Abstract
Poor uterine receptivity leads to implantation defects or failure. Identification of uterine molecules crucial to uterine receptivity and/or embryo implantation provides the opportunity to design a diagnostic screening toolkit for uterine receptivity or targeted drug discovery for treating implantation-based infertility. In this regard, gene-profiling studies performed in humans and rodents have identified numerous genes involved in the transcriptional regulation of uterine receptivity and embryo implantation. In this article, we compared available uterine microarray datasets collected during the time of uterine receptivity and implantation in humans, mice and hamsters to uncover conserved gene sets. We also compared the transcriptome signature of women with unexplained infertility (UIF) and recurrent implantation failure (RIF) to gain insight into genes potentially dysregulated during endometrial receptivity or embryo implantation. Among numerous differentially expressed genes, few were revealed that might have molecular diagnostic screening potential for identifying the uterine receptive state during the time of implantation. Finally, functional annotation of gene sets uncovered altered uterine apoptosis or cell adhesion pathways in women with UIF and RIF, respectively. These conserved or divergent gene sets provide insights into the uterine receptive state for supporting blastocyst implantation.
Keywords: Endometrial receptivity, Implantation site, Gene expression, Unexplained infertility, Recurrent implantation failure
1. Introduction
Blastocyst implantation failure in naturally-occurring and assisted human reproduction occurs in up to 2/3 of all cases, and has been attributed to delayed or failed receptivity. Defective endometrial receptivity is also considered a major cause of unexplained infertility (~10% of reproductive women) and abnormal pregnancies (~25 to 40%).1–6 Past studies have demonstrated that certain morphological parameters and regulation of several uterine genes are associated with successful uterine receptivity and implantation. Predictors of the uterine receptive state are needed to better understand the causes of uterine-based infertility and help women in whom a poor uterine receptive state is considered a limiting factor for blastocyst implantation and pregnancy success. Identification of endometrial molecular signatures will provide the opportunity to design diagnostic screening tests for detecting uterine receptivity status, as well as therapeutic drug discovery for treating implantation-based infertility/pregnancy defects.
Embryo implantation is a multifaceted process beginning with attachment of the blastocyst to the uterine wall. Importantly, embryo implantation occurs during a specific “window” of time when the hormonally prepared uterus becomes receptive and the embryo has reached its proper developmental state.7,8 In women, the uterus becomes receptive ~7 to 10 days after ovulation or the LH surge.9 In mice, the uterus achieves receptivity after a transient pre-implantation estrogen (E2) rise occurring around noon of day 4 of pregnancy.10 Following the short window of implantation, the uterus becomes non-receptive to the implantation-competent blastocyst.7,11 A challenging question has always been how to distinguish the normal or defective uterine receptive state from the non-receptive state.
Since embryo implantation is the initial event defining mammalian pregnancy, it is possible that this event is regulated by conserved gene functions across species. Shared features of embryo implantation in humans and mice include stromal decidualization and hemochorial mode of placentation. Differences in implantation include: (1) hormonal preparation of the receptive uterus, (2) mode of embryo implantation, (3) trophoblast attachment side (polarity) to the luminal epithelium (LE) and (4) timing of decidualization.12 The uterus of humans, rhesus monkeys, pigs, rabbits, guinea pigs and hamsters require only ovarian progesterone (P4) to prepare the uterus for blastocyst implantation, suggesting luteal estrogen may play a permissive role in these species.13–21 However, the uterus of gerbils, rats and mice requires an active role of both P4 and estrogen (E2) to achieve its receptive state.7,10,22 Blastocyst implantation in humans is interstitial (invasive), where the blastocyst completely embeds within the uterine stroma by displacing the underlying epithelium; while mice and hamsters exhibit an eccentric (displacement) type of implantation, where the blastocyst lies within a uterine crypt and causes loss of the underlying epithelial cells.12 Despite these differences, the implantation process in most species involves an initial interaction between the trophectoderm of the blastocyst and the apical surface of the uterine LE.23 Normally, the apical surface of the pre-receptive LE does not allow blastocyst attachment. However, the uterine transition from pre-receptive to receptive state permits fundamental structural and functional changes in epithelial cell organization,24 allowing for successful blastocyst attachment.
Early gene expression analyses and gene targeting technology in mice yielded a substantial amount of information on the importance of individual genes required for uterine receptivity and blastocyst implantation. Such genes included a number of growth factors, cytokines, transcription factors, as well as others.1,25–27 Given the complexity of blastocyst implantation in a receptive uterus, this most likely involves the actions of multiple gene families and gene-environment interactions. Identification of this genetic environment and genetic signaling networks remained a particular challenge for quite sometime, but the development of microarray and RNA-sequencing technologies has helped make such identifications possible.
To gain insight into the necessary genes for embryo implantation in humans, studies have compared the pre-receptive and receptive endometrium of non-conception cycles in order to avoid ethical constraints of collecting endometrial samples from conception cycles in which the embryo is present in the uterus. However, there is a single study to date that inadvertently collected endometrium from a conception cycle to study endometrial gene expressions in humans.28 Studies using mouse and hamster models have added insight into the molecular basis of human implantation because of the existence of some important shared features. Over the past two decades, a variety of platforms have been implemented for measuring gene expression: oligonucleotide chips, cDNA microarrays, serial analysis of gene expression (SAGE) and, more recently, RNAseq. Each technology, utilized by multiple studies, has revealed many differentially expressed genes (DEGs) between: pre-receptive and receptive endometrium of humans,29–34 rhesus monkeys,35–37 rabbits38 and mice39; post-implantation sites and non-implantation sites in hamsters40 and mice.41,42 However, the number of common DEGs is relatively small, as a result of either: (1) conservation of DEGs; (2) differences in experimental design and forms of technology used as older technology examine considerably smaller subsets of genes compared to current technology; and (3) variation in data analysis tools, such as use of various normalization or expression methods may yield different results.
The goal of this review is to provide a comprehensive analysis of available gene expression profiling data sets comparing pre-receptivity, receptivity and post-implantation endometria to elucidate genes needed for receptivity as well as embryo implantation. Data sets were restricted to microarray studies for reasons of: simplicity of comparison and lack of complete RNA-sequencing studies performed on mouse and hamster embryo implantation sites.
2. Endometrial receptivity transcriptomic profiling
2.1. Healthy, natural-cycling women
Many underlying causes of human infertility have been circumvented by in vitro fertilization (IVF) and embryo-transfer techniques. Despite this, implantation rates remain low, likely the result of transferring embryos into non-receptive endometrium. Therefore, several studies have compared the transcriptomics of the human endometrium in different phases of the menstrual cycle, including within the receptivity phase.29–34 These studies demonstrated the existence of differential gene expression patterns in different phases, allowing classification of the endometrium based on its molecular signature. Exploitation of the endometrial signature at the receptivity phase has led to an endometrial receptivity array (ERA) as a clinically-utilized diagnostic tool to differentiate phases of the menstrual cycle, including the window of implantation.43 The ERA has been used to demonstrate a shift in the window of implantation of patients with repeated implantation failure (RIF) and to guide their personalized embryo transfer as a novel therapeutic strategy.44,45 Although improved by this approach, implantation rates of patients with RIF remain suboptimal.
The genes included in the ERA were selected from one study comparing DEG between the pre-receptive and receptive endometrium.46 It is possible that the strength of the ERA could be improved by inclusion of DEGs obtained from other studies comparing the pre-receptive and receptive endometrium. To this end, we have combined the DEGs obtained from five studies (Table 1, green box) with the ERA, resulting in a 1541 non-overlapping ‘Human Endometrial Receptivity’ gene signature of healthy, naturally-cycling women. Of these 1541 DEGs, a total of 241 gene transcripts were shared by two or more microarray studies. We then compared the similarity of the ‘Human Endometrial Receptivity’ transcriptome signature to the endometrial transcriptome of patients with Unexplained Infertility or Recurrent Implantation failure, which is discussed below.
Table 1.
Available microarray data sets used to elucidate important genes during human endometrial receptivity, animal (mouse and hamster) models of embryo implantation to elucidate genes important in these processes as well as to identify shared endometrial DEGs to those reported for patients with unexplained infertility (UIF) and recurrent implantation failure (RIF).
| Study | Species | Tissues Compared | Total Gene | DEG | DEG converted to human official gene symbol | Complete DEG | ||
|---|---|---|---|---|---|---|---|---|
| Pre-receptive | Receptive | 1,541 non- overlapping combined |
Human Endometrial Receptivity |
|||||
| Carson et al., 2002 | Human | LH+2 to +4 | LH+7 to +9 | 12,000 | 699 (329 up, 370 down) | 725 | ||
| Riesewijk et al., 2003 | Human | LH+2 | LH+7 | 12,000 | 211 (153 up, 58 down) | 210 | ||
| Mirkin et al., 2005 | Human | LH+3 | LH+8 | 12,686 | 107 (49 up, 58 down) | 113 | ||
| Kao et al., 2011 | Human | late proliferative | LH+8 to LH+10 | 12,686 | 332 (114 up, 218 down) | 394 | ||
| Chan et al., 2013 | Human | LH+2 | LH+7 | 22,333 | 244 (126 up, 119 down) | 244 | ||
| Endometrial Receptivity Array | ||||||||
| Diaz-Gimeo et al., 2011 | Human | Proliferative (day 8-12 menstrual cycle) |
Receptive (LH+7) | 1,105 | 179 (93 up, 86 down) | 238 | ||
| Pre-receptive (LH+1 to LH+5) |
200 (110 up, 90 down) | |||||||
|
Pre-implantation (pre-receptive) |
Post-Implantation (receptive) |
2,123 non- overlapping combined |
Mouse/ Hamster Embryo Implantation |
|||||
| Xiao et al., 2014 | Mouse | day 4 LE | day 5 LE | 28,853 | 627 (383 up, 244 down) | 719 | ||
| Post-implantation | ||||||||
| Non-Receptive | Receptive | |||||||
| Chen et al., 2006 | Mouse | day 5 LE | day 5 LE | 12,345 | 114 (64 up, 50 down) | 133 | ||
| Reese et al., 2001 | Mouse | day 5 non-implant site | day 5 implant site | 11,987 | 233 (122 up, 111 down) | 314 | ||
| Wei and Herington et al., 2014 | Hamster | day 5 non-implant site | day 5 implant site | 22,690 | 345 (215 up, 130 down) | 513 | ||
| Implantation | ||||||||
| Delayed | Activated | |||||||
| Reese et al., 2001 | Mouse | P4-primed, day 7 uterus |
P4 and E2-primed, day 7 uterus |
11,987 | 591 (214 up, 377 down) | 885 | ||
|
Unexplained Infertility (UIF) |
Fertile | 260 non- overlapping combined |
UIF | |||||
| Altmae et al., 2010 | Human | LH+7 | LH +7 | 44,000 | 260 (145 up, 115 down) | 260 | ||
|
Recurrent Implantation Failure (RIF) |
Fertile | 113 non- overlapping combined |
RIF | |||||
| Tapia-Pizarro et al., 2014 | Human | P+7 | Successful IVF, P+7 Spontaneously Fertile, P+7 |
38,500 | 82 (51 up, 31 down) | 99 | ||
| Tapia et al., 2008 | Human | P+7 | Successful IVF, P+7 Spontaneously Fertile, P+7 |
9,128 | 21 (1 up, 20 down) | 21 | ||
*shared differentially expressed genes (DEGs) between at least two studies. E2= estradiol, IVF = In Vitro Fertilization, LE= luminal epithelium, LH= lutenizing hormone (days since surge), P=progesterone
2.2. Patients with recurrent implantation failure (RIF)
Recurrent implantation failure (RIF) is diagnosed when high-quality embryos fail to implant following several IVF treatment cycles. In the absence of recognizable genital tract, embryonic and endocrine factors, studies have sought to identify genes whose aberrant expression is consistently associated with implantation failure.47 These studies hypothesize that the pattern of endometrial gene expression during the receptive period may differ between women who have had successful versus failed embryo implantation following repeated embryo transfers.47,48
To date, two studies performed transcriptome analysis to identify DEGs in endometrial samples from women with RIF compared to spontaneously fertile women and patients with successful IVF treatment.47,48 Endometrial samples were collected during a receptive period of induced endometrial cycle (used for IVF/embryo transfer) using exogenous E2 and P4. The authors state that the stimulation protocol performed before the endometrial sample collection was the same for all participating women in their studies. Thus, the differential transcript profile in patients with RIF suggests a long-term dysregulation of endometrial gene expression rendering it not suitable for embryo implantation.47 These studies had a total of 6 overlapping DEGs [complement component 4 binding protein, alpha (C4BPA), Clusterin (CLU), Immunoglobulin Heavy Constant Gamma 1 (IGHG1), Microsomal Glutathione S-Transferase 1 (MGST1), Progestagen Associated Endometrial Protein (PAEP) and Ribonucleotide Reductase Catalytic Subunit M1 (RRM1)], all downregulated in patients with RIF.
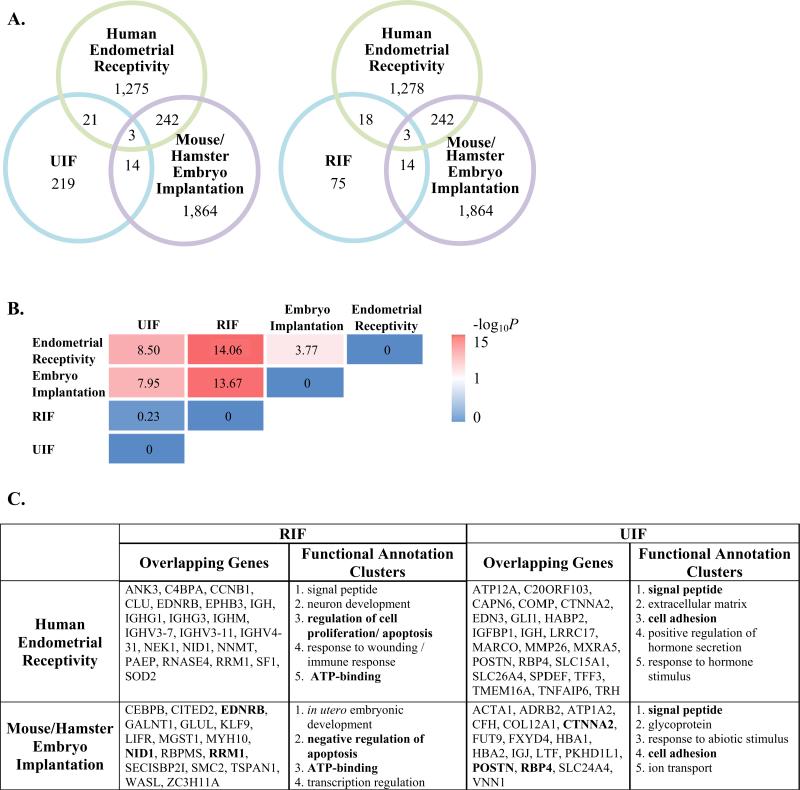
In total, there were 113 non-overlapping DEGs comprising the ‘RIF’ transcriptome signature (Table 1, lower blue box). When comparing the ‘RIF’ and ‘Human Endometrial Receptivity’ profiles, there were 21 shared DEGs (Fig. 1A, listed in panel C). Based on these transcriptomic studies in patients with RIF, it appears that some cases of RIF could be associated with an aberrant gene expression profile. To this end, some of the aberrant gene expression profile is a result of genes associated with endometrial receptivity. Moreover, a subset of these genes has previously been identified as implantation-specific genes from transcriptomic studies performed in animal models of implantation, which will be discussed in more detail below.
Fig. 1.
(A) Venn diagram illustrating the overlap of DEGs between microarray studies (each circle is color coded to microarray data sets described in Table 1). (B) Fisher's exact test was used to examine overlap of DEGs between microarray data sets. The P-value was converted to –log 10. High degrees of significant association are indicated by bright pink. (C) Shared DEGs are listed in the table. Shared DEGs were uploaded into the DAVID bioinformatics resource and clustered based on their known functions. Official gene symbols in bold typeface represent similar shared DEGs in RIF or UIF between human endometrial receptivity and mouse/hamster embryo implantation (see middle of Venn Diagrams, A). Similarly, shared functional annotation clusters are shown in bold.
2.3. Patients with unexplained infertility (UIF)
One likely deficiency found among female infertility of unknown origin may be an intrinsic defect in the expression of crucial genes for implantation.49 Thus, women with ‘unexplained infertility’ have been an attractive study group in the search for target molecules involved in endometrial receptivity and embryo implantation. Specifically, these women have no apparent reason for infertility, having normal ovulatory cycles and hormone profiles, as well as a lack of organ pathology and evidence of male factor infertility.
To the best of our knowledge, only two studies have investigated the endometrial gene expression profiles in women with UIF, compared to fertile controls, during endometrial receptivity.50,51 These studies had no overlapped DEGs, which was attributed to differences in samples utilized (natural cycle LH+7 versus LH+6 to +10),50 though different microarray platforms were utilized as well. To this end, the timing of endometrial biopsy sampling has been shown to be critical for variation between samples.33
Since only Altmae and colleagues reported complete data, their study was used for the comparisons in this review. In total, there were a total of 260 non-overlapping DEGs comprising the ‘UIF’ transcriptome signature (Table 1, upper blue box). When comparing transcriptome profiles of UIF to the ‘Human Endometrial Receptivity’ gene set, there were 24 shared DEGs (Fig. 1A, listed in panel C). Thus, it appears that some of the aberrant gene expression in women with unexplained infertility can be explained in part by endometrial receptivity. It remains to be determined whether any DEGs in women with unexplained infertility overlap with genes needed for embryo implantation; however, this will be discussed below.
Finally, we compared the DEGs from the above RIF microarray studies to the unexplained infertility studies, and found only two shared genes: ASPN, which encodes the protein asporin, and IGH. Though noted for its high level of expression in the uterus,52 the role of ASPN remains to be investigated.
3. Embryo implantation transcriptomic profiling
Genetic studies using animal models have allowed examination into differentially expressed genes during embryo implantation; for ethical reasons, this is not possible in humans. Each animal model has been chosen for its similarity to the process of embryo implantation in humans, though, it remains important to determine whether genes needed for implantation in animal models translate to humans. By examining the transcriptome of multiple models, we can identify genes that are likely to have significant roles in implantation.
3.1. Mouse models of natural implantation
As previously stated, researchers have utilized several different sample collection techniques for their analysis of the mouse implantation transcriptome. First, they may compare the whole segment of implantation site (IS) to the inter- or non-implantation (IIS or NIS) sites. By analyzing the intact IS and NIS, this preserves an undisturbed relationship between the uterine myometrium, stroma, and epithelium. Researchers may remove the blastocyst or embryo, in order to focus just on the maternal transcriptome signature during implantation. Profiling the expression of embryonic factors may identify those that are significant to implantation. Finally, although the different uterine compartments respond to ovarian hormones during uterine preparation for implantation, the functions of these tissues differ. Since the luminal epithelium is the first tissue of contact for the blastocyst, studies have focused on DEG between uterine LE collected from pre-IS versus post-IS, as well as from post-implantation IS vs NIS. Physical disruption of the uterine LE would likely result in different gene expression profiles than those obtained using whole implantation site.
3.2. Mouse models of activated or delayed implantation
In pregnant mice, removal of pre-implantation estrogen secretion by ovariectomy postpones the onset of implantation and induces blastocyst dormancy.7,53 A single injection of estrogen can reactivate the signaling network, resulting in implantation in a P4-primed uterus. Thus, one can compare mice with delayed implantation (P4-only) to mice with E2-induced dissolution of delayed implantation (P4 + E2) to identify implantation-specific genes.42
3.3. Hamster model of natural implantation
The hamster is an animal model where implantation is solely progesterone-dependent,12,19 and thus used to gain understanding of the embryo and uterine events required for implantation. Prior to RNAseq technology, cross-species hybridization microarray technology was used to investigate the transcriptome signature during implantation.40 It is important to note that, the information obtained from these cross-species hybridization microarrays may not identify important and/or rare transcripts due to inadequate gene sequence homology. However, the genes identified by most cross-species microarray probes are those showing high sequence conservation.
A ‘Mouse/Hamster Embryo Implantation’ transcriptome profile was constructed by combining the DEGs obtained from five gene profile studies (Table 1), which included: whole mouse post-implantation IS vs NIS42; mouse preimplantation vs postimplantation LE39; LE from mouse post-implantation IS vs NIS54; and whole hamster post-implantation IS vs NIS.40 Each of these studies collected samples from natural pregnant mice and hamster, respectively. We also chose to examine DEGs from a study comparing active vs delayed implantation uteri.42 We used the available microarray data from these studies and converted either their ‘Affymetrix ID’ or ‘Genbank Accession Number’ to the ‘Official Gene Symbol’, for a more equal comparison to the data from the human microarray studies discussed above and outlined in Table 1. In total, the mouse and hamster microarray studies yielded 2123 non-overlapping DEGs (Table 1, purple box). Of these, 340 transcripts were shared between at least two studies.
4. Comparing the endometrial transcriptomic profiles of receptivity, embryo implantation, RIF and UIF
The number of overlapping DEGs between the ‘Human Endometrial Receptivity’ and ‘Mouse/Hamster Embryo Implantation’ transcriptome profiles compared to ‘UIF’ and ‘RIF’ is illustrated in Fig. 1A. Among the 219 DEGs comprising the endometrial transcriptome signature of patients with unexplained infertility, 24 (11%) and 17 (7.8%) are similar to those gene transcripts associated with endometrial receptivity and embryo implantation, respectively. Surprisingly, 28% (21) and 23% (17) DEGs are shared endometrial genes between women with recurrent implantation failure and those associated with endometrial receptivity and embryo implantation, respectively.
In order to more clearly examine the similarities between the ‘RIF’, ‘UIF’, ‘Human Endometrial Receptivity’ and ‘Mouse/Hamster Embryo Implantation’ transcriptomal signatures, we performed a Fisher's exact test with overlapping and non-overlapping DEGs. As shown in Fig. 1B, we found that the transcriptome profile of women with UIF and women with RIF shows statistically significant (P < 0.0001) association with both transcriptome profiles of human endometrial receptivity and mouse/hamster embryo implantation. Similarly, there is a statistically significant (P < 0.0001) association between the transcriptome signature of human endometrial receptivity and mouse/hamster implantation. Interestingly, the transcriptome profiles of women with UIF and RIF showed no association. This finding was not surprising given that only two genes (ASPN and IGH) were overlapped between the two transcriptomes.
The official gene symbol of overlapping DEGs between the four transcriptional signatures examined are listed in Fig. 1C. Transcripts that are dysregulated in the endometria of women with RIF or UIF, which overlap with those associated with endometrial receptivity and/or embryo implantation could serve as potential therapeutic targets. Researchers are working toward targeted therapeutics for POSTN (periostin),55 which is upregulated in the endometria of UIF.50
Using the DAVID bioinformatics resource, we were able to reveal the enriched functional annotation clusters in each of the four-transcriptome profiles (Fig. 1C). Regulation of apoptosis and ATP-binding are dysregulated biological functions in RIF that are associated with endometrial receptivity and embryo implantation. Similarly, signal peptides and cell adhesion was identified as dysregulated biological functions in UIF that are associated with endometrial receptivity and embryo implantation.
5. Conclusion
5.1. Strategic implications on human reproduction
Uterine receptivity is a brief and distinctive altered state, which allows the blastocyst to implant. Although endometrial receptivity is regulated by ovarian hormones in a species-specific manner,12 it can also be influenced by local activity of the blastocyst.36 Unless we identify a means for detecting the uterine receptive state, improving the pregnancy rate following assisted-reproduction embryo transfer is less likely. Over the past decade, markers for multifactorial uterine receptivity and implantation events have been attempted using single gene expression profile analysis. Although the diagnostic value of the expression of each gene alone is valuable, multigene analysis can increase predictability and potentially be used as a biomarker. Several global gene analyses have been attempted in human, mouse and hamster models in the search for genes that may be involved in inducing receptivity. Although each study has revealed tens or hundreds of genes associated with uterine receptivity and implantation, no attempt has been made to hunt for potential markers by comparing available datasets from collective human, mouse and hamster microarray data sets. Herein, we attempted to provide new information on several genes or pathways that may have functional significance in determining a uterine state that either successfully or fails to support implantation.
5.2. Observations regarding the need for additional research
In the current study, bio-informatics tools used to group genes by their functions implicated different biochemical pathways or processes. Interestingly, comparison of over- or under-expressed genes between human endometrial receptive state or mouse/hamster implantation site and recurrent implantation failure clearly suggests gene regulators of cellular proliferation and apoptosis or ATP-binding is associated with RIF. Similar comparison between human endometrial receptive state or mouse/hamster implantation site from UIF revealed changes in signal peptide and cell adhesion molecules. This finding suggests that UIF and RIF are two events involving defects in different sets of genes, with the exception of two overlapping genes (ASPN and IGH).
In this review, we focused on analyzing and comparing available microarray data sets between receptive and non-receptive states of the human uterus as well as IS and NIS of the mouse and hamster in the quest for human endometrial receptivity molecular marker (s). Analysis of differentially expressed genes among human endometrial receptivity, human UIF and mouse/hamster embryo implantation sites yielded only 3 commonly co-expressed genes. Similar analysis of genes expressed among human endometrial receptivity, human UIF and mouse/hamster embryo implantation sites yielded only 3 co-expressed genes.
As pointed out in this review, advanced technology has emerged to measure gene expression based on next generation sequencing. RNAseq is capable of true genome-wide analysis, sequencing all of the mRNAs present in a sample, while 25% of low-level expressed genes remain undetected in microarray analysis. While microarray studies have yielded similar results to RNAseq, the latter technology reveals several more DEGs. Thus, it may prove beneficial to repeat comparative analysis of transcriptomic studies using RNAseq to refine the ERA, as well as differential transcriptomics in women with RIF. Moreover, validation of these gene predictors using a multi-step approach including genomic, proteomic and tissue array profiling will be necessary for future clinical applications.
Acknowledgements
This work was supported by National Institutes of Health grant HD044741 (BCP), HD080148 (BCP), HD81121 (JR) and research funds (JLH) from the Vanderbilt Office of Clinical and Translational Scientist Development and CTSA award number KL2TR000446 from the National Center for Advancing Translational Sciences.
Footnotes
Authors’ contribution
JLH and BCP conceived and wrote the review. JR assisted writing the review. JLH and YG performed comparative microarray analysis. JLH and YG prepared the figures and tables.
Conflicts of interest
The authors have none to declare.
References
- 1.Lee KY, Jeong JW, Tsai SY, Lydon JP, DeMayo FJ. Mouse models of implantation. Trends Endocrinol Metab. 2007;18:234–239. doi: 10.1016/j.tem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Norwitz ER, Schust DJ, Fisher SJ. Implantation and the survival of early pregnancy. N Engl J Med. 2001;345:1400–1408. doi: 10.1056/NEJMra000763. [DOI] [PubMed] [Google Scholar]
- 3.Sharkey AM, Smith SK. The endometrium as a cause of implantation failure. Best Pract Res Clin Obstet Gynaecol. 2003;17:289–307. doi: 10.1016/s1521-6934(02)00130-x. [DOI] [PubMed] [Google Scholar]
- 4.Simon C, Moreno C, Remohi J, Pellicer A. Cytokines and embryo implantation. J Reprod Immunol. 1998;39:117–131. doi: 10.1016/s0165-0378(98)00017-5. [DOI] [PubMed] [Google Scholar]
- 5.Wilcox AJ, Baird DD, Weinberg CR. Time of implantation of the conceptus and loss of pregnancy. N Engl J Med. 1999;340:1796–1799. doi: 10.1056/NEJM199906103402304. [DOI] [PubMed] [Google Scholar]
- 6.Wilcox AJ, Weinberg CR, O'Connor JF, et al. Incidence of early loss of pregnancy. N Engl J Med. 1988;319:189–194. doi: 10.1056/NEJM198807283190401. [DOI] [PubMed] [Google Scholar]
- 7.Paria BC, Huet-Hudson YM, Dey SK. Blastocyst's state of activity determines the “window” of implantation in the receptive mouse uterus. Proc Natl Acad Sci U S A. 1993;90:10159–10162. doi: 10.1073/pnas.90.21.10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Psychoyos A. Hormonal control of uterine receptivity for nidation. J Reprod Fertil Suppl. 1976:17–28. [PubMed] [Google Scholar]
- 9.Bergh PA, Navot D. The impact of embryonic development and endometrial maturity on the timing of implantation. Fertil Steril. 1992;58:537–542. doi: 10.1016/s0015-0282(16)55259-5. [DOI] [PubMed] [Google Scholar]
- 10.McCormack JT, Greenwald GS. Evidence for a preimplantation rise in oestradiol-17beta levels on day 4 of pregnancy in the mouse. J Reprod Fertil. 1974;41:297–301. doi: 10.1530/jrf.0.0410297. [DOI] [PubMed] [Google Scholar]
- 11.Das SK, Wang XN, Paria BC, et al. Heparin-binding EGF-like growth factor gene is induced in the mouse uterus temporally by the blastocyst solely at the site of its apposition: a possible ligand for interaction with blastocyst EGF-receptor in implantation. Development. 1994;120:1071–1083. doi: 10.1242/dev.120.5.1071. [DOI] [PubMed] [Google Scholar]
- 12.Reese J, Wang H, Ding T, Paria BC. The hamster as a model for embryo implantation: insights into a multifaceted process. Semin Cell Dev Biol. 2008;19:194–203. doi: 10.1016/j.semcdb.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simón C, Cano F, Valuena D, Remohí J, Pellicer A. Clinical evidence for a detrimental effect on uterine receptivity of high serum oestradiol concentrations in high and normal responder patients. Hum Reprod. 1995;10:2432–2437. doi: 10.1093/oxfordjournals.humrep.a136313. [DOI] [PubMed] [Google Scholar]
- 14.Deanesly R. Normal implantation in ovariectomized guinea pigs. Nature. 1960;186:327–328. doi: 10.1038/186327b0. [DOI] [PubMed] [Google Scholar]
- 15.George FW, Wilson JD. Estrogen formation in the early rabbit embryo. Science. 1978;199:200–201. doi: 10.1126/science.579477. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh D, De P, Sengupta J. Luteal phase ovarian oestrogen is not essential for implantation and maintenance of pregnancy from surrogate embryo transfer in the rhesus monkey. Hum Reprod. 1994;9:629–637. doi: 10.1093/oxfordjournals.humrep.a138561. [DOI] [PubMed] [Google Scholar]
- 17.Greenwald GS. Endocrinology of the Pregnant Hamster. Springer US/Plenum Press; New York: 1985. [Google Scholar]
- 18.Harper MJ, Dowd D, Elliott AS. Implantation and embryonic development in the ovariectomized-adrenalectomized hamster. Biol Reprod. 1969;1:253–257. doi: 10.1095/biolreprod1.3.253. [DOI] [PubMed] [Google Scholar]
- 19.Orsini MW, Meyer RK. Effect of varying doses of progesterone on implantation in the ovariectomized hamster. Exp Biol Med. 1962;110:4713–4715. [Google Scholar]
- 20.Perry JS, Heap RB, Amoroso EC. Steroid hormone production by pig blastocysts. Nature. 1973;245:45–47. doi: 10.1038/245045a0. [DOI] [PubMed] [Google Scholar]
- 21.Zegers-Hochschild F, Altieri E. Luteal estrogen is not required for the establishment of pregnancy in the human. J Assist Reprod Genet. 1995;12:224–228. doi: 10.1007/BF02211803. [DOI] [PubMed] [Google Scholar]
- 22.Psychoyos A. Nidation in the rat and the necessary dose of estrogen. C R Hebd Seances Acad Sci. 1961;253:1616–1617. [PubMed] [Google Scholar]
- 23.Carson DD, Bagchi I, Dey SK, et al. Embryo implantation. Dev Biol. 2000;223:217–237. doi: 10.1006/dbio.2000.9767. [DOI] [PubMed] [Google Scholar]
- 24.Kimber SJ, Spanswick C. Blastocyst implantation: the adhesion cascade. Semin Cell Dev Biol. 2000;11:77–92. doi: 10.1006/scdb.2000.0154. [DOI] [PubMed] [Google Scholar]
- 25.Paria BC, Reese J, Das SK, Dey SK. Deciphering the cross-talk of implantation: advances and challenges. Science. 2002;296:2185–2188. doi: 10.1126/science.1071601. [DOI] [PubMed] [Google Scholar]
- 26.Salamonsen LA, Nie G, Dimitriadis E, Robb L, Findlay JK. Genes involved in implantation. Reprod Fertil Dev. 2001;13:41–49. doi: 10.1071/rd00046. [DOI] [PubMed] [Google Scholar]
- 27.Zhang S, Lin H, Kong S, et al. Physiological and molecular determinants of embryo implantation. Mol Aspects Med. 2013;34:939–980. doi: 10.1016/j.mam.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Vaerenbergh I, McIntire R, Van Lommel L, Devroey P, Giudice L, Bourgain C. Gene expression during successful implantation in a natural cycle. Fertil Steril. 2010;93:268.e15–8. doi: 10.1016/j.fertnstert.2009.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carson DD, Lagow E, Thathiah A, et al. Changes in gene expression during the early to mid-luteal (receptive phase) transition in human endometrium detected by high-density microarray screening. Mol Hum Reprod. 2002;8:871–879. doi: 10.1093/molehr/8.9.871. [DOI] [PubMed] [Google Scholar]
- 30.Chan C, Virtanen C, Winegarden NA, Colgan TJ, Brown TJ, Greenblatt EM. Discovery of biomarkers of endometrial receptivity through a minimally invasive approach: a validation study with implications for assisted reproduction. Fertil Steril. 2013;100:810–817. doi: 10.1016/j.fertnstert.2013.04.047. [DOI] [PubMed] [Google Scholar]
- 31.Hu S, Yao G, Wang Y, et al. Transcriptomic changes during the pre-receptive to receptive transition in human endometrium detected by RNA-Seq. J Clin Endocrinol Metab. 2014;99:E2744–E2753. doi: 10.1210/jc.2014-2155. [DOI] [PubMed] [Google Scholar]
- 32.Kao LC, Tulac S, Lobo S, et al. Global gene profiling in human endometrium during the window of implantation. Endocrinology. 2002;143:2119–2138. doi: 10.1210/endo.143.6.8885. [DOI] [PubMed] [Google Scholar]
- 33.Mirkin S, Arslan M, Churikov D, et al. In search of candidate genes critically expressed in the human endometrium during the window of implantation. Hum Reprod. 2005;20:2104–2117. doi: 10.1093/humrep/dei051. [DOI] [PubMed] [Google Scholar]
- 34.Riesewijk A, Martin J, van Os R, et al. Gene expression profiling of human endometrial receptivity on days LH+2 versus LH+7 by microarray technology. Mol Hum Reprod. 2003;9:253–264. doi: 10.1093/molehr/gag037. [DOI] [PubMed] [Google Scholar]
- 35.Ace CI, Okulicz WC. Microarray profiling of progesterone-regulated endometrial genes during the rhesus monkey secretory phase. Reprod Biol Endocrinol. 2004;2:54. doi: 10.1186/1477-7827-2-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghosh D, Sharkey AM, Charnock-Jones DS, Smith SK, Sengupta J. Effect of low-dose mifepristone administration on day 2 after ovulation on transcript profiles in implantation-stage endometrium of rhesus monkeys. Reproduction. 2009;138:357–370. doi: 10.1530/REP-08-0442. [DOI] [PubMed] [Google Scholar]
- 37.Sun XY, Li FX, Li J, et al. Determination of genes involved in the early process of embryonic implantation in rhesus monkey (Macaca mulatta) by suppression subtractive hybridization. Biol Reprod. 2004;70:1365–1373. doi: 10.1095/biolreprod.103.018523. [DOI] [PubMed] [Google Scholar]
- 38.Liu JL, Zhao M, Peng Y, Fu YS. Identification of gene expression changes in rabbit uterus during embryo implantation. Genomics. 2016;107:216–221. doi: 10.1016/j.ygeno.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 39.Xiao S, Diao H, Zhao F, Li R, He N, Ye X. Differential gene expression profiling of mouse uterine luminal epithelium during periimplantation. Reprod Sci. 2014;21:351–362. doi: 10.1177/1933719113497287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lei W, Herington J, Galindo CL, et al. Cross-species transcriptomic approach reveals genes in hamster implantation sites. Reproduction. 2014;148:607–621. doi: 10.1530/REP-14-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma XH, Hu SJ, Ni H, et al. Serial analysis of gene expression in mouse uterus at the implantation site. J Biol Chem. 2006;281:9351–9360. doi: 10.1074/jbc.M511512200. [DOI] [PubMed] [Google Scholar]
- 42.Reese J, Das SK, Paria BC, et al. Global gene expression analysis to identify molecular markers of uterine receptivity and embryo implantation. J Biol Chem. 2001;276:44137–44145. doi: 10.1074/jbc.M107563200. [DOI] [PubMed] [Google Scholar]
- 43.Diaz-Gimeno P, Horcajadas JA, Martinez-Conejero JA, et al. A genomic diagnostic tool for human endometrial receptivity based on the transcriptomic signature. Fertil Steril. 2011;95:50–6060.e1–15. doi: 10.1016/j.fertnstert.2010.04.063. [DOI] [PubMed] [Google Scholar]
- 44.Garrido-Gomez T, Ruiz-Alonso M, Blesa D, Diaz-Gimeno P, Vilella F, Simon C. Profiling the gene signature of endometrial receptivity: clinical results. Fertil Steril. 2013;99:1078–1085. doi: 10.1016/j.fertnstert.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 45.Ruiz-Alonso M, Blesa D, Diaz-Gimeno P, et al. The endometrial receptivity array for diagnosis and personalized embryo transfer as a treatment for patients with repeated implantation failure. Fertil Steril. 2013;100:818–824. doi: 10.1016/j.fertnstert.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 46.Horcajadas JA, Minguez P, Dopazo J, et al. Controlled ovarian stimulation induces a functional genomic delay of the endometrium with potential clinical implications. J Clin Endocrinol Metab. 2008;93:4500–4510. doi: 10.1210/jc.2008-0588. [DOI] [PubMed] [Google Scholar]
- 47.Tapia A, Gangi LM, Zegers-Hochschild F, et al. Differences in the endometrial transcript profile during the receptive period between women who were refractory to implantation and those who achieved pregnancy. Hum Reprod. 2008;23:340–351. doi: 10.1093/humrep/dem319. [DOI] [PubMed] [Google Scholar]
- 48.Tapia-Pizarro A, Figueroa P, Brito J, Marin JC, Munroe DJ, Croxatto HB. Endometrial gene expression reveals compromised progesterone signaling in women refractory to embryo implantation. Reprod Biol Endocrinol. 2014;12:92. doi: 10.1186/1477-7827-12-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tabibzadeh S. Molecular control of the implantation window. Hum Reprod Update. 1998;4:465–471. doi: 10.1093/humupd/4.5.465. [DOI] [PubMed] [Google Scholar]
- 50.Altmae S, Martinez-Conejero JA, Salumets A, Simon C, Horcajadas JA, Stavreus-Evers A. Endometrial gene expression analysis at the time of embryo implantation in women with unexplained infertility. Mol Hum Reprod. 2010;16:178–187. doi: 10.1093/molehr/gap102. [DOI] [PubMed] [Google Scholar]
- 51.Feroze-Zaidi F, Fusi L, Takano M, et al. Role and regulation of the serum- and glucocorticoid-regulated kinase 1 in fertile and infertile human endometrium. Endocrinology. 2007;148:5020–5029. doi: 10.1210/en.2007-0659. [DOI] [PubMed] [Google Scholar]
- 52.Lorenzo P, Aspberg A, Onnerfjord P, Bayliss MT, Neame PJ, Heinegard D. Identification and characterization of asporin. A novel member of the leucine-rich repeat protein family closely related to decorin and biglycan. J Biol Chem. 2001;276:12201–12211. doi: 10.1074/jbc.M010932200. [DOI] [PubMed] [Google Scholar]
- 53.Yoshinaga K, Adams CE. Delayed implantation in the spayed, progesterone treated adult mouse. J Reprod Fertil. 1966;12:593–595. doi: 10.1530/jrf.0.0120593. [DOI] [PubMed] [Google Scholar]
- 54.Chen Y, Ni H, Ma XH, et al. Global analysis of differential luminal epithelial gene expression at mouse implantation sites. J Mol Endocrinol. 2006;37:147–161. doi: 10.1677/jme.1.02009. [DOI] [PubMed] [Google Scholar]
- 55.Lee YJ, Kim IS, Park SA, et al. Periostin-binding DNA aptamer inhibits breast cancer growth and metastasis. Mol Ther. 2013;21:1004–1013. doi: 10.1038/mt.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]