Abstract
Purpose
EGFR mutation is a predictor of epidermal growth factor receptor–tyrosine kinase inhibitor treatment response in patients with non–small-cell lung cancer (NSCLC). However, it remains unclear whether chemotherapy affects EGFR mutation status in NSCLC. We investigated the influence of chemotherapy on EGFR mutations in plasma and tumor tissues from patients with NSCLC.
Patients and Methods
Samples were derived from three cohorts: one, 264 patients with advanced NSCLC who received first-line chemotherapy with matched pre- and postchemotherapy blood samples; two, 63 patients with stages IIb to IIIb disease with pre– and post–neoadjuvant chemotherapy tumor tissues; and three, 79 patients with advanced NSCLC who underwent palliative surgery. EGFR mutation status was determined and analyzed to reveal potential impact of chemotherapy.
Results
In the first cohort, EGFR mutations were detected in 34.5% of the prechemotherapy plasma samples (91 of 264) but in only 23.1% of the postchemotherapy plasma samples (61 of 264). The decrease in EGFR mutation rate was statistically significant (P < .001). Patients whose EGFR mutations switched from positive to negative after chemotherapy had a better partial response (PR) than patients with a reverse change (P = .037). A similar decrease in EGFR mutation rate was observed in tissues after neoadjuvant chemotherapy in the second cohort (34.9% [22 of 63] v 19.0% [12 of 63]; P = .013). In the third cohort, 38.0% of the tumors (30 of 79) showed an intratumor heterogeneity of EGFR mutation, whereas 62.0% (49 of 79) were homogeneous, either with EGFR mutation or no mutation.
Conclusion
Our results suggest that chemotherapy may reduce EGFR mutation frequency in patients with NSCLC, likely the result of a preferential response of subclones with EGFR mutations in tumors with heterogeneous tumor cell populations.
INTRODUCTION
Oral tyrosine kinase inhibitors (TKIs) targeting epidermal growth factor receptor (EGFR) have become an indispensable and important modality for treating advanced non–small-cell lung cancer (NSCLC). Because only a limited number of patients will likely benefit from these agents,1,2 the identification of such patients is urgently needed.
Somatic mutations in the EGFR tyrosine kinase domain have been linked to EGFR-TKI treatment response in patients with advanced NSCLC.3–11 Recent phase III clinical studies of advanced NCSLC have demonstrated that EGFR mutations are the most effective predictor of clinical outcome in response to first-line TKIs.12–16 These mutations have become important biomarkers in determining optimal first-line therapy (chemotherapy or TKI therapy), and the use of these biomarkers is accepted as a paradigm of genotype-based individualized target therapy for patients with NSCLC.
However, the significant predictive value of EGFR mutations observed in first-line TKI treatment has not been maintained in second-line TKI treatment.2,17 Biomarker analysis indicated that EGFR mutations were not associated with the outcomes of TKI treatment in the BR.21 trial2 or in the ISEL (IRESSA Survival Evaluation in Lung Cancer) study, which compared erlotinib or gefitinib with placebo in patients for whom platinum-based chemotherapy had failed.17 Moreover, the rate of tumor response to second-line TKI therapy was lower than that for first-line therapy in patients with EGFR mutations. The reason for the inconsistency in the predictive value of EGFR mutations between first- and second-line treatments is unknown. We postulated that first-line chemotherapy may influence the status of EGFR mutations, and thus, assessment of EGFR mutations using specimens collected at the initial diagnosis might be inadequate for predicting response to EGFR-TKI treatment after chemotherapy.
However, it is difficult to obtain tumor biopsies from patients for whom chemotherapy has failed. Plasma DNA may provide a noninvasive and repeatable source of genotypic information. We and others have previously shown that plasma DNA is a reliable source for EGFR mutation analysis in patients with advanced NSCLC.18–20 The aims of the current study were threefold: first, to compare EGFR mutation status before and after first-line chemotherapy in plasma DNA from patients with advanced NSCLC; second, to identify neoadjuvant chemotherapy–related variation in EGFR mutation in tissue samples from patients with stages IIb to IIIb NSCLC; and third, to explore the potential mechanism of EGFR mutation variation by analyzing the heterogeneity of intratumoral EGFR mutations in tissue samples obtained during palliative surgical resection.
PATIENTS AND METHODS
Study Cohorts
Three cohorts of patients with NSCLC were enrolled onto this study. All patients were treated at the Peking University Cancer Hospital (Beijing, China) between April 1, 2006, and December 31, 2009.
The first cohort consisted of 264 consecutive patients with histologically confirmed stages IIIb to IV NSCLC who had received two cycles of platinum-based first-line chemotherapy (cisplatin and carboplatin plus gemcitabine, vinorelbine and taxanes). Pre- and postchemotherapy peripheral blood were collected from each patient.
The second cohort included patients with locally advanced NSCLC (n = 63) who had received two to four cycles of neoadjuvant chemotherapy (gemcitabine plus cisplatin, vinorelbine plus cisplatin) to confirm chemotherapy-related EGFR mutation status changes observed in the first cohort. Matched biopsy and surgical resection samples were collected before and after neoadjuvant treatment.
The third cohort consisted of 79 patients with stages IIIa to IV NSCLC who had received palliative surgical resection without prior treatment. Tumor cells were microdissected at multiple small regions of the formalin-fixed paraffin-embedded specimens individually and subjected to EGFR mutation analysis.
Patients' clinical information was derived from the clinical database established in 1999. Smoking status was defined as those who had smoked more than 100 lifetime cigarettes and was based on records at patients' first clinic visit. Histologic subtypes were based on WHO criteria.21 Staging was based on the 2009 International Union Against Cancer–American Joint Committee on Cancer–TNM system (version 7).22
The study was reviewed and approved by the Institutional Ethic Committee at Peking University Cancer Hospital. All patients provided written informed consent before samples were collected.
Sample Collection and Processing
Blood was collected in anticoagulated tubes from patients in the first cohort before and after two cycles of first-line chemotherapy. Plasma DNA was extracted according to a method reported previously.18
For the second cohort, tumor tissues were macrodissected to avoid influence of necrotic tissues resulting from neoadjuvant chemotherapy. For the third cohort, one section 15 μm in thickness for each patient case was stained with hematoxylin and eosin and microdissected using the AS Laser Microdissection system (Leica Microsystems, Wetzlar, Germany). Thirty to 40 tumor foci containing approximately 100 cancer cells in each were obtained. E.Z.N.A formalin-fixed paraffin-embedded DNA kits (Omega Bio-Tek, Norcross, GA) were used to extract DNA from the tissues. The quality and concentration of extracted DNA were determined using NanoDrop 2000 (Thermo Fisher Scientific, Wilmington, DE). The extracted DNA was then used for qualitative or semiquantitative EGFR mutation analysis by denaturing high-performance liquid chromatography (DHPLC).
Mutation Analysis
We analyzed all matched samples in the same condition to equalize the detection conditions. The EGFR exon 19 deletion or exon 21 substituted mutations were detected according to the method reported by us previously.18 The amplification refractory mutation system (ARMS), a more sensitive method, was used to re-evaluate the patient cases with EGFR mutation discrepancies before and after chemotherapy or neoadjuvant chemotherapy.
A semiquantitative analysis of mutation abundance was performed by calculating a ratio (M/W) between the peak heights for mutant and wild-type product. M represents mutant peak height, and W represents normal peak height. The analysis was only used in exon 19 mutation examination, not in exon 21 analysis, because mutant peak and wild peak were separate in exon 19 but overlapped in exon 21.
Statistical Analysis
Frequency tabulation and summary statistics were provided to characterize the data distribution. The McNemar test was applied to compare the change of mutation status before and after treatment. The Cochran-Armitage trend test was used to test whether change in mutation status was associated with clinical outcome in terms of partial response (PR), stable disease (SD), or disease progression (PD). The associations of unpaired categorical variables were analyzed using the χ2 test; however, Fisher's exact test was used for small sample sizes (expected value < 5 in any cell of the contingency table). The Wilcoxon rank sum test was applied to compare the mutant abundance between the different mutation groups. Statistical significance was set at a level of .05. Two-sided tests were performed in all settings except for the trend test, in which a one-sided test was applied. All calculations were performed using SAS version 10.0 (SAS Institute, Cary, NC).
RESULTS
Patient Characteristics
Patients' sociodemographic and clinical characteristics are listed in Table 1.
Table 1.
Sociodemographic and Clinical Characteristics by Study Cohort
| Characteristic | Cohort One: First-Line Chemotherapy (n = 264) |
Cohort Two: Neoadjuvant Chemotherapy (n = 63) |
Cohort Three: Palliative Surgery (n = 79) |
|||
|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | |
| Age, years | ||||||
| ≤ 60 | 160 | 60.6 | 42 | 66.7 | 38 | 48.1 |
| > 60 | 104 | 39.4 | 21 | 33.3 | 41 | 51.9 |
| Sex | ||||||
| Male | 144 | 54.5 | 47 | 74.6 | 44 | 55.7 |
| Female | 120 | 45.5 | 16 | 25.4 | 35 | 44.3 |
| Histologic subtype | ||||||
| Adenocarcinoma | 205 | 77.7 | 25 | 39.7 | 58 | 73.4 |
| Nonadenocarcinoma | 59 | 22.3 | 38 | 60.3 | 21 | 26.6 |
| Smoking status | ||||||
| Former smoker | 136 | 51.5 | 33 | 52.4 | 36 | 45.6 |
| Never smoker | 128 | 48.5 | 30 | 47.6 | 43 | 54.4 |
| Stage | ||||||
| IIb to IIIa | 0 | 0.0 | 58 | 92.1 | 25 | 31.6 |
| IIIb to IV | 264 | 100.0 | 5 | 7.9 | 54 | 68.4 |
EGFR Mutation Shift in Plasma DNA Before and After First-Line Chemotherapy
Change in EGFR mutation status in plasma DNA was assessed for the 264 patients who had undergone first-line chemotherapy (cohort one). EGFR mutations were detected by DHPLC and confirmed by ARMS for 91 patients (34.5%) before first-line chemotherapy and for 61 (23.1%) after chemotherapy. There was only one patient case in which different mutation status was determined by the two methods (the patient's EGFR 21 exon mutation switched from positive to negative after chemotherapy based on the DHPLC result, but no switch was observed based on ARMS). The decrease in EGFR mutation rate after chemotherapy was statistically significant (P < .001 [McNemar test]; Table 2). No shift in subtype of mutation was observed after chemotherapy.
Table 2.
Effect of First-Line Chemotherapy on EGFR Mutation Status Before and After Treatment in Plasma Samples From Patients With Stages IIIb to IV NSCLC (n = 264)
| Prechemotherapy | Postchemotherapy |
Total |
||||
|---|---|---|---|---|---|---|
| Wild Type |
Mutated |
|||||
| No. | % | No. | % | No. | % | |
| Wild type | 149 | 56.4 | 24 | 9.1 | 173 | 65.5 |
| Mutated | 54 | 20.5 | 37 | 14.0 | 91 | 34.5 |
| Total | 203 | 76.9 | 61 | 23.1 | 264 | 100.0 |
NOTE. P < .001 (McNemar test).
Abbreviation: NSCLC, non–small-cell lung cancer.
Influence of Tumor Response to First-Line Chemotherapy on EGFR Mutation Shift
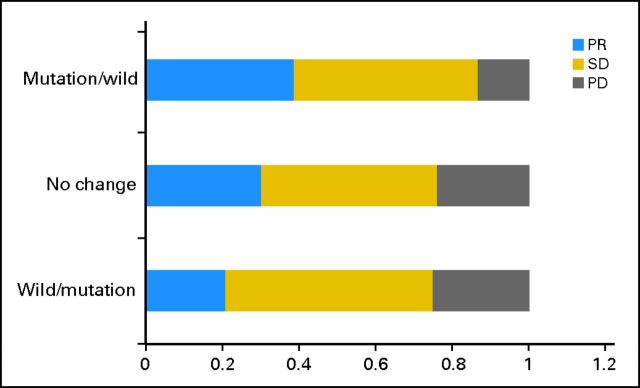
In cohort one, the objective response rate to first-line chemotherapy was 31.1% (82 of 264), without significant statistical association with the pretreatment EGFR mutation status (prechemotherapy: mutant, 31.9% v wild type, 30.6%; P = .84). Among the 264 patients, 54 (20.5%) had mutation before treatment, but this changed to wild type after chemotherapy; 186 (70.4%) remained at mutation or wild type before and after treatment; and 24 (9.1%) had wild type, but this changed to mutation after treatment (Table 2). The PR rates in these three groups were 38.9%, 30.1%, and 20.8%, respectively. Conversely, the PD rates were 13.0%, 23.7%, and 25.0%, respectively. Among patients with discordant EGFR mutation status before and after chemotherapy, decrease in mutation was significantly associated with better clinical response (P = .037 [Cochran-Armitage trend test]; Fig 1; Table 3).
Fig 1.
Response outcome between different subgroups of mutation change in cohort one. The x-axis indicates percentage of different response group; the y-axis indicates three groups according to EGFR mutation variation. PD, progressive disease; PR, partial response; SD, stable disease.
Table 3.
Effect of First-Line Chemotherapy on Change in EGFR Mutation in Plasma Samples Before and After Treatment and Association With Tumor Response (n = 264)
| EGFR Mutation Change From Pre- to Postchemotherapy | Tumor Response |
Total (n = 264) |
||||||
|---|---|---|---|---|---|---|---|---|
| PR |
SD |
PD |
||||||
| No. | % | No. | % | No. | % | No. | % | |
| Mutation → wild type* | 21 | 38.9 | 26 | 48.1 | 7 | 13.0 | 54 | 100.0 |
| No change | 56 | 30.1 | 86 | 46.2 | 44 | 23.7 | 186 | 100.0 |
| Wild → mutation* | 5 | 20.8 | 13 | 54.2 | 6 | 25.0 | 24 | 100.0 |
Abbreviations: PD, progressive disease; PR, partial response; SD, stable disease.
P = .037 (Cochran-Armitage trend test).
EGFR Mutation Shift in Tumor Tissues Before and After Neoadjuvant Chemotherapy
We also assessed change in EGFR mutation status in cohort two (n = 63). Twenty-two patients (34.9%) had EGFR mutations before neoadjuvant chemotherapy; this dropped to 12 (19.0%) after treatment (Table 4). A total of 49 patients (77.8%) retained their initial EGFR status (39 wild type, 10 EGFR mutant); in two patient cases (3.2%), wild type switched to mutation; and in 12 patient cases (19.0%), mutation switched to wild type. The discordant rate of EGFR mutation between pre– and post–neoadjuvant chemotherapy was 22.2%. The decrease in EGFR mutation rate after chemotherapy was statistically significant (P = .013 [McNemar test]; Table 4).
Table 4.
Effect of Neoadjuvant Chemotherapy on EGFR Mutation Status Before and After Treatment in Tissue Samples From Patients With Stages IIb to IIIb NSCLC (n = 63)
| Prechemotherapy | Postchemotherapy |
Total |
||||
|---|---|---|---|---|---|---|
| Wild Type |
Mutated |
|||||
| No. | % | No. | % | No. | % | |
| Wild type | 39 | 61.9 | 2 | 3.2 | 41 | 65.1 |
| Mutated | 12 | 19.0 | 10 | 15.9 | 22 | 34.9 |
| Total | 51 | 81.0 | 12 | 19.0 | 63 | 100.0 |
NOTE. P = .013 (McNemar test).
Abbreviation: NSCLC, non–small-cell lung cancer.
Evaluation of tumor response to neoadjuvant chemotherapy revealed that 38 patients had achieved PR, and 24 had achieved SD. One patient case was not evaluable because of no response data. There was no patient with PD in this cohort. In 14 patients, EGFR mutation status changed before and after treatment, with 12 patients changing from mutation to wild type and two from wild type to mutation. The PR rates for these two groups were 63.6% and 50%, respectively. The difference was not statistically significant (P = .64 [Cochran-Armitage trend test]). Sociodemographic and clinical factors, including chemotherapy regimen, number of cycles, age, sex, smoking status, and histologic subtype, were not associated with EGFR shift (P > .05).
Heterogeneity Analysis of EGFR Mutation in Intratumor Tissue Samples
We performed genetic heterogeneity analysis for samples from 79 patients with stages IIIa to IV NSCLC who had received palliative surgery. Forty-two patient cases were EGFR mutation positive, including 23 with exon 19 mutation (group E19) and 19 with exon 21 mutation (group E21). Of these 42 patient cases with EGFR mutation, a total of 1,331 tumor foci were fractionated, with a median mutation rate of 74.75%. Sixteen of 42 samples consisted of only EGFR-mutated cells, whereas the other 26 samples consisted of cells with both wild-type and mutated EGFR, with the proportion of EGFR-mutant cells ranging from 30% to 90%. Among the 37 patient cases (group W) previously determined in routine detection to be EGFR wild type, 1,175 tumor foci were fractionated for analysis of EGFR mutation, and four (10.8%) showed low mutant frequency (three with EGFR exon 19 mutation, one with EGFR exon 21 mutation) ranging from 7.69% to 20.83%. After combining the 26 mutated patient cases with both wild-type and mutant EGFR cells, and the for nonmutated patient cases with low abundant mutated cells, 38% of the samples (30 of 79) had intratumoral heterogeneity regarding EGFR mutation. The remaining 62% of the samples (49 of 79) contained only EGFR mutation–positive cells or only wild-type cells. In semiquantitative analysis, the mutant abundance values for the three patient cases in group W with low mutation rates were 0.06, 0.08, and 0.22, respectively. On the other hand, the mutant abundance values for the 23 patient cases with exon 19 mutation ranged from 0.4 to 9.5. Not surprisingly, the difference in mutant abundance values was statistically significant (P = .011 [Wilcoxon rank sum test]).
DISCUSSION
Heterogeneity in EGFR gene expression, mutation, or amplification between primary and metastatic tumors has been observed in several small-sample studies, with discordance seen in 25% to 35% of patients.23–25 However, to our knowledge, none of these studies evaluated the influence of chemotherapy on EGFR mutation. Our study provides evidence that first-line chemotherapy may influence EGFR mutation status in both tissue and peripheral blood samples. The decrease in mutation rate was significantly associated with better clinical response (PR v PD).
Recently, several studies have focused on exploring the potential possibility that EGFR mutation detection in circulating tumor DNA from plasma or serum samples had been used as a surrogate of primary tumor tissue samples.18,26–28 The concordance of EGFR mutation status between peripheral blood samples and matched tumor tissue had been reported as varying from 59.1% to 92%, with minimal false-positive rates and variable false-negative rates.18,26–28 In our previous study, 230 plasma samples and matched tumor tissue samples were used for EGFR mutation detection, and concordance was 78%. Recently, we extended the study to include an additional 822 matched plasma and tissue samples, in which 744 patients had advanced NSCLC.29 The mutation concordances of the two kinds of samples were 70% and 78% for total population and patients with advanced disease, respectively, similar to results in our previous report.18 Although the DHPLC method has not been widely used for EGFR mutation analysis, its high sensitivity and specificity have been shown in our and other studies (detection limit approximately 3%-5%).18,30 To minimize the possibility of false results by the DHPLC method, we verified EGFR mutation status using the ARMS method, which is considered more sensitive in mutation analysis; the results were identical, except for one mutation sample deemed negative after chemotherapy by DHPLC but not ARMS. Overall, the DHPLC method for EGFR mutation analysis in plasma is feasible and reproducible.
We showed that the overall incidence of EGFR mutation was lower in plasma DNA after first-line chemotherapy and in tumor tissues after neoadjuvant chemotherapy (Tables 2 and 4). This chemotherapy-related change may partially explain why chemotherapy-resistant tumors are less sensitive to EGFR-TKI treatment than chemotherapy-naive tumors.31 It may also explain why almost all clinical trials involving second-line TKI therapy have failed to show a positive correlation between EGFR mutation and progression-free or overall survival.2,17 EGFR mutation detection at the time of initial diagnosis has failed to reflect exactly mutation status at the initiation of EGFR-TKIs as second- or third-line treatment. Therefore, future prospective trials should consider analyzing biopsies taken immediately before second- or third-line EGFR-TKI therapy. Our study provides an alternative blood-based strategy to assess EGFR mutation status.
Whether EGFR-TKI treatment should precede or be followed by chemotherapy remains an unresolved clinical issue. In a recent multicenter randomized phase III study comparing the order of erlotinib and cisplatin plus gemcitabine in unselective advanced NSCLC, chemotherapy followed by erlotinib resulted in better progression-free and overall survival compared with erlotinib followed by chemotherapy.32 Unfortunately, this study did not address the question of the optimal order of EGFR-TKIs and chemotherapy for patients harboring EGFR mutations. Chin et al33 used an erlotinib-sensitive EGFR-mutant NSCLC cell line to explore whether its prior exposure to platinum agents would affect subsequent response to erlotinib. Their results suggest that first-line chemotherapy in EGFR-mutant NSCLC may reduce the benefit of subsequent EGFR-TKI treatment. Our results are consistent with the observation of Chin et al; decreased EGFR mutation frequency after chemotherapy may reduce the overall clinical benefit of subsequent EGFR-TKI treatment. If these results are confirmed in future prospective and multicenter studies, the therapeutic strategy of using EGFR-TKIs followed by chemotherapy should be considered as an optimal option for patients who harbor EGFR mutations.
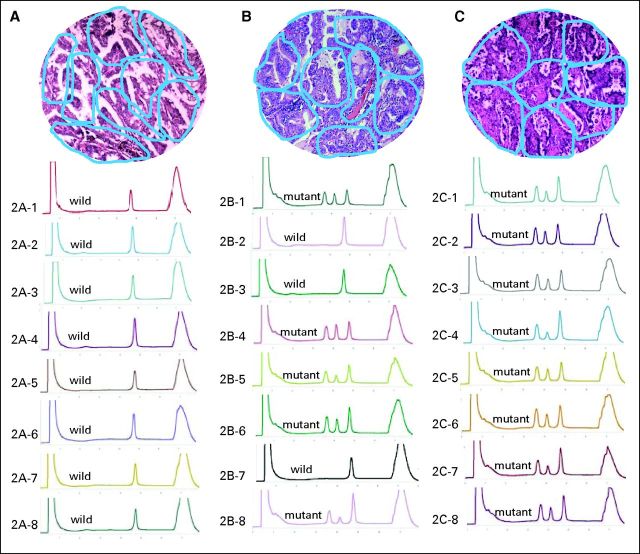
The shift in tumors from EGFR mutation status to wild-type status observed in this study suggests that both mutant and nonmutant cancer cells coexist in the same tumor. To identify intratumor heterogeneity, we microdissected and analyzed EGFR mutation status in more than 2,506 tumor foci of 79 tumors from patients with NSCLC who underwent palliative surgery (Fig 2). Approximately 38% of tumors contained both EGFR-mutant and wild-type foci, similar to the rate reported by Taniguchi et al.34 Recently, the study by Gerlinger et al35 revealed high intratumoral heterogeneity of multiple tumor-suppressor genes converging on loss of function. Conversely, Yatabe et al36 reported a rare occurrence of the heterogeneous distribution of EGFR mutation. However, it should be noted that only 450 tumor micropoints were analyzed in the Yatabe et al study.
Fig 2.
Multiple foci microdissection and analysis of intratumor genetic heterogeneity on EGFR mutation. Blue circles in hematoxylin and eosin staining tissue (10 × 10) represent microdissected foci; DHPLC graph represents corresponding tumor foci mutation status. (A) Tissue with homogeneity of wild-type EGFR; (B) tissue with heterogeneity of EGFR mutation; (C) tissue with homogeneity of mutant EGFR.
It is interesting to note that a majority of EGFR mutation changes after chemotherapy were from mutant state to wild type, suggesting that cancer cells harboring EGFR mutations might be more sensitive to chemotherapy than those without mutation. We further analyzed the relationships between chemotherapy responses and the shift of EGFR mutation status and found patients who achieved PR were more likely to have had EGFR mutation shift than those achieving SD or PD after chemotherapy. This is consistent with the results from the IPASS trial, where the objective response rate was 47.3% in EGFR-mutant patients treated with chemotherapy, significantly higher than the rate in EGFR mutation–negative patients (23.5%).12 Together with previous studies, this study suggests that EGFR mutation shift may be related to the heterogeneity of intratumoral EGFR mutation and to different chemosensitivity levels of mutant and wild-type cells. Interestingly, in our study, four patients (10.8%) had low frequency and abundance of EGFR mutations, which were detected only through microsamples, thus explaining in part why some patients' EGFR mutation status shifted from negative to positive after chemotherapy. To further confirm these results, a study in vivo exploring molecular mechanisms resulting in chemotherapy-related shift of EGFR mutation status is ongoing in our group.
It has been shown that chemotherapy may alter DNA concentration in serum or plasma.37,38 To exclude the influence of DNA concentration change after chemotherapy on the shift of EGFR mutation status, the relationship between them was analyzed. The results revealed that there was no association between change in plasma DNA levels and shift in EGFR gene mutation after chemotherapy, suggesting that the influence of chemotherapy on mutation of the EGFR gene is independent of the change in the level of DNA in the plasma (Appendix; online only).
One of the limitations of the current study is the lack of blood and tissue samples from the same patients. The first cohort provided plasma only, whereas the other two cohorts provided tissue samples only. Nevertheless, we used the plasma samples to show that frequency of EGFR mutation was significantly decreased after two cycles of chemotherapy; we used paired tumor tissues to show that mutation frequency was reduced in the tumors as well; and we used large advanced tumor tissues to show the heterogeneity of EGFR mutations within some of the tumors. Results from the three interconnecting cohorts provided a solid foundation to support our conclusions.
In conclusion, chemotherapy may affect EGFR mutation status in patients with NSCLC, lowering the number of mutations. This observation may be attributable to the heterogeneity of intratumoral EGFR mutations and the different sensitivities of EGFR-mutated and wild-type tumor cells to chemotherapy. These findings should be considered in future studies designed to elucidate the predictive role of EGFR mutation in second-line TKI therapy for patients with NSCLC.
Acknowledgment
We thank Waun Ki Hong, MD, of the University of Texas MD Anderson Cancer Center for his critical comments and suggestions for this article; Xin Shelley Wang, MD, MPH, of The University of Texas MD Anderson Cancer Center for her critical review of this article; and Ning Wang, MD, in the radiologic department of Beijing Cancer Hospital for his assessment of treatment response.
Appendix
Materials and Methods
DNA quantification in plasma.
The DNA level was determined by TaqMan real-time quantitative polymerase chain reaction (PCR) with TaqMan Universal PCR master mix and an ABI StepOne System (Applied Biosystems, Foster City, CA). Each plate consisted of patient samples in triplicate. For construction of the calibration curve on each plate, we used standard Control Human Genomic DNA (Promega, Madison, WI) at 10 ng/μL with appropriate serial dilutions at 10, 5, 2.5, 1.25, and 0.625 ng/μL. For the follow-up study, all consecutive plasma samples for each patient were simultaneously analyzed in the same real-time PCR experiment to allow comparative quantification of samples during the observation time.
Statistical analysis.
Differences in DNA concentration between pre- and post-treatment plasma samples were analyzed using the Wilcoxon matched-pairs signed rank test. Association between DNA concentration and EGFR mutation status in different groups according to chemotherapy response were determined using the nonparameter Kruskal-Wallis test.
Results
Association of circulating cell–free DNA in plasma with EGFR mutation shift during chemotherapy.
We measured circulating cell–free DNA levels in pre- and postchemotherapy plasma from 264 patients with advanced non–small-cell lung cancer. The median DNA level dropped from 163.52 to150.57 ng/mL (P < .001). Median plasma DNA concentration at baseline was significantly higher in patients with stage IV disease than in those with stage IIIb disease (191.69 v 76.83 ng/mL; P = .001). Age, sex, smoking status, and histologic type did not correlate with baseline plasma DNA levels (P > .05).
We used nonparametric Kruskal-Wallis analysis to assess whether changes in EGFR mutation could attribute to the change in the amount of cell-free DNA in plasma after chemotherapy. Chemotherapy-associated shift in EGFR mutation status was not associated with change in the amount of plasma cell–free DNA in patients with partial response (χ2 = 3.278; P = .149), stable disease (χ2 = 2.687; P = .2610), or progressive disease (χ2 = 2.555; P = .279).
Footnotes
Supported by Grants No. 81025012 from the National Natural Sciences Foundation Distinguished Young Scholars Program, No. 81172235 from the National Natural Sciences Foundation General Program, No. 2007-1023 from the Capital Development Foundation, No. 2011-2-22 from the Beijing Health Systems Academic Leader Program, and No. Z090507017709015 from the Science and Technology Project of Beijing.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author(s) indicated no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
Conception and design: Hua Bai, Zhijie Wang, Jie Wang
Financial support: Jie Wang
Administrative support: Jie Wang
Provision of study materials or patients: Zhijie Wang, Keneng Chen, Jun Zhao, Qinghua Zhou
Collection and assembly of data: Hua Bai, Zhijie Wang, Keneng Chen, Jun Zhao, Shuhang Wang, Qinghua Zhou, Minglei Zhuo, Tongtong An, Jianchun Duan, Lu Yang, Meina Wu, Zhen Liang, Xiaozheng Kang
Data analysis and interpretation: Hua Bai, Zhijie Wang, J. Jack Lee, Li Mao, Jie Wang
Manuscript writing: All authors
Final approval of manuscript: All authors
REFERENCES
- 1.Ludovini V, Bianconi F, Pistola L, et al. Optimization of patient selection for EGFR-TKIs in advanced non–small-cell lung cancer by combined analysis of MET, EGFR, KRAS, and PIK3CA mutations. J Clinl Oncol. 2010;28:75–62. doi: 10.1007/s00280-012-1829-7. [DOI] [PubMed] [Google Scholar]
- 2.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 3.Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 4.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 5.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sequist LV, Bell DW, Lynch TJ, et al. Molecular predictors of response to epidermal growth factor receptor antagonists in non–small-cell lung cancer. J Clin Oncol. 2007;25:587–595. doi: 10.1200/JCO.2006.07.3585. [DOI] [PubMed] [Google Scholar]
- 7.Uramoto H, Mitsudomi T. Which biomarker predicts benefit from EGFR-TKI treatment for patients with lung cancer? Br J Cancer. 2007;96:857–863. doi: 10.1038/sj.bjc.6603665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sequist LV, Martins RG, Spigel D, et al. First-line gefitinib in patients with advanced non–small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol. 2008;26:2442–2449. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- 9.Yoshida K, Yatabe Y, Park JY, et al. Prospective validation of prediction of gefitinib sensitivity by epidermal growth factor receptor gene mutation in patients with non-small cell lung cancer. J Thorac Oncol. 2007;2:22–28. [PubMed] [Google Scholar]
- 10.Sunaga N, Tomizawa Y, Yanagitani N, et al. Phase II prospective study of the efficacy of gefitinib for the treatment of stage III/IV non-small cell lung cancer with EGFR mutations, irrespective of previous chemotherapy. Lung Cancer. 2007;56:383–389. doi: 10.1016/j.lungcan.2007.01.025. [DOI] [PubMed] [Google Scholar]
- 11.van Zandwijk N, Mathy A, Boerrigter L, et al. EGFR and KRAS mutations as criteria for treatment with tyrosine kinase inhibitors: Retro- and prospective observations in non-small-cell lung cancer. Ann Oncol. 2007;18:99–103. doi: 10.1093/annonc/mdl323. [DOI] [PubMed] [Google Scholar]
- 12.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 13.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 14.Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): An open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 15.Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): A multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 16.Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): A multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 17.Thatcher N, Chang A, Parikh P, et al. Gefitinib plus best supportive care in previously treated patients with refractory advanced non-small-cell lung cancer: Results from a randomised, placebo-controlled, multicentre study (Iressa Survival Evaluation in Lung Cancer) Lancet. 2005;366:1527–1537. doi: 10.1016/S0140-6736(05)67625-8. [DOI] [PubMed] [Google Scholar]
- 18.Bai H, Mao L, Wang HS, et al. Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in Chinese patients with stages IIIB to IV non–small-cell lung cancer. J Clin Oncol. 2009;27:2653–2659. doi: 10.1200/JCO.2008.17.3930. [DOI] [PubMed] [Google Scholar]
- 19.Moran T, Paz-Ares L, Isla D, et al. High correspondence between EGFR mutations in tissue and in circulating DNA from non-small-cell lung cancer (NSCLC) patients (p) with poor performance status (PS) J Clin Oncol. 2007;25(suppl):540s. abstr 7505. [Google Scholar]
- 20.Chen Z, Feng J, Buzin CH, et al. Analysis of cancer mutation signatures in blood by a novel ultra-sensitive assay: Monitoring of therapy or recurrence in non-metastatic breast cancer. Pub Library Sci. 2009;4:1–14. doi: 10.1371/journal.pone.0007220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brambilla E, Travis WD, Colby TV, et al. The new World Health Organization classification of lung tumours. Eur Respir J. 2001;18:1059–1068. doi: 10.1183/09031936.01.00275301. [DOI] [PubMed] [Google Scholar]
- 22.Detterbeck FC, Boffa DJ, Tanoue LT. The new lung cancer staging system. Chest. 2009;136:260–271. doi: 10.1378/chest.08-0978. [DOI] [PubMed] [Google Scholar]
- 23.Gow CH, Chang YL, Hsu YC, et al. Comparison of epidermal growth factor receptor mutations between primary and corresponding metastatic tumors in tyrosine kinase inhibitor-naïve non-small-cell lung cancer. Ann Oncol. 2009;20:696–702. doi: 10.1093/annonc/mdn679. [DOI] [PubMed] [Google Scholar]
- 24.Bozzetti C, Tiseo M, Lagrasta C. Comparison between epidermal growth factor receptor (EGFR) gene expression in primary non-small cell lung cancer (NSCLC) and in fine-needle aspirates from distant metastatic sites. J Thorac Oncol. 2008;3:18–22. doi: 10.1097/JTO.0b013e31815e8ba2. [DOI] [PubMed] [Google Scholar]
- 25.Italiano A, Vandenbos FB, Otto J. Comparison of the epidermal growth factor receptor gene and protein in primary non-small-cell-lung cancer and metastatic sites: Implications for treatment with EGFR-inhibitors. Ann Oncol. 2006;17:981–985. doi: 10.1093/annonc/mdl038. [DOI] [PubMed] [Google Scholar]
- 26.Kimura H, Suminoe M, Kasahara K, et al. Evaluation of epidermal growth factor receptor mutations in serum DNA as a predictor of response to gefitinib (IRESSA) Br J Cancer. 2007;97:778–784. doi: 10.1038/sj.bjc.6603949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yung TK, Chan KC, Mok TS, et al. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res. 2009;15:2076–2084. doi: 10.1158/1078-0432.CCR-08-2622. [DOI] [PubMed] [Google Scholar]
- 28.Goto K, Ichinose Y, Ohe Y, et al. Epidemal growth factor receptor mutation in circulating free DNA inserum: From IPASS, a phase III study of gefitinib or carboplatin/paclitaxel in non-small cell lung cancer. J Thorac Oncol. 2012;7:115–121. doi: 10.1097/JTO.0b013e3182307f98. [DOI] [PubMed] [Google Scholar]
- 29.Huang Z, Wang Z, Bai H, et al. The detection of EGFR mutation status in plasma is reproducible and can predict the efficacy of EGFR-TKIs dynamically. Thorac Cancer. doi: 10.1111/j.1759-7714.2012.00133.x. doi: 10.1111/j.1759-7714.2012.00133.x. [DOI] [PubMed] [Google Scholar]
- 30.Cohen V, Agulnik JS, Jarry J, et al. Evaluation of denaturing high-performance liquid chromatography as a rapid detection method for identification of epidermal growth factor receptor mutations in non-small-cell lung cancer. Cancer. 2006;107:2858–2865. doi: 10.1002/cncr.22331. [DOI] [PubMed] [Google Scholar]
- 31.Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor (EGFR) mutations in lung cancer. N Engl J Med. 2009;361:958–966. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 32.Gridelli C, Butts C, Ciardiello F, et al. An international, multicenter, randomized phase III study of first-line erlotinib followed by second-line cisplatin/gemcitabine versus first-line cisplatin/gemcitabine followed by second-line erlotinib in advanced non-small-cell lung cancer: Treatment rationale and protocol dynamics of the TORCH trial. Clin Lung Cancer. 2012;9:235–238. doi: 10.3816/CLC.2008.n.037. [DOI] [PubMed] [Google Scholar]
- 33.Chin TM, Quinlan MP, Singh A, et al. Reduced erlotinib sensitivity of epidermal growth factor receptor-mutant non-small cell lung cancer following cisplatin exposure: A cell culture model of second-line erlotinib treatment. Clin Cancer Res. 2008;14:6867–6876. doi: 10.1158/1078-0432.CCR-08-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taniguchi K, Okami J, Kodama K, et al. Intratumor heterogeneity of epidermal growth factor receptor mutations in lung cancer and its correlation to the response to gefitinib. Cancer Sci. 2008;99:929–935. doi: 10.1111/j.1349-7006.2008.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yatabe Y, Matsuo K, Mitsudomi T. Heterogeneous distribution of EGFR mutations is extremely rare in lung adenocarcinoma. J Clin Oncol. 2011;29:2972–2977. doi: 10.1200/JCO.2010.33.3906. [DOI] [PubMed] [Google Scholar]
- 37.Leon SA, Shapiro B, Sklaroff DM, et al. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646–650. [PubMed] [Google Scholar]
- 38.Gautschi O, Bigosch C, Huegli B, et al. Circulating deoxyribonucleic acid as prognostic marker in non–small-cell lung cancer patients undergoing chemotherapy. J Clin Oncol. 2004;22:4157–4164. doi: 10.1200/JCO.2004.11.123. [DOI] [PubMed] [Google Scholar]