Abstract
Mitochondrial DNA (mtDNA) has many similarities with bacterial DNA because of their shared common ancestry. Increasing evidence demonstrates mtDNA to be a potent danger signal that is recognised by the innate immune system and can directly modulate the inflammatory response. In humans, elevated circulating mtDNA is found in conditions with significant tissue injury such as trauma and sepsis and increasingly in chronic organ-specific and systemic illnesses such as steatohepatitis and systemic lupus erythematosus. In this review, we examine our current understanding of mtDNA-mediated inflammation and how the mechanisms regulating mitochondrial homeostasis and mtDNA release represent exciting and previously under-recognised important factors in many human inflammatory diseases, offering many new translational opportunities.
Keywords: mitochondrial DNA, mtDNA, mtDNA-mediated inflammation, inflammatory diseases
Introduction
Mitochondria are intracellular double-membrane-bound organelles (“cellular powerhouses”) with many essential physiological roles in energy production, programmed cell death, calcium homeostasis, and the synthesis of lipids, amino acids, and haem. In addition, they are involved in antibacterial, antiviral, and stress responses to hypoxia and tissue injury 1, 2. Mitochondria are evolutionarily derived from energy-producing alpha-bacteria, engulfed by archezoan cells approximately 2 billion years ago leading to a symbiotic relationship that forms the basis of the eukaryotic cells 3. The mitochondria share several features with bacteria, including the double-membrane structure, a circular genome that replicates independently of nuclear DNA, and the synthesis of N-formylated proteins 4. As the innate immune system recognises conserved bacterial molecules, mitochondrial constituents are similarly immunogenic, acting as damage-associated molecular patterns (DAMPs) when released into the cytosol and extracellular environment, triggering innate immune responses, and promoting inflammation 5. In this review, we focus particularly on the role of mitochondrial DNA (mtDNA) as a specific inflammatory factor, the mechanisms behind its abnormal release, and its effects on downstream inflammatory pathways in human inflammatory diseases.
Elevated circulating mtDNA in human diseases
Freely circulating mtDNA can be detected, and over 60 studies have quantified mtDNA by quantitative polymerase chain reaction (PCR) in plasma and serum in human diseases ( Table 1). In general, they are increased in conditions with acute tissue injury such as trauma, acute myocardial infarction, and sepsis, implicating major cellular stress and uncontrolled cell death as key factors in the release of mtDNA ( Figure 1). In cancer, where its role as “liquid biopsies” is a topic of considerable interest, the pattern is less clear, and relatively lower circulating levels are found in some cancers 6.
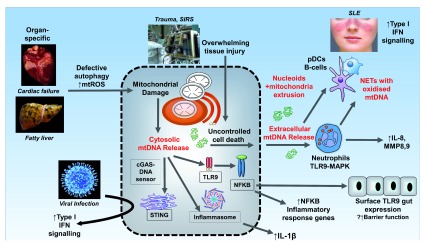
Figure 1. The contribution of mitochondrial DNA to disease pathogenesis.
Medical conditions are in italics. Where and how mitochondria are released are indicated in red. Box in dotted line frames mitochondrial DNA (mtDNA) sensor target. cGAS, cyclic GMP-AMP synthetase; IFN, interferon; IL, interleukin; MAPK, mitogen-activated protein kinase; MMP, matrix metalloproteinase; mtROS, mitochondria-derived reactive oxygen species; NET, neutrophil extracellular trap; NFκB, nuclear factor kappa B; pDC, plasmacytoid dendritic cell; SIRS, systemic inflammatory response syndrome; SLE, systemic lupus erythematosus; STING, stimulator of interferon genes; TLR9, Toll-like receptor 9.
Table 1. Circulating mitochondrial DNA in human disease.
| Disease
category |
Disease | Blood
fraction |
Finding | Reference(s) |
|---|---|---|---|---|
| Trauma | Trauma | Plasma | High mtDNA levels in trauma compared with HCs and correlated
with injury severity |
7 |
| Trauma | Plasma | High mtDNA levels in trauma | 8, 9 | |
| Trauma with MODS | Plasma | Higher levels of mtDNA had higher relative risk for mortality
Higher levels of mtDNA in those with SIRS/MODS compared with those without |
10 | |
| Trauma and severe sepsis | Plasma | mtDNA higher in patients with trauma compared with HCs on day
1 mtDNA correlates with injury severity scores in trauma patients mtDNA higher on day 1 in non-survivors compared with survivors |
11 | |
| Post-traumatic SIRS | Plasma | mtDNA is an independent predictor for post-traumatic SIRS | 12 | |
| Trauma | Plasma | mtDNA higher in trauma patients with correlation with injury
severity |
7 | |
| Trauma (femur
fracture) |
Plasma | mtDNA higher in trauma patients than HCs | 13 | |
| Trauma | Plasma | mtDNA higher in trauma patients compared with HCs at two time
points (pre-hospital and day 1) |
14 | |
| Trauma | Plasma | mtDNA higher in trauma patients than HCs
mtDNA higher in non-survivors compared with survivors |
15 | |
| Sepsis | Severe sepsis | Plasma | mtDNA higher in patients with severe sepsis compared with HCs
No significant difference in mtDNA between non-survivors and survivors in severe sepsis |
11 |
| Severe sepsis in the ED | Plasma | mtDNA higher on admission in severe septic patients than in HCs
mtDNA is higher in non-survivors than in survivors, increases initially and gradually decreases after antimicrobial therapy, and is an independent predictor of fatality |
16 | |
| Sepsis | Plasma | mtDNA higher in septic patients compared with HCs | 17 | |
| Septic shock | Plasma | mtDNA higher in patients with septic shock | 18 | |
| Adult community-
acquired bacterial meningitis |
Plasma | mtDNA levels were higher in patients with aseptic or bacterial
meningitis compared with HCs mtDNA levels fall during course of admission High mtDNA levels associated with poorer outcome in adult community-acquired bacterial meningitis |
19 | |
| Infectious SIRS | Plasma | mtDNA higher in septic patients compared with HCs | 20 | |
| Paediatric sepsis | Plasma | mtDNA higher in septic patients compared with critically ill non-
septic and HC patients |
21 | |
| Severe sepsis in
the ED |
Plasma | No significant difference in mtDNA between sepsis and HC
cohorts |
22 | |
| Critically ill
patients |
ICU patients | Plasma | Increased mtDNA levels associated with medical ICU mortality | 23 |
| Critically ill patients
(in the ICU) |
Plasma | Patients with highest quartile of mtDNA in plasma had higher risk
of dying When stratified by TLR9 expression, only patients with high expression of TLR9 had an association with mortality and mtDNA level |
24 | |
| Out-of-hospital
cardiac arrest |
Plasma | Significantly higher levels in non-survivors than in survivors | 56 a | |
| Liver failure | Acetaminophen-
induced acute liver failure |
Serum | mtDNA higher in acetaminophen-induced acute liver failure
patients compared with HCs mtDNA higher in non-survivors compared with survivors |
25 |
| Acetaminophen-
induced acute liver injury |
Plasma | mtDNA higher in patients with acetaminophen overdose with
abnormal liver function tests compared with HCs and those with acetaminophen overdose but normal liver function tests |
27 | |
| Fulminant liver
failure |
Serum | Higher during acute liver injury | 26 | |
| Heart disease | AMI | Plasma | Significantly higher mtDNA in ST elevation myocardial infarction
patients than in stable angina pectoris patients (reducing rapidly to similar levels 3 days after PCI) |
28 |
| AMI | Plasma | Significantly higher levels in AMI patients compared with HCs
Levels dropped to normal immediately after PCI |
29 | |
| AMI | Plasma | Significantly higher levels in acute AMI patients compared with
HCs on admission |
30 | |
| T2DM with CAD | Plasma | Significantly elevated levels in T2DM compared with HCs
Higher levels in those with diabetes mellitus and CAD compared with those without CAD mtDNA levels correlated with C-reactive protein in patients with CAD |
31 | |
| T2DM with CAD | Plasma | Significantly higher levels in CAD patients with T2DM | 32 | |
| Heart failure | Plasma | Higher levels of mtDNA in heart failure patients compared with
age- and sex-matched HCs; no association with disease severity |
110 | |
| Stroke | Acute ischaemic
stroke |
Plasma | mtDNA levels higher in acute cerebral infarction than in HCs
No significant difference in mtDNA between good versus poor outcome cohorts |
33 |
| Subarachnoid
haemorrhage |
Plasma | No significant difference in mtDNA between subarachnoid
haemorrahge and HC groups Overall plasma mtDNA not a good marker of prognosis |
34 | |
| Intracerebral
haemorrhage |
Plasma | No significant difference in mtDNA between intracerebral
haemorrhage and HC groups No correlation between mtDNA and disease severity |
35 | |
| Malignancy | Breast cancer | Plasma | Reduced levels of mtDNA in benign or malignant breast cancer
compared with HCs |
111 |
| Ovarian cancer | Plasma and
serum |
Plasma: significantly higher levels of mtDNA in ovarian cancer
group compared with HCs and ovarian benign tumour group Serum: no significant difference between groups above |
112 | |
| Testicular germ cell
cancer |
Serum | mtDNA levels were significantly higher in patients with testicular
cancer than in HCs, although it did not correlate with any clinicopathological variable of disease status |
113 | |
| Urological
malignancies |
Serum | mtDNA were significantly higher in “urological malignancies”
(bladder cell, renal cell, and prostate cancer) |
114 | |
| Prostate cancer | Serum | mtDNA could not distinguish between benign prostatic
hypertrophy and prostate cancer Patients with early biochemical recurrence after radical prostatectomy have higher mtDNA levels |
115 | |
| Ewing’s sarcoma | Serum | mtDNA significantly lower in patients with Ewing’s sarcoma
compared with HCs |
116 | |
| Lung cancer | Serum | mtDNA significantly higher in lung cancer patients compared
with those with benign lung diseases and healthy individuals and closely associated with tumour, lymph node, metastasis (TNM) stage |
117 | |
| Advanced prostate
cancer |
Plasma | mtDNA levels are elevated in advanced prostate cancer patients
and are associated with decreased survival |
118 | |
| Adenocarcinoma of
the lung in patients receiving erlotinib |
Plasma | Rise in mtDNA levels in patients with partial response; drop in
mtDNA levels in those with progressive disease or no response No correlation with progression-free survival |
119 | |
| Exposure to
carcinogenic halo-alkane-based pesticides |
Serum | Exposure to these carcinogens was significantly associated with
elevated serum levels of circulating mtDNA (case control study) |
120 | |
| Renal cell
carcinoma |
Plasma | Higher levels in metastatic compared with non-metastatic patients
and controls |
121 | |
| HIV | HIV | Plasma | Higher levels in acute HIV infection, late presenters compared with
long-term non-progressors and HCs Also correlated with viral load |
122 |
| Lipodystrophy
in HIV patients treated with highly active anti-retroviral therapy |
Plasma | Significantly higher levels in HIV-infected versus non-infected
individuals Significantly higher levels in those with lipodystrophy compared with those without lipodystrophy at month 24 |
123 | |
| HIV | Plasma | No significant association between HIV disease status and mtDNA | 124 | |
| Inflammatory
autoimmune conditions |
Rheumatoid arthritis | Plasma | Higher percentage of detectable levels in rheumatoid arthritis
patients compared with controls |
37 b |
| Granulomatosis with
polyangiitis |
Serum | Significantly higher levels in granulomatosis with polyangiitis
patients compared with controls |
38 | |
| Age and
exercise |
Age | Plasma | mtDNA levels increased gradually after the fifth decade of life | 125 |
| Age | Plasma | No association with age but mtDNA associated with HLA-DR | 126 | |
| Aging and “frailty” | Plasma | Aging: no difference in mtDNA between younger and older
subjects Frailty: mtDNA copy number directly correlated with frailty score |
127 | |
| Exercise | Plasma | Reduced mtDNA in response to exercise | 128 | |
| Male volleyball
players |
Plasma | Lower levels in professional volleyball players compared with
healthy non-athlete controls |
129 | |
| Miscellaneous | Corrosive injury
(gastrointestinal ingestion) |
Plasma | Significantly higher mtDNA in mortality group versus survival
group at presentation and after 12 hours |
130 |
| Pulmonary
embolism |
Plasma | Predictor of 15-day mortality | 131 c | |
| Autism | Serum | Significantly higher mtDNA in young autistic children compared
with HCs |
132 | |
| Haemodialysis | Plasma | Significantly higher levels in maintenance haemodialysis patients
compared with HCs |
133 | |
| End-stage renal
failure in Han population |
Plasma | End-stage renal failure patients had higher mtDNA copy number | 134 | |
| Bipolar disorder | Serum | No difference between bipolar disorder and HC groups
Higher levels in bipolar disorder group compared with sepsis |
135 | |
| Low levels of
ionising radiation |
Serum | Higher levels in interventional cardiologists exposed to low levels
of ionising radiation compared with controls |
136 | |
| Friedreich’s ataxia | Plasma | Significantly reduced mtDNA in Friedreich’s ataxia patients
compared with HCs |
137 | |
| Non-haemolytic
transfusion reaction |
Platelet
concentrates |
Higher mtDNA copy number in non-haemolytic transfusion
reaction platelet concentrate versus normal platelet concentrate |
138 |
This table lists studies reporting mitochondrial DNA (mtDNA) analysed by polymerase chain reaction (PCR) on serum or plasma—that is, circulating as a damage-associated molecular pattern (DAMP)—in human diseases. aLetter. bPCR rather than quantitative PCR used. cEarlier study in 2010 not included. AMI, acute myocardial infarction; CAD, coronary artery disease; ED, emergency department; HC, healthy control; HIV, human immunodeficiency virus; HLA-DR, human leukocyte antigen–antigen D related; ICU, intensive care unit; MODS, multiple organ dysfunction syndrome; PCI, percutaneous coronary intervention; SIRS, systemic inflammatory response syndrome; T2DM, type 2 diabetes mellitus; TLR9, Toll-like receptor 9.
Systemic inflammatory response syndrome
Systemic inflammatory response syndrome (SIRS) is a serious condition associated with high mortality, and affected individuals display progressive signs or symptoms of systemic upset reflecting widespread inflammation, often involving multiple organ dysfunction and failure (for example, lungs, kidneys, and brain). SIRS is often a result of major sepsis but also commonly occurs in the context of injury such as trauma. An early study by Lam et al. found that individuals admitted for blunt traumatic injury had increased plasma nuclear DNA and mtDNA levels 7. Subsequently, Hauser et al. made the seminal observation that it is the freely circulating mtDNA following traumatic injury which possesses the distinct ability to trigger and drive the clinical manifestation of SIRS 8. Several studies have confirmed the observation of elevated plasma mtDNA in trauma and SIRS 9– 15. A number of studies have found correlations with injury severity in trauma 7, 11 and higher mtDNA in non-survivors compared with survivors 11, 15. Furthermore, Gu et al. found that elevated plasma mtDNA was an independent predictor of SIRS in trauma patients 12. In sepsis, elevated levels of circulating mtDNA have also been found in multiple studies 11, 16– 21. De Caro et al. found higher mtDNA in the plasma of critically ill paediatric patients who were septic compared with similarly unwell but non-septic patients 21. The one negative study in sepsis may be explained by numerous factors, including a relatively well patient cohort, only one “spot” measurement being taken at presentation, and the potentially confounding factor of cellular content/debris 22. Studies of patients in the intensive care setting have found that higher mtDNA levels are associated with poorer outcomes 23, 24.
Acute single-organ injury: liver, heart, and brain
High levels of mtDNA are present in the serum and plasma of patients with acute injury to a variety of single organs. Acetaminophen overdose induces massive hepatocyte necrosis and in severe cases can lead to multi-organ failure and remains one of the commonest indications for liver transplantation. In fulminant liver failure secondary to acetaminophen overdose, mtDNA in the serum was found to be 30 to 40 times higher than normal, and non-survivors had higher levels than survivors 25; a separate study of drug-induced acute liver failure found serum mtDNA levels to be 10,000-fold higher 26. Serum mtDNA of acetaminophen overdose patients with derangement in the liver enzyme alanine aminotransferase (a marker of hepatocyte damage) is significantly higher than that of overdose patients who had normal liver enzymes 27, suggesting that the extent of mtDNA release into the circulation depends on the extent of hepatocyte necrosis. Similarly, extensive cardiomyocyte necrosis is found in acute myocardial infarction, which is also associated with elevated mtDNA in multiple studies 28– 30 and falls after angioplasty or coronary stent insertion to restore blood flow to the damaged myocardium 28, 29. Patients with diabetes mellitus and coronary artery disease have higher mtDNA levels than those with diabetes but without coronary artery disease 31, 32. mtDNA is also higher in acute cerebral ischaemia, caused by a reduction in cerebral blood flow by embolus or local thrombosis, and plasma levels gradually drop over time after the initial tissue injury 33. Interestingly, studies by the same group relating to plasma mtDNA in subarachnoid haemorrhage and spontaneous intracerebral haemorrhage found no significant difference compared with healthy controls, although both were small studies 34, 35. Higher mtDNA is found in the cerebrospinal fluid of patients with subarachnoid haemorrhage 34 and traumatic brain injury 36 and is associated with worse clinical outcomes. Overall, in these conditions, significant mtDNA release following massive tissue or cellular injury is evident and likely contributes to the uncontrolled inflammatory response 25.
Chronic inflammatory and immune-mediated diseases
The role for mtDNA in immune-mediated inflammatory diseases, unlike conditions relating to injury, is now also emerging. In rheumatoid arthritis, a chronic relapsing autoimmune condition affecting the joints, mtDNA was present in the plasma and synovial fluid of most patients but undetectable in healthy controls 37. Similarly, higher plasma mtDNA is found in granulomatosis with polyangiitis, an autoimmune disease whose features include necrotising granulomatous inflammation and vasculitis 38. Systemic lupus erythematosus (SLE) is a multi-organ autoimmune disease with hallmarks including excessive type I interferon (IFN) and antibodies against nucleic acids. Caielli et al. explored the potential pathogenic importance of oxidised mtDNA in SLE 39. They showed that there is a defect in mitochondrial clearance that leads to abnormal extrusion of oxidised mtDNA, which triggers a subsequent interferogenic response. Elevated anti-mtDNA antibodies were found in a separate study of SLE, particularly in lupus nephritis, where levels correlated with the lupus nephritis activity index better than anti-double-stranded DNA (anti-dsDNA) antibody levels did 40. In a further study of SLE, neutrophil extracellular traps (NETs) released from the inflammatory subset of low-density granulocyte were highly enriched in mtDNA compared with NETs from healthy control neutrophils 41. NETs are networks of extracellular fibres that are primarily composed of DNA and that are strikingly expelled following a form of neutrophil cell death (NETosis) with an aim to control pathogens; however, this study demonstrates that mtDNA-enriched NETs are pro-inflammatory in nature. Similar findings are reported in chronic granulomatous disease in this study. Higher levels of mtDNA have been found in the chronic inflammatory states of HIV (although not in all studies), end-stage renal failure, and diabetes mellitus ( Table 1). In obese individuals with steatohepatitis, mitochondria enclosed in microparticles can also be detected in plasma 42. These findings suggest that mtDNA, otherwise a “self-signal”, may be an active component in the aberrant immune or inflammatory response in chronic diseases and in autoimmunity.
mtDNA contributes to inflammatory response
mtDNA was first directly implicated as a key factor in the development of inflammatory pathology over a decade ago when intra-articular injection of oxidised mtDNA, but not nuclear DNA, triggered inflammatory arthritis in mice 43. There are now numerous studies using in vivo injection of mtDNA to provoke local or systemic inflammation or both 9, 44– 46. Moreover, there are now several in vivo studies to show that genetic deletion or pharmacologic interference of these pathways reduces the inflammatory effect of mtDNA (as will be discussed in the next section). Hence, it is clear that mtDNA release is not an epiphenomenon but directly contributes to the genesis of inflammation ( Figure 1). Current evidence shows that mtDNA-mediated inflammation is predominantly driven by the Toll-like receptor 9 (TLR9), inflammasome, and, more recently, stimulator of interferon genes (STING) pathways.
Toll-like receptor 9
TLR9 is located in the endoplasmic reticulum (ER) of various immune cells and translocates to the endosome upon sensing of hypomethylated DNA with CpG motifs, such as bacterial DNA 47, 48. Given its high frequency of unmethylated CpG dinucleotide repeats, it is postulated that mtDNA mediates inflammation dependent on the TLR9 pathway and potentially exerts a similar effect as on bacterial CpG. TLR9 recognises a variety of types of oligodeoxynucleotides (ODNs); for example, class A ODNs preferentially activate plasmacytoid dendritic cells whilst class B CpG ODNs activate B cells 49. Some of our understanding of how mtDNA may interact with TLR9 is extrapolated from work with class A ODNs, although they do not necessarily have the same effect. After activation of TLR9 by CpG DNA, inflammatory cytokine induction and Th1 immune responses occur 50 and TLR9 is necessary in CpG DNA-driven responses 51. TLR9 ligands can preferentially activate downstream pathways, including pro-inflammatory nuclear factor kappa B (NFκB), nucleotide-bindingdomain and leucine-rich repeat (NLR) pyrin domain containing 3 (NLRP3) inflammasomes, and interferon regulatory factor (IRF)-dependent type 1 IFN, which can upregulate IL-1 receptor antagonist 52, 53.
Most tissue injury models show better outcomes when the tlr9 gene is deleted. Wei et al. recently observed that tlr9 −/− mice have improved survival outcome in a necrotic lung model of cationic nanocarrier-induced necrosis and mtDNA release in vivo 54. Furthermore, the pulmonary inflammation seen after injection of mtDNA was significantly reduced in tlr9 −/− and MyD88 −/− mice, underlining the importance of the TLR9–MyD88 pathway 54. Intravenous injection of mitochondrial debris with substantial amounts of mtDNA into mice induced a systemic inflammatory response in wild-type mice that was significantly attenuated in tlr9 −/− mice 45. Tlr9 −/− mice also have better survival compared with wild-type counterparts in severe renal ischaemia reperfusion injury with associated decreased circulating mtDNA 55. A similar protective effect is seen in tlr9 −/− mice with acute acetaminophen overdose with observed lower serum mtDNA and an absence of lung inflammation in contrast to the findings of wild-type mice 26. Nevertheless, the reduction in mtDNA in tlr9 −/− mice is intriguing and could be explained by the reduced inflammation with lower resultant cellular necrosis. Alternatively, it is possible that TLR9 is somehow involved in mtDNA release into the extracellular circulation. In a recent study using a murine model of non-alcoholic steatohepatitis (NASH), mtDNA from NASH hepatocytes resulted in greater activation of TLR9 than did mtDNA from control livers 42. This suggests that mtDNA that is selectively modified during pathologic disease processes can augment the ensuing inflammatory response. Similarly, the level of TLR9 expression (due to various factors) appears to be important. In those with high mtDNA levels, higher TLR9 expression is associated with increased mortality in the intensive care unit (ICU), as discussed earlier 56.
Neutrophils have received the most attention in studies on mtDNA–TLR9 signalling in several different inflammatory settings. Zhang et al. found that mtDNA activates neutrophil p38 mitogen-activated protein kinase (MAPK) through TLR9 with release of matrix metalloproteinase 8 (MMP8) and MMP9 8, 9, a finding confirmed in a study in which phosphorylated p38 and MMP9 increased after mtDNA treatment of neutrophils 57. A separate study reported similar findings where pre-treatment with TLR9 inhibitor ODN2088 inhibited the activation of p38 MAPK and release of MMP8 54. Gu et al. also found that intratracheal administration of mtDNA provokes lung inflammation through TLR9–p38 MAPK 58. Hip fracture in rats resulted in mtDNA release into the circulation as well as higher TLR9 and NFκB p65 activation and subsequent lung injury 46. The role of other MAPKs such as extracellular signal-regulated kinases (ERKs) and c-Jun N-terminal kinases (JNKs) remains unclear and, to our knowledge, unexamined in this context. These data suggest a pathway where mtDNA activates neutrophils through TLR9 binding and activation of the MAPK pathway with subsequent MMP8 and MMP9 release ( Figure 1).
When mtDNA is considered vis-à-vis the site and location of TLR9 receptor, mtDNA must be either displaced from whole mitochondria and moved into the cytosol or, when extracellular, internalised by some mechanism(s) to act on endosomal TLR9. The endosomal location of TLR9 is most likely a mechanism to avoid unwanted activation 59. It is unclear how extracellular mtDNA is internalised, but possibilities include endocytosis, transmembrane diffusion, phagocytosis, and receptor-mediated endocytosis 60. Transmembrane diffusion is unlikely because of the highly (negatively) charged nature of DNA, which makes it difficult to pass through the cellular membrane. A recent study found that monocyte-derived macrophages can take up whole mitochondria released from necroptosis, suggesting that phagocytosis could be a relevant mechanism 61. The macrophage has a clear role in resolving inflammation by clearing up cellular debris and apoptotic bodies by phagocytosis. When mitochondria are not cleared during non-apoptotic cell death, the macrophage may phagocytose cellular corpses with mtDNA still abundantly present. Typically, apoptotic corpses can suppress the transcription of pro-inflammatory cytokine genes, promote the secretion of anti-inflammatory cytokines by phagocytes, and cause antigen-presenting cells to present dead cell antigen in a manner that promotes immunological tolerance (reviewed by Zitvogel et al. 62) . It will be of interest to consider the fate of mtDNA when macrophages or dendritic cells phagocytose cellular corpses with mtDNA. Does this clear the mtDNA or does it regulate subsequent functions (for example, immune responsiveness) in these cell types? This has yet to be studied in detail. It is also possible that binding to additional cofactors facilitates the internalisation into immune cells, and, in this instance, high-mobility group box 1 (HMGB1) and receptor for advanced glycation end products (RAGE) have been implicated 63. In this study, HMGB1–CpG (class A) complexes resulted in TLR9/RAGE association and recruitment of MyD88 in B cells 63. Here, RAGE was visualised as associating with the DNA and was internalised with some sequestered in endosome-like structures. However, this possible mechanism requires further investigation. It has also been proposed that activation of autoreactive B cells by CpG DNA occurs after B-cell receptor engagement, leading to the delivery of CpG DNA to endosomal TLR9 64.
Although nucleic acid-sensing TLRs on immune cells are found mainly within cells, cell surface expression has also been described. Via flow cytometry, TLR9 has been detected on the surface of resting B lymphocytes 65, 66 and peripheral blood mononuclear cells 67, 68. One functional ex vivo study found primary human and mouse TLR9 surface expression in neutrophils, which are upregulated by a variety of stimuli, including TLR9 agonists 69. However, it remains unclear whether TLR9 is able to signal from the cell surface. In other cell types, TLR9 is also expressed on the cell surface. For example, TLR9 is expressed on both the apical and the basolateral membranes of intestinal epithelial cells, although NFκB is activated only via basolateral stimulation of CpG ligands 70, 71. This is relevant at the gut mucosal interface, as this limits the extent of TLR9 activation at the apical surface, which is in contact with a luminal milieu rich with bacterial DNA. Hence, compromised intestinal barrier integrity and translocation of bacterial CpG from the lumen during gut pathology will lead to basolateral stimulation in this context. Whether mtDNA has a different propensity compared with bacterial CpG to trigger TLR9 depending on epithelial site has not been studied.
The inflammasome
The inflammasomes are targets of mtDNA leading to cleavage and activation of caspase-1 and downstream maturation of interleukin-1β (IL-1β) and IL-18 72. Here, it is the cytosolic release of mtDNA that exerts the dominant effect on inflammasome activation. Of the several inflammasomes described, the NLRP3 inflammasome is the best characterised in this regard. Nakahira et al. showed that depletion of mtDNA reduced IL-1β secretion in macrophages following treatment with known inflammasome triggers lipopolysaccharide (LPS) and ATP 73. Of interest, mitochondria-derived reactive oxygen species (mtROS) is a further key mediator in this process. Pharmacologic induction of mtROS correlates with higher secretion of active IL-1β in an NLRP3- and caspase-1-dependent manner, and treatment with mtROS scavengers suppresses this effect 74. The requirement for mtROS in NLRP3 activation has been confirmed by other studies 73, 75– 77 and may be explained by its oxidising effects on mtDNA. Shimada et al. showed that it is the oxidised form of mtDNA that confers the inflammatogenic potential to mtDNA 75. mtROS enhances not only the oxidative process but also the cytosolic translocation of oxidised mtDNA that then binds directly to NLRP3 75. Non-oxidised mtDNA is insufficient to activate the NLRP3 inflammasome, although it may stimulate IL-1β production via other inflammasomes such as the absent in melanoma 2 (AIM2) 78. Interestingly, genetic deletion of NLRP3 and caspase-1 results in less mtDNA release 73, 77. This suggests a positive-feedback loop, in which activation of the NLRP3 inflammasome by oxidised mtDNA further promotes mtDNA release. The overwhelming or persisting (or both) ROS production by inflammatory cells, for example, is known to damage macromolecules (DNA as well as RNA, lipids, carbohydrates, and proteins) of the surrounding cells. Activated neutrophils produce large amounts of ROS as part of their essential role in host defense 79. Hence, this is a likely major contributory factor to mtDNA damage once the inflammatory process is triggered.
Other factors controlling mitochondria-mediated NLRP3 activation are also relevant. For example, defective autophagy increases caspase-1 activation, IL-1β and IL-18 production, and cytosolic mtDNA translocation in LPS- and ATP-primed macrophages 76. Pharmacological inhibition of mitophagy/autophagy in human macrophages results in the accumulation of damaged mitochondria, ROS generation and IL-1β secretion 74, and increased NLRP3 expression in the presence of LPS 80. Hence, defective autophagy leads to inadequate clearance of damaged mitochondria, priming the internal cellular environment for NLRP3 activation. It is noteworthy that, given the diversity of NLRP3 activators, current literature suggests that the precise mechanism of NLRP3 activation is still under debate 81. Although the role of the inflammasome is often considered separately from TLR9 here, there is evidence that TLR/NFκB activation is a necessary priming step leading to NLRP3 upregulation and subsequent downstream signalling. NFκB-activating stimulus is required for cells to express pro-IL-1β and NLRP3 82. Imeada et al. showed that stimulation of TLR9 by DNA fragments during early acetaminophen-induced cell death can lead to the transcriptional activation of the IL-1β gene, resulting in the formation of pro-IL-1β 83. Using the acetaminophen hepatotoxicity model, they showed that NLRP3 deletion and related inflammasome components ASC and Caspase-1 were protective against induced liver failure 83. A further study, however, did not show any effect of NLRP3 deletion on the outcomes of acetaminophen-induced liver failure 84. Hence, in the context of liver necrosis, the role for NLRP3 inflammasome remains controversial.
STING pathway
The role of mtDNA in innate immunity through the STING pathway has also been a focus of recent studies. STING is a cytosolic protein anchored to the ER 85. STING can be activated either by direct association with dsDNA or by cyclic dinucleotides, which can be derived from intracellular bacteria or viruses or produced by a DNA sensor, cyclic GMP–AMP (cGAMP) synthetase (cGAS) 86. This, in turn, activates IRF3, which ultimately translocates to the nucleus and transcribes type I IFN genes, and also the NFκB pathway 85.
Two independent groups recently discovered that the STING-mediated IFN response can also be activated by mtDNA 87, 88. They first observed that deficiency of apoptotic caspases (3, 7, and 9) resulted in upregulation of type I IFN genes. This response was dependent on Bak/Bax, pro-apoptotic proteins responsible for mitochondrial outer membrane permeabilisation leading to mtDNA release, and the release of cytochrome C, which activates the intrinsic apoptotic pathway. Typically, apoptosis is considered immunologic-silent; for example, it does not trigger an inflammatory response. However, these studies demonstrated that, when caspases (9 and 3/7) responsible for the completion of apoptotic process are inhibited or deleted, cytosolic mtDNA goes on to activate cGAS/STING-mediated type I IFN signalling 87, 88. Hence, these caspases serve as a “brake” on the mtDNA-inflammatory effect during cell death. mtDNA released during cell death has been previously reported to provide a second signal that cooperates with an additional inflammatory signal (for example, LPS) to activate the NLRP3 inflammasome and induce IL-1β production in murine macrophages 75. Further evidence of an mtDNA role in STING-mediated IFN responses comes from West et al. 89. Here, partial deficiency of the mtDNA-binding protein mitochondrial transcription factor A (TFAM) was associated with increased concentrations of cytosolic mtDNA and enhanced type I IFN response, which was attenuated by knockdown of components of the STING pathway.
Aberrant mtDNA–STING signalling has also been implicated in human inflammatory diseases, such as SLE. As discussed earlier, Lood et al. showed that treatment of human neutrophils with SLE-abundant ribonucleoprotein immune complexes induces mtROS, mtDNA oxidation, and translocation of the mitochondria to the plasma membrane 41. Oxidised mtDNA is then released extracellularly as a component of NETs. Transfection of NET-derived mtDNA results in expression of IFN-β in human peripheral mononuclear cells. Systemic injection of oxidised mtDNA increases IFN-stimulated gene expression in the spleen of wild-type but not STING-deficient mice. Similar to inflammasomes, uncontrolled mtROS production promoting cytosolic mtDNA release is important in STING activation and potentially in the case of autoimmunity. These studies highlight the importance of the innate cellular functions to handle mtDNA release during the initiation of cell death, which ultimately will decide whether the ensuing fate will be that of a silent or inflammatory outcome.
Mechanisms for mtDNA release
Two levels of mtDNA release—cytosolic and then extracellular—are critically important steps ( Figure 1). In the former, the mechanism of release of mtDNA from mitochondria relies on the opening of mitochondrial permeability transition (MPT) pores in the inner mitochondrial membrane 90. Inhibition of pore opening with cyclosporine A resulted in lower mtDNA in the cytosol after stimulation with LPS and ATP 73. Ding et al. showed that the induction of ROS using oxidised low-density lipoprotein (ox-LDL) increased mtDNA leakage into the cytosol in a dose-dependent manner, and this effect was ameliorated with blockade of the ox-LDL receptor or a ROS inhibitor 91.
In terms of extracellular release, cellular stress and necrosis are primary factors in the non-discriminant liberation of a host of mitochondrial components such as mtDNA, N-formyl peptides, ATP, TFAM, and mitochondrial lipids. These mitochondrial constituents also exert their respective effects, which are wide-ranging, on key inflammatory pathways (extensively reviewed by Nakahira et al. 81). Aside from this non-selective release after uncontrolled cell death, several studies have suggested additional mechanisms such as necroptosis (or programmed necrosis) 92. Blood transfusion-induced endothelial necroptosis was recently found to increase extracellular mtDNA as a potential mechanism to explain transfusion-related lung injury 93. A recent study suggested that, during necroptosis, mitochondria were released before plasma membrane rupture and then phagocytosed by monocyte-derived macrophages or dendritic cells, triggering an inflammatory response as shown by cytokine production and cell maturation, respectively 61. Thus, ingestion of intact mitochondria may represent a distinct uptake mechanism following necroptosis. In a separate study, platelets were also found to be a source for free extracellular mitochondria release and then to act as an endogenous substrate for bactericidal secreted phospholipase A 2IIA (sPLA 2-IIA) leading to mitochondrial membrane hydrolysis, loss of mitochondrial structural integrity, and mtDNA release 94. Intriguingly, Xin et al. found lower levels of mtROS production when metformin was added to activated platelets, and this was associated with decreased extracellular mtDNA release 95. The authors found lower complex I activity of the platelet mitochondrial respiratory chain and suggested this as a mechanism for the observed suppressed mitochondrial dysfunction.
Whether there is an active element in mtDNA release is an interesting point of consideration. Active cellular transfer of mitochondria from stromal cells to rescue stricken lung alveoli cells in acute lung injury has been demonstrated 96. Extracellular vesicles are important modes of intercellular communication and comprise exosomes (endosomal) and microvesicles (plasma membrane-derived) and are directed by exocytosis. Both chromosomal DNA 97, 98 and mtDNA 99, 100 have been observed in extracellular vesicles; it has been suggested that the transfer of altered mtDNA between cells may play a role in Alzheimer’s disease and skeletal muscle diseases 99, 100. As described earlier, in patients with NASH, a greater percentage of mitochondria was found inside extracellular microparticles and a higher percentage of microparticles contained mitochondria compared with lean subjects 42. Furthermore, subjects with NASH had a higher level of oxidised mtDNA in microparticles. Further clarification is required on the concentration and significance of mtDNA in extracellular vesicles and whether this has different immunostimulatory effects compared with cell-free or surface-bound mtDNA. As previously mentioned, the pro-inflammatory effects of mtDNA are dependent on its oxidisation 75, 101. The highly oxidative extracellular milieu at sites of tissue inflammation in patients with chronic inflammatory disease may overwhelm anti-oxidant systems, further potentiating the inflammatory potential of DAMPs such as mtDNA 5.
mtDNA degradation and clearance
Several well-described clearance mechanisms limit the pro-inflammatory nature of mtDNA. Autophagy as discussed earlier is important 102. Defective autophagy has been implicated in several chronic inflammatory human diseases, including Crohn’s disease 103. A proportion of circulating DNA in the bloodstream appears to cross the kidney barrier and be excreted in the urine 104. Indeed, mtDNA has been detected in the urine at elevated levels in patients with progressive acute kidney injury 105. This may be due to the inflammatory state associated with this condition, the increased clearance with a disturbed kidney barrier, or both. Another possible mechanism of mtDNA clearance is phagocytosis by macrophages in a manner similar to the ingestion of the structurally similar bacterial DNA 106. As described earlier, the outcome of phagocytosis of intact mitochondria may be pro- rather than anti-inflammatory; these divergent effects may also be dependent on the phenotype of the phagocytosing cells (for example, inflammatory versus pro-resolution macrophages/monocytes, neutrophils, and red blood cells) 61, 93.
In general, non-host DNA in the circulation is digested in part by circulating nucleases, and mtDNA may be affected by a similar mechanism 107. Intracellularly, DNases found in the autophagolysosome play a vital role in degrading mtDNA 102, 108. Oka et al. showed that cardiac-specific deletion of DNase II resulted in mtDNA accumulation in cardiomyocytes and the development of heart failure 102. In human umbilical vein endothelial cells, lysosomal DNases protect cells against inflammation from mtDNA damage induced by ox-LDL 91. Here, small interfering RNA (siRNA) knockdown of DNase II amplifies the mtDNA–TLR9-mediated inflammatory response 91. It is unclear whether nucleases have a similar action on mtDNA in the extracellular space or are relevant in the physiological setting, especially when mtDNA is present in microvesicles or housed within intact mitochondria, which protect against DNase II. Intriguingly, DNase pre-treatment abolished renal mitochondrial injury that was observed after injection of mitochondrial debris (including mtDNA) in mice 45. However, the precise role of DNase and its effect on the immunostimulatory effects of mtDNA is likely to be more complex, as illustrated by a recent study which showed that DNase II was required for TLR9 activation by bacterial genomic DNA 109.
Conclusions: translational opportunities for mtDNA-mediated inflammation
mtDNA contributes to inflammation at multiple levels when tissue or cellular homeostasis is perturbed. Damaged mtDNA released into the cytosol has a functional short-range effect on immediate “alarm” systems such as the inflammasome and NFκB. Uncontrolled release of mtDNA into the circulation in conditions with significant tissue injury generates a more systemic effect whilst de-regulation of local mitochondrial homeostatic mechanisms such as autophagy or mtROS detoxification contributes to organ-specific pathology as observed in the heart and liver. Failure of such mechanisms may also give rise to a more wide-ranging consequence (for example, in autoimmune diseases such as SLE).
Our review shows that mtDNA-mediated inflammation is important and relevant to many human inflammatory diseases. However, this remains an underexplored field and more insights will likely emerge in the near future. The current evidence offers a rich realm of translational opportunities to target mtDNA-mediated inflammation. There are many plausible approaches, which include targeting cytosolic mtDNA release (for example, directly at MPT using cyclosporine or by specific mitochondrial anti-oxidant strategies, such as MitoQ10 10 to reduce mtROS), augmenting clearance (for example, using autophagy activators or correcting factors leading to impaired autophagy), diverting the cellular response following mitochondrial damage (for example, induction of pro-apoptotic caspases), and reducing the inflammatory potential of mtDNA (for example, DNases to digest NET-bound mtDNA and reducing oxidation of mtDNA).
Given that mtDNA can be measured and used as a biomarker, it offers a unique opportunity to stratify and identify individuals who may benefit from specific therapeutic targeting of downstream inflammation pathways (for example, TLR9, NLRP3, or STING pathways). As discussed earlier, there are numerous studies in sepsis, trauma, and acute single-organ injury that have already demonstrated that individuals with high mtDNA levels and TLR9 expressions have worse prognosis. Therefore, there are clear groups in which stratification is useful. However, a number of challenges exist to its implementation as a biomarker, such as the variation in which mtDNA is measured (for example, serum versus plasma, mtDNA-specific PCR primers) and reported in the literature. Standardisation of these protocols, including the identification of “normal” and “abnormal” ranges, will be important prior to clinical use. Furthermore, many studies have failed to include clinically relevant predictive statistics; further studies reporting such statistics in a variety of inflammatory conditions are required.
In conclusion, multiple lines of data show that innate responses to mtDNA, which is similar to and evolutionarily derived from bacteria, are hard-wired into our biology and drive the development inflammation with pathologic consequences in many diseases.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Augustine M.K. Choi, Division of Pulmonary and Critical Care Medicine, Joan and Sanford I. Weill Department of Medicine, Weill Cornell Medicine, New York, NY, USA
Kiichi Nakahira, Department of Medicine, Weill Cornell Medicine, New York, NY, USA
Mitchell R. McGill, Department of Pathology and Immunology, Washington University School of Medicine, St Louis, MO, 63110, USA
Antonio Ferrante, Women and Children's Hospital Campus, University of Adelaide, Adelaide, Australia
Funding Statement
This work was supported by Medical Research Council grant G0701898, Crohn’s and Colitis UK M16-1, ECCO IBD Investigator’s Award 2010, Chief Scientist Office ETM/75 award (to G-TH); Edinburgh GI Trustees Grant (2014) (to RKB); Medical Research Society UK Vac-982-2016 (to AT); and Wellcome Trust grant WT096497 (to DAD).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 3 approved]
References
- 1. West AP, Shadel GS, Ghosh S: Mitochondria in innate immune responses. Nat Rev Immunol. 2011;11(6):389–402. 10.1038/nri2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nunnari J, Suomalainen A: Mitochondria: in sickness and in health. Cell. 2012;148(6):1145–59. 10.1016/j.cell.2012.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dyall SD, Brown MT, Johnson PJ: Ancient invasions: from endosymbionts to organelles. Science. 2004;304(5668):253–7. 10.1126/science.1094884 [DOI] [PubMed] [Google Scholar]
- 4. Galluzzi L, Kepp O, Trojel-Hansen C, et al. : Mitochondrial control of cellular life, stress, and death. Circ Res. 2012;111(9):1198–207. 10.1161/CIRCRESAHA.112.268946 [DOI] [PubMed] [Google Scholar]
- 5. Boyapati RK, Rossi AG, Satsangi J, et al. : Gut mucosal DAMPs in IBD: from mechanisms to therapeutic implications. Mucosal Immunol. 2016;9(3):567–82. 10.1038/mi.2016.14 [DOI] [PubMed] [Google Scholar]
- 6. Lee HC, Li SH, Lin JC, et al. : Somatic mutations in the D-loop and decrease in the copy number of mitochondrial DNA in human hepatocellular carcinoma. Mutat Res. 2004;547(1–2):71–8. 10.1016/j.mrfmmm.2003.12.011 [DOI] [PubMed] [Google Scholar]
- 7. Lam NY, Rainer TH, Chiu RW, et al. : Plasma mitochondrial DNA concentrations after trauma. Clin Chem. 2004;50(1):213–6. 10.1373/clinchem.2003.025783 [DOI] [PubMed] [Google Scholar]
- 8. Zhang Q, Raoof M, Chen Y, et al. : Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464(7285):104–7. 10.1038/nature08780 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 9. Zhang Q, Itagaki K, Hauser CJ: Mitochondrial DNA is released by shock and activates neutrophils via p38 map kinase. Shock. 2010;34(1):55–9. 10.1097/SHK.0b013e3181cd8c08 [DOI] [PubMed] [Google Scholar]
- 10. Simmons JD, Lee Y, Mulekar S, et al. : Elevated levels of plasma mitochondrial DNA DAMPs are linked to clinical outcome in severely injured human subjects. Ann Surg. 2013;258(4):591–6; discussion 596–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yamanouchi S, Kudo D, Yamada M, et al. : Plasma mitochondrial DNA levels in patients with trauma and severe sepsis: time course and the association with clinical status. J Crit Care. 2013;28(6):1027–31. 10.1016/j.jcrc.2013.05.006 [DOI] [PubMed] [Google Scholar]
- 12. Gu X, Yao Y, Wu G, et al. : The plasma mitochondrial DNA is an independent predictor for post-traumatic systemic inflammatory response syndrome. PLoS One. 2013;8(8):e72834. 10.1371/journal.pone.0072834 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 13. Hauser CJ, Sursal T, Rodriguez EK, et al. : Mitochondrial damage associated molecular patterns from femoral reamings activate neutrophils through formyl peptide receptors and P44/42 MAP kinase. J Orthop Trauma. 2010;24(9):534–8. 10.1097/BOT.0b013e3181ec4991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Timmermans K, Kox M, Vaneker M, et al. : Plasma levels of danger-associated molecular patterns are associated with immune suppression in trauma patients. Intensive Care Med. 2016;42(4):551–61. 10.1007/s00134-015-4205-3 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 15. Prikhodko AS, Shabanov AK, Zinovkina LA, et al. : Pure Mitochondrial DNA Does Not Activate Human Neutrophils in vitro. Biochemistry (Mosc). 2015;80(5):629–35. 10.1134/S0006297915050168 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 16. Kung CT, Hsiao SY, Tsai TC, et al. : Plasma nuclear and mitochondrial DNA levels as predictors of outcome in severe sepsis patients in the emergency room. J Transl Med. 2012;10:130. 10.1186/1479-5876-10-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bhagirath VC, Dwivedi DJ, Liaw PC: Comparison of the Proinflammatory and Procoagulant Properties of Nuclear, Mitochondrial, and Bacterial DNA. Shock. 2015;44(3):265–71. 10.1097/SHK.0000000000000397 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 18. Timmermans K, Kox M, Scheffer GJ, et al. : Plasma Nuclear and Mitochondrial DNA Levels, and Markers of Inflammation, Shock, and Organ Damage in Patients with Septic Shock. Shock. 2016;45(6):607–12. 10.1097/SHK.0000000000000549 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 19. Lu CH, Chang WN, Tsai NW, et al. : The value of serial plasma nuclear and mitochondrial DNA levels in adult community-acquired bacterial meningitis. QJM. 2010;103(3):169–75. 10.1093/qjmed/hcp201 [DOI] [PubMed] [Google Scholar]
- 20. Garrabou G, Morén C, López S, et al. : The effects of sepsis on mitochondria. J Infect Dis. 2012;205(3):392–400. 10.1093/infdis/jir764 [DOI] [PubMed] [Google Scholar]
- 21. Di Caro V, Walko TD, 3rd, Bola RA, et al. : Plasma Mitochondrial DNA--a Novel DAMP in Pediatric Sepsis. Shock. 2016;45(5):506–11. 10.1097/SHK.0000000000000539 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 22. Yu M: Circulating cell-free mitochondrial DNA as a novel cancer biomarker: opportunities and challenges. Mitochondrial DNA. 2012;23(5):329–32. 10.3109/19401736.2012.696625 [DOI] [PubMed] [Google Scholar]
- 23. Nakahira K, Kyung SY, Rogers AJ, et al. : Circulating mitochondrial DNA in patients in the ICU as a marker of mortality: derivation and validation. PLoS Med. 2013;10(12):e1001577; discussion e1001577. 10.1371/journal.pmed.1001577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Krychtiuk KA, Ruhittel S, Hohensinner PJ, et al. : Mitochondrial DNA and Toll-Like Receptor-9 Are Associated With Mortality in Critically Ill Patients. Crit Care Med. 2015;43(12):2633–41. 10.1097/CCM.0000000000001311 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 25. McGill MR, Staggs VS, Sharpe MR, et al. : Serum mitochondrial biomarkers and damage-associated molecular patterns are higher in acetaminophen overdose patients with poor outcome. Hepatology. 2014;60(4):1336–45. 10.1002/hep.27265 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 26. Marques PE, Amaral SS, Pires DA, et al. : Chemokines and mitochondrial products activate neutrophils to amplify organ injury during mouse acute liver failure. Hepatology. 2012;56(5):1971–82. 10.1002/hep.25801 [DOI] [PubMed] [Google Scholar]
- 27. McGill MR, Sharpe MR, Williams CD, et al. : The mechanism underlying acetaminophen-induced hepatotoxicity in humans and mice involves mitochondrial damage and nuclear DNA fragmentation. J Clin Invest. 2012;122(4):1574–83. 10.1172/JCI59755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bliksoen M, Mariero LH, Ohm IK, et al. : Increased circulating mitochondrial DNA after myocardial infarction. Int J Cardiol. 2012;158(1):132–4. 10.1016/j.ijcard.2012.04.047 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 29. Wang L, Xie L, Zhang Q, et al. : Plasma nuclear and mitochondrial DNA levels in acute myocardial infarction patients. Coron Artery Dis. 2015;26(4):296–300. 10.1097/MCA.0000000000000231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Qin C, Gu J, Liu R, et al. : Release of mitochondrial DNA correlates with peak inflammatory cytokines in patients with acute myocardial infarction. Anatol J Cardiol. 2016. 10.14744/AnatolJCardiol.2016.7209 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 31. Liu J, Cai X, Xie L, et al. : Circulating Cell Free Mitochondrial DNA is a Biomarker in the Development of Coronary Heart Disease in the Patients with Type 2 Diabetes. Clin Lab. 2015;61(7):661–7. 10.7754/Clin.Lab.2014.141132 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 32. Liu J, Zou Y, Tang Y, et al. : Circulating cell-free mitochondrial deoxyribonucleic acid is increased in coronary heart disease patients with diabetes mellitus. J Diabetes Investig. 2016;7(1):109–14. 10.1111/jdi.12366 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 33. Tsai NW, Lin TK, Chen SD, et al. : The value of serial plasma nuclear and mitochondrial DNA levels in patients with acute ischemic stroke. Clin Chim Acta. 2011;412(5–6):476–9. 10.1016/j.cca.2010.11.036 [DOI] [PubMed] [Google Scholar]
- 34. Wang HC, Yang TM, Lin WC, et al. : The value of serial plasma and cerebrospinal fluid nuclear and mitochondrial deoxyribonucleic acid levels in aneurysmal subarachnoid hemorrhage. J Neurosurg. 2013;118(1):13–9. 10.3171/2012.8.JNS112093 [DOI] [PubMed] [Google Scholar]
- 35. Wang HC, Lin YJ, Lin WC, et al. : The value of serial plasma nuclear and mitochondrial DNA levels in acute spontaneous intra-cerebral haemorrhage. Eur J Neurol. 2012;19(12):1532–8. 10.1111/j.1468-1331.2012.03761.x [DOI] [PubMed] [Google Scholar]
- 36. Walko TD, 3rd, Bola RA, Hong JD, et al. : Cerebrospinal fluid mitochondrial DNA: a novel DAMP in pediatric traumatic brain injury. Shock. 2014;41(6):499–503. 10.1097/SHK.0000000000000160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hajizadeh S, DeGroot J, TeKoppele JM, et al. : Extracellular mitochondrial DNA and oxidatively damaged DNA in synovial fluid of patients with rheumatoid arthritis. Arthritis Res Ther. 2003;5(5):R234–40. 10.1186/ar787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Surmiak MP, Hubalewska-Mazgaj M, Wawrzycka-Adamczyk K, et al. : Circulating mitochondrial DNA in serum of patients with granulomatosis with polyangiitis. Clin Exp Immunol. 2015;181(1):150–5. 10.1111/cei.12628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Caielli S, Athale S, Domic B, et al. : Oxidized mitochondrial nucleoids released by neutrophils drive type I interferon production in human lupus. J Exp Med. 2016;213(5):697–713. 10.1084/jem.20151876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang H, Li T, Chen S, et al. : Neutrophil Extracellular Trap Mitochondrial DNA and Its Autoantibody in Systemic Lupus Erythematosus and a Proof-of-Concept Trial of Metformin. Arthritis Rheumatol. 2015;67(12):3190–200. 10.1002/art.39296 [DOI] [PubMed] [Google Scholar]
- 41. Lood C, Blanco LP, Purmalek MM, et al. : Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus-like disease. Nat Med. 2016;22(2):146–53. 10.1038/nm.4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Garcia-Martinez I, Santoro N, Chen Y, et al. : Hepatocyte mitochondrial DNA drives nonalcoholic steatohepatitis by activation of TLR9. J Clin Invest. 2016;126(3):859–64. 10.1172/JCI83885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Collins LV, Hajizadeh S, Holme E, et al. : Endogenously oxidized mitochondrial DNA induces in vivo and in vitro inflammatory responses. J Leukoc Biol. 2004;75(6):995–1000. 10.1189/jlb.0703328 [DOI] [PubMed] [Google Scholar]
- 44. Hu Q, Wood CR, Cimen S, et al. : Mitochondrial Damage-Associated Molecular Patterns (MTDs) Are Released during Hepatic Ischemia Reperfusion and Induce Inflammatory Responses. PLoS One. 2015;10(10):e0140105. 10.1371/journal.pone.0140105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tsuji N, Tsuji T, Ohashi N, et al. : Role of Mitochondrial DNA in Septic AKI via Toll-Like Receptor 9. J Am Soc Nephrol. 2016;27(7):2009–20. 10.1681/ASN.2015040376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gan L, Chen X, Sun T, et al. : Significance of Serum mtDNA Concentration in Lung Injury Induced by Hip Fracture. Shock. 2015;44(1):52–7. 10.1097/SHK.0000000000000366 [DOI] [PubMed] [Google Scholar]
- 47. Latz E, Schoenemeyer A, Visintin A, et al. : TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat Immunol. 2004;5(2):190–8. 10.1038/ni1028 [DOI] [PubMed] [Google Scholar]
- 48. Leifer CA, Kennedy MN, Mazzoni A, et al. : TLR9 is localized in the endoplasmic reticulum prior to stimulation. J Immunol. 2004;173(2):1179–83. 10.4049/jimmunol.173.2.1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Moseman EA, Liang X, Dawson AJ, et al. : Human plasmacytoid dendritic cells activated by CpG oligodeoxynucleotides induce the generation of CD4 +CD25 + regulatory T cells. J Immunol. 2004;173(7):4433–42. 10.4049/jimmunol.173.7.4433 [DOI] [PubMed] [Google Scholar]
- 50. Hemmi H, Takeuchi O, Kawai T, et al. : A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408(6813):740–5. 10.1038/35047123 [DOI] [PubMed] [Google Scholar]
- 51. Bauer S, Kirschning CJ, Häcker H, et al. : Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc Natl Acad Sci U S A. 2001;98(16):9237–42. 10.1073/pnas.161293498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sasai M, Linehan MM, Iwasaki A: Bifurcation of Toll-like receptor 9 signaling by adaptor protein 3. Science. 2010;329(5998):1530–4. 10.1126/science.1187029 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 53. Petrasek J, Dolganiuc A, Csak T, et al. : Type I interferons protect from Toll-like receptor 9-associated liver injury and regulate IL-1 receptor antagonist in mice. Gastroenterology. 2011;140(2):697–708.e4. 10.1053/j.gastro.2010.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wei X, Shao B, He Z, et al. : Cationic nanocarriers induce cell necrosis through impairment of Na +/K +-ATPase and cause subsequent inflammatory response. Cell Res. 2015;25(2):237–53. 10.1038/cr.2015.9 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 55. Bakker PJ, Scantlebery AM, Butter LM, et al. : TLR9 Mediates Remote Liver Injury following Severe Renal Ischemia Reperfusion. PLoS One. 2015;10(9):e0137511. 10.1371/journal.pone.0137511 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 56. Arnalich F, Codoceo R, López-Collazo E, et al. : Circulating cell-free mitochondrial DNA: a better early prognostic marker in patients with out-of-hospital cardiac arrest. Resuscitation. 2012;83(7):e162–3. 10.1016/j.resuscitation.2012.03.032 [DOI] [PubMed] [Google Scholar]
- 57. Sudakov NP, Popkova TP, Katyshev AI, et al. : Level of Blood Cell-Free Circulating Mitochondrial DNA as a Novel Biomarker of Acute Myocardial Ischemia. Biochemistry (Mosc). 2015;80(10):1387–92. 10.1134/S000629791510020X [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 58. Gu X, Wu G, Yao Y, et al. : Intratracheal administration of mitochondrial DNA directly provokes lung inflammation through the TLR9-p38 MAPK pathway. Free Radic Biol Med. 2015;83:149–58. 10.1016/j.freeradbiomed.2015.02.034 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 59. Barton GM, Kagan JC: A cell biological view of Toll-like receptor function: regulation through compartmentalization. Nat Rev Immunol. 2009;9(8):535–42. 10.1038/nri2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ziello JE, Huang Y, Jovin IS: Cellular endocytosis and gene delivery. Mol Med. 2010;16(5–6):222–9. 10.2119/molmed.2009.00101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Maeda A, Fadeel B: Mitochondria released by cells undergoing TNF- α-induced necroptosis act as danger signals. Cell Death Dis. 2014;5:e1312. 10.1038/cddis.2014.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zitvogel L, Kepp O, Kroemer G: Decoding cell death signals in inflammation and immunity. Cell. 2010;140(6):798–804. 10.1016/j.cell.2010.02.015 [DOI] [PubMed] [Google Scholar]
- 63. Tian J, Avalos AM, Mao SY, et al. : Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat Immunol. 2007;8(5):487–96. 10.1038/ni1457 [DOI] [PubMed] [Google Scholar]
- 64. Viglianti GA, Lau CM, Hanley TM, et al. : Activation of autoreactive B cells by CpG dsDNA. Immunity. 2003;19(6):837–47. 10.1016/S1074-7613(03)00323-6 [DOI] [PubMed] [Google Scholar]
- 65. Dasari P, Nicholson IC, Hodge G, et al. : Expression of toll-like receptors on B lymphocytes. Cell Immunol. 2005;236(1–2):140–5. 10.1016/j.cellimm.2005.08.020 [DOI] [PubMed] [Google Scholar]
- 66. Baiyee EE, Flohe S, Lendemans S, et al. : Expression and function of Toll-like receptor 9 in severely injured patients prone to sepsis. Clin Exp Immunol. 2006;145(3):456–62. 10.1111/j.1365-2249.2006.03160.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Eaton-Bassiri A, Dillon SB, Cunningham M, et al. : Toll-like receptor 9 can be expressed at the cell surface of distinct populations of tonsils and human peripheral blood mononuclear cells. Infect Immun. 2004;72(12):7202–11. 10.1128/IAI.72.12.7202-7211.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Saikh KU, Kissner TL, Sultana A, et al. : Human monocytes infected with Yersinia pestis express cell surface TLR9 and differentiate into dendritic cells. J Immunol. 2004;173(12):7426–34. 10.4049/jimmunol.173.12.7426 [DOI] [PubMed] [Google Scholar]
- 69. Lindau D, Mussard J, Wagner BJ, et al. : Primary blood neutrophils express a functional cell surface Toll-like receptor 9. Eur J Immunol. 2013;43(8):2101–13. 10.1002/eji.201142143 [DOI] [PubMed] [Google Scholar]
- 70. Lee J, Mo JH, Shen C, et al. : Toll-like receptor signaling in intestinal epithelial cells contributes to colonic homoeostasis. Curr Opin Gastroenterol. 2007;23(1):27–31. 10.1097/MOG.0b013e3280118272 [DOI] [PubMed] [Google Scholar]
- 71. Ewaschuk JB, Backer JL, Churchill TA, et al. : Surface expression of Toll-like receptor 9 is upregulated on intestinal epithelial cells in response to pathogenic bacterial DNA. Infect Immun. 2007;75(5):2572–9. 10.1128/IAI.01662-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gurung P, Lukens JR, Kanneganti TD: Mitochondria: diversity in the regulation of the NLRP3 inflammasome. Trends Mol Med. 2015;21(3):193–201. 10.1016/j.molmed.2014.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Nakahira K, Haspel JA, Rathinam VA, et al. : Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12(3):222–30. 10.1038/ni.1980 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 74. Zhou R, Yazdi AS, Menu P, et al. : A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469(7329):221–5. 10.1038/nature09663 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 75. Shimada K, Crother TR, Karlin J, et al. : Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012;36(3):401–14. 10.1016/j.immuni.2012.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 76. Zhang Z, Xu X, Ma J, et al. : Gene deletion of Gabarap enhances Nlrp3 inflammasome-dependent inflammatory responses. J Immunol. 2013;190(7):3517–24. 10.4049/jimmunol.1202628 [DOI] [PubMed] [Google Scholar]
- 77. Won JH, Park S, Hong S, et al. : Rotenone-induced Impairment of Mitochondrial Electron Transport Chain Confers a Selective Priming Signal for NLRP3 Inflammasome Activation. J Biol Chem. 2015;290(45):27425–37. 10.1074/jbc.M115.667063 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 78. Dombrowski Y, Peric M, Koglin S, et al. : Honey bee ( Apis mellifera) venom induces AIM2 inflammasome activation in human keratinocytes. Allergy. 2012;67(11):1400–7. 10.1111/all.12022 [DOI] [PubMed] [Google Scholar]
- 79. Holmström KM, Finkel T: Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat Rev Mol Cell Biol. 2014;15(6):411–21. 10.1038/nrm3801 [DOI] [PubMed] [Google Scholar]
- 80. Ding Z, Liu S, Wang X, et al. : LOX-1, mtDNA damage, and NLRP3 inflammasome activation in macrophages: implications in atherogenesis. Cardiovasc Res. 2014;103(4):619–28. 10.1093/cvr/cvu114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Nakahira K, Hisata S, Choi AM: The Roles of Mitochondrial Damage-Associated Molecular Patterns in Diseases. Antioxid Redox Signal. 2015;23(17):1329–50. 10.1089/ars.2015.6407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bauernfeind FG, Horvath G, Stutz A, et al. : Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J Immunol. 2009;183(2):787–91. 10.4049/jimmunol.0901363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Imaeda AB, Watanabe A, Sohail MA, et al. : Acetaminophen-induced hepatotoxicity in mice is dependent on Tlr9 and the Nalp3 inflammasome. J Clin Invest. 2009;119(2):305–14. 10.1172/JCI35958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Williams CD, Antoine DJ, Shaw PJ, et al. : Role of the Nalp3 inflammasome in acetaminophen-induced sterile inflammation and liver injury. Toxicol Appl Pharmacol. 2011;252(3):289–97. 10.1016/j.taap.2011.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ishikawa H, Barber GN: STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455(7213):674–8. 10.1038/nature07317 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 86. Barber GN: STING-dependent cytosolic DNA sensing pathways. Trends Immunol. 2014;35(2):88–93. 10.1016/j.it.2013.10.010 [DOI] [PubMed] [Google Scholar]
- 87. Rongvaux A, Jackson R, Harman CC, et al. : Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell. 2014;159(7):1563–77. 10.1016/j.cell.2014.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 88. White MJ, McArthur K, Metcalf D, et al. : Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell. 2014;159(7):1549–62. 10.1016/j.cell.2014.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 89. West AP, Khoury-Hanold W, Staron M, et al. : Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015;520(7548):553–7. 10.1038/nature14156 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 90. Patrushev M, Kasymov V, Patrusheva V, et al. : Mitochondrial permeability transition triggers the release of mtDNA fragments. Cell Mol Life Sci. 2004;61(24):3100–3. 10.1007/s00018-004-4424-1 [DOI] [PubMed] [Google Scholar]
- 91. Ding Z, Liu S, Wang X, et al. : Oxidant stress in mitochondrial DNA damage, autophagy and inflammation in atherosclerosis. Sci Rep. 2013;3:1077. 10.1038/srep01077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kaczmarek A, Vandenabeele P, Krysko DV: Necroptosis: the release of damage-associated molecular patterns and its physiological relevance. Immunity. 2013;38(2):209–23. 10.1016/j.immuni.2013.02.003 [DOI] [PubMed] [Google Scholar]
- 93. Mangalmurti N, Qing D, Hotz M, et al. : Mitochondrial DNA Released Following Necroptosis Accumulates on RBCs. Am J Respir Crit Care Med. 2016;193:A4309 Reference Source [Google Scholar]
- 94. Boudreau LH, Duchez AC, Cloutier N, et al. : Platelets release mitochondria serving as substrate for bactericidal group IIA-secreted phospholipase A 2 to promote inflammation. Blood. 2014;124(14):2173–83. 10.1182/blood-2014-05-573543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Xin G, Wei Z, Ji C, et al. : Metformin Uniquely Prevents Thrombosis by Inhibiting Platelet Activation and mtDNA Release. Sci Rep. 2016;6:36222. 10.1038/srep36222 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 96. Islam MN, Das SR, Emin MT, et al. : Mitochondrial transfer from bone-marrow-derived stromal cells to pulmonary alveoli protects against acute lung injury. Nat Med. 2012;18(5):759–65. 10.1038/nm.2736 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 97. Waldenström A, Gennebäck N, Hellman U, et al. : Cardiomyocyte microvesicles contain DNA/RNA and convey biological messages to target cells. PLoS One. 2012;7(4):e34653. 10.1371/journal.pone.0034653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Balaj L, Lessard R, Dai L, et al. : Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun. 2011;2:180. 10.1038/ncomms1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Guescini M, Guidolin D, Vallorani L, et al. : C2C12 myoblasts release micro-vesicles containing mtDNA and proteins involved in signal transduction. Exp Cell Res. 2010;316(12):1977–84. 10.1016/j.yexcr.2010.04.006 [DOI] [PubMed] [Google Scholar]
- 100. Guescini M, Genedani S, Stocchi V, et al. : Astrocytes and Glioblastoma cells release exosomes carrying mtDNA. J Neural Transm (Vienna). 2010;117(1):1–4. 10.1007/s00702-009-0288-8 [DOI] [PubMed] [Google Scholar]
- 101. Pazmandi K, Agod Z, Kumar BV, et al. : Oxidative modification enhances the immunostimulatory effects of extracellular mitochondrial DNA on plasmacytoid dendritic cells. Free Radic Biol Med. 2014;77:281–90. 10.1016/j.freeradbiomed.2014.09.028 [DOI] [PubMed] [Google Scholar]
- 102. Oka T, Hikoso S, Yamaguchi O, et al. : Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 2012;485(7397):251–5. 10.1038/nature10992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Boyapati R, Satsangi J, Ho G: Pathogenesis of Crohn's disease. F1000Prime Rep. 2015;7:44. 10.12703/P7-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Botezatu I, Serdyuk O, Potapova G, et al. : Genetic analysis of DNA excreted in urine: a new approach for detecting specific genomic DNA sequences from cells dying in an organism. Clin Chem. 2000;46(8 Pt):1078–84. [PubMed] [Google Scholar]
- 105. Whitaker RM, Stallons LJ, Kneff JE, et al. : Urinary mitochondrial DNA is a biomarker of mitochondrial disruption and renal dysfunction in acute kidney injury. Kidney Int. 2015;88(6):1336–44. 10.1038/ki.2015.240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Stacey KJ, Sweet MJ, Hume DA: Macrophages ingest and are activated by bacterial DNA. J Immunol. 1996;157(5):2116–22. [PubMed] [Google Scholar]
- 107. Lo YM, Zhang J, Leung TN, et al. : Rapid clearance of fetal DNA from maternal plasma. Am J Hum Genet. 1999;64(1):218–24. 10.1086/302205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Okabe Y, Kawane K, Akira S, et al. : Toll-like receptor-independent gene induction program activated by mammalian DNA escaped from apoptotic DNA degradation. J Exp Med. 2005;202(10):1333–9. 10.1084/jem.20051654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Chan MP, Onji M, Fukui R, et al. : DNase II-dependent DNA digestion is required for DNA sensing by TLR9. Nat Commun. 2015;6:5853. 10.1038/ncomms6853 [DOI] [PubMed] [Google Scholar]
- 110. Dhondup Y, Ueland T, Dahl CP, et al. : Low Circulating Levels of Mitochondrial and High Levels of Nuclear DNA Predict Mortality in Chronic Heart Failure. J Card Fail. 2016;22(10):823–8. 10.1016/j.cardfail.2016.06.013 [DOI] [PubMed] [Google Scholar]
- 111. Kohler C, Radpour R, Barekati Z, et al. : Levels of plasma circulating cell free nuclear and mitochondrial DNA as potential biomarkers for breast tumors. Mol Cancer. 2009;8:105. 10.1186/1476-4598-8-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Zachariah RR, Schmid S, Buerki N, et al. : Levels of circulating cell-free nuclear and mitochondrial DNA in benign and malignant ovarian tumors. Obstet Gynecol. 2008;112(4):843–50. 10.1097/AOG.0b013e3181867bc0 [DOI] [PubMed] [Google Scholar]
- 113. Ellinger J, Albers P, Müller SC, et al. : Circulating mitochondrial DNA in the serum of patients with testicular germ cell cancer as a novel noninvasive diagnostic biomarker. BJU Int. 2009;104(1):48–52. 10.1111/j.1464-410X.2008.08289.x [DOI] [PubMed] [Google Scholar]
- 114. Ellinger J, Müller DC, Müller SC, et al. : Circulating mitochondrial DNA in serum: a universal diagnostic biomarker for patients with urological malignancies. Urol Oncol. 2012;30(4):509–15. 10.1016/j.urolonc.2010.03.004 [DOI] [PubMed] [Google Scholar]
- 115. Ellinger J, Müller SC, Wernert N, et al. : Mitochondrial DNA in serum of patients with prostate cancer: a predictor of biochemical recurrence after prostatectomy. BJU Int. 2008;102(5):628–32. 10.1111/j.1464-410X.2008.07613.x [DOI] [PubMed] [Google Scholar]
- 116. Yu M, Wan YF, Zou QH: Cell-free circulating mitochondrial DNA in the serum: a potential non-invasive biomarker for Ewing's sarcoma. Arch Med Res. 2012;43(5):389–94. 10.1016/j.arcmed.2012.06.007 [DOI] [PubMed] [Google Scholar]
- 117. Hou YL, Chen JJ, Wu YF, et al. : Clinical significance of serum mitochondrial DNA in lung cancer. Clin Biochem. 2013;46(15):1474–7. 10.1016/j.clinbiochem.2013.04.009 [DOI] [PubMed] [Google Scholar]
- 118. Mehra N, Penning M, Maas J, et al. : Circulating mitochondrial nucleic acids have prognostic value for survival in patients with advanced prostate cancer. Clin Cancer Res. 2007;13(2 Pt 1):421–6. 10.1158/1078-0432.CCR-06-1087 [DOI] [PubMed] [Google Scholar]
- 119. Huang CY, Chen YM, Wu CH, et al. : Circulating free mitochondrial DNA concentration and its association with erlotinib treatment in patients with adenocarcinoma of the lung. Oncol Lett. 2014;7(6):2180–4. 10.3892/ol.2014.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Budnik LT, Kloth S, Baur X, et al. : Circulating mitochondrial DNA as biomarker linking environmental chemical exposure to early preclinical lesions elevation of mtDNA in human serum after exposure to carcinogenic halo-alkane-based pesticides. PLoS One. 2013;8(5):e64413. 10.1371/journal.pone.0064413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Lu H, Busch J, Jung M, et al. : Diagnostic and prognostic potential of circulating cell-free genomic and mitochondrial DNA fragments in clear cell renal cell carcinoma patients. Clin Chim Acta. 2016;452:109–19. 10.1016/j.cca.2015.11.009 [DOI] [PubMed] [Google Scholar]
- 122. Cossarizza A, Pinti M, Nasi M, et al. : Increased plasma levels of extracellular mitochondrial DNA during HIV infection: a new role for mitochondrial damage-associated molecular patterns during inflammation. Mitochondrion. 2011;11(5):750–5. 10.1016/j.mito.2011.06.005 [DOI] [PubMed] [Google Scholar]
- 123. Dai Z, Cai W, Hu F, et al. : Plasma Mitochondrial DNA Levels as a Biomarker of Lipodystrophy Among HIV-infected Patients Treated with Highly Active Antiretroviral Therapy (HAART). Curr Mol Med. 2015;15(10):975–9. 10.2174/1566524016666151123114401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Lauring AS, Lee TH, Martin JN, et al. : Lack of evidence for mtDNA as a biomarker of innate immune activation in HIV infection. PLoS One. 2012;7(11):e50486. 10.1371/journal.pone.0050486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Pinti M, Cevenini E, Nasi M, et al. : Circulating mitochondrial DNA increases with age and is a familiar trait: Implications for "inflamm-aging". Eur J Immunol. 2014;44(5):1552–62. 10.1002/eji.201343921 [DOI] [PubMed] [Google Scholar]
- 126. Verschoor CP, Loukov D, Naidoo A, et al. : Circulating TNF and mitochondrial DNA are major determinants of neutrophil phenotype in the advanced-age, frail elderly. Mol Immunol. 2015;65(1):148–56. 10.1016/j.molimm.2015.01.015 [DOI] [PubMed] [Google Scholar]
- 127. Jylhävä J, Nevalainen T, Marttila S, et al. : Characterization of the role of distinct plasma cell-free DNA species in age-associated inflammation and frailty. Aging Cell. 2013;12(3):388–97. 10.1111/acel.12058 [DOI] [PubMed] [Google Scholar]
- 128. Shockett PE, Khanal J, Sitaula A, et al. : Plasma cell-free mitochondrial DNA declines in response to prolonged moderate aerobic exercise. Physiol Rep. 2016;4(1): pii: e12672. 10.14814/phy2.12672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Nasi M, Cristani A, Pinti M, et al. : Decreased Circulating mtDNA Levels in Professional Male Volleyball Players. Int J Sports Physiol Perform. 2016;11(1):116–21. 10.1123/ijspp.2014-0461 [DOI] [PubMed] [Google Scholar]
- 130. Chou CC, Fang HY, Chen YL, et al. : Plasma nuclear DNA and mitochondrial DNA as prognostic markers in corrosive injury patients. Dig Surg. 2008;25(4):300–4. 10.1159/000152846 [DOI] [PubMed] [Google Scholar]
- 131. Sabatino L, Botto N, Borghini A, et al. : Development of a new multiplex quantitative real-time PCR assay for the detection of the mtDNA 4977 deletion in coronary artery disease patients: a link with telomere shortening. Environ Mol Mutagen. 2013;54(5):299–307. 10.1002/em.21783 [DOI] [PubMed] [Google Scholar]
- 132. Zhang B, Angelidou A, Alysandratos KD, et al. : Mitochondrial DNA and anti-mitochondrial antibodies in serum of autistic children. J Neuroinflammation. 2010;7:80. 10.1186/1742-2094-7-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Cao H, Ye H, Sun Z, et al. : Circulatory mitochondrial DNA is a pro-inflammatory agent in maintenance hemodialysis patients. PLoS One. 2014;9(12):e113179. 10.1371/journal.pone.0113179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Zhang Y, Zhao Y, Wen S, et al. : Associations of mitochondrial haplogroups and mitochondrial DNA copy numbers with end-stage renal disease in a Han population. Mitochondrial DNA A DNA Mapp Seq Anal. 2016:1–7. 10.1080/24701394.2016.1177038 [DOI] [PubMed] [Google Scholar]
- 135. Stertz L, Fries GR, Rosa AR, et al. : Damage-associated molecular patterns and immune activation in bipolar disorder. Acta Psychiatr Scand. 2015;132(3):211–7. 10.1111/acps.12417 [DOI] [PubMed] [Google Scholar]
- 136. Borghini A, Mercuri A, Turchi S, et al. : Increased circulating cell-free DNA levels and mtDNA fragments in interventional cardiologists occupationally exposed to low levels of ionizing radiation. Environ Mol Mutagen. 2015;56(3):293–300. 10.1002/em.21917 [DOI] [PubMed] [Google Scholar]
- 137. Dantham S, Srivastava AK, Gulati S, et al. : Plasma circulating cell-free mitochondrial DNA in the assessment of Friedreich’s ataxia. J Neurol Sci. 2016;365:82–8. 10.1016/j.jns.2016.04.016 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 138. Yasui K, Matsuyama N, Kuroishi A, et al. : Mitochondrial damage-associated molecular patterns as potential proinflammatory mediators in post-platelet transfusion adverse effects. Transfusion. 2016;56(5):1201–12. 10.1111/trf.13535 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation