Significance
Postnatal growth retardation is a Down syndrome (DS) phenotype, but its mechanism is unknown. Our previous studies have shown that Huntingtin-associated protein 1 (Hap1) is important for the postnatal growth of mice. Here we report that Hap1 binds DDB1- and CUL4-associated factor 7 (Dcaf7) competitively in the cytoplasm with dual-specificity tyrosine phosphorylation-regulated kinase 1A (DYRK1A), a protein implicated in DS. Loss of Hap1 in the hypothalamus, which can impair postnatal growth, is associated with increased levels of DYRK1A. Transgenic DS mice overexpressing DYRK1A show reduced Hap1–Dcaf7 association in the hypothalamus. Furthermore, the overexpression of DYRK1A in the hypothalamus led to delayed growth in postnatal mice, suggesting that DYRK1A regulates the Hap1–Dcaf7 interaction and postnatal growth and that targeting Hap1 or Dcaf7 may ameliorate growth retardation in DS.
Keywords: Hap1, DYRK1A, Dcaf7, WDR68, Down syndrome
Abstract
Huntingtin-associated protein 1 (Hap1) is known to be critical for postnatal hypothalamic function and growth. Hap1 forms stigmoid bodies (SBs), unique neuronal cytoplasmic inclusions of unknown function that are enriched in hypothalamic neurons. Here we developed a simple strategy to isolate the SB-enriched fraction from mouse brain. By analyzing Hap1 immunoprecipitants from this fraction, we identified a Hap1-interacting SB component, DDB1 and CUL4 associated factor 7 (Dcaf7)/WD40 repeat 68 (WDR68), whose protein level and nuclear translocation are regulated by Hap1. Moreover, we found that Hap1 bound Dcaf7 competitively in cytoplasm with dual-specificity tyrosine phosphorylation-regulated kinase 1A (DYRK1A), a protein implicated in Down syndrome (DS). Depleting Hap1 promoted the DYRK1A–Dcaf7 interaction and increased the DYRK1A protein level. Transgenic DS mice overexpressing DYRK1A showed reduced Hap1–Dcaf7 association in the hypothalamus. Furthermore, the overexpression of DYRK1A in the hypothalamus led to delayed growth in postnatal mice, suggesting that DYRK1A regulates the Hap1–Dcaf7 interaction and postnatal growth and that targeting Hap1 or Dcaf7 could ameliorate growth retardation in DS.
Huntingtin-associated protein 1 (Hap1) is a cytoplasmic protein highly expressed in neurons of the CNS (1, 2). Hap1 was initially recognized for its binding to polyglutamine-expanded huntingtin (Htt), the protein responsible for Huntington’s disease (3). Because Hap1 is expressed at various levels in different types of neurons (1, 4–8), it is likely involved in cell type-specific functions of the brain. For example, Hap1 is abundantly expressed in the hypothalamus, and its expression in the hypothalamus influences the central control of feeding (9); as a direct result, mice lacking Hap1 are malnourished and die in the early postnatal period (5, 10, 11). The function of Hap1 is also age dependent and appears to be important for postnatal neurogenesis in the paraventricular nuclei, the lateral hypothalamus (5, 11), and the dentate gyrus of the hippocampus (7).
The specific function of Hap1 in the hypothalamus may be associated with unique cytoplasmic structures called “stigmoid bodies” (SBs), which are formed by Hap1 in the hypothalamus (12, 13) and appear to be non–membrane-bound, ovoid to circular in shape, and 0.5–3 µm in diameter (14, 15). Widely regarded as a determinant marker for SBs, Hap1 is capable of sequestering several important disease-related proteins into SB-like inclusions in cell cultures, including Abelson helper integration site 1 (Ahi1), androgen receptor, TBP, and ataxin-3 (6, 16–18). However, in vivo evidence for such sequestration exists only for Ahi1, which is responsible for specific forms of Joubert syndrome. Other proteins, such as SorLA/LR11, sortilin, 5-HT7 receptor, and 14-3-3, are also reported to be associated with SBs (19–21). Although SBs are abundant in the hypothalamus, their functions remain elusive, despite speculation that they might be involved in sex-steroid maturation (15). We believe that identifying new protein components of the SBs will help shed light on the specific function of Hap1 in the hypothalamus.
In the current study, we started out by developing a strategy to allow the identification of proteins interacting with Hap1 in hypothalamic SBs. Through mass spectrometry analysis followed by validation of candidate proteins, we identified DDB1 and CUL4 associated factor 7 (Dcaf7)/WD40 repeat 68 (WDR68) as a protein that interacts with Hap1 and a component of the SB. Dcaf7 contains WD40-repeat (WDR) domains, is highly conserved from animals to plants, and is involved in a variety of cellular processes. It binds DYRK1A, a dual-specificity tyrosine phosphorylation-regulated kinase that is encoded by a gene located on chromosome 21 and is overexpressed in Down syndrome (DS) (22). DYRK1A is a major candidate gene in producing features of DS, and transgenic mice overexpressing DYRK1A show cognitive and behavioral alterations similar to those in persons with DS (23, 24). Also, recent studies show that administration of epigallocatechin-3-gallate (EGCG), which antagonizes DYRK1A, can improve the cognitive function of individuals with DS (25, 26).
We found that Hap1 controls the protein level of Dcaf7 and inhibits its nuclear translocation. More importantly, Hap1 and DYRK1A competitively bind Dcaf7 in the cytoplasm, and DYRK1A overexpression in the hypothalamus of DS mice leads to a decreased association between Hap1 and Dcaf7. Furthermore, we found that mice injected in the hypothalamus with Adeno-associated viruses (AAV) expressing DYRK1A gained less body weight during the postnatal period; this result could explain, in part, the growth retardation phenotype in DS. Thus, our work offers insight into the mechanism underlying the postnatal growth retardation in DS and also opens up the possibility of targeting Hap1 or Dcaf7 as a treatment for DS.
Results
A Strategy to Enrich SBs from Mouse Brain.
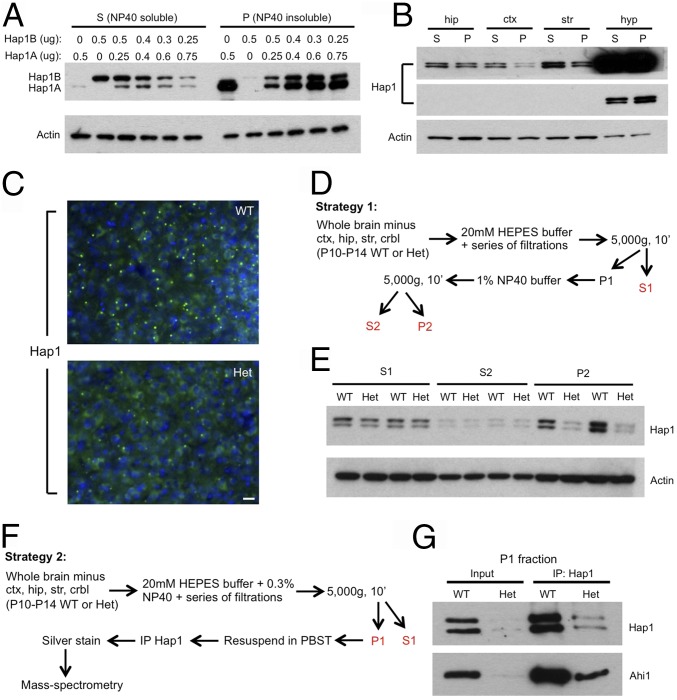
Because the SB is a Hap1-containing nonmembranous cytoplasmic structure (15) and because transfected Hap1A, one of the two murine Hap1 isoforms, can initiate the formation of this structure in HEK293 cells (12), we set out to see whether we could enrich SBs using the detergent Nonidet P-40. With increasing ratios of Hap1A to Hap1B transfected into HEK293 cells, we found more Hap1 was retained in the Nonidet P-40–insoluble (P) fraction (Fig. 1A), indicating that the detergent-insoluble pellet indeed contained inclusions formed by Hap1A. To test the applicability of this strategy in vivo, we analyzed the relative abundance of Hap1 in Nonidet P-40–soluble (S) and –insoluble (P) fractions from mouse hippocampus, cortex, striatum, and hypothalamus. The highest P/S ratio of Hap1 was seen in the hypothalamus (Fig. 1B), in agreement with the finding that SBs are more abundant in the hypothalamus than in other tissues (5, 10, 14). Thus, these results showed that the use of a detergent-based strategy to enrich SBs in mouse brain was feasible.
Fig. 1.
A strategy to isolate the SB-enriched fraction from mouse brain. (A) Different ratios of Hap1A to Hap1B were transfected into HEK293 cells, and the lysate was separated into Nonidet P-40–soluble (S) and Nonidet P-40–insoluble (P) fractions. Western blotting shows that, as the ratio of Hap1A to Hap1B increases, more Hap1 is found in the P fraction. (B) Western blotting of Hap1 on S and P fractions from the hippocampal (hip), cortical (ctx), striatal (str), and hypothalamic (hyp) tissues isolated from postnatal day 18 WT mouse brain. Both longer (Upper) and shorter (Middle) exposures for Hap1 are shown. (C) Immunostaining of Hap1 in postnatal day 14 WT and Hap1-KO (Het) mouse hypothalamus demonstrates markedly fewer Hap1 puncta in Het hypothalamus. (Scale bar: 20 µm.) (D) A strategy to isolate the SB-enriched fraction. (E) Western blotting of Hap1 on WT and Het mouse brain fractions obtained using strategy 1. (F) A strategy to use the SB-enriched fraction for identifying novel proteins associated with Hap1-contianing SB. (G) IP of Hap1 in P1 fractions from postnatal day 11 WT and Het mouse brain derived with strategy 2.
Heterozygous Hap1-KO (Het) mice are suggested to have fewer Hap1-containing SBs (10). Our staining also showed a decrease in the number of Hap1-containing SBs in the hypothalamus of Het mouse brain (Fig. 1C). Next, we used a detergent-based strategy to isolate the SB-enriched fraction from mouse brain (excluding regions that express a low amount of Hap1) with help from a previous protocol (Fig. 1D) (27). Compared with Het control mice, WT mice had significantly more Hap1 in the P2 fraction, even though there was no obvious difference in soluble Hap1 level in the two groups (Fig. 1E); our results support an enrichment of Hap1-containing SBs in the P2 fraction in the hypothalamus of WT mice. Notably, the S2 fraction after 1% Nonidet P-40 treatment contained very little Hap1, suggesting that the P1 fraction is already enriched in Hap1-containing SBs that are resistant to further Nonidet P-40 extraction. Thus, the P1 fraction was used to identify Hap1 partners in our simplified strategy 2 (Fig. 1F). Indeed, the enrichment of Hap1-containing SBs in the P1 and P2 fractions was confirmed by staining Hap1 on a glass slide loaded with these fractions (Fig. S1). After resuspension in PBS and Triton X-100 (PBST), the P1 fraction was subjected to immunoprecipitation (IP) by a monoclonal antibody against Hap1. The efficiency of IP was tested via Western blotting, in which high levels of Hap1 and Ahi1, a protein that is known to bind tightly to Hap1 (6), were found in the WT sample as compared with the Het sample (Fig. 1G). With these steps we successfully developed a protocol to isolate the SB-enriched fraction, which can be used to identify novel SB-associated proteins, from mouse brain.
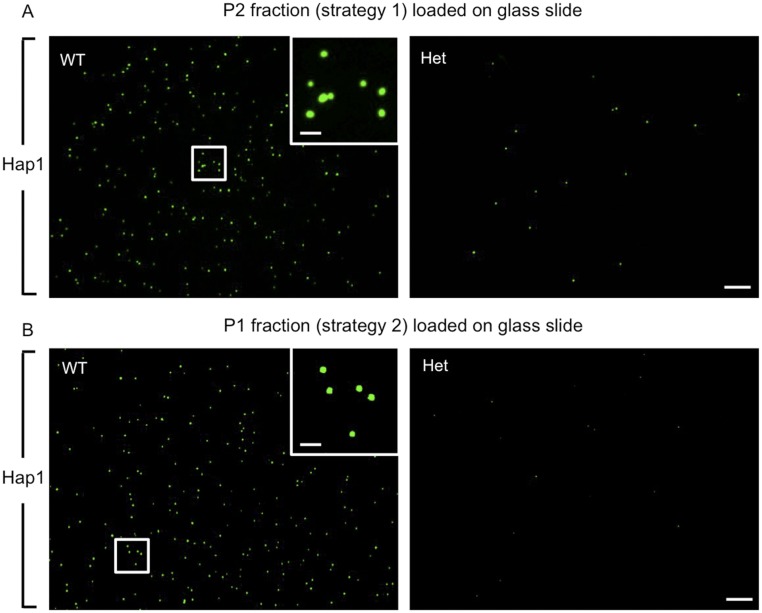
Fig. S1.
Hap1 immunostaining of the SB-enriched fractions from mouse brains. Postnatal day 11 WT and Het P2 fractions derived with strategy 1 (Fig. 1D) (A) or P1 fractions derived with strategy 2 (Fig. 1F) (B) were loaded on glass slides and immunostained with a Hap1 antibody. Numerous Hap1 puncta were observed in WT fractions, but markedly fewer could be seen in Het fractions. (Scale bars: 20 µm.) Insets are enlarged images of the boxed areas. (Scale bars: 5 µm.)
Dcaf7 Interacts with Hap1 and Is a Component of the SB.
Next, we ran the Hap1 immunoprecipitants from WT and Het mice on a gel and performed silver staining. By comparing the banding of WT and Het samples, we identified one prominent band, A, and another weak band, B, that were more enriched in the WT sample, beyond the known bands for Hap1 and Ahi1 (Fig. 2A). The protein constitutions of the two bands were uncovered by mass spectrometry analysis (Fig. 2B). Two major proteins—a growth cone-associated protein (Gap43) (16) and a highly conserved WD40 repeat-containing scaffolding protein important for craniofacial development, Dcaf7/WDR68 (28)—were found in band A, whereas five isoforms of 14-3-3 protein were detected in band B. Because an interaction between 14-3-3 protein and Hap1 had been reported previously (20), we focused on verifying the interactions of Hap1 with the other two proteins, Gap43 and Dcaf7.
Fig. 2.
Identification of Dcaf7 as a component of the SB. (A) Silver staining revealed two bands, A and B, that are more prominent in Hap1 immunoprecipitants from the WT P1 fraction. (B) Mass spectrometry analysis of bands A and B uncovered candidate proteins associated with the SB. (C and D) Western blotting of Gad43 and Dcaf7 (arrows) coimmunoprecipitated by Hap1 in P1 fractions obtained using strategy 2 (Fig. 1F) (C) or P2 fractions obtained using strategy 1 (Fig. 1D) (D) from WT and Het mouse brains. (E) Western blotting of Ahi1 and Dcaf7 coimmunoprecipitated by Hap1 in the S1 fraction of WT mouse brain. (F) Coimmunostaining of Hap1 and Dcaf7 in postnatal day 14 WT mouse hypothalamus confirming the presence of Dcaf7 in SBs. (Scale bar: 20 µm.) (G) Immunohistochemical staining of Dcaf7 in postnatal day 18 WT mouse hypothalamus and cortex. Note that Dcaf7 is more enriched in SBs in hypothalamic neurons than in cortical neurons. (Scale bars: 10 µm.)
Western blotting clearly showed that Dcaf7 is specifically associated with Hap1 in the P1 (strategy 2; Fig. 1F) and P2 (strategy 1; Fig. 1D) fractions, whereas Gap43, although more enriched in the WT Hap1 immunoprecipitant, was pulled down much less efficiently by Hap1 antibody, likely in a nonspecific fashion because the control IgG also could pull down a comparable amount of Gap43 from the WT sample (Fig. 2 C and D). We therefore focused on the interaction between Dcaf7 and Hap1, which was found to exist not only in detergent-insoluble P fractions but also in the soluble S1 fraction in which Hap1 also binds tightly with Ahi1 (Fig. 2E). To confirm that Dcaf7 is indeed localized in the SB, we performed coimmunostaining of Hap1 and Dcaf7 in sections of WT mouse hypothalamus and found both proteins were present in SBs (Fig. 2F). In Het hypothalamus, Dcaf7 appears largely diffuse and does not form puncta by itself because of the loss of Hap1 (Fig. S2), indicating that Hap1 has a central role in recruiting different components of the SB. Indeed, we saw more abundant Dcaf7-containing SBs in the hypothalamus than in the cortex (Fig. 2G), as is consistent with the greater abundance of Hap1 in the hypothalamus (Fig. 1B). Overall, we identified Dcaf7 as an Hap1-interacting protein and a component of the SB.
Fig. S2.
Dcaf7 does not form obvious puncta in Hap1 Het-KO mouse hypothalamus. Coimmunoprecipitation of Hap1 and Dcaf7 in postnatal day 14 Hap1 Het-KO mouse hypothalamus displays prominently diffuse staining for both proteins, indicating that Dcaf7 does not form puncta when Hap1 is reduced. (Scale bar: 10 µm.)
Binding of Hap1 Inhibits the Nuclear Translocation of Dcaf7.
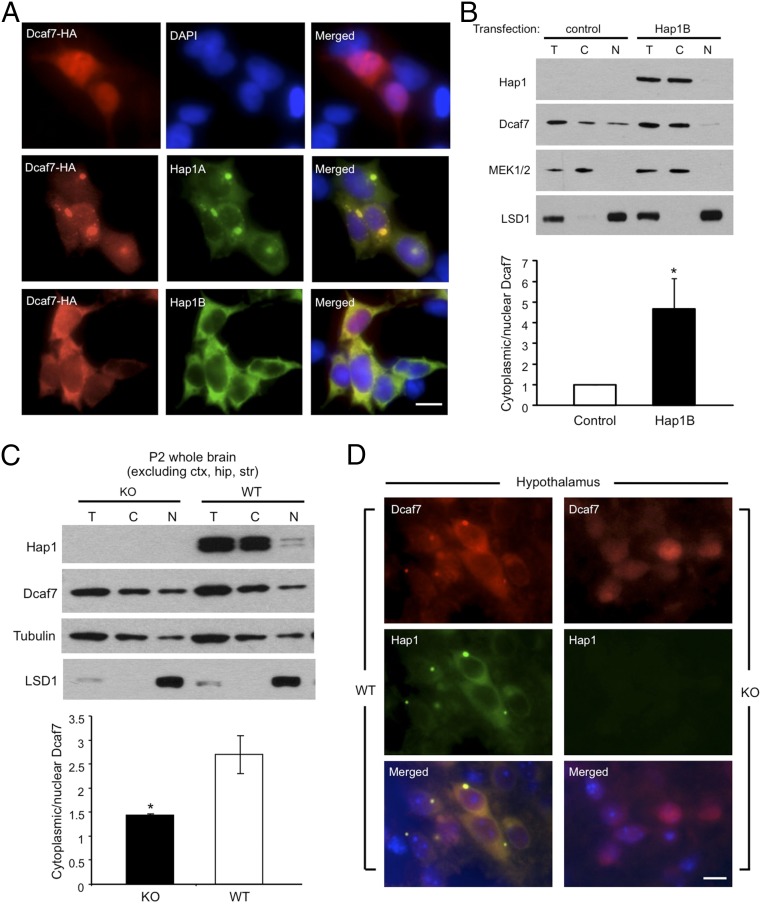
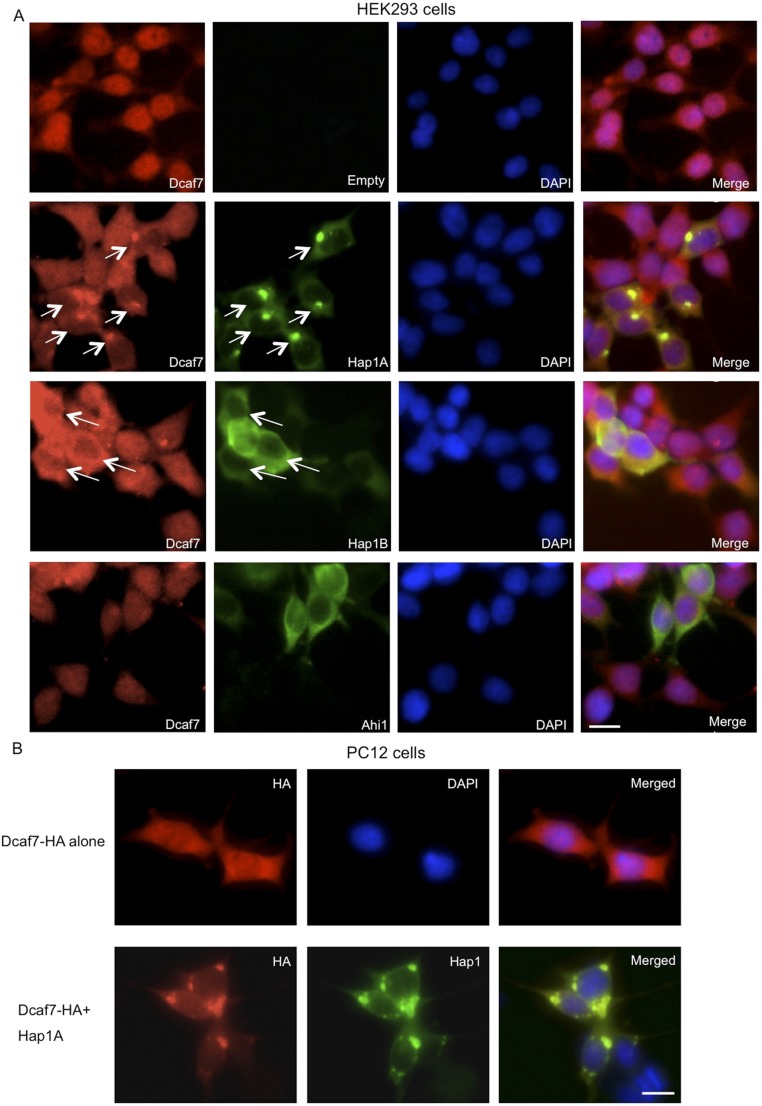
Dcaf7 localization to be more nuclear in the cortex appears to be more nuclear in the cortex than in the hypothalamus (Fig. 2G), raising the possibility that Hap1 may regulate the subcellular distribution of Dcaf7. To test this idea, we transfected plasmid Dcaf7-HA into HEK293 cells that do not express Hap1. When transfected alone, Dcaf7 exhibited a more nuclear localization; however, when cotransfected with either Hap1A or Hap1B, Dcaf7 became noticeably more cytoplasmic (Fig. 3A). Transfection of Hap1A or Hap1B, but not Ahi1, also redirected endogenous Dcaf7 into the cytoplasm (Fig. S3A), suggesting that Hap1 alone is able to direct the cytoplasmic distribution of Dac7. Subcellular fractionation of HEK293 cells and Western blotting using MAPK/ERK kinase 1/2 (MEK1/2) as a cytoplasmic marker and lysine-specific histone deethylase 1 (LSD1) as a nuclear marker validated a more cytoplasmic distribution of endogenous Dcaf7 in the presence of transfected Hap1B (Fig. 3B). Using additional cultured cell lines, we also showed that Hap1A was able to sequester Dcaf7 into cytoplasmic inclusions resembling SBs in vivo in both HEK293 and PC12 cells (Fig. 3A and Fig. S3).
Fig. 3.
Hap1 sequesters Dcaf7 in the cytoplasm. (A) Immunostaining of Dcaf7 when transfected alone or cotransfected with Hap1A or Hap1B into HEK293 cells. (Scale bar: 8 µm.) (B) Subcellular fractionation of HEK293 cells transfected with Hap1B or a control vector. Western blotting (Upper) and quantification (Lower) of total (T), cytoplasmic (C), and nuclear (N) fractions indicate a significant enrichment of Dcaf7 in the cytoplasm when Hap1B is present. Ratios were normalized to control level. n = 4 each. (C) Subcellular fractionation of P2 WT and Hap1-KO mouse brains. Western blotting (Upper) and quantification (Lower) show a decreased cytoplasmic/nuclear ratio of Dcaf7 in Hap1-KO brains. n = 3 each. (D) Coimmunostaining of Dcaf7 and Hap1 in postnatal day 2 WT and Hap1-KO mouse hypothalamus displays differential distributions of Dcaf7. (Scale bar: 10 µm.) *P < 0.05.
Fig. S3.
Hap1 sequesters Dcaf7 in the cytoplasm. (A) Transfection of Hap1A or Hap1B, but not Ahi1, in HEK293 cells redirects a great portion of endogenous Dcaf7 to the cytoplasm. Arrows indicate cells with redistributed Dcaf7. (B) Cotransfection of Dcaf7-HA and Hap1A in PC12 cells shows punctate cytoplasmic staining for both, whereas transfecting Dcaf7-HA alone gives diffuse staining of Dcaf7 with more nuclear signal. (Scale bars: 10 µm.)
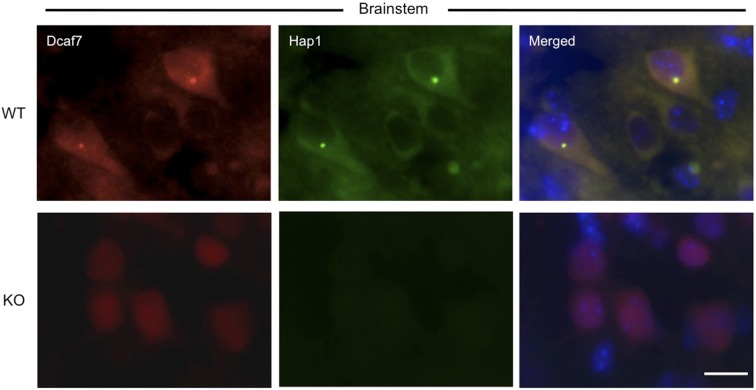
If Hap1 indeed regulates the subcellular distribution of Dcaf7, we reasoned that we should see a perturbed distribution pattern of Dcaf7in Hap1-KO mouse brain. Therefore, we conducted fractionation of P2 WT and Hap1-KO mouse brains and found that the loss of Hap1 led to more nuclear translocation of Dcaf7 (Fig. 3C). Because the mixed tissue types used for Western blotting express Hap1 at different levels, we used immunocytochemistry to examine Dcaf7 distribution in the hypothalamus, which is enriched in Hap1, and observed a shift in Dcaf7 localization leaning toward the nucleus, as well as a loss of cytoplasmic Dcaf7 puncta, when Hap1 is depleted in KO mice (Fig. 3D). Similarly, in the brainstem, which also expresses Hap1 at a high level, loss of Hap1 results in the nuclear distribution of Dcaf7 in neuronal cells (Fig. S4).
Fig. S4.
Dcaf7 localizes more into the nucleus in Hap1-KO mouse brainstem. Coimmunostaining of Dcaf7 and Hap1 in postnatal day WT and Hap1-KO mouse brainstems shows less cytoplasmic and more nuclear immunoreactivity of Dcaf7 in the KO tissue. (Scale bar: 15 µm.)
Hap1 Stabilizes the Protein Level of Dcaf7 by Inhibiting Its Proteasome Degradation.
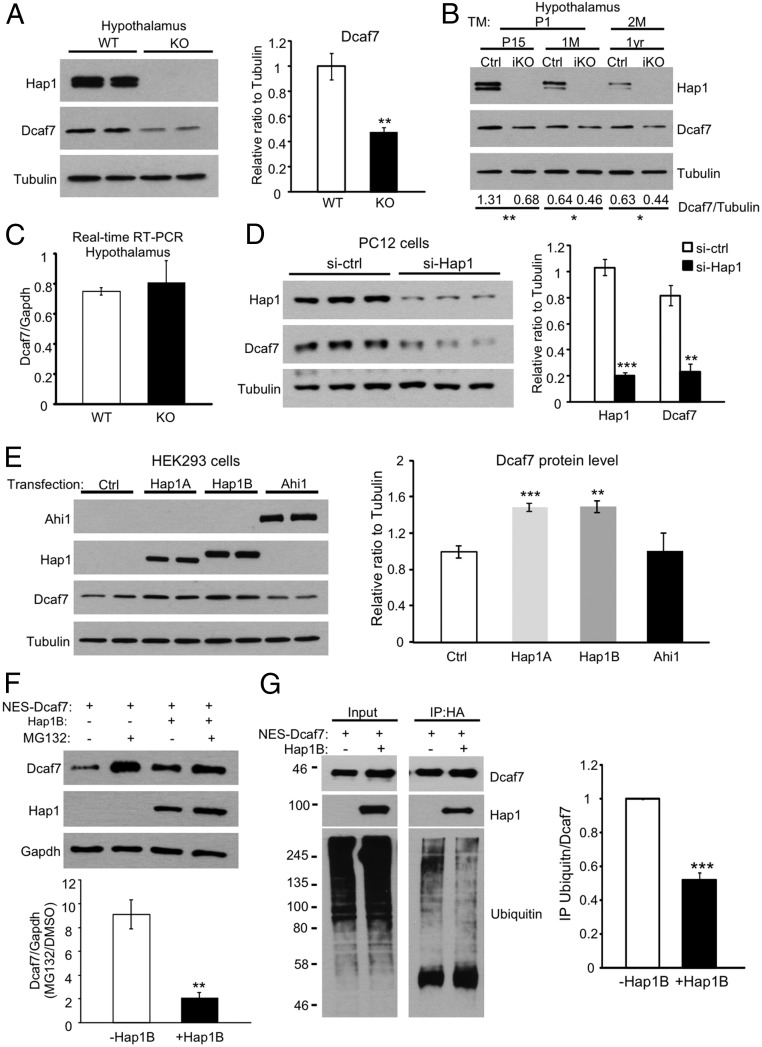
Because Hap1 regulates the protein level of its most prominent binding partner, Ahi1 (6), we next examined whether Hap1 may also play a role in Dcaf7 protein turnover. In Hap1-KO mouse hypothalamus, Dcaf7 was reduced drastically, by more than 50% (Fig. 4A). No difference in Dcaf7 was seen in Hap1-KO cortex, in which very little Hap1 was expressed (Fig. S5A). In 15-d/1-mo-old or 1-y-old conditional Hap1-KO mice in which Hap1 was deleted via tamoxifen injection at postnatal day 1 or 2 mo of age, decreased levels of Dcaf7 in the hypothalamus were also observed (Fig. 4B). However, Dcaf7 mRNA levels remained unaltered in KO hypothalamus (Fig. 4C), suggesting that Hap1 stabilizes Dcaf7 at the protein level. In addition, knocking down Hap1 in PC12 cells via siRNA also induced a dramatic reduction in the Dcaf7 protein level (Fig. 4D).
Fig. 4.
Hap1 stabilizes Dcaf7 in the cytoplasm by reducing its proteasome degradation. (A) Western blotting shows a significant reduction in Dcaf7 protein level in Hap1-KO mouse hypothalamus. Ratios were normalized to WT. n = 4 each. (B) Decreased Dcaf7 levels also were observed in the hypothalami of postnatal day 15/1-mo-old induced Hap1-KO (iKO) mice injected with tamoxifen for 3 d beginning at postnatal day 1 or of 1-y-old iKO mice injected with tamoxifen for 5 d beginning at 2 mo of age. The ratios of Dcaf7 to tubulin were obtained from three independent experiments and are shown beneath the blots. (C) Real-time RT-PCR found no difference in Dcaf7 mRNA levels in hypothalami from postnatal day 2 WT and Hap1-KO mice. n = 4 per group. (D) Knockdown of Hap1 using siRNA in PC12 cells drastically down-regulated Dcaf7. Ratios were normalized to the control level. n = 3 per group. (E) Transfections of Hap1A or Hap1B, but not Ahi1, increased the endogenous Dcaf7 levels in HEK293 cells. Ratios were normalized to the control level. n = 4 per group. (F) HEK293 cells treated with 5 µM MG132 for 16 h showed more NES-Dcaf7 accumulation without Hap1B than when Hap1B was cotransfected. n = 3 per group. (G) Hap1B cotransfection with NES-Dcaf7-HA reduces the polyubiquitination of Dcaf7. NES-Dacf7-HA was immunoprecipitated by anti-HA. Ratios were normalized to the NES-Dcaf7-HA–only group. n = 3 per group. *P < 0.05; **P < 0.01; ***P < 0.001.
Fig. S5.
The protein level of Dcaf7 is correlated with Hap1 expression and is degraded by the proteasome. (A) The Dcaf7 level is not changed in Hap1-KO mouse cortex, in which Hap1 expression is very low. (B) Hap1A and Hap1B increase the half-life of endogenous Dcaf7 in HEK293 cells. n = 6 for each group. (C) HEK293 cells were treated with DMSO, 5 µM MG132 (proteasome inhibitor), or 50 nM BFA, an autophagy inhibitor, for 16 h before being lysed to examine the protein levels of endogenous Dcaf7. Quantification of the Western blotting result is shown on the right. MG132, but not BFA, markedly stabilizes Dcaf7 compared with DMSO treatment. *P < 0.05; **P < 0.01.
Next, we transfected Hap1A, Hap1B, or Ahi1 into HEK239 cells and found that Hap1, but not Ahi1, increased the protein level of Dcaf7 (Fig. 4E). Both Hap1A and Hap1B extended the half-life of endogenous Dcaf7 in HEK293 cells (Fig. S5B). To find out which pathway was primarily used to degrade Dcaf7, we treated HEK293 cells with either the proteasome inhibitor MG132 or the autophagy inhibitor bafilomycin A1 (BFA). Only MG132 treatment significantly stabilized the level of Dcaf7, suggesting that Dcaf7 is degraded mainly via the ubiquitin-proteasome system (UPS) (Fig. S5C). To examine whether Hap1 inhibits the proteasome degradation of Dcaf7, we compared the levels of nuclear export signal (NES)-tagged Dcaf7-HA after MG132 treatment with or without Hap1B cotransfection. We used NES-Dcaf7-HA so that we would assess only the degradation of Dcaf7 in the cytoplasm where Hap1 localizes. The result demonstrated that the proteasome degradation of Dcaf7 over the course of 16 h was remarkably decreased in the presence of Hap1B (Fig. 4F). In addition, Hap1B also reduced the polyubiquitination of immunoprecipitated NES-Dcaf7-HA (Fig. 4G). Thus, we concluded that Hap1 not only sequesters but also stabilizes Dcaf7 in the cytoplasm by inhibiting its degradation by the UPS. Because Hap1 can protect internalized receptors from lysosomal degradation (11, 29–31), Hap1 may stabilize its binding partners from subsequent degradation through either the lysosome or UPS.
Hap1 Competes for Dcaf7 Binding with DYRK1A, Levels of Which Are Increased in Hap1-KO Mouse Brains.
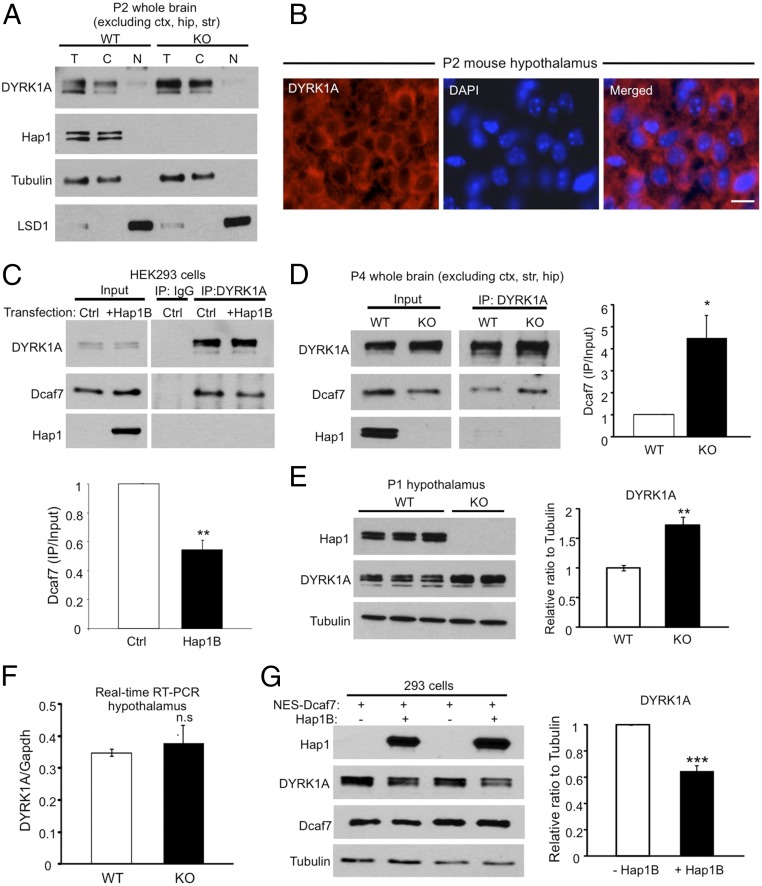
We know that Dcaf7 also binds DYRK1A (32), a kinase that phosphorylates a wide range of proteins involved in metabolism, synaptic function, and neurodegeneration (33). Of greatest interest, DYRK1A maps to human chromosome 21 and perhaps is implicated in DS (34–36). We were interested in whether Hap1 and DYRK1A interact with Dcaf7 competitively or synergistically. First, however, we wanted to examine the subcellular localization of DYRK1A, because studies have demonstrated both cytoplasmic and nuclear distribution of endogenous DYRK1A in neuronal cells, whereas ectopically expressed DYRK1A is found mainly in the nucleus (37–41). We found via subcellular fractionation and immunostaining that endogenous DYRK1A is localized predominantly in the cytoplasm in mouse brain in vivo and in cultured HEK293 and PC12 cells (Fig. 5 A and B and Fig. S6 A and B). Next, we evaluated the interaction between Dcaf7 and DYRK1A in HEK293 cells with or without Hap1. It is known that both Hap1A and Hap1B form heterodimers to interact with other proteins. Because Hap1A transfection leads to the formation of insoluble puncta in transfected cells, whereas Hap1B alone remains soluble (12), we used Hap1B transfection to examine the interaction of soluble Hap1 with other proteins. The result indicated attenuated binding between Dcaf7 and DYRK1A in the presence of Hap1B, suggesting competition between Hap1 and DYRK1A for Dcaf7 binding (Fig. 5C). Because Hap1 is strictly a cytoplasmic protein in neurons, this competition most likely happens in the neuronal cytoplasm; thus, it is reasonable to assume that the association of Dcaf7 and DYRK1A may be enhanced in the Hap1-KO mouse brain. Indeed, we saw a potentiated Dcaf7–DYRK1A interaction in Hap1-KO brain, despite a decrease in the protein level of Dcaf7 (Fig. 5D).
Fig. 5.
DYRK1A and Hap1 compete for Dcaf7 binding in the cytoplasm, and the protein level of DYRK1A is increased in Hap1-KO mouse brains. (A) Fractionation of postnatal day 2 mouse brains reveals a preponderantly cytoplasmic distribution of DYRK1A. T, total; C, cytoplasmic; N, nuclear. (B) Immunostaining of DYRK1A in postnatal day 2 mouse hypothalamus also demonstrates cytoplasmic localization of DYRK1A. (Scale bar: 10 µm.) (C) Hap1B expression significantly reduces the interaction between DYRK1A and Dcaf7 in HEK293 cells. Ratios were normalized to control level. n = 3 each. (D) The association between DYRK1A and Dcaf7 is dramatically enhanced in Hap1-KO mouse brains. Ratios were normalized to the WT level. n = 3 per group. (E) The protein level of DYRK1A is increased in postnatal day 1 Hap1-KO hypothalamus. Ratios were normalized to WT level. n = 4 each. (F) Real-time RT-PCR found no difference in DYRK1A mRNA levels in postnatal day 2 WT and Hap1-KO mouse hypothalami. n = 4 per group. (G) The expression of transfected NES-Dcaf7 was adjusted at similar levels in groups with or without Hap1B cotransfection in HEK293 cells. The presence of Hap1B reduces the level of endogenous DYRK1A. Ratios were normalized to the NES-Dcaf7–only group. n = 3 per group. *P < 0.05; **P < 0.01; ***P < 0.001, n.s., not significant.
Fig. S6.
DYRK1A is predominantly localized in the cytoplasm of HEK293 and PC12 cells, and its level is increased in induced Hap1-KO mouse hypothalamus. (A and B) Coimmunostaining of DYRK1A and Dcaf7 in HEK293 (A) and PC12 cells (B) demonstrates a cytoplasmic distribution of DYRK1A in both cell types, whereas Dcaf7 is distributed more evenly throughout the whole cell. (Scale bars: 10 µm.) (C) Increased DYRK1A levels were observed in the hypothalami of postnatal day 15/1-mo-old induced Hap1-KO (iKO) mice injected with tamoxifen for 3 d beginning at postnatal day 1 or of 3-mo-old iKO mice injected with tamoxifen for 5 d beginning at 2 mo of age. The ratios of Decaf7 to tubulin were obtained from three independent experiments and are shown beneath the blots. *P < 0.05; **P < 0.01.
Surprisingly, when protein levels of DYRK1A were assessed, we saw an increase of about 70% in Hap1-KO mouse brains compared with WT controls (Fig. 5E). The up-regulation occurs posttranscriptionally, because mRNA levels of DYRK1A were comparable in WT and KO mice (Fig. 5F). In conditional Hap1-KO mice in which Hap1 had been depleted via postnatal tamoxifen injection (11), increased levels of DYRK1A were also observed (Fig. S6C). Because of the enhanced association between Dcaf7 and DYRK1A in Hap1-KO brain, we hypothesized that the binding of Dcaf7 might be important for DYRK1A stability. To test this hypothesis without confounding factors, such as nuclear transportation and changes in the level of Dcaf7 protein, we expressed equal amounts of NES-Dcaf7 in HEK293 cells with or without transfected Hap1B and measured the levels of endogenous DYRK1A. DYRK1A levels were reduced in groups coexpressing Hap1B, probably because a portion of NES-Dcaf7 was bound to Hap1B (Fig. 5G).
Overexpression of DYRK1A Impairs Hap1–Dcaf7 Binding and Causes Body Weight Reduction in Mice.
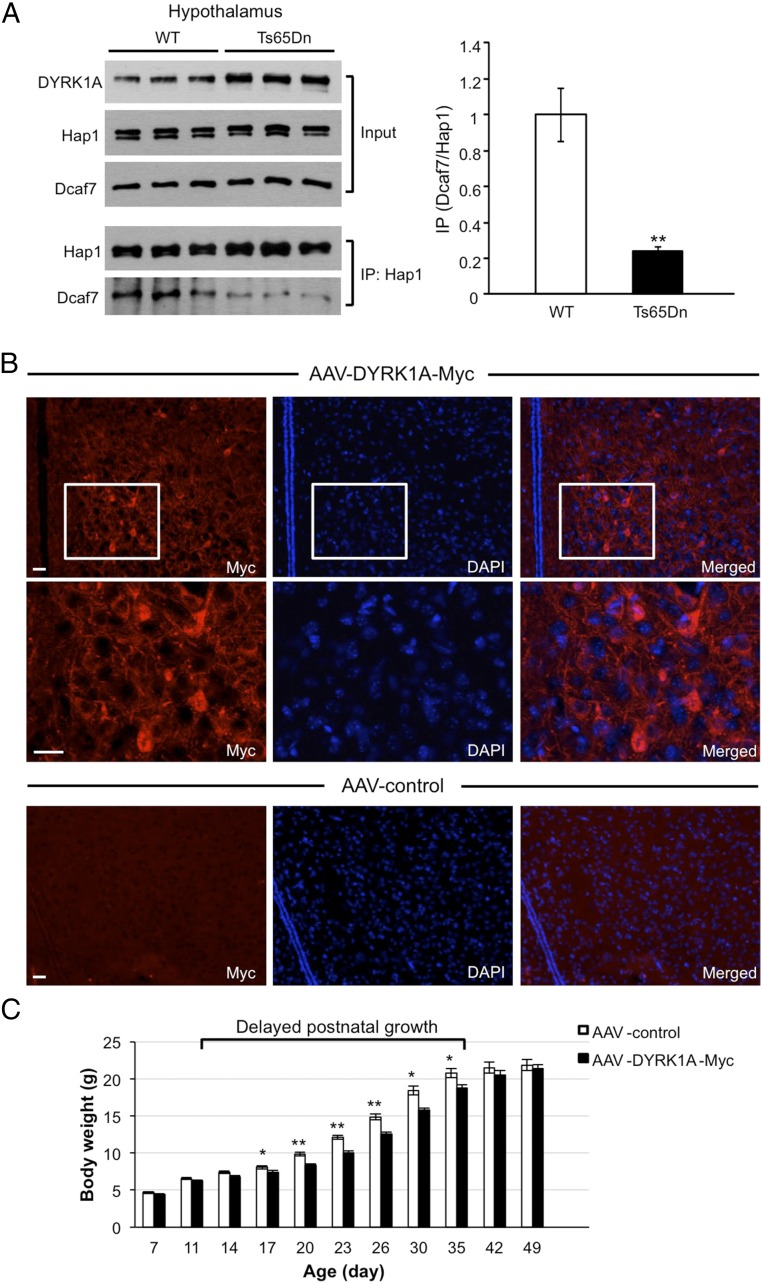
The increased DYRK1A level in the Hap1-KO mouse brain was particularly interesting because DYRK1A also is overexpressed in the brains of individuals with DS, and this overexpression may explain some of the neuropathological traits of this syndrome (22, 42). To validate the competition between Hap1 and DYRK1A for binding with Dcaf7 in a different and more disease-relevant model, we examined the association of Hap1 and Dcaf7 in a DS mouse model, Ts65Dn, which carries a partial trisomy of mouse chromosome 21, including the DYRK1A gene (43, 44). In the hypothalamus of Ts65Dn mice overexpressing DYRK1A, the interaction between Hap1 and Dcaf7 was dramatically diminished (Fig. 6A). Ts65Dn mice exhibit neurodevelopmental delay and reduced body weight similar to that seen in individuals with DS (45, 46). Because Hap1-KO mice suffer from growth retardation (5, 10), we proposed that in DS and its mouse models elevated DYRK1A pulls Dcaf7 away from Hap1, thereby influencing the normal function of Hap1 and thus contributing to the growth defect. To test this notion, we injected AAV-expressing DYRK1A-Myc into mouse hypothalamus (a major brain region in which Hap1 exerts its feeding-regulation function) at postnatal day 1 or 2 (9). The expression of the viral vector could be verified via immunostaining of the Myc tag (Fig. 6B). We then measured the body weight of injected mice and found weight gain was significantly reduced starting from postnatal days 14–17 in mice injected with AAV-DYRK1A as compared with the mice injected with the AAV control (Fig. 6C). Because recombinant AAV does not integrate into the host cell genome, the overexpression of DYRK1A in the injected region was not sustained over a long time. Also, the rapid and dramatic increase in brain volume and neurogenesis that occur during postnatal development (47, 48) could dilute the effect of AAV-transduced neurons.
Fig. 6.
Overexpression of DYRK1A in mouse hypothalamus reduces the Hap1–dcaf7 interaction and leads to reduced body weight gain. (A) IP of Hap1 in the DYRK1A-overexpressing DS mouse model. Ts65Dn hypothalamus reveals a decreased association of Hap1 and Dcaf7. (B) Immunostaining of Myc in postnatal day 20 WT mouse hypothalamus that had been injected with AAV-DYRK1A-Myc or AAV-control at postnatal day 2. Enlarged images of the boxes are shown below. (Scale bars: 15 µm.) (C) Body weight recording indicates that DYRK1A overexpression in the hypothalamus reduced the postnatal weight gain of WT mice. n = 11–20. *P < 0.05; **P < 0.01.
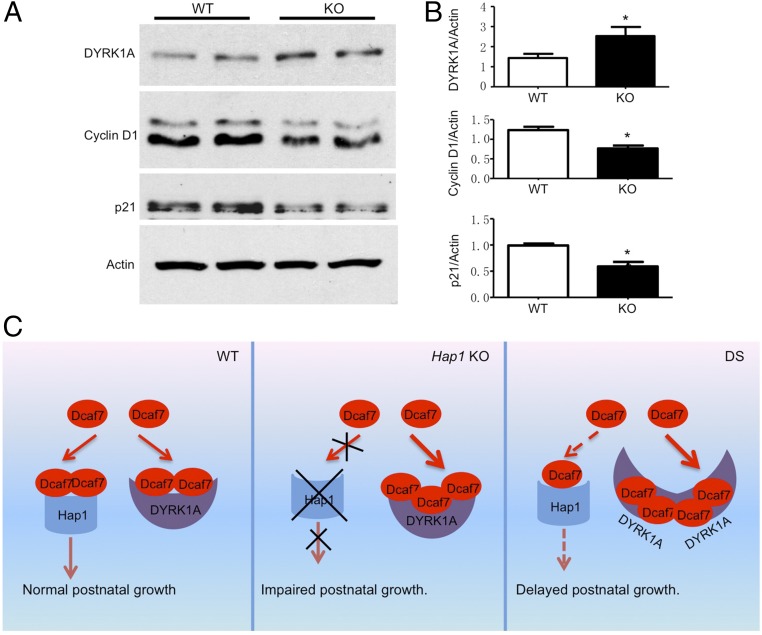
The importance of the Hap1–Dcaf7–DYRK1A interaction in postnatal growth can be tested further by examining the cellular function related to this interaction. Our data have shown that loss of Hap1 can lead to enhanced interaction between Dcaf7 and DYRK1A, resulting in an increase in DYRK1A. It has been reported that increased DYRK1A can affect cell-cycle regulation and neuronal differentiation by reducing cyclin D1 and the cyclin-dependent kinase inhibitor p21 (49, 50), two important proteins that promote neuronal differentiation (50–54). Thus, we isolated hypothalamic tissues from WT and Hap1-KO postnatal pups and performed Western blotting with anti-cyclin D1 and anti-p21. We found that cyclin D1 and p21 were indeed decreased in the Hap1-KO hypothalamic tissues (Fig. 7 A and B). This finding also supports the role of altered Hap1–Dcaf7–DRK1A interactions in the delayed growth of postnatal mice. Based on these findings, we propose that Dcaf7 binds both Hap1 and DYRK1A in the cytoplasm. The Hap1–Dcaf7 interaction may be critical for postnatal growth, and the loss of Hap1 in KO mice impairs postnatal growth. In DS, overexpressed DYRK1A reduces the Hap1–Dcaf7 interaction in the hypothalamus, also affecting postnatal growth (Fig. 7C).
Fig. 7.
(A) Western blotting analysis and quantitative analysis of DYRK1A, Cyclin D1, and p21 levels in the hypothalamus of WT and Hap1-KO mouse hypothalami. (B) The relative levels of DYRK1A, Cyclin D1, and p21 on the Western blots. Actin was used as loading control. n = 3 per group. *P < 0.05. (C) A schematic of imbalanced Hap1–Dcaf7 and DYRK1A–Dcaf7 interactions in Hap1-KO mouse and DS hypothalamic neurons. (Left) In WT hypothalamic neurons, Dcaf7 binds both Hap1 and DYRK1A in the cytoplasm. The Hap1–Dcaf7 interaction may be critical for postnatal growth. (Center) In a Hap1-KO hypothalamic neuron, more Dcaf7 binds to DYRK1A, increasing the DYRK1A protein level. Absence of the Hap1–Dcaf7 complex may impair postnatal growth. (Right) In a DS hypothalamic neuron, an elevated level of DYRK1A leads to more Dcaf7 being bound to DYRK1A and less to Hap1, possibly resulting in delayed postnatal growth.
Discussion
Our previous findings and those of others have shown that Hap1 is highly expressed in the hypothalamus and that the loss of Hap1 affects postnatal growth (5, 10, 11). However, the mechanisms behind Hap1’s function in the hypothalamus and the relevance to human diseases of the defective postnatal growth mediated by the loss of Hap1 remain unclear. The findings in our current study suggest that the interaction of Hap1 with Dcaf7 in the hypothalamus is important for postnatal growth and that overexpressed DYRK1A could affect this interaction, thereby affecting postnatal growth in DS.
The interaction between Dcaf7 and Hap1 is strongly supported by their coexistence in hypothalamic SBs and their coimmunoprecipitation and also by the regulation of Dcaf7 stability at the protein level by Hap1. We show that Hap1 not only stabilizes the protein level of Dcaf7 but also sequesters Dcaf7 from entering the nucleus. In the Hap1-KO mouse hypothalamus, Dcaf7 is down-regulated, and its distribution becomes more nuclear. Whether its loss of function in the cytoplasm or gain of function in the nucleus promotes the deleterious effects of Hap1 deletion is unknown. Characterization of Dcaf7 function in mammals is limited, so we can only make inferences from studies in other organisms or cell cultures. For example, in zebrafish (Danio rerio), nuclear localization of Dcaf7 is required for craniofacial development (47, 48). It is logical to think that the subcellular localization of Dcaf7 may depend largely on the expression profiles of its cell type-specific interacting proteins as well as on the timing of its expression during development. Thus, it would be interesting to investigate whether the accumulation of Dcaf7 in the nuclei of Hap1-null hypothalamic neurons might be detrimental to the cell, e.g., by causing transcriptional dysregulation. Moreover, knocking down Dcaf7 in COS7 cells has been reported to induce apoptosis (32). Thus, future studies are needed to investigate the effects of Dcaf7 ablation on Hap1 function, SB formation, neuronal survival, and mouse behavior.
The more important finding in our study is that DYRK1A can regulate the interaction of Dcaf7 and Hap1. The interaction and relative abundance of Dcaf7 and DYRK1A are known to influence the localization of these two proteins (32, 48). Because craniofacial maldevelopment is a characteristic common to all individuals with DS (55), and the subcellular distribution of DYRK1A varies in different species and cell types, it is possible that DYRK1A may localize predominantly to the cytoplasm of cranial neural crest cells, and its overexpression in DS could cause more Dcaf7 to be retained in the cytoplasm, leading to impaired signaling in craniofacial development.
Our study highlights a physiological balance between Hap1–Dcaf7 and DYRK1A–Dcaf7 interactions and the perturbation of this balance in DS. Because we have demonstrated that both Hap1 and DYRK1A localize primarily to the cytoplasm of mouse hypothalamic neurons, where they compete for Dcaf7 binding, changes in the expression levels of either protein would disturb this balance and likely cause harm to neurons. DYRK1A levels are increased in individuals with DS; our findings indicate that this increase should tip the balance toward enhanced DYRK1A–Dcaf7 binding and weakened Hap1–Dcaf7 binding. As a result, the function of Hap1 may be attenuated.
We must point out that the wide range of symptoms and behavioral changes in DS are likely caused by the abnormal expression of a large array of genes and their interactions resulting from an extra copy of chromosome 21. DYRK1A is a major candidate gene for DS and likely is responsible for selective DS phenotypes. To look for DS symptoms that could be related to the regulation of DYRK1A on the Dcaf7–Hap1 interaction, we focused on growth retardation, which is a cardinal feature of DS (56) and is the most noticeable phenotype in Hap1-KO mice (5, 10). Both individuals with DS and Hap1-KO mice exhibit impaired growth velocity after birth; the difference, however, is that in DS, growth velocity is most reduced between 6 mo and 3 y of age (56, 57), whereas Hap1-KO mice are born at normal weight but display severe failure in body weight gain shortly after birth (5, 10). These differences may reflect species differences in terms of maturation and life span to a certain extent, but are more likely explained by the differential consequences of partial loss of Hap1 function and a complete elimination of Hap1. The interactions of Hap1 with other proteins may mediate age-dependent functions, because loss of Hap1 at different ages causes completely different phenotypes (7). Also, triplication of an entire chromosome 21 or segment of this chromosome could affect multiple genes involved in growth control, so that the growth retardation in DS may be mediated by a combination of mechanisms.
The generation and characterization of different mouse models of DS may shed light on the causative role of dosage-sensitive genes in distinct DS phenotypes and aid in therapeutic development for DS (58). The most widely used DS mouse model, Ts65Dn mice, are trisomic for more than half of the mouse orthologs of genes on human chromosome 21 (59), whereas Ts1Cje mice carry a smaller segmental triplication included in Ts65Dn trisomy (60). That these two models display DS-characteristic growth retardation suggests that certain genes within the triplicated region are responsible, at least in part, for the phenotype (45, 46, 61). DYRK1A transgenic (Tg) mouse models were also made in an effort to reveal its role in DS. Cognitive deficits were noted in two models overexpressing DYRK1A, one under the sheep metallothionein 1a (sMT-1a) promoter (24) and the other in a BAC under the human DYRK1A promoter (23). Although original reports of these two models did not mention changes in body weight, a recent study showed that DYRK1A BAC Tg mice are lean and resistant to diet-induced obesity (62). The trend of decreased body weight starts from the earliest time point measured, i.e., 6 wk after birth. Because it is unclear how transgenic DYRK1A is expressed in the hypothalamus in these mouse models, it would be important to assess the phenotypes of mice that express a high level of DYRK1A in the hypothalamus. To this end, we performed viral injection of DYRK1A into newborn mouse hypothalami and saw a reduction in weight gain in the injected mice. Although this reduction is consistent with delayed postnatal growth in DS, it is less severe than in Hap1-KO mice and individuals with DS. It is possible that viral vector injection allows only restricted and temporary expression of transgenic DYRK1A. Another possibility is that overexpression of other genes in chromosome 21 in DS may be required to stabilize a high level of DYRK1A in the hypothalamus. Furthermore, because of the difficulty in determining feeding behavior and metabolic functions of the limited number of pups injected postnatally with AAV-DRYK1A-Myc, it remains to be investigated whether the reduced body weight in these pups is caused by alterations in feeding activities or by metabolic changes, either of which could result from the dysfunction of hypothalamic neurons.
Because the function of DYRK1A may vary in different brain regions, we believe that hypothalamic DYRK1A is important for normal body growth. We also propose that diminished Hap1–Dcaf7 interaction is a likely mechanism for DYRK1A-mediated growth retardation in DS. Downregulating DYRK1A levels might not be an ideal solution, because DYRK1A acts on a large number of substrate proteins, including transcription factors, splicing factors, and synaptic proteins (42). If the level of Hap1–Dcaf7 interaction indeed mediates the function of Hap1 in food intake, enhancement of this interaction by up-regulating either Hap1 or Dcaf7 could provide promising therapeutic options for treating stunted growth in DS and other growth disorders.
Materials and Methods
Mice were housed in the Division of Animal Resources at Emory University on a 12-h light/dark cycle with lights on at 7 AM and lights off at 7 PM. All procedures were approved by the Animal Care and Use Committee at Emory University (2002557) (X.-J.L.) or The Johns Hopkins University (M015M347) (R.H.R.). Generation of germline Hap1-KO mice and tamoxifen-inducible Cre-ER/floxed-Hap1 mice was described previously (5, 11). Brain tissues of Ts65Dn mice were provided by R.H.R. Additional information is provided in SI Materials and Methods.
SI Materials and Methods
Reagents and Antibodies.
DMEM and FCS were obtained from Life Technologies. Trypsin, BSA, 3,3′-diaminobenzidine (DAB), MG132, and BFA were all from Sigma. Cell-culture dishes, coverslips, plates, and flasks were purchased from Corning and Nunc, Inc. The Vectastain Elite ABC Kit was from Vector Laboratories. Pierce protein A/G magnetic beads were from Thermo Fisher Scientific. Guinea pig antibody to Hap1 was generated in X.-J.L. laboratory at Emory University and was used for Western blotting (1:10,000) and immunofluorescence (1:1,000). Mouse anti-Hap1 (1B6) from Novus was used for IP. Rabbit antibody to Ahi1 was generated in our previous work (6) and was used for Western blotting (1:2,000). Mouse anti-DRRK1A (M01; Abnova) was used for Western blotting (1:10,000) and immunofluorescence (1:200). The following were used for Western blotting: β-actin (1:20,000), γ-tubulin (1:20,000; T-6557; Sigma), GAPDH (1:10,000; GA1R; Thermo Fisher Scientific), ubiquitin (1:5,000; P4D1; Cell Signaling), MEK1/2 (1:1,000; L38C12; Cell Signaling), P21 (1:50; NBP2-29463; Novus), LSD1 (1:3,000; 2139; Cell Signaling), and cyclin D1 (1:1,000; 2978; Cell Signaling). Myc (1:5,000; 9B11; Cell Signaling) and HA (1:2,000; C29F4; Cell Signaling) were used for immunofluorescence. Rabbit anti-Dcaf7 (EPR8712; Abcam) was used for Western blotting (1:10,000), immunofluorescence (1:2,000), and immunohistochemistry (1:10,000). HRP-tagged, biotinylated, or fluorescent secondary antibodies were obtained from Jackson ImmunoResearch Laboratories.
Enrichment of SBs from the Mouse Brain.
For strategy 1 (Fig. 1D), a WT or Het mouse was killed at postnatal day 11–14, and the whole brain was obtained. The cortex, hippocampus, striatum, and cerebellum were removed, and the remaining tissues enriched in SBs were homogenized in 1.5 mL Hepes buffer (20 mM Hepes, 11 mM K-acetate, 5 mM Na-acetate, 2 mM MgCl2, 1 mM DTT) containing Halt protease inhibitor mixture and phosphatase inhibitors. The homogenate was transferred to a glass Dounce homogenizer and was disrupted with six strokes. Then it was filtered through a 20-µm filter followed by a 10-µm and a 5-µm polycarbonate filter (Millipore) inside a filter holder (Millipore) driven by a syringe to remove cell debris and most of the nuclei. Centrifugation was applied at 5,000 × g for 10 min at 4 °C to separate the S1 supernatant and P1 pellet fractions. The P1 fraction then was lysed with 1 mL 1% Nonidet P-40 buffer, and the lysate was centrifuged at 5,000 × g for 10 min at 4 °C to separate the S2 and P2 fractions. The P2 fraction was resuspended in 500 mL PBST (PBS + 0.1% Triton X-100). For strategy 2 (Fig. 1F), 0.3% Nonidet P-40 was added to the initial Hepes buffer, and the P1 fraction was resuspended in 500 mL PBST for follow-up analysis.
To examine the abundance and quality of SBs in the P2 (strategy 1) or P1 (strategy 2) fractions, a portion of the fraction resuspended in PBST was loaded on a gelatin-coated glass slide. We allowed 10 min for SBs to sink and attach to the surface before removing the liquid and then proceeded with immunofluorescent staining procedures to visualize Hap1+ SBs.
Subcellular Fractionation.
For HEK293 cells, cells were washed with PBS and pelleted at 500 × g for 5 min at 4 °C. Two packed cell volumes (PCVs) of the hypotonic buffer [10 mM Hepes (pH 7.9), 10 mM KCl, 1.5 mM MgCl2, 1 mM DTT] containing Halt protease inhibitor mixture and phosphatase inhibitors were added, and after 10 min incubation on ice, the homogenate was transferred to a glass Dounce homogenizer and disrupted with 10 strokes. Centrifugation was applied to the homogenate at 1,000 × g for 5 min at 4 °C. The supernatant was removed and spun again to yield the cytoplasmic fraction (C). The pellet representing the nuclear fraction (N) was washed once with 5–10 PCVs of the hypotonic buffer and was extracted with radioimmunoprecipitation (RIPA) buffer [50 mM Tris (pH 8.0), 150 mM NaCl, 1 mM EDTA (pH 8.0), 1 mM EGTA (pH 8.0), 0.1% SDS, 0.5% DOC, and 1% Triton X-100].
For mouse brain fractionation, a postnatal day 2 WT or Hap1-KO mouse whole brain excluding the cortex, hippocampus, and striatum was homogenized in Hepes buffer (20 mM Hepes, 11 mM K-acetate, 5 mM Na-acetate, 2 mM MgCl2, 1 mM DTT) containing Halt protease inhibitor mixture and phosphatase inhibitors. The homogenate was transferred to a glass Dounce homogenizer and disrupted with 10 strokes. After 15-min incubation on ice, the homogenate was centrifuged at 1,000 × g for 10 min at 4 °C. The S1 supernatant was collected, and the P1 pellet was resuspended in Hepes buffer and loaded onto a 1.8-M sucrose layer for centrifugation at 71,000 × g for 90 min at 4 °C. The P2 pellet represents the nuclear fraction (N), and the S2 supernatant was combined with the S1 supernatant to yield the cytoplasmic fraction (S).
Western Blot Analysis.
Mouse brain tissues or harvested cells were lysed in ice-cold RIPA buffer containing Halt protease inhibitor mixture and phosphatase inhibitors. The lysates were incubated on a rocker at 4 °C for 30 min, sonicated, and centrifuged at top speed for 10 min. The supernatants were collected and subjected to SDS/PAGE. The proteins on the gel were transferred to a nitrocellulose membrane. The blot was blocked with 5% (wt/vol) milk/PBS for 1 h at room temperature and incubated with a primary antibody in 3% (wt/vol) BSA/PBS overnight at 4 °C. After three washes in PBS, the blot was incubated with HRP-conjugated secondary antibodies in 5% (wt/vol) milk/PBS for 1 h at room temperature. The blot then was washed three times in PBS and developed using ECL Prime (GE Healthcare).
IP.
For IP in HEK293 cells or cells from the hypothalamus, cells grown in a 10-cm plate or the hypothalamic tissues were homogenized with Nonidet P-40 buffer [50 mM Tris (pH 7.4), 50 mM NaCl, 0.1% Triton X-100, 0.5% Nonidet P-40] containing Halt protease inhibitor mixture using a glass Dounce homogenizer. The lysate was centrifuged at 20,000 × g at 4 °C for 5 min, and 500 μg of the supernatant was used for IP. Samples were adjusted to 500 μL in volume and were incubated with a primary antibody at 4 °C overnight. Next, 15 μL of protein G beads were added for an additional hour to pull down the antibody–protein complex. Beads were spun down at 2,000 × g at room temperature for 30 s and were washed three times with lysis buffer. After final wash, SDS loading buffer was added to the samples, and the immunoprecipitants were boiled and resolved by electrophoresis for Western blot analysis. To immunoprecipitate Hap1 from fractioned mouse brains, 300 μg of the desired fraction was taken, and the total volume was adjusted to 500 μL. For fractions enriched in SBs, at least 2 h preclearing with the magnetic beads was required to reduce nonspecific bindings by the beads. Mouse anti-Hap1 antibody (5 μL; 1B6; Novus) then was added to incubate with the lysate at 4 °C overnight. Twenty-five microliters of the magnetic beads were added for 45 min to pull down Hap1. The beads containing Hap1 immunoprecipitants were collected using a magnetic particle concentrator (Dynal Biotech).
To identify Hap1-interacting proteins, Hap1 immunoprecipitants were resolved in SDS/PAGE and examined via silver staining. The bands that were more abundant in Hap1 immunoprecipitants than in IgG immunoprecipitants were excised to identify their nature by reverse-phase LC-MS/MS at the Emory Core facility. Acquired MS/MS spectra were searched against the mouse reference database of the National Center for Biotechnology Information.
Immunofluorescent and Immunohistochemical Microscopy.
For immunofluorescent staining of cultured cells, cells were washed once with PBS and were fixed with 4% (wt/vol) paraformaldehyde (PFA) for 15 min. Fixed cells were washed three times with PBS and were blocked with 3% (wt/vol) BSA + 5% (vol/vol) normal donkey serum/PBST (PBS + 0.2% Triton X-100) for 1 h at room temperature. Primary antibodies were diluted in blocking buffer and incubated with the cells at 4 °C overnight followed by three washes with PBS and incubation with fluorescent secondary antibodies and nuclear dye. After three washes, the fluorescence was examined under an Axiovert 200M microscope (Zeiss) with a digital camera (Orca-100; Hamamatsu Photonics) and OpenLAB software (Improvision). Immunofluorescent staining of brain sections was performed as described previously (7). Briefly, mice were deeply anesthetized and were perfused with saline followed by 4% (wt/vol) PFA fixation. Brains were postfixed overnight in the same fixative and then were switched to 30% (wt/vol) sucrose at 4 °C. After the brains were completely sunk in the sucrose solution, they were sectioned at 15 μm with a cryostat at −19 °C and were mounted onto gelatin-coated slides. The tissues on the slides were washed, blocked, and immunostained with antibodies using the method described above for cultured cells.
For immunohistochemical staining, brain sections first were incubated with 3% (vol/vol) H2O2 in distilled H2O for 20 min. After three washes in PBS, blocking and primary antibody incubation were performed as for immunofluorescent staining. The sections were washed three times and then were incubated with a biotinylated secondary antibody for 1 h at room temperature. After three washes, ABC reagent (prepared 30 min before use) was added for 30 min at room temperature. DAB then was used after three washes to develop signals. As soon as desired signal was reached, sections were quickly immersed in PBS to stop the reaction. The sections then were placed in a 37 °C incubator overnight, dehydrated in 70% (×2), 95% (×2), and 100% (×2) (vol/vol) alcohol, cleared in xylene (×2), and then mounted for observation under an Axio Imager A2 microscope (Zeiss).
Silver Staining.
Silver staining was performed as described previously (63). Briefly, the protein gel was fixed in 40% (vol/vol) ethanol, 10% (vol/vol) acetic acid, and 50% (vol/vol) H2O for 1 h followed by washing with several changes of water for a total of 1 h. The gel was sensitized in 0.02% sodium thiosulfate for 1 min and washed three times in H2O (20 s each washing) followed by incubation for 20 min in 4 °C 0.1% silver nitrate solution. After three 20-s washings in H2O, the gel was transferred to a new staining tray and was washed for another minute before being developed in 3% (wt/vol) sodium carbonate (0.05% formaldehyde added just before use). Developer solution was changed immediately when it turned yellow. Staining was terminated in 5% (vol/vol) acetic acid when sufficient. The gel was left at 4 °C in 1% acetic acid for storage.
RNA Isolation, RT-PCR, and Real-Time PCR.
RNA from mouse hypothalamus was isolated by the RNeasy Lipid Tissue Mini Kit (74804; QIAGEN). The RNA concentration was determined by a Synergy H4 reader, and an equal amount of RNA was used for cDNA synthesis with the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Real-time PCR was performed using SYBR Green Real-Time PCR Master Mix (Applied Biosystems) in an Eppendorf Mastercycler RealPlex instrument. The following primers were used: Dcaf7 forward: 5′-AGA ACA CCT TTG ACC ACC CGT ACC-3′, reverse: 5′-AGC CAC TTG TTG CCA GGA GGT CTG-3′; DYRK1A forward: 5′-TAC GCA GGA TTC TAT GGA GGT TGG-3′, reverse: 5′-ACC ACT GGG CGG TTC TGA TAG G-3′; Gapdh forward: 5′-AAC TTT GTC AAG CTC ATT TCT GGT-3′, reverse: 5′-GGT TTC TTA CTC CTT GGA GCC ATG -3′.
Tamoxifen Induction in Mice.
Tamoxifen induction in mice was performed as described previously (11). Briefly, tamoxifen (T5648; Sigma) was dissolved in ethanol at 20 mg/mL and stored at −20 °C before use. To induce Hap1 depletion, a calculated amount of tamoxifen was mixed with corn oil, and ethanol was removed by a vacuum centrifuge. One-day-old mouse pups were injected s.c. with TM/40 (1.1 mg/40 g body weight) for three consecutive days. Adult mice received i.p. injections of tamoxifen (4 mg/40 g body weight) for five consecutive days.
Preparation of Adenoviral Hap1 siRNA and AAV-DYRK1A-Myc.
Adenoviral siRNAs targeting Hap1 or a scramble sequence were prepared as in our previous study (64). Viral stocks were adjusted to 1 × 108 viral particles/μL before use. PC12 cells were incubated with adenoviral Hap1 siRNA at a multiplicity of infection of 50 for 48 h. Mouse DYRK1A cDNA was fused with an Myc tag in the C terminus and subcloned into a pAAV-MCS vector (Cell Biolabs). The Ubiquitin C (UBC) promoter sequence was inserted into the construct to replace the original promoter in the vector. AAV-DYRK1A-Myc (serotype 9) was packaged and amplified by the Viral Vector Core at Emory University. AAV-GFP also was packaged and used as a control.
Stereotaxic Viral Injection.
A 1- to 2-d-old mouse pup was placed in plastic wrap and immersed up to the neck in crushed ice and water (2–3 °C) for 7–10 min. The pup then was stabilized on a platform with ice underneath while being injected with 1 μL of AAV-DYRK1A-Myc or AAV-control viral particles (1 × 1012 particles/mL) into the hypothalamus (at the midline coordinates of 1.5 mm rostral to the lambda and 3.5 mm below the skull) over 2 min from a 5-μL Hamilton syringe and 33-gauge needle. The needle was kept in place for another 2 min before withdrawal. The pup then was transferred to a clean cage with a nest placed on a heated pad (33 °C) to recover completely from hypothermia before returning to the home cage.
Statistical Analysis.
All data are expressed as mean ± SEM. Statistical significance was determined by two-tailed Student’s t test or two-way ANOVA, followed by post hoc t tests using Prism 5.0 software. A value of P < 0.05 was considered statistically significant.
Acknowledgments
We thank the viral vector core at Emory University for preparing AAV viruses, Duc M. Duong at the Center for Neurodegenerative Diseases, Emory University for assistance in mass spectrometry analysis, Dr. Stephanie Sherman for advice, Cheryl Strauss for critical reading of this manuscript, and Benjamin Devenney for tissue harvest from Ts65Dn mice. This work was supported by NIH Grants NS036232 and NS041449 (to X.-J.L.), AG031153 and NS0405016 (to S.L.), and DH038384 (to R.H.R.) and by National Natural Science Foundation of China Grant 91332206 (to X.-J.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1614893114/-/DCSupplemental.
References
- 1.Li XJ, et al. Huntingtin-associated protein (HAP1): Discrete neuronal localizations in the brain resemble those of neuronal nitric oxide synthase. Proc Natl Acad Sci USA. 1996;93(10):4839–4844. doi: 10.1073/pnas.93.10.4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gutekunst CA, et al. The cellular and subcellular localization of huntingtin-associated protein 1 (HAP1): Comparison with huntingtin in rat and human. J Neurosci. 1998;18(19):7674–7686. doi: 10.1523/JNEUROSCI.18-19-07674.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li XJ, et al. A huntingtin-associated protein enriched in brain with implications for pathology. Nature. 1995;378(6555):398–402. doi: 10.1038/378398a0. [DOI] [PubMed] [Google Scholar]
- 4.Dragatsis I, Dietrich P, Zeitlin S. Expression of the Huntingtin-associated protein 1 gene in the developing and adult mouse. Neurosci Lett. 2000;282(1-2):37–40. doi: 10.1016/s0304-3940(00)00872-7. [DOI] [PubMed] [Google Scholar]
- 5.Li SH, et al. Lack of huntingtin-associated protein-1 causes neuronal death resembling hypothalamic degeneration in Huntington’s disease. J Neurosci. 2003;23(17):6956–6964. doi: 10.1523/JNEUROSCI.23-17-06956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheng G, et al. Huntingtin-associated protein 1 interacts with Ahi1 to regulate cerebellar and brainstem development in mice. J Clin Invest. 2008;118(8):2785–2795. doi: 10.1172/JCI35339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiang J, Yan S, Li SH, Li XJ. Postnatal loss of hap1 reduces hippocampal neurogenesis and causes adult depressive-like behavior in mice. PLoS Genet. 2015;11(4):e1005175. doi: 10.1371/journal.pgen.1005175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin EJ, et al. Analysis of Huntingtin-associated protein 1 in mouse brain and immortalized striatal neurons. J Comp Neurol. 1999;403(4):421–430. [PubMed] [Google Scholar]
- 9.Sheng G, et al. Hypothalamic huntingtin-associated protein 1 as a mediator of feeding behavior. Nat Med. 2006;12(5):526–533. doi: 10.1038/nm1382. [DOI] [PubMed] [Google Scholar]
- 10.Chan EY, et al. Targeted disruption of Huntingtin-associated protein-1 (Hap1) results in postnatal death due to depressed feeding behavior. Hum Mol Genet. 2002;11(8):945–959. doi: 10.1093/hmg/11.8.945. [DOI] [PubMed] [Google Scholar]
- 11.Xiang J, et al. Huntingtin-associated protein 1 regulates postnatal neurogenesis and neurotrophin receptor sorting. J Clin Invest. 2014;124(1):85–98. doi: 10.1172/JCI69206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li SH, Gutekunst CA, Hersch SM, Li XJ. Association of HAP1 isoforms with a unique cytoplasmic structure. J Neurochem. 1998;71(5):2178–2185. doi: 10.1046/j.1471-4159.1998.71052178.x. [DOI] [PubMed] [Google Scholar]
- 13.Fujinaga R, et al. Anti-human placental antigen complex X-P2 (hPAX-P2) anti-serum recognizes C-terminus of huntingtin-associated protein 1A common to 1B as a determinant marker for the stigmoid body. Histochem Cell Biol. 2007;128(4):335–348. doi: 10.1007/s00418-007-0315-5. [DOI] [PubMed] [Google Scholar]
- 14.Shinoda K, Mori S, Ohtsuki T, Osawa Y. An aromatase-associated cytoplasmic inclusion, the “stigmoid body,” in the rat brain: I. Distribution in the forebrain. J Comp Neurol. 1992;322(3):360–376. doi: 10.1002/cne.903220306. [DOI] [PubMed] [Google Scholar]
- 15.Shinoda K, Nagano M, Osawa Y. An aromatase-associated cytoplasmic inclusion, the “stigmoid body,” in the rat brain: II. Ultrastructure (with a review of its history and nomenclature) J Comp Neurol. 1993;329(1):1–19. doi: 10.1002/cne.903290102. [DOI] [PubMed] [Google Scholar]
- 16.Takeshita Y, Fujinaga R, Zhao C, Yanai A, Shinoda K. Huntingtin-associated protein 1 (HAP1) interacts with androgen receptor (AR) and suppresses SBMA-mutant-AR-induced apoptosis. Hum Mol Genet. 2006;15(15):2298–2312. doi: 10.1093/hmg/ddl156. [DOI] [PubMed] [Google Scholar]
- 17.Prigge JR, Schmidt EE. HAP1 can sequester a subset of TBP in cytoplasmic inclusions via specific interaction with the conserved TBP(CORE) BMC Mol Biol. 2007;8:76. doi: 10.1186/1471-2199-8-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takeshita Y, et al. Interaction of ataxin-3 with huntingtin-associated protein 1 through Josephin domain. Neuroreport. 2011;22(5):232–238. doi: 10.1097/WNR.0b013e32834505f4. [DOI] [PubMed] [Google Scholar]
- 19.Gutekunst CA, et al. Stigmoid bodies contain type I receptor proteins SorLA/LR11 and sortilin: New perspectives on their function. J Histochem Cytochem. 2003;51(6):841–852. doi: 10.1177/002215540305100615. [DOI] [PubMed] [Google Scholar]
- 20.Rong J, et al. 14-3-3 protein interacts with Huntingtin-associated protein 1 and regulates its trafficking. J Biol Chem. 2007;282(7):4748–4756. doi: 10.1074/jbc.M609057200. [DOI] [PubMed] [Google Scholar]
- 21.Muneoka KT, Takigawa M. 5-Hydroxytryptamine7 (5-HT7) receptor immunoreactivity-positive 'stigmoid body'-like structure in developing rat brains. Int J Dev Neurosci. 2003;21(3):133–143. doi: 10.1016/s0736-5748(03)00029-7. [DOI] [PubMed] [Google Scholar]
- 22.Hämmerle B, Elizalde C, Galceran J, Becker W, Tejedor FJ. The MNB/DYRK1A protein kinase: Neurobiological functions and Down syndrome implications. J Neural Transm Suppl. 2003;67:129–137. doi: 10.1007/978-3-7091-6721-2_11. [DOI] [PubMed] [Google Scholar]
- 23.Ahn KJ, et al. DYRK1A BAC transgenic mice show altered synaptic plasticity with learning and memory defects. Neurobiol Dis. 2006;22(3):463–472. doi: 10.1016/j.nbd.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 24.Altafaj X, et al. Neurodevelopmental delay, motor abnormalities and cognitive deficits in transgenic mice overexpressing Dyrk1A (minibrain), a murine model of Down’s syndrome. Hum Mol Genet. 2001;10(18):1915–1923. doi: 10.1093/hmg/10.18.1915. [DOI] [PubMed] [Google Scholar]
- 25.De la Torre R, et al. Epigallocatechin-3-gallate, a DYRK1A inhibitor, rescues cognitive deficits in Down syndrome mouse models and in humans. Mol Nutr Food Res. 2014;58(2):278–288. doi: 10.1002/mnfr.201300325. [DOI] [PubMed] [Google Scholar]
- 26.de la Torre R, et al. TESDAD study group Safety and efficacy of cognitive training plus epigallocatechin-3-gallate in young adults with Down’s syndrome (TESDAD): A double-blind, randomised, placebo-controlled, phase 2 trial. Lancet Neurol. 2016;15(8):801–810. doi: 10.1016/S1474-4422(16)30034-5. [DOI] [PubMed] [Google Scholar]
- 27.Torre ER, Coleman S, Yi H, Gutekunst CA. A protocol for isolation and biochemical characterization of stigmoid bodies from rat brain. J Neurosci Methods. 2003;125(1-2):27–32. doi: 10.1016/s0165-0270(03)00026-8. [DOI] [PubMed] [Google Scholar]
- 28.Maekawa S, Iino S, Miyata S. Molecular characterization of the detergent-insoluble cholesterol-rich membrane microdomain (raft) of the central nervous system. Biochim Biophys Acta. 2003;1610(2):261–270. doi: 10.1016/s0005-2736(03)00023-3. [DOI] [PubMed] [Google Scholar]
- 29.Li Y, Chin LS, Levey AI, Li L. Huntingtin-associated protein 1 interacts with hepatocyte growth factor-regulated tyrosine kinase substrate and functions in endosomal trafficking. J Biol Chem. 2002;277(31):28212–28221. doi: 10.1074/jbc.M111612200. [DOI] [PubMed] [Google Scholar]
- 30.Kittler JT, et al. Huntingtin-associated protein 1 regulates inhibitory synaptic transmission by modulating gamma-aminobutyric acid type A receptor membrane trafficking. Proc Natl Acad Sci USA. 2004;101(34):12736–12741. doi: 10.1073/pnas.0401860101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rong J, et al. Regulation of intracellular trafficking of huntingtin-associated protein-1 is critical for TrkA protein levels and neurite outgrowth. J Neurosci. 2006;26(22):6019–6030. doi: 10.1523/JNEUROSCI.1251-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyata Y, Nishida E. DYRK1A binds to an evolutionarily conserved WD40-repeat protein WDR68 and induces its nuclear translocation. Biochim Biophys Acta. 2011;1813(10):1728–1739. doi: 10.1016/j.bbamcr.2011.06.023. [DOI] [PubMed] [Google Scholar]
- 33.Tejedor FJ, Hämmerle B. MNB/DYRK1A as a multiple regulator of neuronal development. FEBS J. 2011;278(2):223–235. doi: 10.1111/j.1742-4658.2010.07954.x. [DOI] [PubMed] [Google Scholar]
- 34.Guimerá J, et al. A human homologue of Drosophila minibrain (MNB) is expressed in the neuronal regions affected in Down syndrome and maps to the critical region. Hum Mol Genet. 1996;5(9):1305–1310. doi: 10.1093/hmg/5.9.1305. [DOI] [PubMed] [Google Scholar]
- 35.Song WJ, et al. Isolation of human and murine homologues of the Drosophila minibrain gene: Human homologue maps to 21q22.2 in the Down syndrome “critical region”. Genomics. 1996;38(3):331–339. doi: 10.1006/geno.1996.0636. [DOI] [PubMed] [Google Scholar]
- 36.Shindoh N, et al. Cloning of a human homolog of the Drosophila minibrain/rat Dyrk gene from “the Down syndrome critical region” of chromosome 21. Biochem Biophys Res Commun. 1996;225(1):92–99. doi: 10.1006/bbrc.1996.1135. [DOI] [PubMed] [Google Scholar]
- 37.Martí E, et al. Dyrk1A expression pattern supports specific roles of this kinase in the adult central nervous system. Brain Res. 2003;964(2):250–263. doi: 10.1016/s0006-8993(02)04069-6. [DOI] [PubMed] [Google Scholar]
- 38.Wegiel J, et al. Cell type- and brain structure-specific patterns of distribution of minibrain kinase in human brain. Brain Res. 2004;1010(1-2):69–80. doi: 10.1016/j.brainres.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 39.Hämmerle B, et al. Expression patterns and subcellular localization of the Down syndrome candidate protein MNB/DYRK1A suggest a role in late neuronal differentiation. Eur J Neurosci. 2003;17(11):2277–2286. doi: 10.1046/j.1460-9568.2003.02665.x. [DOI] [PubMed] [Google Scholar]
- 40.Murakami N, Bolton D, Hwang YW. Dyrk1A binds to multiple endocytic proteins required for formation of clathrin-coated vesicles. Biochemistry. 2009;48(39):9297–9305. doi: 10.1021/bi9010557. [DOI] [PubMed] [Google Scholar]
- 41.Becker W, et al. Sequence characteristics, subcellular localization, and substrate specificity of DYRK-related kinases, a novel family of dual specificity protein kinases. J Biol Chem. 1998;273(40):25893–25902. doi: 10.1074/jbc.273.40.25893. [DOI] [PubMed] [Google Scholar]
- 42.Park J, Song WJ, Chung KC. Function and regulation of Dyrk1A: Towards understanding Down syndrome. Cell Mol Life Sci. 2009;66(20):3235–3240. doi: 10.1007/s00018-009-0123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davisson MT, et al. Segmental trisomy as a mouse model for Down syndrome. Prog Clin Biol Res. 1993;384:117–133. [PubMed] [Google Scholar]
- 44.Reeves RH, et al. A mouse model for Down syndrome exhibits learning and behaviour deficits. Nat Genet. 1995;11(2):177–184. doi: 10.1038/ng1095-177. [DOI] [PubMed] [Google Scholar]
- 45.Belichenko PV, et al. Synaptic structural abnormalities in the Ts65Dn mouse model of Down Syndrome. J Comp Neurol. 2004;480(3):281–298. doi: 10.1002/cne.20337. [DOI] [PubMed] [Google Scholar]
- 46.Fuchs C, Ciani E, Guidi S, Trazzi S, Bartesaghi R. Early-occurring proliferation defects in peripheral tissues of the Ts65Dn mouse model of Down syndrome are associated with patched1 over expression. Lab Invest. 2012;92(11):1648–1660. doi: 10.1038/labinvest.2012.117. [DOI] [PubMed] [Google Scholar]
- 47.Nissen RM, Amsterdam A, Hopkins N. A zebrafish screen for craniofacial mutants identifies wdr68 as a highly conserved gene required for endothelin-1 expression. BMC Dev Biol. 2006;6:28. doi: 10.1186/1471-213X-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang B, et al. Wdr68 requires nuclear access for craniofacial development. PLoS One. 2013;8(1):e54363. doi: 10.1371/journal.pone.0054363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen JY, Lin JR, Tsai FC, Meyer T. Dosage of Dyrk1a shifts cells within a p21-cyclin D1 signaling map to control the decision to enter the cell cycle. Mol Cell. 2013;52(1):87–100. doi: 10.1016/j.molcel.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soppa U, et al. The Down syndrome-related protein kinase DYRK1A phosphorylates p27(Kip1) and Cyclin D1 and induces cell cycle exit and neuronal differentiation. Cell Cycle. 2014;13(13):2084–2100. doi: 10.4161/cc.29104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Georgopoulou N, et al. BM88 is a dual function molecule inducing cell cycle exit and neuronal differentiation of neuroblastoma cells via cyclin D1 down-regulation and retinoblastoma protein hypophosphorylation. J Biol Chem. 2006;281(44):33606–33620. doi: 10.1074/jbc.M602689200. [DOI] [PubMed] [Google Scholar]
- 52.Yano M, Okano HJ, Okano H. Involvement of Hu and heterogeneous nuclear ribonucleoprotein K in neuronal differentiation through p21 mRNA post-transcriptional regulation. J Biol Chem. 2005;280(13):12690–12699. doi: 10.1074/jbc.M411119200. [DOI] [PubMed] [Google Scholar]
- 53.Diolaiti D, et al. Functional cooperation between TrkA and p75(NTR) accelerates neuronal differentiation by increased transcription of GAP-43 and p21(CIP/WAF) genes via ERK1/2 and AP-1 activities. Exp Cell Res. 2007;313(14):2980–2992. doi: 10.1016/j.yexcr.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 54.Hung SP, et al. Genistein-induced neuronal differentiation is associated with activation of extracellular signal-regulated kinases and upregulation of p21 and N-cadherin. J Cell Biochem. 2005;96(5):1061–1070. doi: 10.1002/jcb.20626. [DOI] [PubMed] [Google Scholar]
- 55.Fischer-Brandies H, Schmid RG, Fischer-Brandies E. Craniofacial development in patients with Down’s syndrome from birth to 14 years of age. Eur J Orthod. 1986;8(1):35–42. doi: 10.1093/ejo/8.1.35. [DOI] [PubMed] [Google Scholar]
- 56.Myrelid A, Gustafsson J, Ollars B, Annerén G. Growth charts for Down’s syndrome from birth to 18 years of age. Arch Dis Child. 2002;87(2):97–103. doi: 10.1136/adc.87.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pueschel SM, Rothman KJ, Ogilby JD. Birth weight of children with Down’s syndrome. Am J Ment Defic. 1976;80(4):442–445. [PubMed] [Google Scholar]
- 58.Salehi A, Faizi M, Belichenko PV, Mobley WC. Using mouse models to explore genotype-phenotype relationship in Down syndrome. Ment Retard Dev Disabil Res Rev. 2007;13(3):207–214. doi: 10.1002/mrdd.20164. [DOI] [PubMed] [Google Scholar]
- 59.Sturgeon X, Gardiner KJ. Transcript catalogs of human chromosome 21 and orthologous chimpanzee and mouse regions. Mamm Genome. 2011;22(5-6):261–271. doi: 10.1007/s00335-011-9321-y. [DOI] [PubMed] [Google Scholar]
- 60.Sago H, et al. Ts1Cje, a partial trisomy 16 mouse model for Down syndrome, exhibits learning and behavioral abnormalities. Proc Natl Acad Sci USA. 1998;95(11):6256–6261. doi: 10.1073/pnas.95.11.6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Belichenko PV, Kleschevnikov AM, Salehi A, Epstein CJ, Mobley WC. Synaptic and cognitive abnormalities in mouse models of Down syndrome: Exploring genotype-phenotype relationships. J Comp Neurol. 2007;504(4):329–345. doi: 10.1002/cne.21433. [DOI] [PubMed] [Google Scholar]
- 62.Song WJ, et al. Phosphorylation and inactivation of glycogen synthase kinase 3β (GSK3β) by dual-specificity tyrosine phosphorylation-regulated kinase 1A (Dyrk1A) J Biol Chem. 2015;290(4):2321–2333. doi: 10.1074/jbc.M114.594952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mortz E, Krogh TN, Vorum H, Görg A. Improved silver staining protocols for high sensitivity protein identification using matrix-assisted laser desorption/ionization-time of flight analysis. Proteomics. 2001;1(11):1359–1363. doi: 10.1002/1615-9861(200111)1:11<1359::AID-PROT1359>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 64.McGuire JR, Rong J, Li SH, Li XJ. Interaction of Huntingtin-associated protein-1 with kinesin light chain: Implications in intracellular trafficking in neurons. J Biol Chem. 2006;281(6):3552–3559. doi: 10.1074/jbc.M509806200. [DOI] [PubMed] [Google Scholar]