Significance
Natural killer (NK) cells play a key role in early viral control of CMV replication and in shaping the adaptive immune response. Despite an early control, CMV persists by exploiting host immune inhibitory pathways. Here, we describe a previously unidentified pathway wherein the cytokine Epstein–Barr virus-induced 3 (EBI3) affects the establishment of mouse cytomegalovirus (MCMV) latency. We also show that both human and mouse NK cells express EBI3 and the EBI3 receptor, gp130, after stimulation. MCMV-infected EBI3-deficient mice showed decreased IL-10 production by NK cells, increased dendritic cell maturation and activation of CD8+ T cells, and significantly diminished viral loads in the salivary glands and oral lavage. Together, our results provide insight into how CMV establishes latent infection.
Keywords: natural killer cell, EBI3, cytomegalovirus
Abstract
Natural killer (NK) cells are key mediators in the control of cytomegalovirus infection. Here, we show that Epstein–Barr virus-induced 3 (EBI3) is expressed by human NK cells after NKG2D or IL-12 plus IL-18 stimulation and by mouse NK cells during mouse cytomegalovirus (MCMV) infection. The induction of EBI3 protein expression in mouse NK cells is a late activation event. Thus, early activation events of NK cells, such as IFNγ production and CD69 expression, were not affected in EBI3-deficient (Ebi3−/−) C57BL/6 (B6) mice during MCMV infection. Furthermore, comparable levels of early viral replication in spleen and liver were observed in MCMV-infected Ebi3−/− and wild-type (WT) B6 mice. Interestingly, the viral load in salivary glands and oral lavage was strongly decreased in the MCMV-infected Ebi3−/− B6 mice, suggesting that EBI3 plays a role in the establishment of MCMV latency. We detected a decrease in the sustained IL-10 production by NK cells and lower serum levels of IL-10 in the MCMV-infected Ebi3−/− B6 mice. Furthermore, we observed an increase in dendritic cell maturation markers and an increase in activated CD8+ T cells. Thus, EBI3 dampens the immune response against MCMV infection, resulting in prolonged viral persistence.
Natural killer (NK) cells play an essential role in host defense against viral infections, particularly herpesviruses, such as cytomegalovirus (CMV) (1). During infection, NK cell activation is tightly controlled by the integration of signals derived from activating and inhibitory receptors, through the interaction with target or accessory cells, and from cytokine receptors. Several activating NK receptors exist, including the activating killer cell Ig-like receptors (KIRs) in humans, the activating Ly49 receptors in rodents, NKG2D, the natural cytotoxicity receptors (i.e., NKp30, NKp44, and NKp46), and the activating Fc receptor CD16 (2). The activating receptors recognize either stress-induced ligands on viral-infected cells, virus-encoded proteins, or Ig-coated cells. Signals from the activating receptors promote cytoskeletal rearrangements and proliferation, as well as secretion of cytolytic granules and cytokines (2). The inhibitory receptors Ly49 and KIR recognize polymorphic major histocompatibility complex class I ligands that can dampen or prevent the NK cells from attacking self (2).
NK cell-mediated control of viral infections has been studied extensively in mice infected with mouse CMV (MCMV). NK cells contribute directly to the early control of MCMV infection by eliminating the virus-infected cells. In C57BL/6 (B6) mice, Ly49H+ NK cells recognize MCMV-infected cells expressing the virus-encoded protein m157. This antigen-specific recognition leads to NK cell activation (3), as well as expansion and differentiation of memory NK cells (4), which is dependent on the DAP12 adapter protein, the costimulatory receptor DNAM-1, and the proinflammatory cytokine IL-12 (4–6). The DAP10 adapter protein and the cytokines IL-33 and IL-18 are required for optimal expansion of Ly49H+ NK cells, but not for memory NK cell differentiation (7–9). In addition, optimal activation of both Ly49H+ and Ly49H− NK cells and production of IFNγ during MCMV infection is critically dependent on both IL-12 and IL-18 (9, 10). In addition to mediating early control of MCMV infection, NK cells also play a role in shaping the subsequent adaptive immune responses. Crosstalk between NK cells and dendritic cells (DCs) early during MCMV infection affects the outcome of the T-cell responses. IL-10 secreted by various immune cells, including NK cells, dampens the T-cell response by negatively affecting the maturation of DCs, and in the absence of IL-10 secretion of IFNγ and TNFα by NK cells enhances the maturation of DCs, which boosts the T-cell response (11).
The cytokine Epstein–Barr virus-induced 3 (EBI3) was first identified in B cells infected with Epstein–Barr virus (12), but several other cells from the immune system have also been found to express and secrete EBI3, including activated DCs, regulatory T cells, and regulatory B cells (13–15). EBI3 belongs to the IL-12 family of cytokines that consists of the four heterodimeric cytokines IL-12 (p35/p40), IL-23 (p19/p40), IL-27 (p28/EBI3), and IL-35 (p35/EBI3), which signal through unique pairings of the five receptor chains IL-12Rβ1, IL-12Rβ2, IL-23R, gp130, and WSX-1 (16). IL-27 and IL-35 lack disulfide linkage and pair poorly and are therefore less stable and secreted in much lower amounts than the disulfide-linked family members IL-12 and IL-23 (16). It has been well-documented that IL-12 and IL-23 function as proinflammatory cytokines. However, studies with IL-27 and IL-35 have been complicated by their instability in solution and the lack of specific reagents. IL-27 has been proposed to possess both proinflammatory and anti-inflammatory properties in that it can promote Th1 polarization (17) but also stimulate the production of IL-10 (18, 19). IL-35 appears to possess anti-inflammatory properties with the predominant mechanism being suppression of T-cell proliferation and conversion of naive T cells into IL-10–producing T cells (13). It has been speculated that EBI3 can be secreted and function as a homodimer; however, this remains to be elucidated (15, 16). Here we identify activated human and mouse NK cells as producers of EBI3 and show that EBI3 promotes the persistence of MCMV infection.
Results
Gene Expression Analysis of NKL Cells Stimulated Through Activating Receptors.
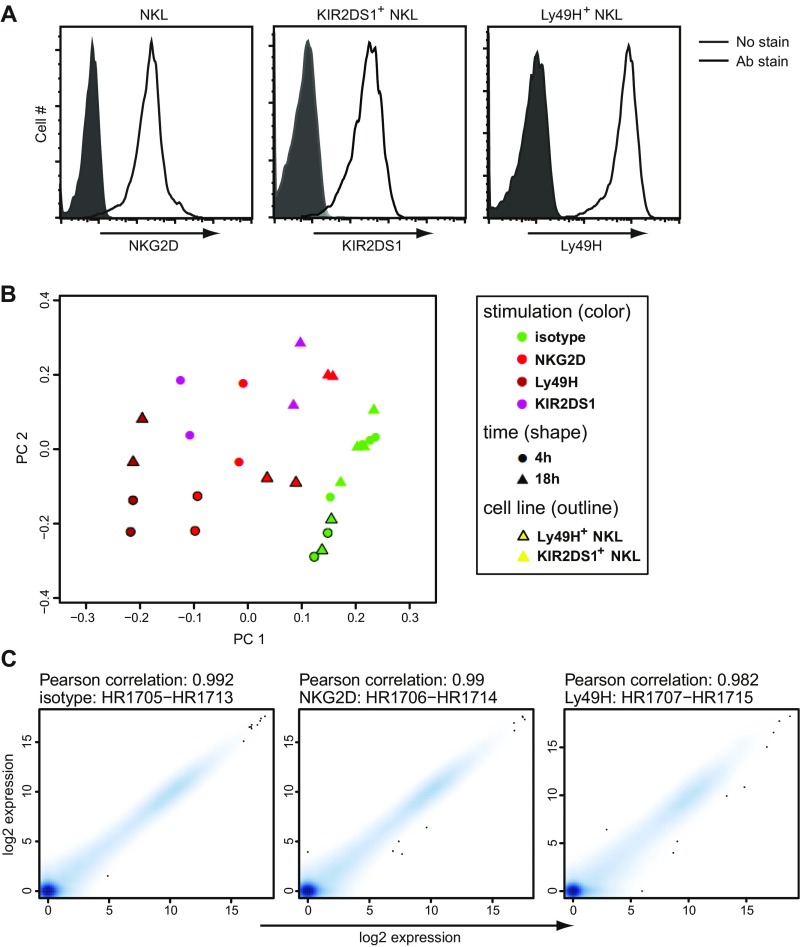
We investigated the global gene expression profile of human NK cells stimulated through various activating receptors to identify genes involved in the regulation of NK cell functions. We used the human-transformed NK cell line, NKL, stably transduced with either the activating mouse Ly49H receptor or the activating human KIR2DS1 receptor (Fig. S1A). NKL cells, which constitutively express the activating NKG2D receptor (Fig. S1A), were stimulated with saturating amounts of plate-bound isotype-matched control Ig or receptor-specific monoclonal antibodies (mAbs), and the RNA was extracted and used for deep sequencing. Before analysis the quality of the gene expression data was verified using principal component analysis (PCA) and by calculating the Pearson correlation coefficient for the experimental replicates (Fig. S1 B and C).
Fig. S1.
(A) NKG2D, KIR2DS1, and Ly49H cell-surface expression of transduced NKL cells was verified by flow cytometry. (B) The PCA plot was generated using the Bioconductor package made4 (version 1.40.0) (32) and calculated after removing nonexpressed genes and scaling the log2 expression values to 0 mean and unit variance. (C) The replicate plots were calculated as Pearson correlations of the log2 expression values of all measured genes and plotted using the smoothScatter function of R. (B and C) The individual datasets represent independent experiments with NKL cells.
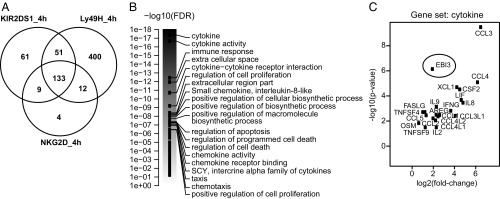
In our gene expression analysis of the activated NKL cells, we scrutinized genes that were up- and down-regulated with at least a 1.5-fold change relative to the control samples. A total of 254, 158, or 596 genes were identified to be up- and down-regulated 4 h after KIR2DS1, NKG2D, or Ly49H stimulation, respectively (Fig. 1A and Dataset S1). We examined the 133 genes that were commonly regulated by all three stimulations (Fig. 1A) by gene set enrichment analysis (GSEA). GSEA is a pathway-based analysis that detects changes in expression of genes in entire pathways or gene sets and is therefore more robust than analyses based upon individual genes. GSEA identified 20 gene sets and pathways that were significantly different relative to the control samples (Fig. 1B). The gene set with the highest enrichment score identified by GSEA was “cytokines,” which included many genes known to be expressed by activated NK cells, i.e., IFNG, FASLG, XCL1, CCL1, CCL3, CCL4, and CCL5 (2, 20, 21) (Fig. 1C). Interestingly, we detected an up-regulation of EBI3 in the activated NKL cells (Fig. 1C). EBI3 expression has recently been shown to be up-regulated in human NK cells in response to Fc receptor activation, but only in the presence of IL-12 (22), confirming our results that activated NK cells can express EBI3; however, to our knowledge no previous studies have examined EBI3 protein expression and secretion by human and mouse NK cells. Furthermore, the effect of EBI3 on NK cell function in vivo is unknown.
Fig. 1.
Gene expression analysis of NKL cells stimulated through activating receptors. (A) RNA was isolated from NKL cells stimulated with the indicated plate-bound Abs for 4 h and then analyzed by deep sequencing. Shown is the number of genes that were up- and down-regulated with at least a 1.5-fold change in the receptor-stimulated NKL cells relative to the control-treated cells (n = 2–4 independent experiments). (B and C) GSEA was performed on the deep-sequencing data. (B) Shown are the strongest associated gene sets/pathways when analyzing genes that were significantly differentially expressed between NKG2D-stimulated NKL cells versus control-treated cells at 4 h. The scale is given as DAVID-reported –log10 (false discovery rate) value, i.e., the gene-set score. The score of all gene sets is shown, but only the top 20 are labeled. (C) Shown is a volcano plot of the genes contained in the “cytokine” gene set, which had the strongest enrichment score in B. The x axis indicates a log2 fold-change, with positive values corresponding to genes for which expression is up-regulated.
EBI3 Protein Expression and Secretion Is Increased in Human NK Cells in Response to Receptor- and Cytokine-Mediated Stimulation.
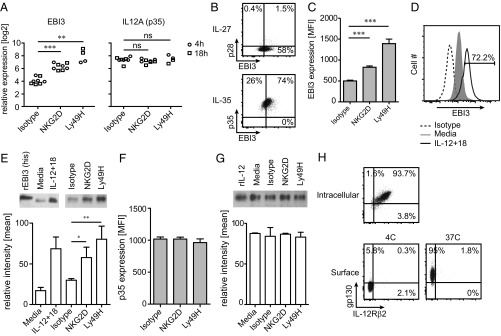
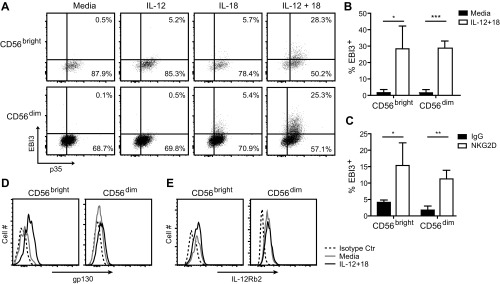
NKL cells constitutively express EBI3, which was significantly increased upon receptor-mediated stimulation (Fig. 2A). IL12A (i.e., p35) was also constitutively expressed in NKL cells, but its expression was not increased after receptor-mediated stimulation (Fig. 2A). In agreement with the gene expression data, the majority of or all NKL cells express EBI3 and p35, respectively (Fig. 2B), and the amount of intracellular EBI3, but not p35, proteins was increased following receptor-mediated stimulation for 24 h (Fig. 2 C and F, respectively). In addition, the intracellular EBI3 protein level was increased in response to IL-12 plus IL-18 treatment for 24 h (Fig. 2D). EBI3 and p35 protein was found in the supernatant of NKL cells after culture, but only EBI3 secretion was increased after stimulation (Fig. 2 E and G), which was consistent with the intracellular protein levels observed. To confirm that EBI3 is also expressed by primary human NK cells, we examined the intracellular EBI3 protein level in resting and activated CD56dim (i.e., mature) and CD56bright (i.e., immature) NK cells derived from healthy human blood donors. Resting NK cells did not express EBI3 (Fig. S2 A and B), suggesting that the constitutive expression of EBI3 observed in NKL cells is due to cell transformation and/or the in vitro culture conditions. Treatment with IL-12 alone for 24 h did not induce EBI3 protein expression, and only a slight increase was observed after treatment with IL-18 alone (Fig. S2A). However, treatment with IL-12 in combination with IL-18 for 24 h led to a robust induction of EBI3 protein expression in both CD56bright and CD56dim NK cells (Fig. S2 A and B). Priming with IL-2 for 24 h did not induce EBI3 protein expression; however, a significant increase was observed after stimulation of IL-2–primed NK cells with plate-bound anti-NKG2D mAb for 24 h (Fig. S2C). In contrast to EBI3 expression, the majority of resting NK cells constitutively expressed p35 (Fig. S2A). We did not detect any protein expression of p28 (Fig. 2B). NKL cells stained positive for both gp130 and IL-12Rβ2 as detected by intracellular protein staining (Fig. 2H, “intracellular”). Furthermore, gp130 was detected at the cell surface of NKL cells, but only after incubating the cells with the anti-gp130 Ab at 37 °C (Fig. 2H, “surface”), suggesting that the gp130 receptor is rapidly recirculated at the cell surface of the NKL cells. Resting primary human NK cells expressed low levels of gp130 at the cell surface, but the expression was increased in response to IL-12 plus IL-18 treatment, with the strongest induction observed in the CD56bright NK cells (Fig. S2D). Furthermore, the majority of CD56bright NK cells and all of the CD56dim NK cells constitutively expressed IL-12Rβ2 as detected by intracellular protein staining (Fig. S2E). No cell-surface staining of IL-12Rβ2 was detected by flow cytometry (Fig. 2H, “surface”).
Fig. 2.
EBI3 protein expression is increased in response to receptor- and cytokine-mediated stimulation. (A) Gene expression data for EBI3 and p35 in NKL cells stimulated with the indicated plate-bound Abs for 4 h (circle) or 18 h (square) were obtained from the deep-sequencing analysis (n = 2–4 independent experiments; statistical analysis is shown for 4-h samples). (B) Intracellular EBI3, p28, and p35 protein expression was examined by flow cytometry in resting NKL cells. Data are representative of three independent experiments. (C and D) Intracellular EBI3 protein expression was measured by flow cytometry in NKL cells stimulated with (C) plate-bound Abs or (D) IL-12+IL-18 for 24 h. (C) Data show mean ± SD from three independent experiments. (D) Data are representative of three independent experiments. (E) NKL cells were stimulated with plate-bound Abs, media, or IL-12+IL-18 for 24 h, and EBI3 was detected in the supernatant by Western blot analysis. The graphs show mean ± SD from two or three independent experiments. (F) Intracellular p35 protein expression was measured by flow cytometry in NKL cells stimulated with plate-bound Abs for 24 h. (G) NKL cells were stimulated with media or plate-bound Abs for 24 h, and p35 was detected in the supernatant by Western blot analysis. The graphs show mean ± SD from three independent experiments. (H) Surface and intracellular protein expression of gp130 and IL-12Rβ2 was examined by flow cytometry in resting NKL cells. Data are representative of three independent experiments. Statistical analysis was performed by two-tailed unpaired Student’s t test (*P < 0.05, **P < 0.01, and ***P < 0.001).
Fig. S2.
EBI3 and gp130 protein expression is increased in human primary NK cells in response to receptor- and cytokine-mediated stimulation. Human primary CD56bright and CD56dim NK cells were isolated from blood obtained from healthy blood donors and (A, B, D, and E) either left unstimulated or stimulated with IL-12 and/or IL-18 for 24 h or (C) primed with IL-2 for 24 h before being stimulated with plate-bound isotype-matched control Ig or anti-NKG2D mAb for 24 h. (A–C) Intracellular EBI3 and p35 protein expression was analyzed by flow cytometry. (A) Data are representative of three different donors. (B and C) Data show mean ± SD from three different donors. (D) Surface gp130 expression and (E) intracellular IL-12Rβ2 expression was examined by flow cytometry. Data are representative of three different donors. Statistical analysis was performed by two-tailed unpaired Student’s t test (*P < 0.05, **P < 0.01, and ***P < 0.001).
MCMV Infection Induces EBI3 Expression in Mouse NK Cells.
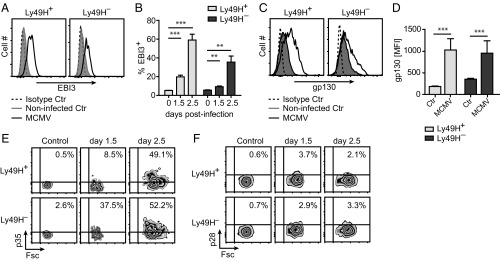
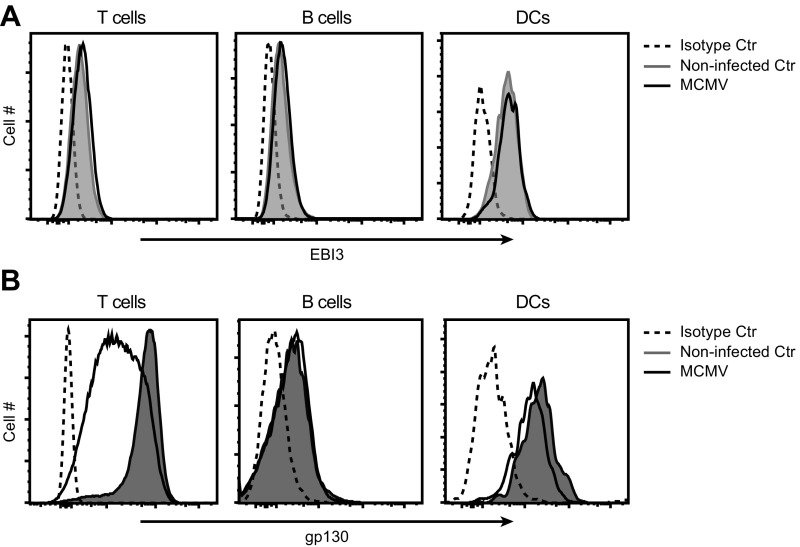
Based on our results with human NK cells and the NKL cells transduced to express Ly49H, we examined if MCMV infection could be used as an in vivo model system to study the functional role of EBI3. During MCMV infection we detected an increase in the intracellular EBI3 protein level in splenic NK cells (Fig. 3 A and B), whereas no difference was detected in T cells, B cells, and DCs from the same samples (Fig. S3A). The increase in EBI3 protein expression in mouse NK cells was strongest at day 2.5 postinfection (p.i.) (Fig. 3B) with the highest levels observed in MCMV-specific Ly49H+ (Fig. 3 A and B). The cell-surface expression of gp130 was increased on splenic NK cells in response to MCMV infection (Fig. 3 C and D), which indicates a possibility for an autocrine effect. This increase in gp130 surface expression was specific for NK cells, as the surface expression on T cells, B cells, and DCs was either decreased or unchanged (Fig. S3B). p35, but not p28, protein expression was increased in splenic NK cells in response to MCMV infection (Fig. 3 E and F), and like EBI3, the strongest induction of p35 protein expression was observed at day 2.5 p.i. (Fig. 3E).
Fig. 3.
MCMV infection induces EBI3 protein expression in mouse NK cells. Intracellular (A and B) EBI3, (E) p35, and (F) p28 protein expression was examined by flow cytometry in splenic Ly49H+ and Ly49H– NK cells from noninfected and infected (day 1.5 and day 2.5) WT B6 mice. (A, E, and F) Data are representative of four mice for each time point from two independent experiments. (B) Data show mean ± SD from four or six mice for each time point from two independent experiments. (C and D) Surface expression of gp130 was examined by flow cytometry in splenic Ly49H+ and Ly49H– NK cells from noninfected and MCMV-infected (day 2) WT B6 mice. (C) Data are representative of six mice from two independent experiments. (D) Data show mean ± SD from six mice from two independent experiments. Statistical analysis was performed by two-tailed unpaired Student’s t test (**P < 0.01 and ***P < 0.001).
Fig. S3.
(A) Intracellular EBI3 protein expression was examined by flow cytometry in splenic T cells, B cells, and DCs from noninfected and MCMV-infected (day 2.5) WT B6 mice. (B) gp130 surface expression was examined by flow cytometry in splenic T cells, B cells, and DCs from noninfected and MCMV-infected (day 2) WT B6 mice. (A and B) Data are representative of four or six mice from two independent experiments.
EBI3-Deficient Mice Show Decreased MCMV Replication in the Salivary Glands and Oral Lavage.
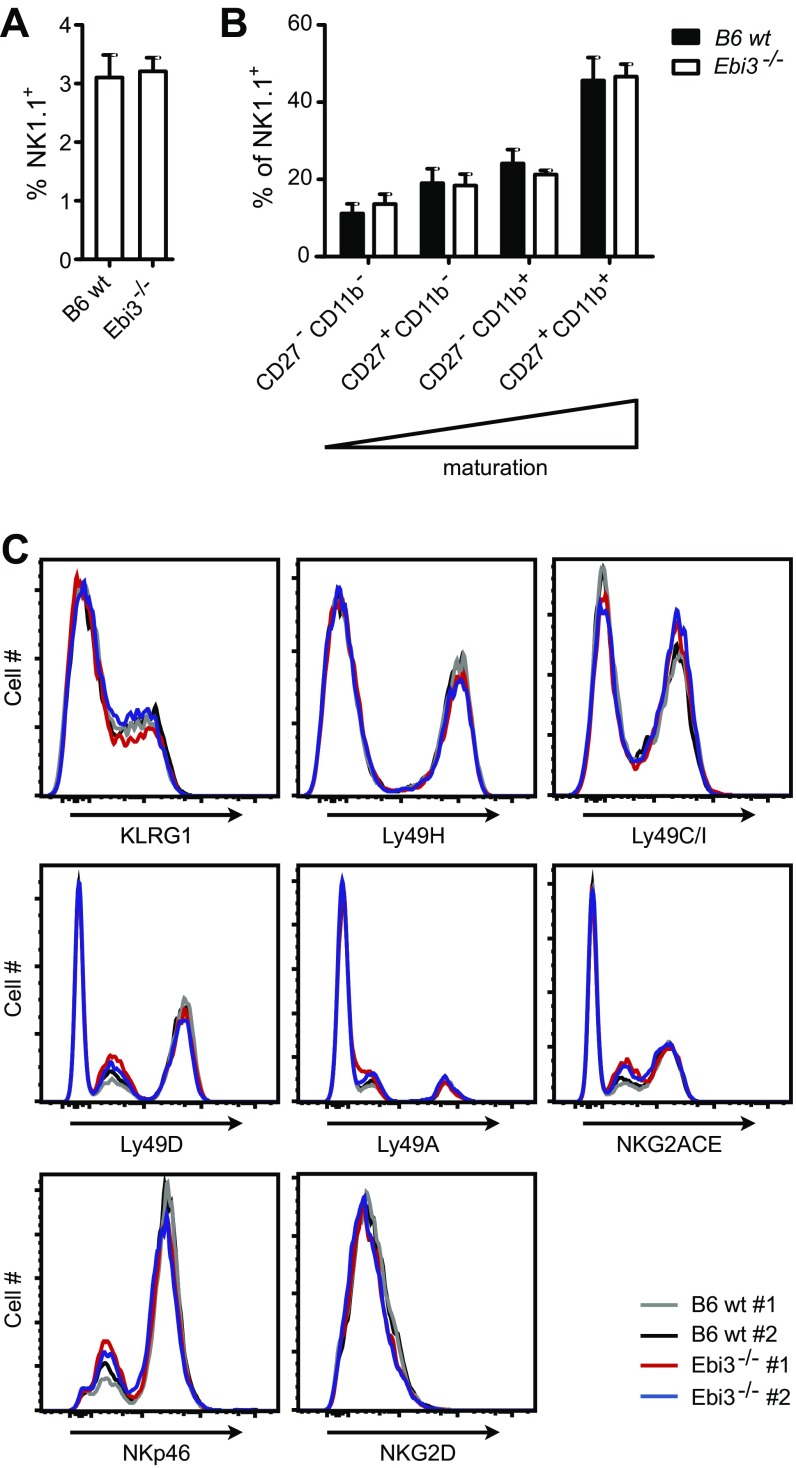
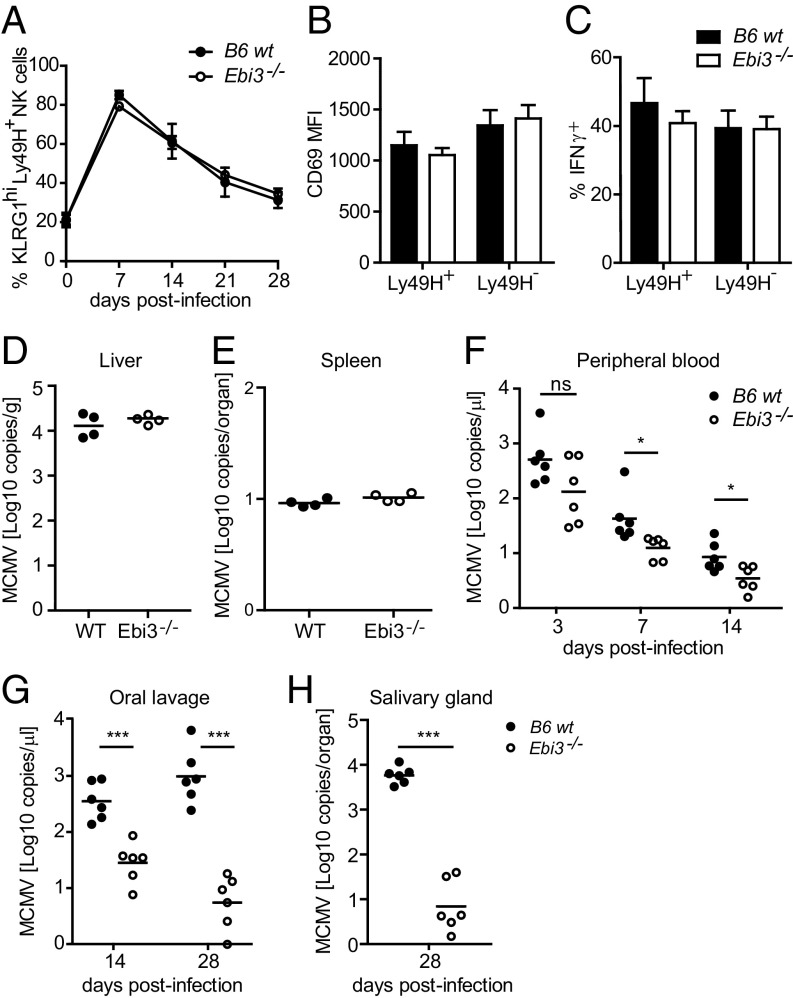
To examine the functional role of EBI3 in vivo, we infected wild-type (WT) and EBI3-deficient (Ebi3−/−) C57BL/6 (B6) mice with MCMV. The Ebi3−/− B6 mice lack exons 2–5 of the Ebi3 gene, corresponding to amino acids 24–228 of the EBI3 protein (23), which includes the functional fibronectin type 3 domain found at amino acids 128–216 (24). Thus, the truncated version of EBI3 likely to be present in the deficient mice would be nonfunctional. No difference was observed between the two mouse strains with regard to the percentages of splenic NK cells and the immature and mature NK cell subsets (Fig. S4 A and B). Furthermore, the splenic NK cells showed similar expression levels of several NK receptors, including KLRG1, Ly49H, Ly49C/I, Ly49D, Ly49A, NKG2,A,C,E, NKp46, and NKG2D (Fig. 4A and Fig. S4C). Thus, EBI3 deficiency is not associated with any phenotypic or maturational defects in the NK cells. During MCMV infection, the early activation of splenic NK cells, measured by CD69 expression and IFNγ production at day 1.5 p.i., was comparable between the WT and Ebi3−/− B6 mice (Fig. 4 B and C, respectively). Furthermore, no difference in the viral load in liver and spleen from the mice was observed at day 4 p.i. (Fig. 4 D and E). We also observed a similar expansion of peripheral blood KLRG1hi Ly49H+ NK cells in the MCMV-infected mice (Fig. 4A). However, the viral load was significantly (P < 0.05) decreased in the blood in the MCMV-infected Ebi3−/− B6 mice at day 7 and day 14 p.i. (Fig. 4F). Interestingly, we found that MCMV was cleared most efficiently in the salivary glands and oral lavage of the Ebi3−/− B6 mice (Fig. 4 G and H), which are important sites for virus persistence and dissemination (25).
Fig. S4.
(A) The percentage of splenic TCRβ− NK1.1+ cells and (B) the maturation stages from WT and Ebi3−/− B6 mice were examined by flow cytometry. n = 5 for each mouse strain from two independent experiments. Data show mean ± SD. (C) Surface levels of the indicated NK cell receptors on splenic TCRβ− NK1.1+ cells from WT and Ebi3−/− B6 mice were examined by flow cytometry. Data shown are from two mice from each strain and are representative of five mice from two independent experiments.
Fig. 4.
EBI3-deficient mice show decreased MCMV replication in the salivary glands and oral lavage. (A) Expansion of peripheral KLRG1hi Ly49H+ NK cells from WT or Ebi3−/− B6 mice was measured by flow cytometry at days 0, 7, 14, 21, and 28 p.i. n = 6 for each mouse strain and time point from two independent experiments (mean ± SD). (B) CD69 expression and (C) IFNγ production was measured by flow cytometry in splenic Ly49H+ and Ly49H– NK cells from WT or Ebi3−/− B6 mice at day 1.5 post-MCMV infection. n = 4 for each mouse strain from two independent experiments. Data show mean ± SD. MCMV titer in WT or Ebi3−/− B6 mice was determined by real-time PCR in (D) liver and (E) spleen at day 4 p.i. in (F) peripheral blood at days 3, 7, and 14 p.i., in (G) saliva (by oral lavage) at day 14 and day 28 p.i., and (H) in salivary glands at day 28 p.i. n = 4 or 6 for each mouse strain and time point from two independent experiments. Statistical analysis was performed by two-tailed unpaired Student’s t test (*P < 0.05 and ***P ≤ 0.0003).
EBI3 Promotes IL-10 Production by NK Cells and Negatively Affects the Maturation of DCs and Activation of CD8+ T Cells During MCMV Infection.
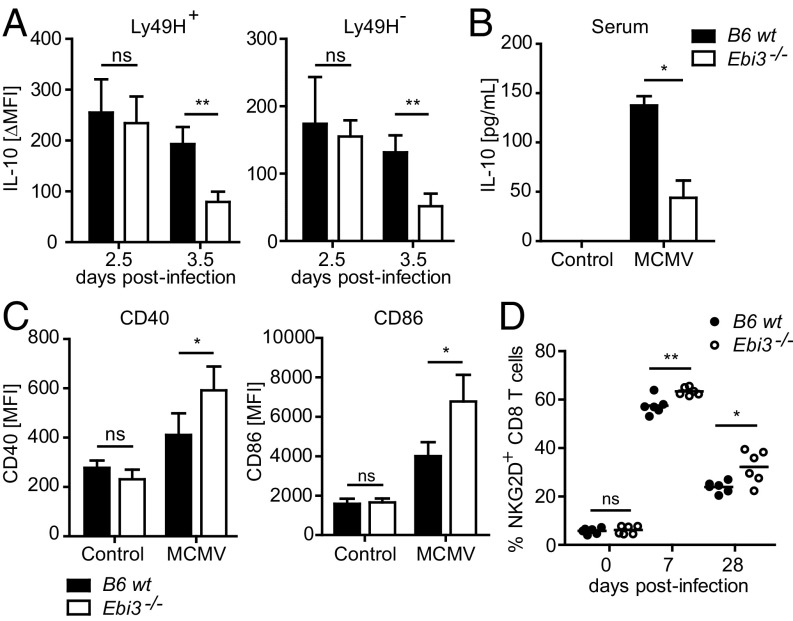
Several cells in the immune system, including NK cells, produce IL-10 early after MCMV infection. The early production of IL-10 promotes virus replication in the salivary glands by negatively affecting the maturation of DCs, leading to poor priming of T cells (26, 27). We found that splenic Ly49H+ and Ly49H– NK cells from the mice produced similar levels of IL-10 at day 2.5 post-MCMV infection (Fig. 5A). However, the IL-10 production was significantly decreased at day 3.5 p.i. in the Ebi3−/− B6 mice (Fig. 5A), suggesting that EBI3 plays an essential role in the sustained, but not initial, production of IL-10 by NK cells during MCMV infection. The serum level of IL-10 peaks at day 5 p.i. during MCMV infection (26). We found that the levels of IL-10 in sera were significantly decreased in the MCMV-infected Ebi3−/− B6 mice (Fig. 5B), indicating that the overall production of IL-10 was affected in the mice. We further examined the impact of EBI3 deficiency on DC maturation and T-cell activation, both of which are affected by IL-10 during MCMV infection (26, 27). We observed higher levels of the maturation markers CD86 and CD40 on splenic DCs derived from the Ebi3−/− B6 mice at day 5 p.i. (Fig. 5C). Furthermore, the percentage of activated peripheral CD8+ T cells (i.e., NKG2D-positive CD8+ T cells) was increased in the MCMV-infected Ebi3−/− B6 mice (Fig. 5D). Thus, together these results indicate that EBI3 promotes persistent MCMV infection, presumably in part by sustaining IL-10 production, which negatively affects the maturation of DCs and the activation of T cells.
Fig. 5.
EBI3 promotes IL-10 production by NK cells and negatively affects the maturation of DCs and activation of CD8+ T cells during MCMV infection. (A) IL-10 expression was measured by flow cytometry in splenic Ly49H+ and Ly49H– NK cells from WT or Ebi3−/− B6 mice at day 2.5 and 3.5 p.i. n = 4 for each mouse strain from two independent experiments. Shown are the mean ± SD of the induced IL-10 mean fluorescence intensity (MFI) in MCMV-infected compared with noninfected control mice. (B) IL-10 was measured in sera from MCMV-infected (day 5) or noninfected WT or Ebi3−/− B6 mice by ELISA. Data are representative of four mice for each time point from two independent experiments. (C) CD40 and CD86 expression was measured by flow cytometry on splenic DCs from MCMV-infected (day 5) or noninfected control WT or Ebi3−/− B6 mice. n = 4 for each mouse strain from two independent experiments (mean ± SD). (D) NKG2D expression was measured on peripheral CD8+ T cells from WT or Ebi3−/− B6 mice by flow cytometry at days 0, 7, and 28 p.i. n = 6 for each mouse strain and time point from two independent experiments. Statistical analysis was performed by two-tailed unpaired Student’s t test (*P < 0.05 and **P < 0.01).
Discussion
In this study, we show that both human and mouse NK cells express EBI3 protein after stimulation. Furthermore, we describe a previously unidentified pathway wherein EBI3 affects the establishment of MCMV latency. Interestingly, mice deficient in EBI3 showed almost no viral replication in salivary glands and oral lavage, which are the main sites for viral persistence and dissemination (25), whereas the early viral replication in the spleen and liver was comparable between the EBI3-deficient and WT mice. MCMV-infected mice displayed lower levels of IL-10 in the absence of EBI3, an effect that was observed in splenic NK cells at day 3.5 p.i. and in the serum at day 5 p.i. As reported previously, production of IL-10 early during MCMV infection is important for limiting DC maturation and T-cell activation to prevent harmful immune-mediated tissue damage in the host (26, 27). During MCMV infection, IL-10 production by NK cells and other immune cells can suppress the maturation DCs, leading to poor priming of CD4+ T cells (26). Furthermore, NK cell-mediated IL-10 production during MCMV infection regulates CD8+ T-cell activation, where a blockade of IL-10 increases the CD8+ T-cell response against MCMV (27). We detected an increase in both the maturation markers of DCs and the percentage of activated CD8+ T cells in the MCMV-infected EBI3-deficient mice, indicating that the observed decrease in IL-10 production in the EBI3-deficient mice was able to enhance the subsequent T-cell response. In this study we found that EBI3 affects the production of IL-10 by NK cells during MCMV infection, as measured by direct ex vivo intracellular staining for IL-10 protein. However, it remains to be determined whether the production of IL-10 by other cell subsets, such as myeloid cells and CD4+ T cells, is affected by EBI3 during MCMV infection. Although beyond the scope of this article, it is also possible that EBI3 plays an additional role(s) in MCMV latency that is independent of IL-10. Production of IL-10 early during MCMV infection depends on the magnitude of viral replication (28). Following a low-dose MCMV infection, viral replication is controlled rapidly within a couple of days and only low levels of IL-10 are produced to limit the immune response. In contrast, during a high-dose MCMV infection, which leads to sustained and elevated levels of viral replication, sustained and higher amounts of IL-10 are needed to limit the immune response and prevent tissue damage (28). Our data suggest that EBI3 is essential to sustain IL-10 production during a high-dose MCMV infection.
EBI3 can interact with p28 to form IL-27 heterodimers or with p35 to form IL-35 heterodimers (16). We detected p35, but never p28, protein expression in human and mouse NK cells. Both EBI3 and p35 were constitutively secreted by the NKL cells. Therefore, in our experimental settings activated NK cells do not produce or secrete IL-27 heterodimers. For several reasons we were not able to distinguish between IL-35 heterodimers and EBI3 homodimers and therefore were unable to determine which of the two species are predominantly formed and secreted by the activated NK cells. First, there are no blocking Abs that can distinguish between EBI3 and IL-35. Second, IL-35 is very unstable in solution (16), which makes the detection of small amounts by ELISA or immunoprecipitation difficult or impossible. Finally, mice deficient in p35 also lack IL-12, a cytokine essential for NK cell activation and proliferation in response to MCMV infection (6, 10). EBI3 protein and the gp130 receptor was up-regulated in splenic NK cells during MCMV infection, whereas no up-regulation was observed in T cells, DCs, or B cells at the time points examined in this study. However, the basal or background level differed between the various cell types examined. We cannot distinguish between the role of EBI3 derived from NK cells versus other cell types during MCMV infection as there presently is no conditional knockout of Ebi3 to definitively address this issue.
The immune system has established multiple layers of control to ensure effective protection against viral infections, but at the same time to keep the immune system in check to avoid excessive inflammation and autoimmunity. NK cells play a key role in the early control of CMV replication and in modulating the adaptive immune response against the virus (1). Despite the cooperative work between NK cells and T and B cells, CMV can establish persistent infections in mice and humans by exploiting host immune inhibitory pathways to modulate the virus–host balance toward its own benefit (25). Induction of the regulatory cytokine IL-10 during infection represents one such immune inhibitory pathway (29). Our results suggest that the induction of EBI3 represents an inhibitory pathway that can be exploited by CMV to establish latent infection. Whether EBI3 can affect the persistent infection of other herpesviruses remains to be elucidated. However, it is noteworthy that Epstein–Barr virus, another virus that can establish latency by exploiting the IL-10 inhibitory pathway (25), is also a strong inducer of EBI3 expression (12).
Materials and Methods
WT and EBI3-deficient (B6.129 × 1Ebi3tm1Rsb/J) mice on a C57BL/6 background were maintained and used in accordance with guidelines of the University of California at San Francisco (UCSF) Institutional Animal Care and Use Committee. Peripheral blood mononuclear cells (PBMCs) were isolated from blood obtained from the Stanford Blood Center or the Blood Centers of the Pacific under an Institutional Review Board approved protocol (IRB# 10–00265) by density gradient centrifugation using Ficoll-Paque PLUS (GE Healthcare Bio-Sciences AB).
Reagents, vendors, and protocols are included in SI Materials and Methods. Details of the mice and cells used, as well as details of the methods used including MCMV infection, in vitro stimulation of NK cells, real-time PCR, flow cytometry, Western blot analysis, IL-10 ELISA, and deep sequencing are presented in SI Materials and Methods. Additional questions pertaining to methods, protocols, and reagents are available upon request.
SI Materials and Methods
Mice and MCMV Infection.
WT and EBI3-deficient (B6.129 × 1Ebi3tm1Rsb/J) mice on a C57BL/6 background were maintained. Mice between 7–10 wk of age were infected with 104 pfu of the MCMV Smith strain by intraperitoneal injection.
Cells.
The KIR2DS1+ NKL cells were generated by stable transduction of the human NK cell line, NKL, with a pMxs-puro retroviral vector containing cDNA of human KIR2DS1 (GenBank accession no. X89892), a kind gift from Roberto Biassoni, Istituto Giannina Gaslini, Genoa, Italy. The KIR2DS1+ NKL cells were selected with puromycin and subsequently sorted to generate a homogenous KIR2DS1-expressing cell line. The Ly49H+ NKL cells were generated by stable transduction of NKL cells with a pMX retroviral vector, a kind gift from Hisashi Arase, Osaka University, Osaka, containing cDNA of the mouse B6 allele of Klra8 (GenBank accession no. U12889). The Ly49H+ NKL cells were single-cell–cloned to generate a homogenous Ly49H-expressing cell line. NKL cells were maintained in RPMI-1640 medium (Corning Cellgro, Mediatech Inc.) containing 10% (vol/vol) FBS (Thermo Scientific), 2 mM l-glutamine (UCSF cell culture facility), and penicillin (100 IU/mL) and streptomycin (100 μg/mL) (Corning Cellgro), supplemented with 200 U/mL human IL-2 (generously provided by Prometheus Laboratories, Inc.). PBMCs were isolated from blood obtained from the Stanford Blood Center or the Blood Centers of the Pacific by density gradient centrifugation using Ficoll-Paque PLUS (GE Healthcare Bio-Sciences AB). NK cells were purified from PBMCs using an NK cell isolation kit (Miltenyi Biotec GmbH) (>90% purity) or by sorting CD3–CD56+ NK cells (>99% purity). The primary human NK cells were cultured in RPMI-1640 medium containing 10% FBS, 2 mM l-glutamine, penicillin, and streptomycin.
In Vitro Stimulation of NKL Cells and Primary Human NK Cells.
Plate-bound Ab stimulations.
Nunc maxisorp ELISA plates (Thermo Fisher Scientific) were washed twice in PBS and then coated with 1 µg/mL anti-NKG2D mAb (149810, R&D Systems), 5 µg/mL anti-NKG2D mAb (1D11, BioLegend), 5 µg/mL anti-KIR2D mAb (clone HP3E4, BD Biosciences), 5 µg/mL anti-Ly49H (3D10, eBioscience), 1 or 5 µg/mL control mouse IgG1 (MOPC-21, UCSF Monoclonal Antibody Core), or 5 µg/mL control mouse IgM (11E10, eBioscience) in PBS for 24 h at 4 °C. The ELISA plates were washed twice in PBS and blocked in complete culture medium for 10 min at room temperature (RT) before 150,000 cells were added to each well in the ELISA plate. NKL cells were stimulated with control IgG1, control IgM, anti-NKG2D, anti-KIR2D, or anti-Ly49H plate-bound mAb for 4 h or 18 h (for deep sequencing) or for 24 h (for flow cytometry and Western blot analysis) at 37 °C and 5% CO2. Primary human NK cells were primed in 200 U/mL human IL-2 for 24 h and then stimulated with plate-bound control IgG1 or anti-NKG2D mAb for 24 h (flow cytometry) at 37 °C and 5% CO2.
Cytokine stimulations.
NKL cells and primary human NK cells were stimulated with 20 ng/mL human IL-18 (R&D Systems) and/or 10 ng/mL human IL-12 (R&D Systems) for 24 h (for flow cytometry and Western blot analysis) at 37 °C and 5% CO2. For the EBI3, p35, and p28 measurements by flow cytometry, GolgiSTOP and GolgiPLUG (BD Bioscience, final dilution 1:1,500 and 1:1,000, respectively) were added for the last 4 h (NKL cells) or 6 h (primary human NK cells) of stimulation.
Real-Time PCR.
For MCMV quantification, oral lavage was collected by washing the mouse sublingual cavity with sterile saline, and 1-μL samples were used for quantitative PCR (qPCR) analysis. DNA was isolated from peripheral blood and homogenates of salivary glands by using a ReliaPrep Blood gDNA Miniprep System kit (Promega), and 1-μL samples were used for qPCR analysis. The relative copy numbers of MCMV IE1 were determined by quantitative PCR analysis with SYBR green master mix reagent (Invitrogen) using standard conditions and the following primer pair: MCMV-IE1_Forward—5′-AGCCACCAACATTGACCACGCAC-3′ and MCMV-IE1_Reverse—5′-GCCCCAACCAGGACA CACAACTC-3′. Mice were monitored daily for weight loss and survival.
Flow Cytometry.
The mAbs used for cell-surface staining were the following: FITC-conjugated anti-mouse MHC class II (M5/114.15.2, eBioscience); FITC-conjugated anti-Ly49D (4E5, BioLegend); FITC-conjugated anti-mouse NKG2A,C,E (20d5, eBioscience); FITC-conjugated anti-mouse CD69 (HI.2F3, BD Biosciences); FITC- or PE-conjugated anti-Ly49H (3D10, BioLegend); PE-conjugated anti-mouse CD27 (M-T271, BioLegend); PE-conjugated anti-mouse NKp46 (29A1.4, BioLegend); PE-conjugated anti-mouse NKG2D (CX5, eBioscience); PE-conjugated anti-mouse CD86 (GL1, BioLegend); PE-conjugated anti-mouse NKG2D (CX5, eBioscience); PE-conjugated anti-mouse gp130 (4H1B35, BioLegend); PE-conjugated anti-human gp130 (2E1B02, BioLegend); APC-conjugated anti-mouse CD11b (M1/70, BioLegend); APC-conjugated anti-mouse CD40 (3/23, BioLegend); APC-conjugated anti-mouse KLRG1 (2FI/KLRG1, BioLegend); APC-conjugated anti-human and -mouse IL-12Rβ2 (305719, R&D Systems); APC-conjugated anti-human KIR2DL1/DS1 (11PB6, Miltenyi Biotec); APC-conjugated anti-human NKG2D (149810, R&D Systems); PerCP-Cy5.5-conjugated anti-mouse NK1.1 (PK136, BioLegend); PerCP-Cy5.5-conjugated anti-human CD56 (HCD56, BioLegend); AF700-conjugated anti-mouse TCRβ (H57-597, BioLegend); AF700-conjugated anti-human CD3 (HIT3a, BioLegend); PE-Cy7–conjugated anti-mouse CD11c (HL3, BD Biosciences); PE-Cy7–conjugated anti-mouse TCRβ (H57-597, BioLegend); APC-Cy7-conjugated anti-mouse TCRβ (H57-597, BioLegend); vF421-conjugated anti-mouse CD8 (53-6.7, Tonbo Biosciences); PB-conjugated anti-Ly49A (A1/Ly49A, BioLegend); and PB-conjugated anti-B220 (R43-6B2, BioLegend). The mAbs used for intracellular staining werethe following: eFlour660-conjugated anti-human p28 (3D1p28, eBioscience); PE-conjugate anti-human EBI3 (B032F6, BioLegend); APC-conjugated anti-human and mouse p35 (27537, R&D Systems); APC-conjugated anti-human and mouse IL-12Rβ2; APC-conjugated anti-mouse IFNγ (XMG1.2, BioLegend); PE-conjugated anti-mouse IL-10 (JES5-16E3, BioLegend); PE-conjugated anti-mouse EBI3 (355022, R&D Systems); AF647-conjugated anti-mouse p28 (516903, BioLegend). For surface staining, cells were incubated with the indicated mAbs or isotype-matched control antibodies (Jackson ImmunoResearch, BD Biosciences, eBioscience, or BioLegend) for 20 min on ice. When applicable, mouse Fc receptors were blocked with anti-CD16 + CD32 mAb 2.4G2 before staining. When indicated, NKL cells were stained by incubating the cells with mAbs for 1 h at 37 °C with 5% CO2. For intracellular staining, cells were fixed and permeabilized using the Cytofix/Cytoperm kit (BD Biosciences) according to the manufacturer’s protocol and subsequently stained with the indicated mAbs or isotype-matched control antibodies (BD Biosciences, eBioscience, or BioLegend) for either 20 min on ice or 30 min at RT. Samples were acquired on a LSRII (BD Biosciences) and analyzed with FlowJo software (Tree Star). Live single cells were gated based on forward (Fsc) and side light-scatter (Ssc) profiles. Human NK cells were gated as CD3−CD56+ cells. Mouse NK cells, B cells, T cells, and DCs were gated as TCRβ− NK1.1+ cells, TCRβ− B220+ cells, TCRβ+ NK1.1− cells, and TCRβ−NK1.1−B220−MHC class II+CD11chi cells, respectively.
IL-10 ELISA.
Blood was obtained from MCMV-infected (day 5) and noninfected control mice by cardiac puncture and was allowed to clot in serum separator tubes for 30 min at RT. Sera were collected after centrifugation (13,000 × g for 10 min at RT) and the IL-10 concentration was determined using an OptEIA Mouse IL-10 ELISA kit (BD Biosciences) according to the manufacturer’s instructions.
Western Blot Analysis.
Supernatants from stimulated NKL cells were concentrated 10× using 10K Amicon Ultra centrifugal filters (Millipore). The supernatants for p35 Western blot analysis were precleared with goat anti-mouse IgG Dynabeads by rotating for 30 min at 4 °C, and the Dynabeads were removed by magnet before concentration. The concentrated supernatants were then diluted in RIPA buffer (150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH 8), and the samples were incubated at 70 °C for 10 min in NuPAGE SDS Sample Buffer (Life Technologies) containing NuPAGE Reducing Agent (Life Technologies). Proteins were separated on a 4–12% Bis–Tris NuPAGE gel (Life Technologies) in MES or MOPS NuPAGE Running Buffer (Life Technologies) containing NuPAGE Antioxidant (Life Technologies) at 200 V for 90 min at 4 °C. Human recombinant EBI3 (ab83026, Abcam) or human recombinant IL-12 (R&D Systems) was included as positive control. Samples were transferred to Immobilon PVDF membranes (Millipore) in NuPAGE transfer buffer (Life Technologies) containing NuPAGE antioxidant and 10% methanol for 1 h at 20 V at RT and blocked in 5% milk in Tris-buffered saline with Tween 20 (TBST) buffer (0.01 M Tris, pH 8, 0.15 M NaCl, 0.5% Tween-20) for 1 h at RT. Membranes were probed with rabbit anti-human EBI3 polyclonal antibody (H-110, Santa Cruz Biotechnology, 1:400 dilution) or mouse anti-human/mouse IL-12/IL-35 p35 monoclonal Ab (27537, R&D Systems, 1:500 dilution) in TBST + 2% BSA+ 0.05% azide buffer overnight at 4 °C, followed by incubation with HRP-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch) or HRP-conjugated donkey anti-mouse IgG (Jackson ImmunoResearch) in TBST + 2% BSA buffer) for 1 h at RT and then visualized by using the Amersham ECL Prime WB detection reagent (GE Healthcare). The mean intensity of the bands was calculated using Photoshop by subtracting the histogram mean for the background from the mean obtained for each sample.
Deep Sequencing.
Total RNA was isolated from NKL cells stimulated with plate-bound Ab for 4 h or 18 h using an RNeasy Kit (Qiagen). Libraries were constructed according to standard Illumina protocols. The integrity and quality of RNA and cDNA were monitored by using an Agilent Bioanalyzer 2100. Ultrahigh-throughput sequencing was performed on the Illumina HiSeq for 2 × 100-bp paired-end reads. Fastqc was used for read quality control, and fastq quality filter (-q 30 -p 85) and fastq quality trimmer (-t 30 -l 40) were used for read filtering. All reads where then aligned with tophat2 and the GRCh37_r69 human reference genome, using the default setting except for allowing segment-mismatches (2), segment-length (25), and mate-std-dev (395) (30). Read counts were obtained with Htseq running in intersection-nonempty mode. Read counts where normalized using the calcNormFactors function of the edgeR software package together with log2 transformation. Comparative analysis was performed using linear models on these normalized values as recommended by Dillies et al. (31). The individual datasets represent independent experiments with NKL cells. The GSEA was performed using the online analysis tool DAVID (version 6.7) together with all gene sets from the Gene Ontology (GOTERM_MF_FAT definition).
Supplementary Material
Acknowledgments
We thank the L.L.L. laboratory for comments and discussions; Viola Lam and Tsukasa Nabekura for assistance with MCMV infections; Melissa Lodoen for generating the Ly49H+ NKL cells; Sandra Laurence Lopez-Verges for generating the KIR2DS1+ NKL cells; Roberto Biassoni for providing the cDNA of human KIR2DS1; Hisashi Arase for providing the pMXs retroviral vector; Prometheus Laboratories, Inc., for providing human IL-2; and Novo Nordisk, Inc., for providing the deep-sequencing platform. L.L.L. is an American Cancer Society Professor and funded by National Institutes of Health Grants AI066897 and AI068129 and the Parker Institute for Cancer Immunotherapy. H.J. is supported by a Lundbeck Foundation Postdoctoral Fellowship. In addition, the work was supported by NIH Grants U19AI057229, U19AI100627, R33CA183654, R33CA0183692, R01GM10983601, R01CA184968, R01CA19665701, R21CA183660, R01NS08953301, 5UH2AR067676, and R01HL120724; the Northrop-Grumman Corporation; Novartis Grant CMEK162AUS06T; Pfizer Grant 123214; Juno Therapeutics Grant 122401; Department of Defense Grants OC110674 and W81XWH-14-1-0 180; Gates Foundation Grant OPP1113682; and Food and Drug Administration Grant BAA-15-00121.
Footnotes
Conflict of interest statement: L.L.L. and the University of California, San Francisco have licensed intellectual property rights regarding NKG2D for commercial applications.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700231114/-/DCSupplemental.
References
- 1.Lanier LL. Evolutionary struggles between NK cells and viruses. Nat Rev Immunol. 2008;8(4):259–268. doi: 10.1038/nri2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lanier LL. Up on the tightrope: Natural killer cell activation and inhibition. Nat Immunol. 2008;9(5):495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002;296(5571):1323–1326. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- 4.Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature. 2009;457(7229):557–561. doi: 10.1038/nature07665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nabekura T, et al. Costimulatory molecule DNAM-1 is essential for optimal differentiation of memory natural killer cells during mouse cytomegalovirus infection. Immunity. 2014;40(2):225–234. doi: 10.1016/j.immuni.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun JC, et al. Proinflammatory cytokine signaling required for the generation of natural killer cell memory. J Exp Med. 2012;209(5):947–954. doi: 10.1084/jem.20111760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orr MT, et al. Ly49H signaling through DAP10 is essential for optimal natural killer cell responses to mouse cytomegalovirus infection. J Exp Med. 2009;206(4):807–817. doi: 10.1084/jem.20090168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nabekura T, Girard J-P, Lanier LL. IL-33 receptor ST2 amplifies the expansion of NK cells and enhances host defense during mouse cytomegalovirus infection. J Immunol. 2015;194(12):5948–5952. doi: 10.4049/jimmunol.1500424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madera S, Sun JC. Cutting edge: Stage-specific requirement of IL-18 for antiviral NK cell expansion. J Immunol. 2015;194(4):1408–1412. doi: 10.4049/jimmunol.1402001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orange JS, Wang B, Terhorst C, Biron CA. Requirement for natural killer cell-produced interferon gamma in defense against murine cytomegalovirus infection and enhancement of this defense pathway by interleukin 12 administration. J Exp Med. 1995;182(4):1045–1056. doi: 10.1084/jem.182.4.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vivier E, et al. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331(6013):44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devergne O, et al. A novel interleukin-12 p40-related protein induced by latent Epstein-Barr virus infection in B lymphocytes. J Virol. 1996;70(2):1143–1153. doi: 10.1128/jvi.70.2.1143-1153.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collison LW, et al. The inhibitory cytokine IL-35 contributes to regulatory T-cell function. Nature. 2007;450(7169):566–569. doi: 10.1038/nature06306. [DOI] [PubMed] [Google Scholar]
- 14.Wang R-X, et al. Interleukin-35 induces regulatory B cells that suppress autoimmune disease. Nat Med. 2014;20(6):633–641. doi: 10.1038/nm.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wirtz S, et al. EBV-induced gene 3 transcription is induced by TLR signaling in primary dendritic cells via NF-kappa B activation. J Immunol. 2005;174(5):2814–2824. doi: 10.4049/jimmunol.174.5.2814. [DOI] [PubMed] [Google Scholar]
- 16.Vignali DAA, Kuchroo VK. IL-12 family cytokines: Immunological playmakers. Nat Immunol. 2012;13(8):722–728. doi: 10.1038/ni.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pflanz S, et al. IL-27, a heterodimeric cytokine composed of EBI3 and p28 protein, induces proliferation of naive CD4+ T cells. Immunity. 2002;16(6):779–790. doi: 10.1016/s1074-7613(02)00324-2. [DOI] [PubMed] [Google Scholar]
- 18.Awasthi A, et al. A dominant function for interleukin 27 in generating interleukin 10-producing anti-inflammatory T cells. Nat Immunol. 2007;8(12):1380–1389. doi: 10.1038/ni1541. [DOI] [PubMed] [Google Scholar]
- 19.Stumhofer JS, et al. Interleukins 27 and 6 induce STAT3-mediated T cell production of interleukin 10. Nat Immunol. 2007;8(12):1363–1371. doi: 10.1038/ni1537. [DOI] [PubMed] [Google Scholar]
- 20.Robertson MJ. Role of chemokines in the biology of natural killer cells. J Leukoc Biol. 2002;71(2):173–183. [PubMed] [Google Scholar]
- 21.Eischen CM, Schilling JD, Lynch DH, Krammer PH, Leibson PJ. Fc receptor-induced expression of Fas ligand on activated NK cells facilitates cell-mediated cytotoxicity and subsequent autocrine NK cell apoptosis. J Immunol. 1996;156(8):2693–2699. [PubMed] [Google Scholar]
- 22.Campbell AR, et al. Gene expression profiling of the human natural killer cell response to Fc receptor activation: Unique enhancement in the presence of interleukin-12. BMC Med Genomics. 2015;8:66. doi: 10.1186/s12920-015-0142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nieuwenhuis EES, et al. Disruption of T helper 2-immune responses in Epstein-Barr virus-induced gene 3-deficient mice. Proc Natl Acad Sci USA. 2002;99(26):16951–16956. doi: 10.1073/pnas.252648899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campbell ID, Spitzfaden C. Building proteins with fibronectin type III modules. Structure. 1994;2(5):333–337. doi: 10.1016/s0969-2126(00)00034-4. [DOI] [PubMed] [Google Scholar]
- 25.Stack G, Stacey MA, Humphreys IR. Herpesvirus exploitation of host immune inhibitory pathways. Viruses. 2012;4(8):1182–1201. doi: 10.3390/v4081182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mandaric S, et al. IL-10 suppression of NK/DC crosstalk leads to poor priming of MCMV-specific CD4 T cells and prolonged MCMV persistence. PLoS Pathog. 2012;8(8):e1002846. doi: 10.1371/journal.ppat.1002846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee S-H, Kim K-S, Fodil-Cornu N, Vidal SM, Biron CA. Activating receptors promote NK cell expansion for maintenance, IL-10 production, and CD8 T cell regulation during viral infection. J Exp Med. 2009;206(10):2235–2251. doi: 10.1084/jem.20082387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tarrio ML, et al. Proliferation conditions promote intrinsic changes in NK cells for an IL-10 response. J Immunol. 2014;193(1):354–363. doi: 10.4049/jimmunol.1302999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Humphreys IR, et al. Cytomegalovirus exploits IL-10-mediated immune regulation in the salivary glands. J Exp Med. 2007;204(5):1217–1225. doi: 10.1084/jem.20062424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim D, et al. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dillies M-A, et al. French StatOmique Consortium A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief Bioinform. 2013;14(6):671–683. doi: 10.1093/bib/bbs046. [DOI] [PubMed] [Google Scholar]
- 32.Culhane AC, Thioulouse J, Perrière G, Higgins DG. MADE4: An R package for multivariate analysis of gene expression data. Bioinformatics. 2005;21(11):2789–2790. doi: 10.1093/bioinformatics/bti394. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.