided fundamental insights into brain development, shedding light on genes, signaling pathways, and general rules of brain formation. It is not incidental to point out that obvious species-specific differences exist in many aspects between mice and humans (gestation, morphology, and gene expression regulation in time and space). Thus, additional model systems are needed to uncover specific functions of a gene in human development (2, 3). This task is also driven by the need to investigate neurological diseases, such as HD, in a model that more closely resembles human biology.
Here, we decided to take advantage of human embryonic stem (hES) cells to develop a model to study the roles of selected transcription factors (TFs) in human striatal development and as a strategy to increase recovery of authentic MSNs for transplantation purposes. During brain development, a set of TFs are expressed in different regions and times and cooperate to establish a dorsal–ventral and medial–lateral positional identity in progenitor cells and to specify neuronal terminal differentiation. In particular, in the developing telencephalon, two TFs play a key role in contributing to the formation of the striatum: the GS Homeobox 2 (Gsx2) and Early B-cell factor 1 (Ebf1).
Gsx2 is expressed in the ventral ventricular zone (VZ) of the telencephalon, where it is involved in maintaining the identity of early striatal progenitors, and it is required for promoting a striatal fate (4–8). Recently, two studies have reported about the role of Gsx2 in mouse neural stem cells, showing that Gsx2 regulates progenitor proliferation and differentiation (9, 10). Nonetheless, these studies focused on Gsx2 function in mouse neurospheres and in adult neural stem cells models that could bear different signatures with respect to human embryonic ventral progenitors. Ebf1 is a helix–loop–helix TF that has been shown to control cell differentiation in the murine embryonic striatum (11–13), but it has never been studied in a human model system of striatal development.
We have previously demonstrated that human ventral telencephalic progenitors can be generated from hES cells by using a Shh treatment coupled with Wnt inhibition (14, 15). These progenitors eventually differentiate into mature, electrophysiologically active neurons. However, the protocol yielded cultures containing Darpp32+–Ctip2+ cells never exceeding 10–15%. We therefore wished to establish a hES cell-based inducible gain-of-function (iGOF) system whereby TFs expressed in the developing striatum can be harnessed to improve MSN differentiation and to study human striatal development. We uncovered roles for Gsx2 and Ebf1 during human striatal specification and differentiation, in particular in cell-cycle regulation. Moreover, we report that a specific temporal window of Gsx2 and Ebf1 overexpression in hES cells achieves high yields of MSNs in vitro, expressing Darpp32 and Ctip2, and that these cells can be found in vivo after transplantation. We show that these hES-derived striatal progenitors can be transplanted in animal models and can differentiate and integrate into the host, extending fibers over a long distance.
Results
Generation of Inducible hES Cell Lines.
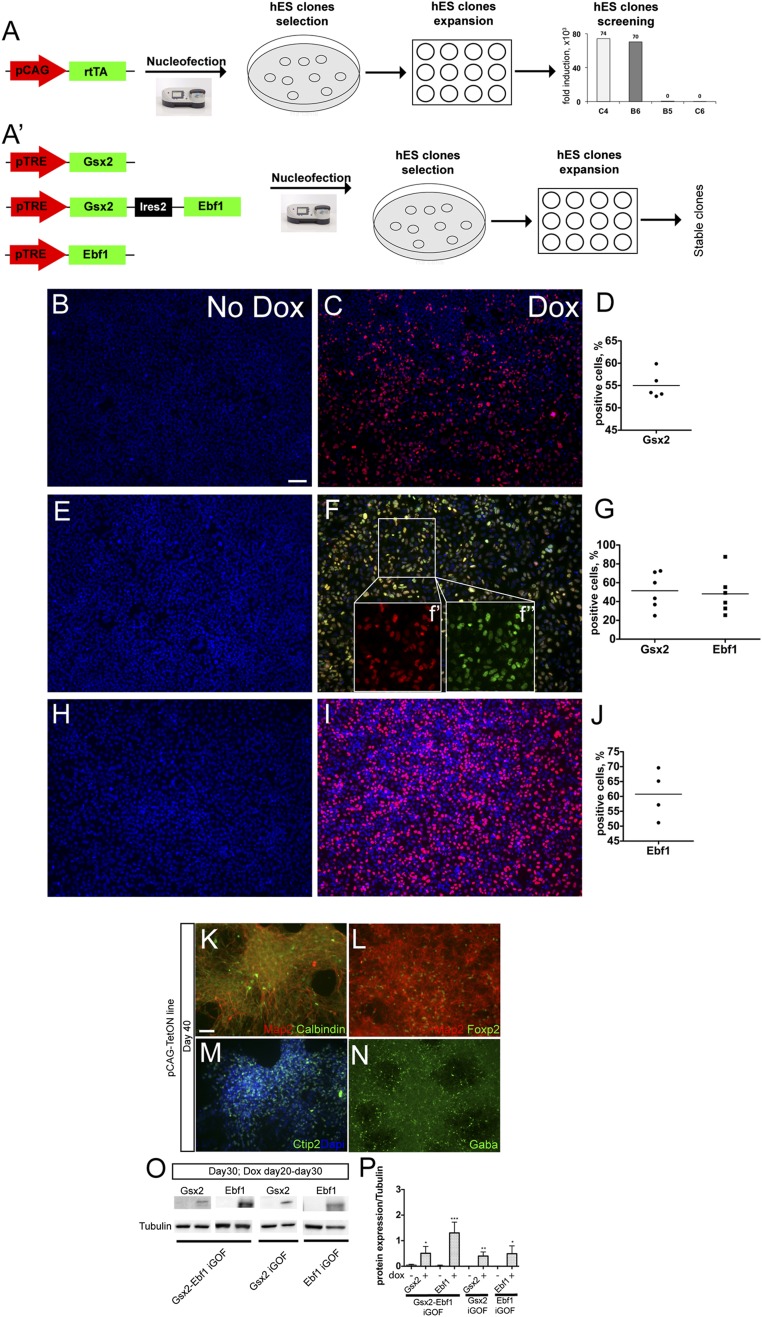
To shed light on the transcriptional program that drives human striatal differentiation, we decided to develop an inducible overexpression system in hES H9 cells. To this goal, we modified a commercially available TetON construct (Clontech) by moving the TetON cassette into a chicken beta-actin promoter with CMV enhancer-based plasmid (pCAG) (Methods) to avoid silencing effects (16). This construct was introduced by nucleofection (Fig. S1A) in hES H9 cells (p40–p50) along with a linear construct carrying a gene encoding for puromycin resistance. After selection, several stable hES cell clones were picked, amplified, and tested for inducibility by using a pTRE-Luciferase construct. We selected four clones that showed no basal Luciferase activity and high induction after 48 h of doxycycline treatment. We amplified and characterized the clones C4 and B6 that showed the highest Luciferase expression after transient transfection (Fig. S1A, chart). They responded promptly to differentiation stimuli similarly to the original H9 cells (Fig. S1 K–N). Next, we constructed three conditional vectors with the pTRE promoter, regulating Gsx2 (pTRE-Gsx2), Gsx2 alongside with Ebf1 by means of an IRES2 sequence (pTRE-Gsx2-Ebf1), and Ebf1 alone (pTRE-Ebf1). Using these three constructs, we carried out nucleofections in the hES-inducible clones (Fig. S1A′). After selection, several stable hES cell clones were picked, amplified, and tested for Gsx2, Gsx2–Ebf1, and Ebf1 expression. Four Gsx2, one Gsx2–Ebf1, and two Ebf1 overexpressing clones were chosen for the next experiments. We quantified Gsx2, Gsx2–Ebf1, and Ebf1 overexpression in the inducible hES cell clones after 72 h of doxycycline treatment (Fig. S1 B–J). Quantification of Gsx2+ cells after 72 h of doxycycline induction in Gsx2 iGOF showed 55 ± 3% expression (Fig. S1D). Quantification of Gsx2+ and Ebf1+ cells in Gsx2–Ebf1 iGOF showed 51 ± 19% and 48 ± 22% expression, respectively (Fig. S1G), with virtually all of the cells coexpressing Gsx2 and Ebf1 (Fig. S1 F′ and F′′). Finally, quantification of Ebf1+ cells in Ebf1 iGOF showed 60 ± 8% expression (Fig. S1J). We next used Western Blot analysis to perform a second quantification experiment during neuronal differentiation, at day30, after 10 d of doxycycline treatment (Fig. S1O) in the three iGOF lines. Western Blot quantification (Fig. S1P) showed up-regulation of Gsx2 (12-fold in Gsx2–Ebf1 and 124-fold in Gsx2 iGOF) and Ebf1 (243-fold in Gsx2–Ebf1 and 267-fold in Ebf1 iGOF) in the three lines compared with basal culture conditions (no doxycycline).
Fig. S1.
Inducible line generation, differentiation potential, and quantification of transgene expression in inducible lines. (A) Schematic representation of nucleofection of a pCAG-TeON-3G plasmid in hES H9 cells, puromycin selection, isolation of resistant clones, and screening of the best responsive clones after pTRE-Luciferase transient transfections. Chart shows quantification of Luciferase activity in selected clones after doxycycline treatment for 24 h, compared with basal levels. (A’) Schematic representation of nucleofection of pTRE-Gsx2, pTRE-Gsx2-Ebf1, and pTRE-Ebf1 plasmids in Tet-ON3G hES cells, puromycin selection, isolation of resistant clones, and selection of the best expressing clones. (B–J) Representative immunofluorescent images and their quantifications of stable hES cell lines for Gsx2 (B–D), Gsx2–Ebf1 (E–G), and Ebf1 (H–J) after 72 h of doxycycline treatment in pluripotency conditions. Quantifications showed transgene expression of 55 ± 3% (Gsx2 in Gsx2 iGOF), 51 ± 19%; 48 ± 22% (Gsx2 and Ebf1 in Gsx2–Ebf1 iGOF, respectively), and 60 ± 8% (Ebf1 in Ebf1 iGOF). Data are presented as means (black lines) and distribution of single experiments (dots). (Scale bar, 100 μm.) (K–N) Representative images of neuronal monolayers generated from Gsx2 and Gsx2–Ebf1 iGOF lines in basal (no doxycycline) conditions, showing that the genetic modifications did not alter neuronal differentiation potential. Immunofluorescence images for (K) Map2 (red) and Calbindin (green) expression, (L) Map2 (red) and Foxp2 (green) expression, (M) Ctip2 (green) expression, and (N) Gaba (green) expression. (Scale bar, 75 μm.) (O) Western blot analyses of Gsx2, Gsx2 and Ebf1, and Ebf1 overexpression after 10 d of doxycycline treatment during the day 20–30 differentiation time window used in the study. (P) Quantification of Western blots shown in O. Error bars show SD. TF overexpression data are normalized over Tubulin levels. *P < 0.05, **P < 0.005, ***P < 0.001.
Gsx2 and Ebf1 Regulation of Patterning Genes.
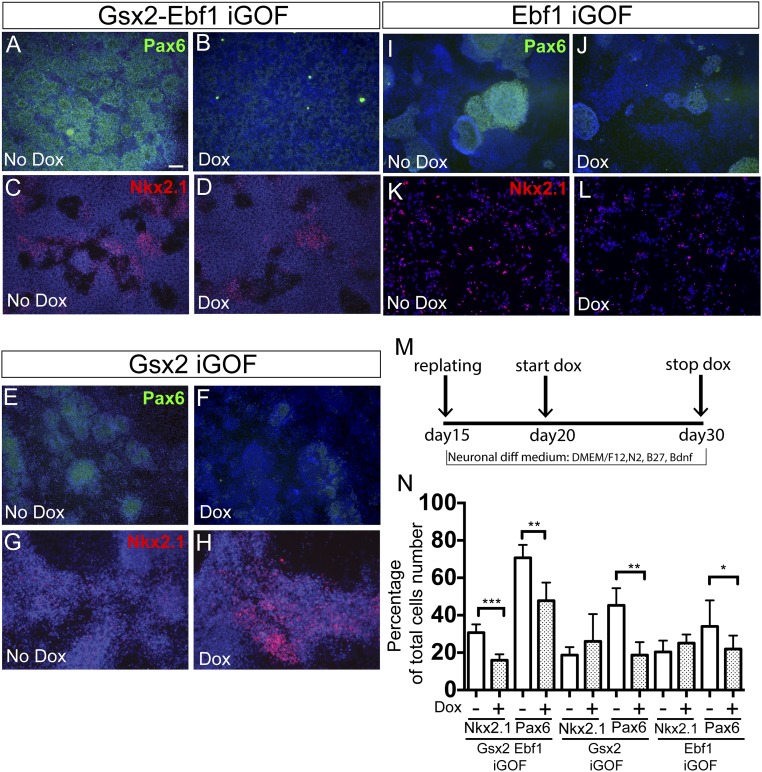
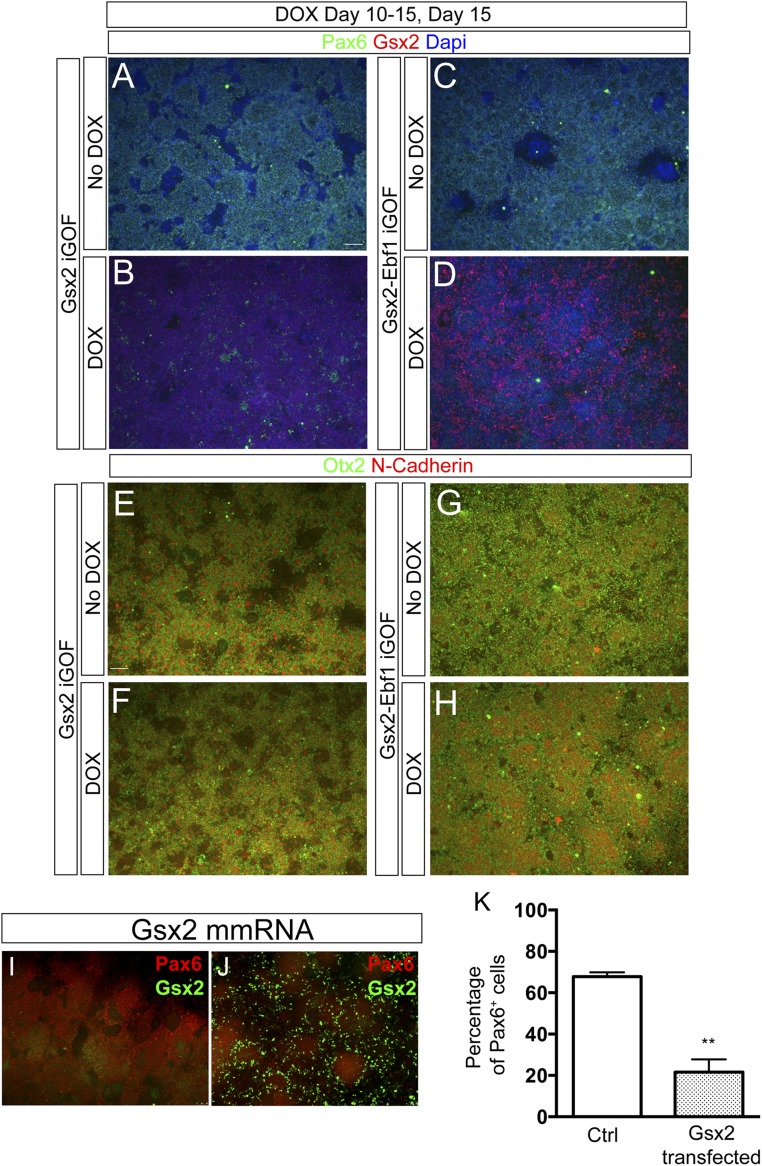
The patterning activity of Gsx2 during ventral telencephalic development has been extensively studied in mouse models (4–7, 17). However, no information is available about its roles during human development. To determine the effects of Gsx2 and Ebf1 overexpression in human neural progenitors, we used a specific protocol that we previously showed to have the potential to generate, first, ventral telencephalic progenitors and, then, mature MSNs after 80 d in vitro (14, 15). However, the protocol yields cultures containing Darpp32+/Ctip2+ cells never exceeding 10–15%. We therefore wished to implement this protocol by establishing a hES cell-based iGOF system whereby TFs expressed in the developing striatum can be used to increase MSN yield. Thus, we decided to overexpress Gsx2, Gsx2–Ebf1, and Ebf1 in different temporal windows during hES neural differentiation: day 10–15, day 15–20, and day 20–30. To test this TF-mediated specification, we first analyzed regional patterning in the hES-derived neural progenitors. We found that Gsx2, Gsx2–Ebf1, and Ebf1 iGOF down-regulated Pax6, a dorsal cortical marker, at day 30 (Fig. 1 A, B, E, F, I, and J, and quantification in Fig. 1N) and at day15 (Fig. S2 A–D), corresponding to the end of the doxycycline treatments. Because Pax6 is also an important early neuroectodermal marker in humans (3), we sought to determine if Gsx2 overexpression could compromise the process of neural induction in hES cells. To test this possibility, Gsx2 expression was induced during the day 10–15 time window, the earliest period used in this study. Importantly, Gsx2 activation did not down-regulate Otx2 and N-Cadherin (two early neural plate markers) expression at day15, the end of the doxycycline treatment (Fig. S2 E–H), suggesting that the cells correctly went through neural induction.
Fig. 1.
Gsx2 and Ebf1 roles during patterning of telencephalic progenitors. (A–D) Gsx2–Ebf1 iGOF line down-regulates Pax6 and Nkx2.1 expression during the day 20–30 developmental window. Instead, Gsx2 and Ebf1 single lines down-regulated only Pax6 (E–L). Representative immunofluorescence images for Pax6 (green) and Nkx2.1 (red) expression. (Scale bar, 100 μm.) (M) Schema illustrating the experimental design. (N) Quantification analysis for Pax6 and Nkx2.1 expressing cells; n = 3 biological replicates. For Pax6 analysis, n = 8 (no dox) and n = 10 (dox). *P < 0.05, **P < 0.01, ***P < 0.003 two-tailed t test analysis. Data are presented as means ± SD.
Fig. S2.
Early neuroectoderm differentiation in Gsx2 and Gsx2–Ebf1 line. (A–D) Immunostaining for Pax6 (green) and Gsx2 (red) at day 15 showing Pax6 down-regulation of Gsx2 in Gsx2 and Gsx2–Ebf1 iGOF lines after 5 d of doxycycline treatment. (E–H) Immunostaining for Otx2 (green) and N-Cadherin (red) of Gsx2 in Gsx2 and Gsx2–Ebf1 iGOF lines after 5 d of doxycycline treatment, showing a correct neural induction. (Scale bar, 100 μm.) (I–K) Representative immunofluorescence images for Pax6 (red) and Gsx2 (green) in control (I) and Gsx2-transfected cells (J). (K) Quantification of Pax6+ cells in the two cell populations. **P < 0.01, two-tailed t test analysis. Data are represented as means ± SD; n = 3 technical replicates.
Next, we performed immunostaining for Nkx2.1, a marker expressed in proliferative cells of the MGE, in striatal interneurons and in Ctip2+ cells of the mature striatum (and in the hypothalamus). Because at this time point (day 30) most cells are still proliferating and we do not usually detect Ctip2 expression, the down-regulation of Nkx2.1 that we found in the double Gsx2–Ebf1 iGOF line (Fig. 1 C, D, G, H, K, and L, and quantification in Fig. 1N) suggests a suppression of an MGE fate.
Next, to move forward a transient and nonintegrating system, we generated a modified mRNA (mmRNA) for Gsx2. We transfected this mmRNA into H9 hES-derived neural progenitor cells from day 20 to day 25 of differentiation using the same protocol used for the iGOF lines. As shown in Fig. S2 I–K, Gsx2 overexpressing cells reduced Pax6 expression similarly to that found in the Gsx2 iGOF line (threefold decrease in both overexpressing systems).
Together, the data indicate that in hES cells that are undergoing neuronal conversion, Gsx2 and Ebf1 overexpression suppresses the dorsal marker Pax6 and the MGE marker Nkx2.1 while maintaining typical neuroepithelial markers (Otx2, N-Cadherin).
Given the in vivo expression of Gsx2 in progenitor cells and Ebf1 in early postmitotic neurons, we next investigated if and how Gsx2 and Ebf1 overexpression modified cell proliferation.
Gsx2 and Ebf1 Regulate Cell-Cycle Kinetics.
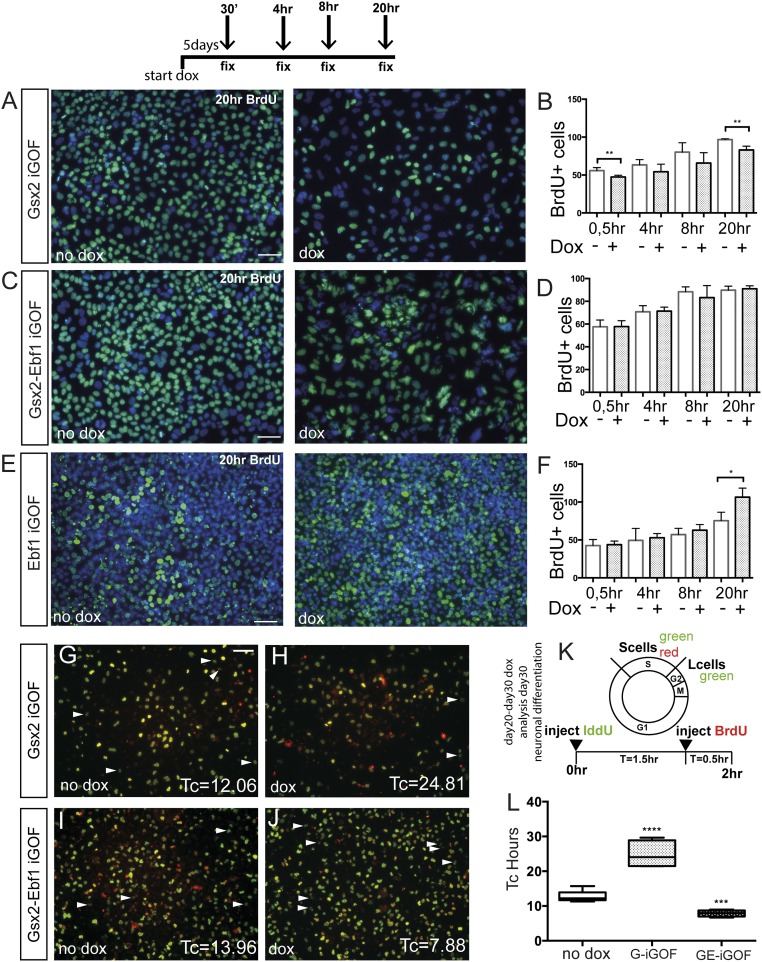
Regulation of cell proliferation in the developing telencephalon is a tightly regulated process, and it is essential to produce the correct number of postmitotic neurons. To examine the effects of Gsx2 and Ebf1 overexpression in human progenitor cells, we first performed a cumulative BrdU analysis in the hES cell lines inducible for Gsx2 and Gsx2–Ebf1. After treating the cells with doxycycline for 5 d, we administered BrdU for 30 min and 4, 8, and 20 h. We found that the Gsx2 iGOF line showed a reduced BrdU incorporation compared with the untreated cells (Fig. 2A and quantification in Fig. 2B). In contrast, the Gsx2–Ebf1 double inducible line showed a similar BrdU incorporation propensity compared with the control line, suggesting that cell-cycle alteration by Gsx2 was rescued by Ebf1 (Fig. 2 C and D). Finally, we performed the same analysis also in the Ebf1 iGOF line, finding that at 20 h there was a significant increase in BrdU incorporation compared with the control line, the opposite phenotype found in Gsx2 iGOF cells (Fig. 2 E and F). These data suggested an involvement of Gsx2 and Ebf1 in cell-cycle regulation.
Fig. 2.
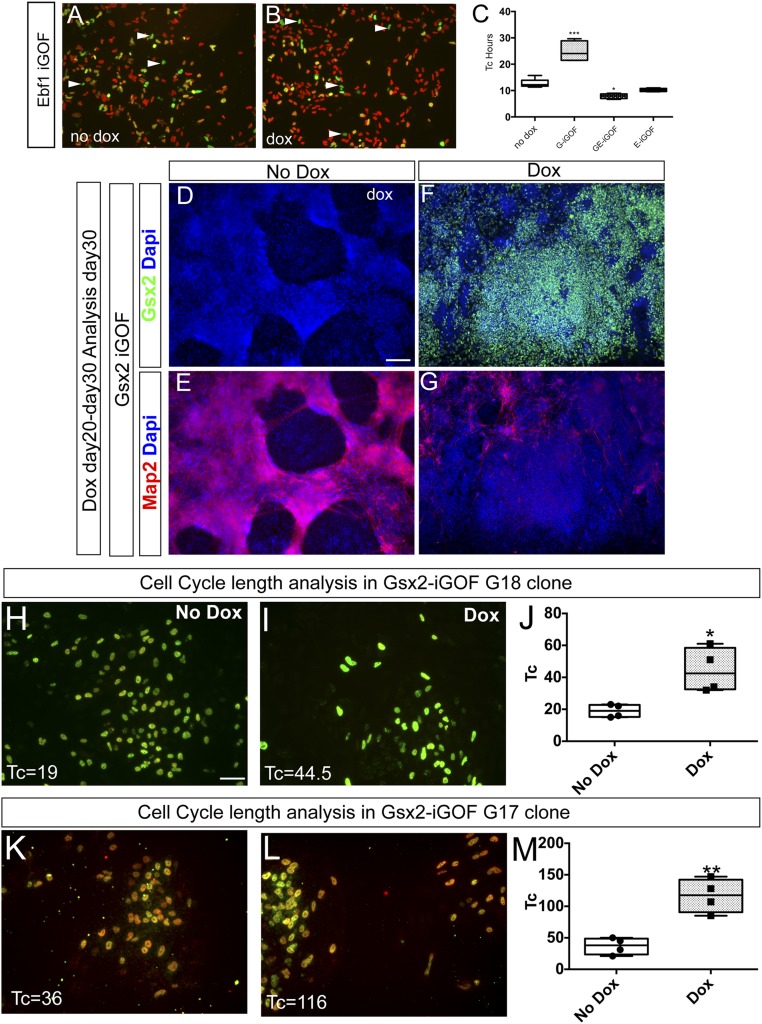
Gsx2 and Ebf1 modulate cell-cycle kinetics. (A, C, and E) Representative images of a BrdU cumulative labeling experiment in Gsx2 (A), Gsx2–Ebf1 (C), and Ebf1 (E) iGOF lines in culture condition allowing pluripotency. BrdU was added to the culture media at 0.5, 4, 8, and 20 h. (B, D, and F) Quantification of BrdU+ cells at the different time points. Data are represented as means ± SD; two-tailed t test analysis. *P < 0.05, **P < 0.01. (Scale bar, 75 μm.) (G–J) Representative images of neuronal progenitor cell-cycle length analysis using BrdU/IdU colabeling. Day 30 hES cell-derived neural progenitors, treated for 10 d with doxycycline, were exposed to IdU at T 0 h and with BrdU at T 1.5 h (see experimental design in K). Arrowheads point to cells that left the S-phase at T 1.5 h (Lcells, green only cells), whereas yellow cells are still in the S-phase at T 2 h. (Scale bar, 75 μm.) (L) Quantification of Tc estimation from BrdU/IdU analysis of Gsx2 and Gsx2–Ebf1 iGOF. Box shows the median and the 25th and 75th percentiles. The whiskers of the graph show the largest and smallest values. ***P < 0.0005, ****P < 0.0001.
To test this hypothesis also in hES-derived neural progenitor cells (the biological context that more closely resembles the developing embryonic human brain), we administered doxycycline from day 20 to day 30 of the neuronal differentiation protocol and analyzed cell-cycle kinetics by a BrdU/IddU double labeling paradigm (18, 19) (Methods for details and Fig. 2K for experimental design). We first tested this method in our hES cell lines, in basal conditions (no doxycycline), with culture conditions permitting pluripotency, finding a cell-cycle time (Tc) of 19.4 ± 4.4 h, comparable to previous published data (20). Next, we analyzed the Tc of day 30 hES-derived neural progenitor cells (Fig. 2 G–L), and we found results in agreement with the BrdU cumulative analysis performed in Fig. 2 A–E. Estimation of control cell (no doxycycline) Tc was 12 ± 1 h, whereas the Gsx2 overexpressing cells showed a Tc of 24 ± 4 h. Next, we analyzed the contribution of Ebf1 by measuring Tc in Gsx2–Ebf1 iGOF cells, finding a value of 7 ± 1 h, suggesting that Ebf1 could override a Gsx2-mediated increase of cell-cycle length. In agreement with these findings, when we examined the single Ebf1 iGOF line, we found that this line showed a cell cycle similar to the double Gsx2–Ebf1 iGOF (Fig. S3 A–C).
Fig. S3.
Gsx2 and Ebf1 regulate differentiation and cell-cycle progression. (A–C) BrdU-IddU assay showing Ebf1 role in Gsx2-Ebf1 double line (see Fig. 2). *P < 0.05; ***P < 0.0005. (D–G) Immunostaining of day 30 neuronal monolayers generated from the Gsx2 iGOF line for Gsx2 (D–F) and Map2 (E–G), showing reduced Map2 expression in cells overexpressing Gsx2 after 10 d of doxycycline (compare E and G). (H–M) Cell-cycle length analysis in two additional clones inducible for Gsx2 (G17 and G18). (H, I, K, and L) immunostaining for BrdU and IddU using two antibodies recognizing BrdU-IddU (green) or only BrdU (red) following the design showed in Fig. 3K. (J and M) Estimation of cell-cycle length in G18 (J) and G17 (M) clones showing results (Tc increase) similar to Fig. 2 for the Gsx2 iGOF clone used throughout the study (G4). Box shows the median and the 25th and 75th percentiles. The whiskers of the graph show the largest and smallest values. *P < 0.02, **P < 0.002.
To rule out the possibility that Gsx2 iGOF cells were undergoing differentiation (and thus incorporating less BrdU), we analyzed Map2 expression at day30, the same time point used for the previous analysis. Gsx2 iGOF showed a marked reduction of Map2+ cells (Fig. S3 D–G), in agreement with the previous cell-cycle analysis data and further suggesting that Gsx2+ cells could not exit the cell cycle. Moreover, we investigated this Gsx2-mediated cell-cycle regulation also in another two hES inducible clones (G18 and G17; Fig. S3 H–M), finding similar results.
Together, these results suggest that Gsx2 regulates cell-cycle progression in human neural progenitor cells.
Gsx2 Constitutive Overexpression Modifies Proliferative Characteristics and Differentiation Potential of hES-Derived Neuroepithelial Stem Cells.
To test if this Gsx2-driven cell-cycle regulation is telencephalic-dependent or represents a general role, we decided to test its overexpression in long-term self-renewing neuroepithelial stem (LT-NES) cells. LT-NES cells represent an excellent model for studying human neuroepithelial cell biology (21). They are hES cell-derived neural progenitors with an anterior hindbrain identity. Here, we decided to take advantage of this cell population and its regional identity to gain insights into the cell-cycle regulation by Gsx2.
We generated an LT-NES cell line overexpressing Gsx2 by nucleofection of a pCAG-Gsx2-IRES-Puromicyn vector and isolation of stable, positive clones. We characterized different clones, finding identical phenotypes across the different lines. A control cell line was also generated by using a pCAG-EGFP-IRES-Puromycin vector, and we found identical self-renewal capacity and differentiation potential compared with the unmodified cell line.
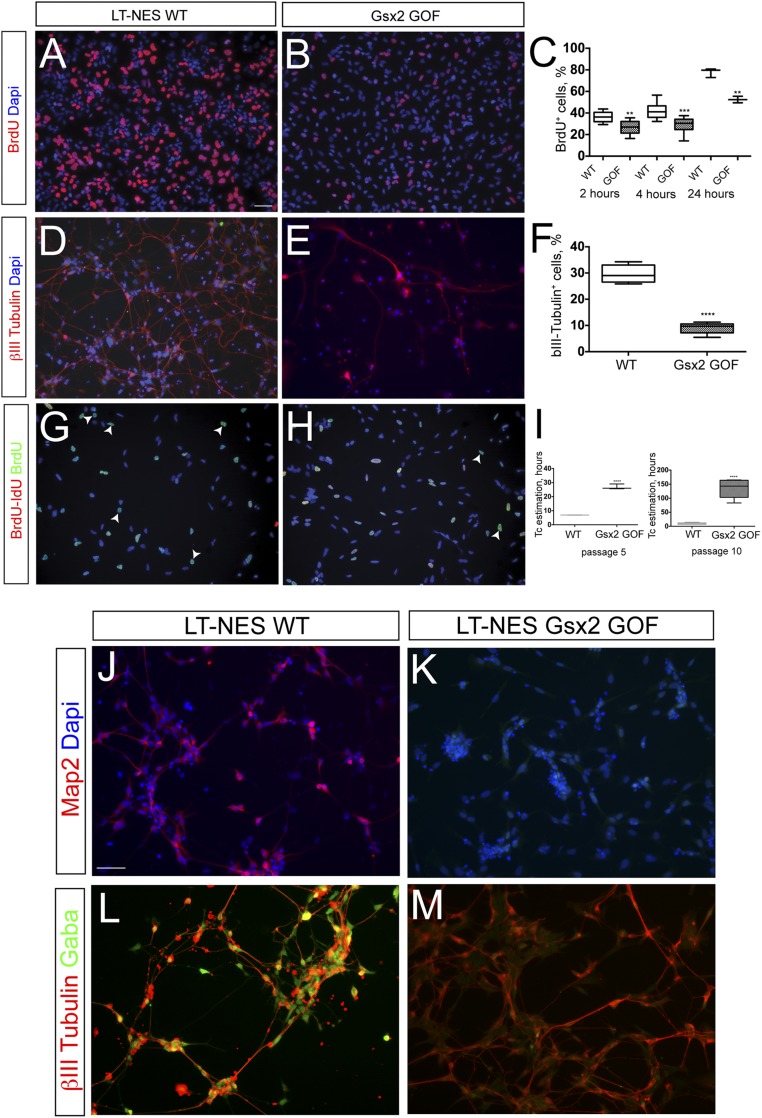
First, we decided to analyze the effects of Gsx2 overexpression during proliferation of LT-NES cells by means of BrdU studies. We first performed a BrdU pulse of 2 h, and we found a decrease in BrdU incorporation in LT-NES cells overexpressing Gsx2 (LT-NES-Gsx2), compared with the control cell line (form 36.3 ± 5.1% to 26.8 ± 5.9%, P < 0.005; Fig. S4 A–C). A similar proliferative defect was found after BrdU pulses of 4 and 24 h (Fig. S4C). We reasoned that this decrease in BrdU incorporation could be linked to an increase in cell differentiation or to an increase in cell-cycle length, which leads to a reduction in the number of times the cells pass through the S phase, thus reducing BrdU incorporation. Thus, we performed differentiation experiments and cell-cycle length studies to distinguish between these two possibilities.
Fig. S4.
Gsx2 constitutive overexpression decreases proliferation and differentiation. (A and B) Representative images of BrdU-labeled LT-NES cells after a 2-h pulse of BrdU in the control line (A) and in one constitutively overexpressing Gsx2 (B). (C) Graph illustrates the percentage of BrdU positive cells of total population in WT and Gsx2 GOF LT-NES cells after 2, 4, and 24 h of BrdU exposure (2 h, 36.3 ± 5.1% and 26.8 ± 6%; 4 h, 41.9 ± 7.7% and 29.2 ± 6.6%; 24 h, 87.1 ± 6.4% and 58.9 ± 8.7%, respectively). In the time points analyzed, **P < 0.005 and ***P < 0.0005 (unpaired, two-tailed t test analysis) by comparing WT and Gsx2 GOF. Box shows the median and the 25th and 75th percentiles. The whiskers of the graph show the largest and smallest values. (D and E) Representative images of neuronal monolayers generated from LT-NES WT and Gsx2 GOF lines. Seven days after growth factor withdrawal, LT-NES WT cells readily differentiated into βIII-Tubulin+ neurons (D, red signal), whereas LT-NES Gsx2 GOF cells did not (E, red signal). (F) Graph illustrates quantification of βIII-Tubulin+ cells of the total population in the two lines, by cell counts of 10× fields, normalized to cell nuclei. Box shows the median and the 25th and 75th percentiles. The whiskers of the graph show the largest and smallest values. ****P < 0.0005. (G and H) Analysis of cell-cycle characteristics using IdU/BrdU colabeling. Monoclonal antibodies specific for both BrdU and IdU (red) and BrdU alone (green) are used to identify cells in the S-fraction [red and green double-labeled cells, (Scells)] and cells leaving the S phase (green only cells, Lcells, arrowheads in G and H). (I) Tc estimation from BrdU/IdU analysis of WT and Gsx2 GOF line. The LT-NES Gsx2 GOF line increases dramatically after a few passages (P0–P5–P10). Box shows the median and the 25th and 75th percentiles. The whiskers of the graph show the largest and smallest values. (Scale bar, 75 μm.) (J and K) Representative images of neuronal monolayers generated from WT and Gsx2 GOF cells at day 10 of differentiation after Fgf2 and Egf withdrawal: The mature neuronal marker Map2 was absent in LT-NES-Gsx2 cells. (L and M) Early passage LT-NES-Gsx2 cells express the early neuronal marker βIII-Tubulin (red) but failed to express the lineage-specific marker Gaba (green).
First, we differentiated the cells for 10 d, and we analyzed the expression of the early neuronal marker βIII-Tubulin. We found that the number of newly formed neurons was decreased in LT-NES-Gsx2 compared with the control cell line (form 28.9 ± 7.8% to 15.1 ± 1.8%, P < 0.05; Fig. S4 D–F), in agreement with the results previously found in the Gsx2 iGOF hES line (Fig. S3 D–G). A similar result was found when studying the expression of a more mature neuronal marker such as Map2 (Fig. S4 J and K). Moreover, even when it was possible to detect βIII-Tubulin expression in LT-NES-Gsx2 cells (at early passages), more mature and lineage-specific markers, such as GABA, were absent (Fig. S4 L and M), further suggesting that Gsx2 overexpression impairs neuronal differentiation and maturation. Next, we asked if the decrease in BrdU incorporation was caused by a cell-cycle disregulation. To this goal, we performed an analysis of cell-cycle characteristics using the BrdU/IddU double labeling paradigm to estimate the cell-cycle length in the two cell populations, the LT-NES-Gsx2 and LT-NES-EGFP cells. We found that Gsx2 overexpression caused a significant increase in total cell-cycle length (Tc) compared with the control cell line (Fig. S4 G–I). This increase was even more pronounced after a few passages (Fig. S4I, see increment between p5 and p10, from 26.88 ± 1.84 h to 135.30 ± 33.79 h, P < 0.005), suggesting that Gsx2 overexpression has a cumulative effect during time. The control cell line, during the same time period, did not show a statistically significant increase in Tc (from 6.9 ± 0.1 to 9.5 ± 3, P > 0.1; Fig. S4I).
In conclusion, these data demonstrated that a constitutive Gsx2 overexpression was detrimental for proper neuronal differentiation and maturation, even in a nontelencephalic compartment, corroborating the results obtained in the hES inducible lines. Thus, to summarize the results shown in Fig. 2 and Fig. S4, Gsx2 has a major role in regulating proliferation, by lengthening the cell cycle in a context-independent manner.
Ebf1 Promotes Neuronal Differentiation and Maturation.
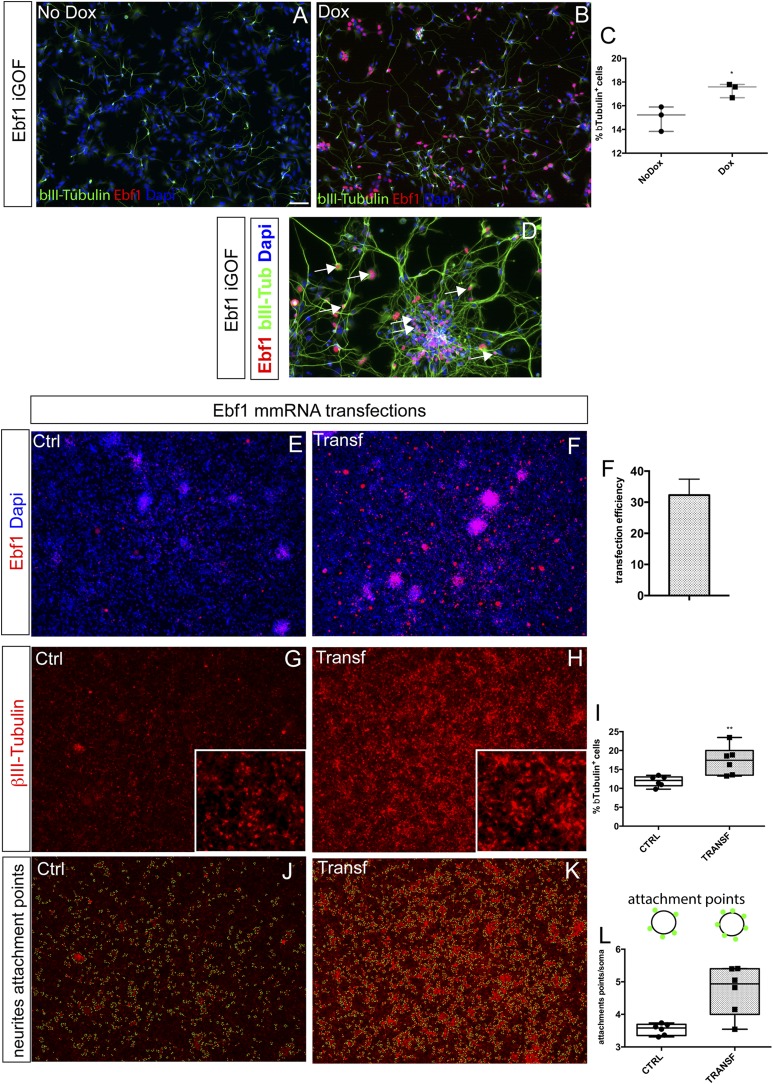
The foregoing data demonstrate that Gsx2 has important roles in regulating cell-cycle progression, whereas Ebf1 expression probably enhances differentiation. To investigate the specific role of Ebf1 in human neural progenitor cells, we first studied the Ebf1 iGOF line. After overexpressing Ebf1 (by doxycycline treatment) in the day 20–30 temporal window of hES neuronal differentiation, we compared βIII-Tubulin expression with control (no doxycycline) cells. Ebf1 overexpression resulted in a significant increase in the number of βIII-Tubulin+ cells (Fig. S5 A–D, quantification in Fig. S5C). Next, to further test the Ebf1 role in increasing neuronal differentiation, we transfected an mmRNA for Ebf1 (Miltenyi Biotec) in unmodified H9 hES-derived neural progenitor cells (exposed to the same protocol used for the iGOF lines). First, we tested transfection efficiency by staining for Ebf1 after 2 consecutive days of mmRNA delivery, finding a transfection efficiency of 32 ± 5% (Fig. S5 E and F). Next, we investigated βIII-Tubulin expression after 5 consecutive days of transfections (day 30). We found 17 ± 3% of cells expressing βIII-Tubulin compared with 11 ± 1% of untreated cells (Fig. S5 G and H, and quantification in Fig. S5I, P < 0.01, n = 3, unpaired t test). We then investigated if Ebf1 overexpression had an effect on neurite length or complexity. Interestingly, by using NeurphologyJ analysis (22), we found an increase of attachment points (Fig. S5 J–L) on neuronal soma (from 3.5 ± 0.1% to 4.7 ± 0.7% in transfected cells, normalized over total soma number, P < 0.05, n = 3, unpaired t test). These data strongly suggest that Ebf1 has a role as a neuronal differentiation player during hES differentiation.
Fig. S5.
Ebf1 enhances neuronal differentiation in hES-derived neural progenitors. (A and B) Representative images of day 30 neuronal monolayers generated from Ebf1 iGOF hES line in basal conditions (A) and after 10 d of doxycycline treatment (B) showing bIII-Tubulin (green) and Ebf1 (red) expression. (C) Graph shows quantification of βIII-Tubulin+ cells in the two conditions. Center lines show the average; whiskers indicate minimum and maximum. (D) Higher magnification of Ebf1+bIIITubulin+ cells in Ebf1 iGOF cells. (E–I) Ebf1 mmRNA transfections of H9 hES-derived neural progenitor cells. (D and E) Representative images of day 30 neuronal monolayers generated from H9 hES cells transfected with 200 ng of Ebf1 mmRNA, showing Ebf1 overexpression (red). (F) Quantification of Ebf1+ cells 48 h after transfection. (G and H) Representative images of βIII-Tubulin expression of neuronal monolayers in control cells (G) and after 5 d of Ebf1 mmRNA transfections (H). (I) Quantification of βIII-Tubulin+ neurons. Center lines show the average; whiskers indicate minimum and maximum. (J–L) Neurite attachment point quantification of βIII-Tubulin staining performed in G and H. (L) Graph shows attachment point quantification by NeurphologyJ software. Box shows the median and the 25th and 75th percentiles. The whiskers of the graph show the largest and smallest values. (Scale bar, 100 μm.) *P < 0.05, **P < 0.005.
Gsx2 and Ebf1 Overexpression Differentially Regulates Early Neuronal Differentiation.
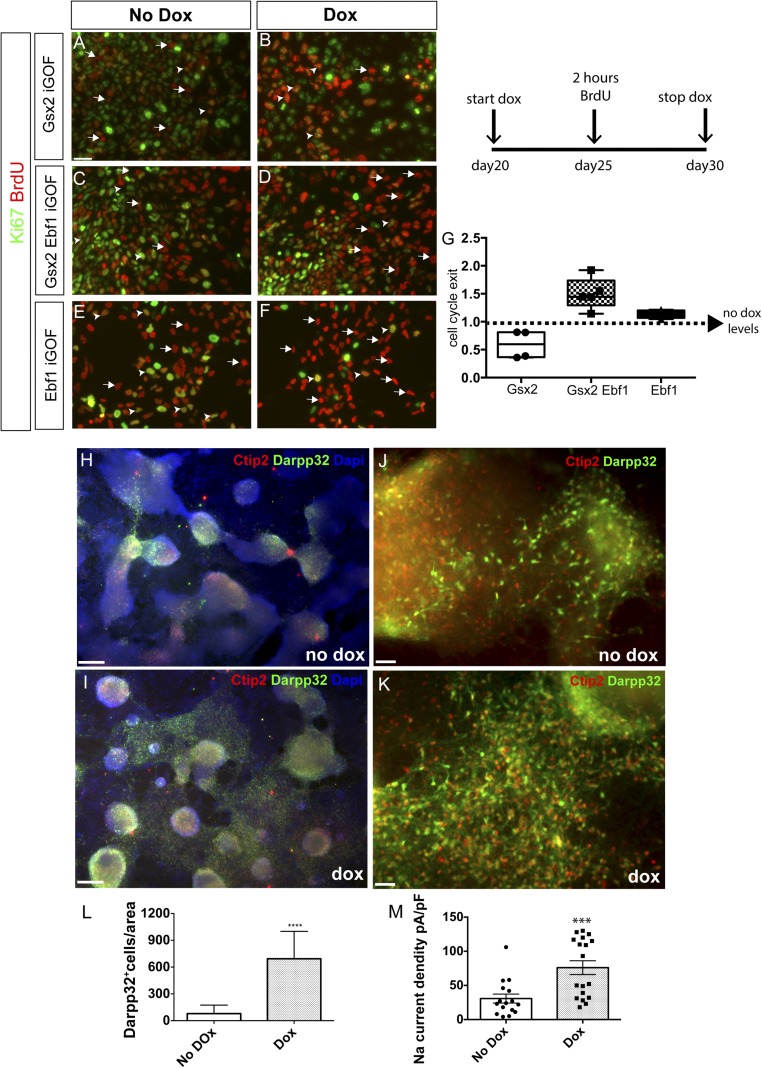
Taking into account the different proliferative responses of hES-derived neural progenitors to Gsx2 and Ebf1 overexpression and the increased neurogenesis after Ebf1 overexpression, we decided to investigate the tendency of Ebf1 and Gsx2–Ebf1 iGOF lines toward differentiation. First, we monitored neuronal differentiation during the differentiation process, finding better neuronal morphology in the two lines after doxycycline treatment. To quantify this differentiation propensity, we performed cell-cycle exit studies, by administering for 2 h BrdU at day 25 of neuronal maturation in a day 20–30 temporal window of doxycycline treatment. The cells were then fixed at day 30 and analyzed for BrdU and Ki67 expression (see schema in Fig. 3). Cell-cycle exit index was calculated by dividing the total number of BrdU+ Ki67− cells by the total number of BrdU+ cells. As shown in Fig. 3 A–F and quantified in Fig. 3G, the three cell lines showed different phenotypes. Gsx2 overexpressing cells were more likely to remain in the cell cycle (50.2 ± 29.7% reduction of cell-cycle exit over no-doxycycline cells; no doxycycline levels were arbitrarily set to 100; P < 0.005, n = 3; Fig. 3 A and B, quantification in Fig. 3G), in agreement with the data presented in Fig. S3 D–G. Ebf1 incorporation caused increased differentiation output in Gsx2–Ebf1 iGOF line (150 ± 28% increase in cell-cycle exit over no-doxycycline cells; no doxycycline levels were arbitrarily set to 100; P < 0.05, n = 3; Fig. 3 C and D, quantification in Fig. 3G). Finally, Ebf1 single iGOF overexpressing cells were 113 ± 7% more likely to exit the cell cycle (no doxycycline levels were arbitrarily set to 100%; P < 0.05, n = 3; Fig. 3 E and F, chart in Fig. 3G).
Fig. 3.
Gsx2 and Ebf1 differentially regulate cell-cycle exit and promote striatal differentiation. (A–F) Representative images of cell-cycle exit studies following the experimental design depicted in Top Right. Arrows point to BrdU+Ki67− cells (that exited cell cycle); arrowheads point to BrdU+Ki67+ cells (still proliferating). (Scale bar, 75 μm.) (G) Quantification of cell-cycle exit in Gsx2, Gsx2–Ebf1, and Ebf1 lines after 10 d of doxycycline treatments compared with basal conditions (no doxycycline, dotted line). Box shows the median and the 25th and 75th percentiles. The whiskers of the graph show the largest and smallest values. **P < 0.005. (H–K) Representative images of neuronal monolayers generated from the Gsx2–Ebf1 iGOF line: immunofluorescence for Ctip2 (red) and Darpp32 (green) at day 80 of striatal differentiation, in day 20–30 doxycycline-treated (I and K) and nontreated cells (H and J). H and I, 5× magnification; J and K, 20× magnification. (L) Quantification of Darpp32+ cells by automated cell counts of 10× fields normalized to the area occupied by nuclear counterstaining. The images in the figure represent reproducible results from four out of five differentiation experiments reaching day 80. [Scale bar, 250 μm (J and K) and 75 μm (H and I).] ****P < 0.0001. (M) Sodium current density of neuronal monolayer cultures at day 100 of differentiation in control and doxycycline-treated conditions. Data are represented as means ± SD. Individual round and squared dots represent individual recorded cells. Sodium current density was significantly higher (***P < 0.001) in Gsx2–Ebf1 overexpressing cells.
Again, these results were in line with the hypothesis of Gsx2 retaining neural progenitor cells in an undifferentiated state and Ebf1 controlling cell-cycle exit and progenitor maturation.
Gsx2–Ebf1 Overexpression Induces MSN Differentiation from hES Cells.
To determine the striatal differentiation potential of hES cells overexpressing Gsx2–Ebf1 in the day 20–30 developmental window, we conducted long-term differentiation experiments and analyzed the cells at day 60 and day 80.
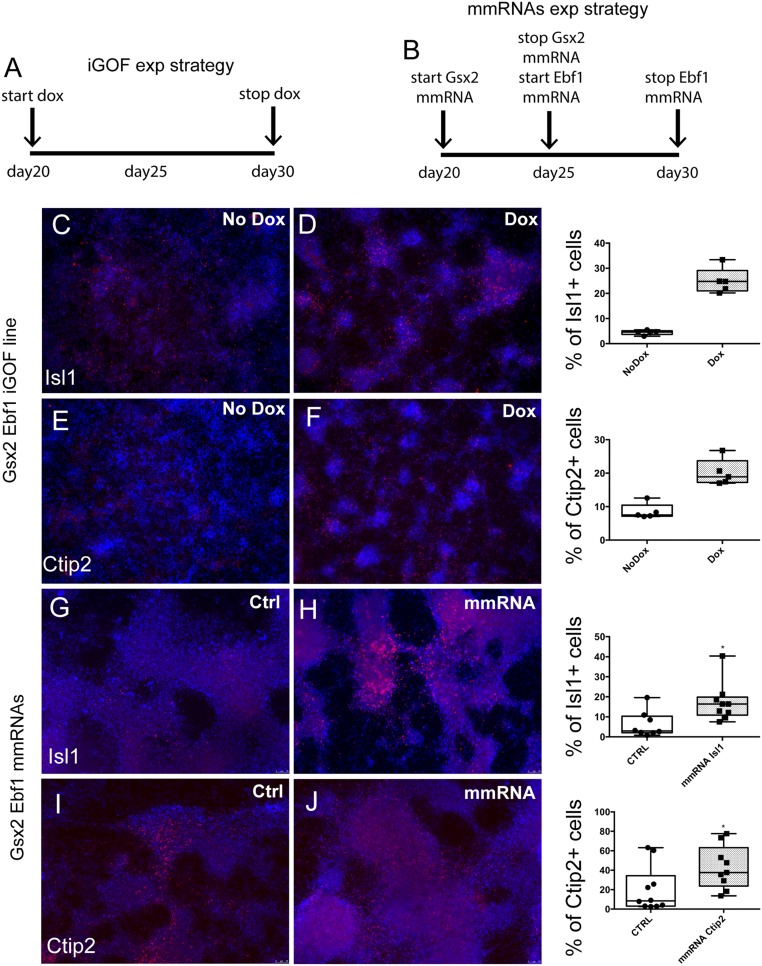
First, we evaluated the number of cells expressing the striatal neuronal markers Isl1 and Ctip2 at day 60 of differentiation. Isl1+ cells increased from 4.4 ± 0.9% in control cells (no doxycycline) to 25 ± 5% in Gsx2–Ebf1 overexpressing cells (P < 0.00005, n = 3; Fig. S6 C and D). Ctip2+ cells increased from 8.5 ± 2.3% in control cells (no doxycycline) to 20 ± 3.9% in Gsx2–Ebf1 overexpressing cells (P < 0.0005, n = 3; Fig. S6 E and F). To further validate these findings using a different system and to move toward a nonintegrating system, we also performed transfection experiments in the RC17 hES cell line using mmRNAs for Gsx2 and Ebf1. Following the experimental strategy shown in Fig. S6B, Isl1+ cells increased from 6.2 ± 2.2% in nontransfected cells to 17.2 ± 3.2% in cells transfected sequentially with Gsx2 and Ebf1 (Fig. S6 G and H). At day 60 of differentiation, Ctip2+ cells increased from 20.10 ± 7.4% in nontransfected cells to 42.9 ± 7.5% in cells transfected sequentially with Gsx2 and Ebf1 (Fig. S6 I and J).
Fig. S6.
Gsx2 and Ebf1 overexpression induces Isl1 and Ctip2. (A and B) The two different experimental designs in iGOF line (A) and by using mmRNAs (B). We performed daily transfections for Gsx2 mmRNA from day 20 to day 25 and then for Ebf1 mmRNA from day 26 to day 30. (C–J) Representative images of neuronal monolayers generated from Gsx2–Ebf1 iGOF line (C–F) and after Gsx2 and Ebf1 mmRNA sequential transfections (G–J). Graphs illustrate quantification of Isl1+ and Ctip2+ cells of the total cell population in the two experimental approaches by cell counts of 10× fields, normalized to cell nuclei. Box shows the median and the 25th and 75th percentiles. The whiskers of the graph show the largest and smallest values. *P < 0.05; black squares show the individual measurements, unpaired t test analysis.
Next, we analyzed the neuronal population at day 80 of differentiation by studying Darpp32 and Ctip2 expression. Initially, we quantified the generated striatal neurons by expressing the density of Ctip2+/Darpp32+ area per arbitrary surface area (Fig. 3 H and I), finding a higher efficiency of Darpp32+/Ctip2+ neuron generation in the iGOF line compared with the control line (from 3.8 ± 3.1% to 38.8 ± 13.7%). Then we focused on the number of Darpp32+ cells by performing automating soma cell counting (by using the NeurphologyJ ImageJ plugin; Methods for quantification details), and we found a higher number of Darpp32+ cells per unit area in the iGOF line than in control cells (from 79.5 ± 26.3 in basal condition to 693 ± 76 in iGOF; number of cells per area; Methods for quantification details, n = 3; Fig. 3 J and K, quantification in Fig. 3L; n = 3).
Finally, we studied if the Gsx2–Ebf1 combination could confer functional electrophysiological properties to the differentiated neurons. Although passive properties did not change significantly between doxycycline-treated and nontreated cells, we found interesting results studying sodium currents. In particular, Na+ current density was significantly higher in doxycycline-treated cells (from 30.7 ± 6.6 pA/pF in control cells to 76.1 ± 10.1 pA/pF in Gsx2–Ebf1 overexpressing cells, P < 0.001; Fig. 3M).
Gsx2–Ebf1 iGOF Cells Survive and Differentiate in Vivo After Transplantation.
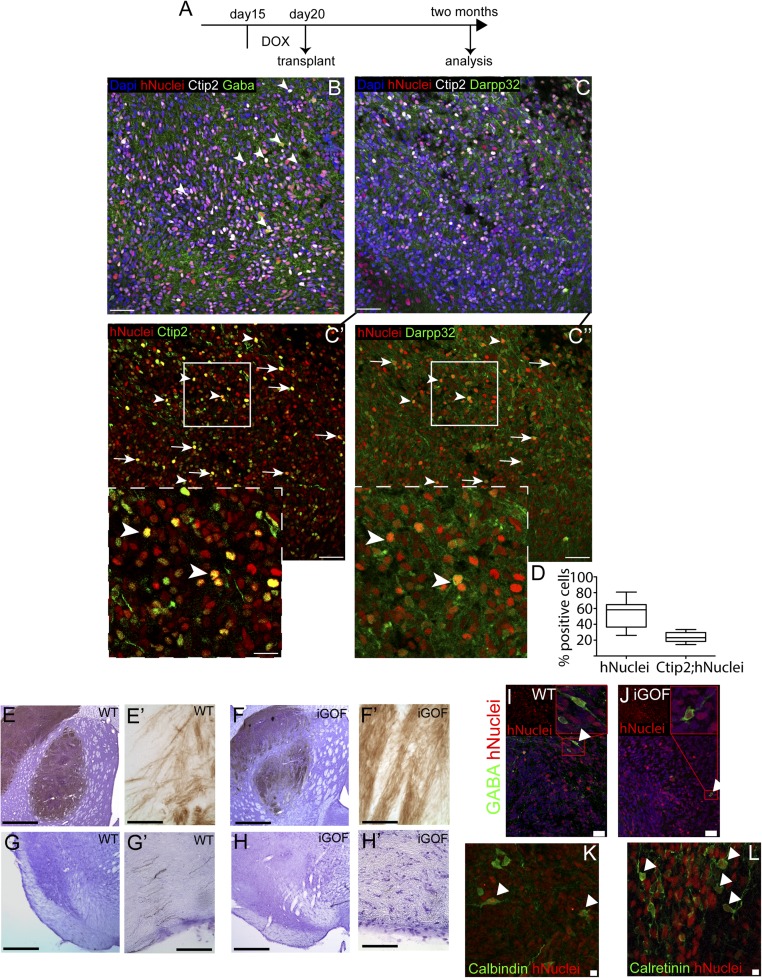
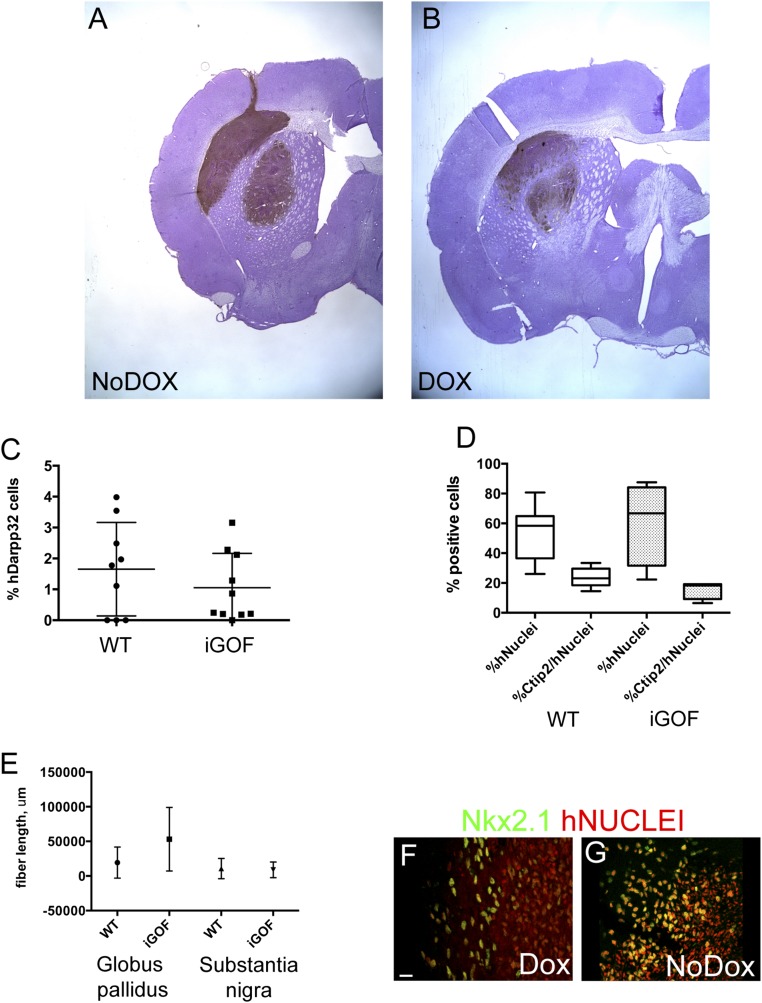
Next, we wanted to assess long-term survival and differentiation of Gsx2–Ebf1 iGOF cells after transplantation in the striatum of QA-lesioned, athymic adult rats. The transplanted animals were followed up to 2 mo and then killed for immunohistochemical analysis. To this goal, we decided to induce Gsx2 and Ebf1 expression from day 15–20 of neuronal differentiation and perform the transplant at day 20 (Fig. 4A). This time point was chosen according to previous studies performed in our laboratory, showing an increase in cell survival when cells were transplanted at day 20 compared with day 30. Two months after transplantation, we found many human nuclei+ cells in the transplanted site (Fig. 4 B and C, red cells), suggesting optimal survival (average of 53 ± 16% human nuclei+ cells; Fig. 4D). We then analyzed the expression of markers of mature striatal neurons: Ctip2, GABA, and Darpp32. Interestingly, Ctip2 and GABA were largely present in the lesioned transplanted site (human nuclei+ cells) (Fig. 4B, arrowheads point to examples of Ctip2+/hNuclei+ cells). In addition, immunostaining for Darpp32 and Ctip2 showed similar results (Fig. 4C), with these two striatal markers expressed in the site of transplantation. To further investigate the coexpression of Ctip2 and Darpp32 in human nuclei+ cells, we analyzed the immunostaining for Ctip2/human nuclei (Fig. 4C’) and Darpp32/human nuclei (Fig. 4C’’) on the same section shown in Fig. 4C. Insets in Fig. 4 C’ and C’’ show representative human nuclei+ cells expressing both Ctip2 and Darpp32 markers. We quantified the cells that were human nuclei/Ctip2 double-positive, and we found 23 ± 6% of cells expressing both markers. When we quantified control (No Dox) cells, we found similar results (Fig. S7 C and D for Darpp32 and Ctip2 quantifications, respectively). Future studies will need to address the role of the in vivo environment in differentiating hES cells. These results suggest that Gsx2–Ebf1 iGOF cells were able to differentiate into striatal neurons in vivo as in vitro.
Fig. 4.
Gsx2–Ebf1 overexpressing cells maturate in vivo into MSNs and extend axonal projections into distant targets. (A) Experimental design for hES cell-derived neural progenitor transplantation in QA-lesioned athymic rats, after 5 d of doxycycline treatment. (B–C’’) Representative images of grafted cells 2 mo after transplantation, assayed for human nuclei marker and specific MSNs markers. Insets in C’ and C’’ are magnifications of regions depicted in C’ and C’’. Arrowheads point to human cells expressing both Ctip2 and Darpp32. Arrows point to grafted human cells expressing either Ctip2 or Darpp32. [Scale bar, 35 μm (B–C’’) and 15 μm (Insets).] (D) Quantification of human cells in the grafted site (hNuclei+ cells) and of Ctip2+ cells in the hNuclei+ population. Box shows the median and the 25th and 75th percentiles. The whiskers of the graph show the largest and smallest values. [Scale bar, 75 μm (B, C, C’, and C’’) and 30 μm (Insets in C’ and C’’).] (E–H’) DAB-developed sections stained for human NCAM antibody showing the neuronal outgrowth of intrastriatal transplants of hES-derived striatal progenitor cells. E–H and G’ and H’ were counterstained with Cresyl-Violet to show the surrounding tissue. (I–J) Examples of hNuclei+GABA+ cells found both in WT (I) and iGOF cells (J). [Scale bar, 250 μm (E, F, G, and H) and 25 μm (E’, F’, G’, and H’).] (K and L) Examples of hNuclei cells expressing the local interneurons markers Calbindin (K) and Calretinin (L). Calbindin+ cells displayed a morphology reminiscent of the typical fusiform shape of human interneurons. Calretinin+ cells exhibited ovoid somata, as expected for human striatal interneurons. [Scale bar, 25 μm (I and J) and 5 μm (K and L).]
Fig. S7.
(A and B) Low-magnification images of graft size in NoDox (A) and Dox (B) conditions, as assayed by hNCAM staining, 2 mo posttransplantation. Quantification of (C) Darpp32 cells of human origin in the grafts, (D) Ctip2 cells of human origin in the grafts, and (E) fiber length as assayed by hNCAM expression in two targets of striatal projections. (F and G) Nxx2.1 staining 2 mo after transplantation in the graft region, showing Nkx2.1+ cells, in agreement with the interneuron population shown in Fig. 4 K and L. (Scale bar, 75 μm.)
Long-Distance Axonal Outgrowth and Local Circuitry Integration of hES-Derived Striatal Neurons After Transplantation.
We next performed a histological analysis of long-distance, target-specific outgrowth by using a human-specific antibody for NCAM (hNCAM). When grafted to the lesioned striatum, both WT and iGOF cells showed hNCAM-rich grafts (Fig. 4 E and E’ as well as F and F’ and Fig. S7 A and B for low-magnification pictures showing graft size). These grafts were able to extend axons to the substantia nigra (Fig. 4 G and G’ as well as H and H’), a specific striatal target. We also quantified fiber length (Methods) in iGOF and control cells (Fig. S7E), finding similar results. Moreover, we could also observe the presence of human cells expressing the neurotransmitter GABA (Fig. 4 I and J). Lastly, these hES-derived progenitors were also able to differentiate into local circuitry interneurons, as shown by the presence of human cells expressing Calbindin (Fig. 4K), Calretinin (Fig. 4L), and Nkx2.1 (Fig. S7 F and G).
In summary, we provide evidence of an efficient integration in the host neuronal circuitry with human axonal extension to striatal-specific targets as the substantia Nigra.
Discussion
This study aimed to achieve two goals: (i) to study Gsx2 and Ebf1 function during human ventral telencephalic development, and (ii) to improve MSN differentiation from hES cells by transcriptional specification. In both efforts, we have succeeded in applying an iGOF system for forcing TF expression in defined temporal windows and in combining this approach with a morphogens-driven ventral telencephalic specification.
In making progress toward the first aim, we have demonstrated a dual role for Gsx2 in embryonic human neural progenitors. First, it imparts a regional identity by directly or indirectly down-regulating Pax6 expression. We found this effect during different time windows of Gsx2 induction, suggesting a time-independent primary function for this TF. It is also important to note that Gsx2 iGOF cells responded properly to neural induction extrinsic signals as evidenced by the correct expression of early neural plate markers such as Otx2 and N-Cadherin. Second, Gsx2 has a major role in regulating proliferation, by lengthening the cell cycle in a context-independent manner: We found similar results in cells as different as LT-NES, self-renewing hES cells, and hES-derived neural progenitors. To begin with, we show that in LT-NES cells, a model of human neuroepithelial cells, constitutive Gsx2 overexpression caused a progressive increase in cell-cycle length during passages, leading to a proliferation block and to differentiation impairment.
Interestingly, Gsx2 time-restricted overexpression showed the same consequences on cell-cycle regulation, suggesting that this is a key Gsx2 role in neuronal progenitors. Moreover, because LT-NES cells have a ventral anterior hindbrain identity and hES-derived neural progenitors express more anterior markers, such as Otx2, this Gsx2 activity on cell-cycle regulation is context-independent and likely reflects a primary role. Interestingly, it is well accepted that during mouse development cell-cycle lengthening is correlated with enhanced neurogenesis (23). Our data about Gsx2-regulated cell-cycle lengthening are somehow in contrast, as we found a reduction in differentiation. It is probable that Gsx2 retains human neural progenitor cells in a condition that prevents excessive proliferation and differentiation, with implications for the generation of the correct number of differentiated progeny during human development.
A recent paper has evidenced that in adult neural stem cells Gsx2 overexpression promotes the transition from quiescent to activated neural stem cells (9). Nonetheless, they also pointed out how a high level of Gsx2 blocks the lineage progression toward transit amplifying progenitors, a more differentiated cell population. Our findings obtained in human neural progenitors are in line with the suggestion that fine-tuned Gsx2 levels must be reached to promote neuronal differentiation. The ventral mouse and human telencephalon express at high-level Gsx2 in the VZ (4), including the LGE proliferative region, and these expression data likely reflect the roles played by this TF. Later during development, the Gsx2 expression is reduced in both the number of Gsx2+ cells and the intensity levels (5), suggesting that its expression must be down-regulated over time to allow neuronal maturation.
Thus, our data point to a role for Gsx2 in restraining cell-cycle progression in neural progenitors while instructing a regional ventral phenotype. Of note, the differentiation defect observed in Gsx2 iGOF cells was rescue by Ebf1: the corecruitment of Gsx2 and Ebf1 caused a more efficient neuronal differentiation, while preserving the regional patterning activity of Gsx2, as shown by the Pax6 down-regulation. Interestingly, even if these two TFs are not expressed in the same region and time during development, their combination in hES cell-derived neural progenitors allowed a proper cell-cycle progression and neuronal differentiation, while maintaining a patterning activity (Pax6 down-regulation). We show here that Ebf1, by using iGOF lines or mmRNA transfections, can enhance neuronal differentiation in hES-derived neuronal populations, in terms of neuronal numbers and morphological characteristics.
In this work, we also identified a temporal window for an efficient iGOF transcriptional activation or mmRNAs transfections leading to improved human neural progenitor patterning and differentiation toward MSNs. In the last few years, the use of specific extrinsic signals in combination with the dual SMAD inhibition strategy resulted in the development of protocols for the derivation of many central and peripheral nervous system lineages from hES and iPS cells (24). Here we show that TFs with a different expression pattern and timing can be combined to efficiently differentiate hES H9 cells toward a striatal phenotype. This study has then provided a working system for combining extrinsic (morphogens) and intrinsic (TFs) players to manipulate hES or iPS cell fates. In particular, by combining a ventral-inducer like Gsx2 and a neuronal differentiation-effector as Ebf1 we could shift the differentiation outcome toward MSNs. Of interest for future studies in stem-cell therapies for HD, we show that, upon transplantation in HD rat models, Gsx2–Ebf1 iGOF cells can survive, differentiate, and express key striatal markers such as Ctip2 and Darpp32. Moreover, we show that striatal-patterned hES cells can project axons over long distances in the adult brain (of clinical importance), providing appropriate innervation of striatal GABA targets as the substantia nigra.
These findings in combination with the ability to use mmRNAs for nonintegrating transient gene expression might further pave the way for a rational modulation of cell fates, especially in clinical settings.
We also show that hES cells can be harnessed to model human embryonic development and neuronal differentiation by inducible expression of key developmental TFs. This technique allows mimicking and testing the temporal widows of TF activation during human embryonic development.
Methods
ES and LT-NES Cell Culture.
The hES H9 cell line (Wicell) was cultured on Matrigel (BD, Becton Dickinson) or Matrix (Cell Guidance System). Pluripro (Cell Guidance System) medium was changed daily. Cells were passaged enzymatically with Accutase (Invitrogen) every 3 d. LT-NES cells were derived as described in ref. 21 and maintained in DMEM/F12 (Life Technologies) supplemented with N2 1:100 (Life Technologies), B27 1:1,000 (Life Technologies), and 10 ng/mL Fgf2 and Egf (Peprotech). LT-NES neuronal differentiation was triggered by removing Fgf2 and Egf from the medium. Mycoplasma contamination was checked every 3 mo.
Neuronal Differentiation.
hES cells were plated for neuronal induction as described in ref. 25. Briefly, cells were plated at a density of 0.7 × 105 cells per cm−2 on Matrigel-coated dishes in Pluripro medium supplemented with 10 µM ROCK inhibitor (Y-2763226, Sigma). Cell cultures were expanded for 3 d until they were nearly confluent. The starting differentiation medium included DMEM/F12 (Life Technologies) with N2 and B27 (Life Technologies), supplemented with 5 µM Dorsomorphin (Sigma) or 500 nM LDN 193189 (Sigma) and 10 µM SB431542 (Tocris), which were used until day 12. Every 2 d, the medium was replaced with new medium. Starting on day 5, 200 ng·mL−1 SHHC-25II (R&D) and 100 ng·mL−1 DKK-1 (Peprotech) were added to the culture and maintained for 3 wk. After the appearance of rosettes (around day 15), the entire cell population was detached using Accutase (Millipore) and replated at a cell density of 2.5 × 104 cells per cm−2 on dishes coated with Matrigel (BD, Becton Dickinson). The cells were maintained in terminal differentiation medium, which was composed of N2 medium supplemented with B27 and 30 ng·mL−1 BDNF, until the end of differentiation.
Generation of hES H9 Inducible Lines.
To generate an inducible hES cell line, we first modified a pCMV-TetON-3G (Clontech) by removing the TetOn-3G cassette by digestion with EcoRI and HindIII (Biolabs). Then, we removed the CRE cassette of a pCAG-CRE vector (Addgene) and inserted the gel-purified TetON-3G cassette to generate a pCAG-TetON-3G vector. Next, we inserted in the pTRE3G-IRES responsive vector (Clontech) Gsx2 alone (in the first MCS), Gsx2 (in the first MCS) together with Ebf1 (in the second MCS), and Ebf1 alone (in the first MCS). Gsx2 cDNA was a gift from Kenneth Campbell, Cincinnati Children's Hospital Medical Center, Cincinnati, Ebf1 cDNA was a gift from Giacomo Consalez, Division of Neuroscience, San Raffaele Scientific Institute, Milan.
The hES H9 cell line was cultured as described. We used 8 × 106 cells for introducing the constructs by Nucleofection (Lonza) using a mouse ES cell nucleofection kit and electroporation protocol B16. We used 7 μg of pCAG-TetON-3G in the first round and 7 μg of pTRE-Gsx2 or pTRE-Gsx2-Ebf1 in the second round, together with 700 ng of linear resistant marker (Clontech, Puromycin during the first round and Hygromycin during the second round). Cells were then plated in two Matrigel-coated 6-cm dishes with Pluripro medium supplemented with Rock inhibitor (Y-2763226). After 72 h, antibiotics (Puromycin during the first round and Hygromycin during the second round) were added to the medium for positive selection. Following ∼2 wk in selection medium, hES cell colonies were carefully selected and expanded in Matrigel-coated 48-well plates. Clones were then expanded and tested for transgene expression after 48 h of doxycycline treatment. During the first round, the clones were screened by transient transfections with a pTRE-Luciferase vector (Clontech). During the second round, the clones were screened by 48 h of doxycycline treatment, and immunofluorescence analysis was performed for Gsx2 and Gsx2–Ebf1.
Immunofluorescence.
Cells were fixed in 4% (vol/vol) paraformaldehyde (PFA) for 15 min at room temperature (RT) and washed 3× with PBS. Cells were then permeabilized with 0.5% Triton (Sigma) and blocked with 10% normal goat serum (Vector) for 1 h at RT. Next, cells were incubated overnight at 4 °C with the following primary antibodies and dilutions: anti-OCT4, 1:100 (Santa Cruz); anti-OTX2, 1:500 (Chemicon); anti-PAX6, 1:2,000 (Hybridoma Bank); anti-NESTIN, 1:200 (R&D); anti–βIII-Tubulin, 1:1,000 (Promega); anti-MAP2, 1:500 (BD Bioscience); anti-CALBINDIN, 1:200 (Swant); anti-GABA, 1:500 (Sigma); anti-CTIP2, 1:500 (Abcam); anti-DARPP32, 1:200 (Epitomics); anti-GSX2 (Millipore); hNuclei (Chemicon); and Calbindin, Calretinin, and Parvalbumin (Swant). After three washes in PBS 0.1% Triton, appropriate secondary antibodies conjugated to Alexa fluorophores 488 or 568 (Molecular Probes, Invitrogen) were diluted 1:500 in blocking solution and mixed with Hoechst 33258 (5 µg·mL−1; Molecular Probes, Invitrogen) to counterstain the nuclei. Images were acquired with a Leica DMI 6000B microscope (5×, 10×, and 20× objectives) and analyzed with LAS-AF imaging software and then processed using Adobe Photoshop, only to adjust contrast for optimal RGB rendering with the same procedure in doxycycline-treated and untreated cells.
Histological specimens from transplantations were examined using a Leica TCS SP5 confocal microscope. Confocal images were taken at a resolution of 1024 × 1024 dpi and 150 Hz speed, and each focal plane was 1 μm thick. Laser intensity, gain, and offset were maintained constant in each analysis. Three animals for each transplant type were analyzed. hNCAM fiber quantification was performed using Spaceballs (MBFbiosciences).
Cell-Cycle Analysis with IdU and BrdU.
IddU (Sigma) was first added in the culture medium for 1.5 h followed by BrdU (Sigma) for 30 min. Cells were then fixed at the end of the BrdU treatment. For IdU/BrdU double labeling, primary antibodies used were mouse anti-BrdU/IdU (which recognizes both BrdU and IdU, clone B44, 1:100; BD), and rat anti-BrdU (clone BU1/75, 1:100; Abcam). After 4% PFA fixation, cells are first treated with 0.2N HCl for 5 min at RT and then with 2N HCl for 20 min at 37° for BrdU/IdU immunofluorescence. Cell-cycle lengths (estimation) were calculated as previously described (18): Cells labeled initially with IdU and leaving S-phase during the interval between IdU and BrdU were labeled with IdU but not BrdU (leaving fraction).
Cumulative BrdU Labeling.
BrdU is added to the cell culture medium for different time windows in different wells. BrdU immunofluorescence is performed as described above for the cell-cycle analysis.
Cell-Cycle Exit Study.
iGOF cell lines were treated with doxycycline from day 20 to day 30 of neuronal differentiation. At day 25 cells were exposed to BrdU for 2 h to label cells in the S-phase of the cell cycle. Neuronal differentiation was carried on until day30, when cells were fixed and processed as described above for BrdU immunofluorescence. Cells were also stained for Ki67 to label all proliferating cells at day 30. Cell-cycle exit index was calculated by dividing the total number of BrdU+ Ki67− cells by the total number of BrdU+ cells.
mmRNA Transfections.
The transfection mix was prepared according to the manual of the StemMACS mRNA Reprogramming Kit (Miltenyi Biotec) using the StemMACS mRNA Transfection Reagent and StemMACS mRNA Transfection Buffer. We used 200 ng mmRNA of GSX2 and EBF1 (gently provided by Miltenyi Biotec) daily for 5 consecutive days. As a transfection control, 100 ng of nuclear GFP (Miltenyi Biotec) was used the first day of transfection to monitor the transfection efficiency.
Transplantations.
Athymic NIH-FOXN1 Nude rats (Charles River) of 200–250 g were lesioned 8 d before transplantation with quinolinic acid (QA). The lesion was generated by monolateral injection of 210 nmol of freshly made QA in the right striatum using the following stereotaxic coordinates: AP, +0.6; L, ±2.8; V, 5.0. We injected 1 M PBS in the left striatum. Gsx2–Ebf1 iGOF cells were differentiated as described above. Cells were treated with doxycycline from day 20 to day 30 of differentiation to induce Gsx2 and Ebf1 expression. At day 30, cells were detached with Accutase supplemented with N2 1:100 for 20–30 min at 37 °C. Cells were then resuspended to obtain a single cell suspension at a concentration of 50 × 103 cells per μL and then transplanted in complete medium by bilateral stereotaxic transplantation in lesioned adult athymic rats using the following coordinates: AP, +0.9; L, +3.1/–3.1; DV, 5.0. A total of 2 × 105 cells (4 μL) per injection site were delivered by a single injection. Two months after transplantation, the animals were killed, transcardially perfused, and the brains cryosectioned for immunohistochemical analyses. Animal experiments were carried out according to the National regulatory requirements and the Institutional Animal Care and Use Committee (IACUC). The experimental protocol has been approved by the Ethics Committee of the San Raffaele Scientific Institute and by the Italian Ministry of Health (Protocol no. 722 approved on January 12th, 2016).
Patch-Clamp Recordings and Data Analysis.
Whole-cell patch-clamp recordings were performed at RT in voltage and current-clamp configuration. During recordings cells were visualized using an inverted microscope (Nikon Eclipse TE200) and maintained in a solution containing (in mM) NaCl (140), KCl (3), glucose (10), Hepes (10), MgCl2 (1), and CaCl2 (2) at pH 7.4 with NaOH. Pipettes were produced from borosilicate glass capillary tubes (Hilgenberg GmbH) by means of a horizontal puller (P-97, Sutter instruments), and their resistance was 2–4 MΩ, when filled with (in mM) CsCl (135), NaCl (3), Hepes (10), EGTA (10), CaCl2 (0.5), and MgCl2 (1) at pH 7.3 with CsOH. To isolate the sodium current, cells were recorded using an extracellular solution containing (in mM) NaCl (140), KCl (3), TEA-Cl (10), Hepes (10), 4-AP (5), MgCl2 (1), and CaCl2 (1) at pH 7.4 with NaOH, and pipettes were filled with a solution containing (in mM) CsCl (120), NaCl (10), TEA-Cl (20), Hepes (10), EGTA (10), and MgCl2 (2) at pH 7.3 with CsOH. Recordings were performed with an AXOPATCH 200B amplifier (Molecular Devices) and digitized with a DigiData1322A (Molecular Devices). Data were acquired using the software Clampex (Molecular Devices), sampled at 50 kHz and filtered at 10 kHz. The series resistance was minimized and monitored throughout the experiment.
Analysis was performed with Clampfit (Molecular Devices) and Origin 6.0 (Microcal Software Inc.). Statistics reported are mean ± SEM, unless otherwise specified. Statistical tests were performed using Instat (GraphPad Software). Two-tailed P values were used throughout.
Statistical Analysis.
Statistical tests were performed with PRISM software (GraphPad, version 6). Statistical significance was tested with the unpaired (nonparametric) t test as reported in each figure and legend. All results were expressed as means ± SD. The sample size was chosen based on our preliminary studies and on the variability across differentiations. Given that the long-term differentiation experiments (80 d) are susceptible to variability, we decided to perform five different biological experiments (Fig. 3) to address this issue. No data points were excluded from the reported analyses. Differentiation experiments were excluded when a poor neural induction was obtained (low Otx2, N-cadherin, and Pax6 expression).
The majority of the cell counting experiments (Figs. 1–4) were performed using specific softwares (CellProfiler, fully automatic, or ITCN in ImageJ, partially automatic); therefore, they were performed blindly. The remaining cell counts were performed manually; no blinding was performed.
Acknowledgments
We thank Alice Abbondanza for help with cell cultures; Oliver Brustle and Philippe Koch for their help in the generation of LT-NES cells; Paola Conforti and other members of our group for fruitful comments and technical assistance; WiCell for providing the H9 cell line; Roslin Cells for the RC17 cell line; and the families of HD patients for their continuous support. This work was supported by Marie Curie Reintegration Grant 268248 (to A.F.), CHDI Foundation Grant A7333 (to E. Cattaneo), Programmi di Ricerca Scientifica di rilevanza Nazionale Grant 2008JKSHKN_001 (to E. Cattaneo), and NeurostemcellRepair Grant NSCR 602278 (to E. Cattaneo).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1611473114/-/DCSupplemental.
References
- 1.Calabresi P, Picconi B, Tozzi A, Ghiglieri V, Di Filippo M. Direct and indirect pathways of basal ganglia: A critical reappraisal. Nat Neurosci. 2014;17(8):1022–1030. doi: 10.1038/nn.3743. [DOI] [PubMed] [Google Scholar]
- 2.Finger S, Heavens RP, Sirinathsinghji DJ, Kuehn MR, Dunnett SB. Behavioral and neurochemical evaluation of a transgenic mouse model of Lesch-Nyhan syndrome. J Neurol Sci. 1988;86(2-3):203–213. doi: 10.1016/0022-510x(88)90099-8. [DOI] [PubMed] [Google Scholar]
- 3.Zhang X, et al. Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell. 2010;7(1):90–100. doi: 10.1016/j.stem.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corbin JG, Gaiano N, Machold RP, Langston A, Fishell G. The Gsh2 homeodomain gene controls multiple aspects of telencephalic development. Development. 2000;127(23):5007–5020. doi: 10.1242/dev.127.23.5007. [DOI] [PubMed] [Google Scholar]
- 5.Pei Z, et al. Homeobox genes Gsx1 and Gsx2 differentially regulate telencephalic progenitor maturation. Proc Natl Acad Sci USA. 2011;108(4):1675–1680. doi: 10.1073/pnas.1008824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toresson H, Potter SS, Campbell K. Genetic control of dorsal-ventral identity in the telencephalon: Opposing roles for Pax6 and Gsh2. Development. 2000;127(20):4361–4371. doi: 10.1242/dev.127.20.4361. [DOI] [PubMed] [Google Scholar]
- 7.Waclaw RR, Wang B, Pei Z, Ehrman LA, Campbell K. Distinct temporal requirements for the homeobox gene Gsx2 in specifying striatal and olfactory bulb neuronal fates. Neuron. 2009;63(4):451–465. doi: 10.1016/j.neuron.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yun K, Potter S, Rubenstein JL. Gsh2 and Pax6 play complementary roles in dorsoventral patterning of the mammalian telencephalon. Development. 2001;128(2):193–205. doi: 10.1242/dev.128.2.193. [DOI] [PubMed] [Google Scholar]
- 9.López-Juárez A, et al. Gsx2 controls region-specific activation of neural stem cells and injury-induced neurogenesis in the adult subventricular zone. Genes Dev. 2013;27(11):1272–1287. doi: 10.1101/gad.217539.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Méndez-Gómez HR, Vicario-Abejón C. The homeobox gene Gsx2 regulates the self-renewal and differentiation of neural stem cells and the cell fate of postnatal progenitors. PLoS One. 2012;7(1):e29799. doi: 10.1371/journal.pone.0029799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garel S, Marín F, Grosschedl R, Charnay P. Ebf1 controls early cell differentiation in the embryonic striatum. Development. 1999;126(23):5285–5294. doi: 10.1242/dev.126.23.5285. [DOI] [PubMed] [Google Scholar]
- 12.Lobo MK, Karsten SL, Gray M, Geschwind DH, Yang XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci. 2006;9(3):443–452. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- 13.Lobo MK, Yeh C, Yang XW. Pivotal role of early B-cell factor 1 in development of striatonigral medium spiny neurons in the matrix compartment. J Neurosci Res. 2008;86(10):2134–2146. doi: 10.1002/jnr.21666. [DOI] [PubMed] [Google Scholar]
- 14.Delli Carri A, et al. Human pluripotent stem cell differentiation into authentic striatal projection neurons. Stem Cell Rev. 2013;9(4):461–474. doi: 10.1007/s12015-013-9441-8. [DOI] [PubMed] [Google Scholar]
- 15.Delli Carri A, et al. Developmentally coordinated extrinsic signals drive human pluripotent stem cell differentiation toward authentic DARPP-32+ medium-sized spiny neurons. Development. 2013;140(2):301–312. doi: 10.1242/dev.084608. [DOI] [PubMed] [Google Scholar]
- 16.Liew CG, Draper JS, Walsh J, Moore H, Andrews PW. Transient and stable transgene expression in human embryonic stem cells. Stem Cells. 2007;25(6):1521–1528. doi: 10.1634/stemcells.2006-0634. [DOI] [PubMed] [Google Scholar]
- 17.Waclaw RR, Wang B, Campbell K. The homeobox gene Gsh2 is required for retinoid production in the embryonic mouse telencephalon. Development. 2004;131(16):4013–4020. doi: 10.1242/dev.01272. [DOI] [PubMed] [Google Scholar]
- 18.Martynoga B, Morrison H, Price DJ, Mason JO. Foxg1 is required for specification of ventral telencephalon and region-specific regulation of dorsal telencephalic precursor proliferation and apoptosis. Dev Biol. 2005;283(1):113–127. doi: 10.1016/j.ydbio.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 19.Shibui S, Hoshino T, Vanderlaan M, Gray JW. Double labeling with iodo- and bromodeoxyuridine for cell kinetics studies. J Histochem Cytochem. 1989;37(7):1007–1011. doi: 10.1177/37.7.2659659. [DOI] [PubMed] [Google Scholar]
- 20.Becker KA, et al. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J Cell Physiol. 2006;209(3):883–893. doi: 10.1002/jcp.20776. [DOI] [PubMed] [Google Scholar]
- 21.Koch P, Opitz T, Steinbeck JA, Ladewig J, Brüstle O. A rosette-type, self-renewing human ES cell-derived neural stem cell with potential for in vitro instruction and synaptic integration. Proc Natl Acad Sci. 2009;106(9):3225–3230. doi: 10.1073/pnas.0808387106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho SY, et al. NeurphologyJ: An automatic neuronal morphology quantification method and its application in pharmacological discovery. BMC Bioinformatics. 2011;12:230. doi: 10.1186/1471-2105-12-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salomoni P, Calegari F. Cell cycle control of mammalian neural stem cells: Putting a speed limit on G1. Trends Cell Biol. 2010;20(5):233–243. doi: 10.1016/j.tcb.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 24.Tabar V, Studer L. Pluripotent stem cells in regenerative medicine: Challenges and recent progress. Nat Rev Genet. 2014;15(2):82–92. doi: 10.1038/nrg3563. [DOI] [PMC free article] [PubMed] [Google Scholar]