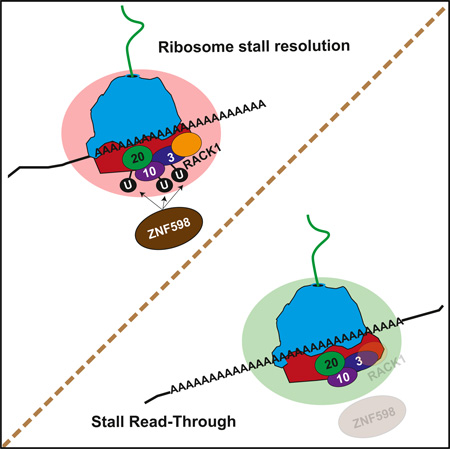
SUMMARY
Ribosomes that experience terminal stalls during translation are resolved by ribosome-associated quality control (QC) pathways that oversee mRNA and nascent chain destruction and recycle ribosomal subunits. The proximal factors that sense stalled ribosomes and initiate mammalian ribosome-associated QC events remain undefined. We demonstrate that the ZNF598 ubiquitin ligase and the 40S ribosomal protein RACK1 help to resolve poly(A)-induced stalled ribosomes. They accomplish this by regulating distinct and overlapping regulatory 40S ribosomal ubiquitylation events. ZNF598 primarily mediates regulatory ubiquitylation of RPS10 and RPS20, whereas RACK1 regulates RPS2, RPS3, and RPS20 ubiquitylation. Gain or loss of ZNF598 function or mutations that block RPS10 or RPS20 ubiquitylation result in defective resolution of stalled ribosomes and subsequent readthrough of poly(A)-containing stall sequences. Together, our results indicate that ZNF598, RACK1, and 40S regulatory ubiquitylation plays a pivotal role in mammalian ribosome-associated QC pathways.
Graphical Abstract
INTRODUCTION
The ribosome has emerged as a quality control (QC) hub that facilitates removal of both defective mRNAs and nascent chains that arise from errors in transcription, mRNA processing, or translation (Brandman and Hegde, 2016; Lykke-Andersen and Bennett, 2014; Rodrigo-Brenni and Hegde, 2012; Shoemaker and Green, 2012). Various impediments encoded in mRNA sequences or nascent chains can result in terminally stalled elongating ribosomes that require ribosome-associated QC factors for effective stall resolution and recycling of ribosomal subunits (Bengtson and Joazeiro, 2010; Brandman et al., 2012; Doma and Parker, 2006; Ito-Harashima et al., 2007; Kuroha et al., 2010; Letzring et al., 2010). How terminally stalled ribosomes are initially sensed and the upstream events that facilitate recruitment of a ribosome-associated QC complex (RQC) and RNA decay factors remain uncharacterized.
Genetic screens in S. cerevisiae identified the 40S ribosomal protein Asc1 (RACK1 in mammals) and the ubiquitin ligase Hel2 (ZNF598 in mammals) as factors required for turnover of model reporter proteins containing polybasic sequences (Brandman et al., 2012; Kuroha et al., 2010). Unlike the accumulation of truncated proteins arising from stall products observed upon genetic disruption of the RQC factor Ltn1, deletion of ASC1 or HEL2 resulted in readthrough of the stall-inducing sequence and the production of full-length reporter protein (Brandman et al., 2012; Kuroha et al., 2010). Similarly, loss of Asc1 results in read-through of CGA repeat stall sequences in yeast (Letzring et al., 2013). These observations suggest that Asc1 and Hel2 may be early-acting factors in ribosome-associated QC pathways. However, the precise role for Asc1 or Hel2 in regulating ribosome-associated QC is unclear.
Although ribosome-associated QC pathways are beginning to be well defined in S. cerevisiae, relatively little is known about the corresponding pathways in mammalian cells. We recently discovered that activation of the unfolded protein response and translation elongation inhibition stimulates the site-specific regulatory ubiquitylation of 40S ribosomal proteins (RRub) (Higgins et al., 2015). These findings suggested that a subset of RRub events may play a role in regulating ribosome-associated QC. Discovery of the cellular factors that catalyze or otherwise mediate individual RRub events is critical to understanding the mechanism by which RRub regulates ribosome-associated functions and overall cellular physiology.
Here we identify ZNF598 and RACK1 as two factors that regulate specific RRub events. We show that both loss of ZNF598 function and mutations within the 40S proteins RPS10 and RPS20, which block regulatory ubiquitylation, result in readthrough of stall sequences. We show that ribosome-associated RACK1 regulates a distinct but overlapping subset of RRub events compared with ZNF598. These results establish ZNF598, RACK1, and RRub as critical early-acting factors that regulate mammalian ribosome-associated QC pathways.
RESULTS
Establishment of a Mammalian Cell-Based Assay to Study Ribosome-Associated Quality Control
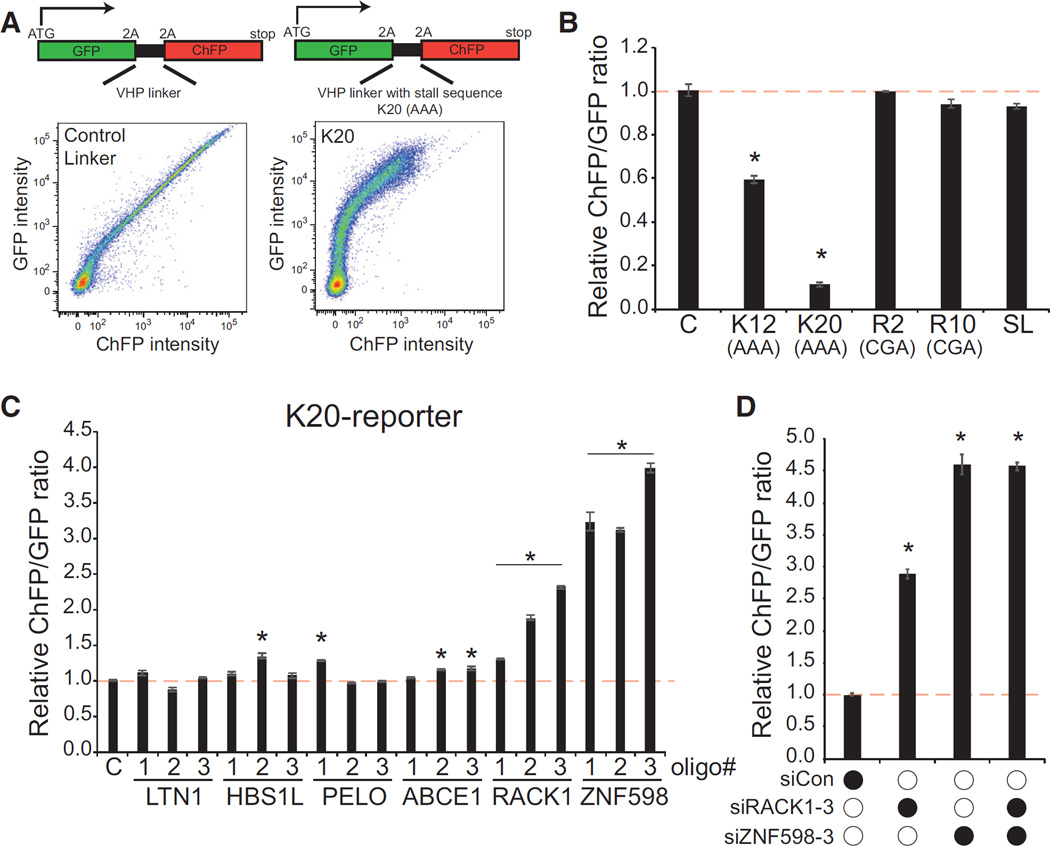
To define the factors and mechanisms that govern mammalian ribosome-associated QC pathways, we utilized a dual fluorescence reporter system in which GFP and cherry fluorescent protein (ChFP) are expressed in-frame from a single mRNA that is separated by a linker sequence encoding a FLAG-tagged, independently folding villin headpiece (VHP) that has been used previously to study ribosome-associated QC (Shao and Hegde, 2014; Shao et al., 2013; Figure 1A). Ribosomal 2A skipping sequences flank the linker region, allowing GFP and ChFP to be independent markers of translation prior to or after the linker region. Introduction of this reporter plasmid into mammalian cells resulted in a linear correlation between cellular GFP and ChFP fluorescence over four orders of magnitude (Figure 1A). Several sequence elements that have been shown previously to induce ribosome stalling and initiate ribosome-associated QC events in S. cerevisiae were introduced into the VHP linker region (Doma and Parker, 2006; Ito-Harashima et al., 2007; Kuroha et al., 2010; Letzring et al., 2010). Of the stall sequences examined, only a stretch of consecutive lysine codons (encoded by AAA) resulted in robust repression of downstream ChFP translation compared with GFP (Figure 1B). Interestingly, ChFP expression downstream of poly(A)-containing sequences was more prevalent in cells with higher overall expression levels (Figure 1A). Titrating the amount of transfected reporter plasmid revealed that ChFP expression was augmented to a greater extent compared with GFP expression when larger DNA amounts were transfected (Figure S1A). These observations suggest that some limiting factor needed to resolve stalled ribosomes becomes saturated at higher reporter expression levels. Our demonstration that the K20 sequence represses downstream translation is consistent with observations in S. cerevisiae where polybasic sequences result in ribosome stalling and destruction of the nascent chain (Kuroha et al., 2010). However, the mechanism for stalling appears to be distinct from S. cerevisiae because poly-arginine codons (CGA) failed to repress ChFP expression in mammalian cells. These observations suggest that sequence elements that nucleate ribosome stalling and QC events differ between yeast and mammalian cells. Despite this, the poly(A) sequence robustly and reproducibly represses downstream ChFP expression and can be used as a cell-based assay to examine mammalian ribosome-associated QC function.
Figure 1. Poly(A) Sequence-Induced Translational Repression Is Mediated by ZNF598 and RACK1.
(A) Schematic of dual fluorescence translation stall reporters. Plasmids expressing a reporter without a stall-inducing sequence or one containing 20 consecutive lysine codons (AAA, K20) in the linker region were transfected into 293T cells. The resulting cellular GFP and ChFP levels are depicted in the fluorescence-activated cell sorting (FACS) plots.
(B) Reporter plasmids containing either no stall sequence (C) or the indicated stall sequence were transfected into 293T cells and analyzed by flow cytometry. The relative ChFP:GFP ratio is depicted for each reporter. K12, 12 lysine codons (AAA); K20, 20 lysine codons (AAA); R2, two arginine codons (CGA); R10, ten arginine codons (CGA); SL, stem loop.
(C) 293T cells were transfected with three separate siRNA oligos targeting the indicated genes and subsequently transfected with the K20-containing dual fluorescence reporter. The relative ChFP:GFP ratio is depicted. C, control scrambled siRNA.
(D) 293T cells were transfected with control siRNA or siRNA targeting RACK1 or ZNF598 either individually or in combination. The resulting relative ChFP:GFP ratio from the K20 reporter plasmid is depicted. Error bars represent SEM for three separate transfections and flow cytometry measurements. *p < 0.01 using Student’s t test compared with control transfections. See also Figures S1 and S2.
Knockdown of RACK1 or ZNF598 Results in Readthrough of poly(A) Stall-Inducing Sequences
Having established a mammalian ribosome-associated QC assay, we set out to test whether human homologs of identified S. cerevisiae ribosome-associated QC factors play similar roles in mammalian cells. Knockdown of Ltn1 or the ribosome splitting factors Hbs1L, Pelota, or ABCE1 did not result in robust or consistent recovery of ChFP expression downstream of a poly(A) stall sequence compared with GFP (Figure 1C; Figure S1B). These results suggest that loss of Ltn1 or individual ribosome splitting factors does not robustly effect mRNA translation downstream of poly(A)-induced stalled ribosomes. Knockdown of RACK1 or ZNF598 resulted in a robust elevation of ChFP levels expressed downstream of the poly(A) sequence, suggesting that loss of RACK1 or ZNF598 function results in readthrough of stall-inducing mRNA sequences (Figure 1C). Knockdown of ZNF598 reproducibly elevated ChFP expression from the K20 reporter to a larger extent than observed after RACK1 knockdown, and combined knockdown of RACK1 and ZNF598 did not have an additive effect (Figures 1C and 1D). This result suggests that RACK1 and ZNF598 are acting within the same pathway to enable readthrough of stall-inducing sequences.
ZNF598 Functions within the Ribosome-Associated QC Pathway in a Manner Distinct from Ltn1 and NEMF
Elegant biochemical studies have demonstrated that Ltn1 and NEMF act subsequent to splitting of stalled 80S complexes to facilitate ubiquitylation of the trapped peptidyl-linked tRNA species (Shao and Hegde, 2014; Shao et al., 2013). To directly examine the protein products from poly(A)-mediated stalling events and to determine whether ZNF598 acts similarly to Ltn1 and NEMF, we visualized the central VHP translation product that contains the poly-lysine sequence by immunoblotting. Knockdown of either Ltn1 or NEMF resulted in the accumulation of stall translation products that are slightly smaller than the expected molecular weight of either the VHP-polyK product or the VHP-polyK-ChFP product that would result from a failed 2A-mediated skipping event (Figure S2A). Knockdown of ZNF598 resulted in the robust accumulation of full-length VHP-polyK and VHP-polyK-ChFP products, indicating that loss of ZNF598 allowed for readthrough of stall sequences that would normally result in the production of defective translation stall products (Figure S2A). These results are consistent with the hypothesis that ZNF598 acts prior to Ltn1, NEMF, and ribosome splitting during ribosome-associated QC events.
Although we consistently observed that loss of ZNF598 function increased ChFP expression from the K20 reporter, ChFP levels increased to only ~25% of the levels observed with a reporter lacking a stall-inducing sequence (Figure S1C). Previous studies have documented that poly(A) sequences can induce frameshifting events during translation (Arthur et al., 2015). Thus, it is possible that the dual fluorescence reporter may be underrepresenting the extent of readthrough upon loss of ZNF598 function because stall readthrough events in only one reading frame would result in ChFP production. To directly examine whether loss of ZNF598 results in stall readthrough in multiple frames, we mutated the second ribosomal 2A skipping sequence that separates the VHP-polyK translation product from ChFP to allow for more facile differentiation of frameshifted translation products from readthrough events in the correct frame. Knockdown of ZNF598 resulted in accumulation of both VHP-polyK-ChFP and smaller VHP-containing products that are the result of frameshifting events (Figure S2B). Quantification revealed nearly twice as much accumulation of the smaller products compared with the larger VHP-polyK-ChFP. These results indicate that loss of ZNF598 results in readthrough of poly(A)-induced stall events in multiple reading frames and that ChFP expression from our reporter underrepresents the extent of stall readthrough. Further, it appears that loss of ZNF598 allows for nearly complete readthrough of poly(A)-induced translation stalls.
Mutations in RPS20 or RPS10 that Block Regulatory Ubiquitylation Result in Defective Resolution of Stalled Ribosomes
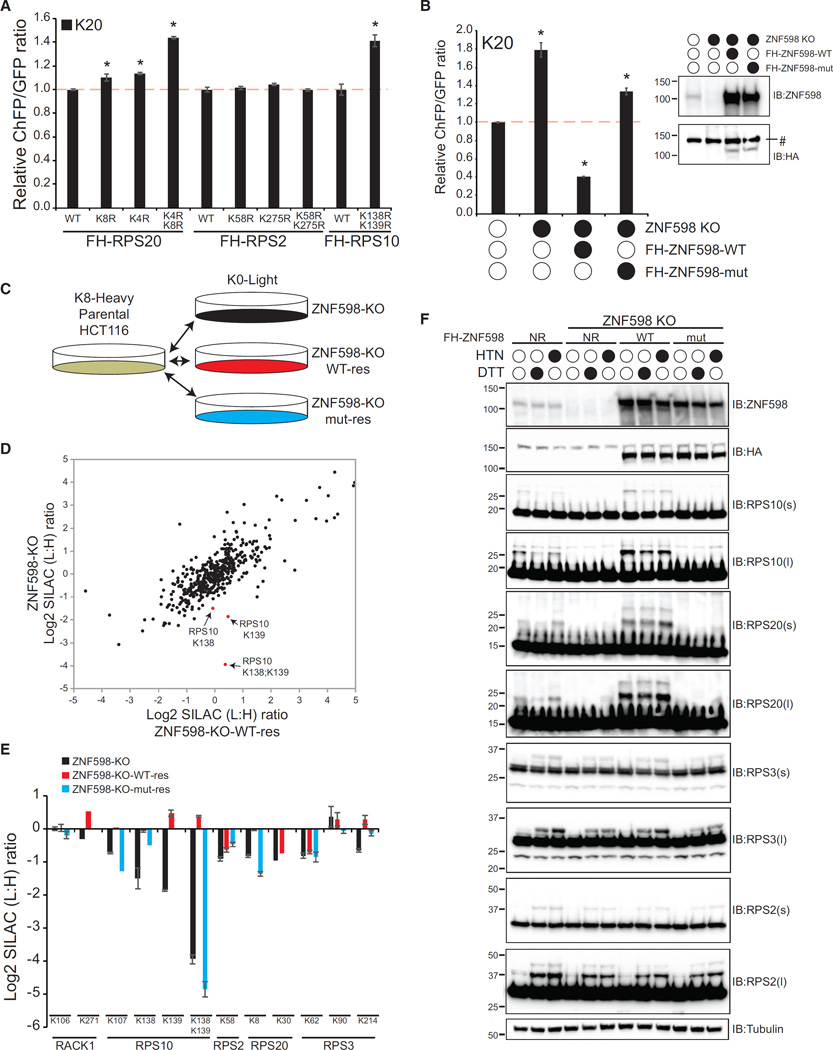
Examination of previous ubiquitin proteomics experiments (Higgins et al., 2015) identified specific lysine residues in RPS2, RPS3, RPS10, and RPS20 that became ubiquitylated upon treatment with the translation elongation inhibitor anisomycin (Figure S3A). Having validated our cell-based mammalian ribosome-associated QC assay, we tested whether some of the specific RRub events we observed to be stimulated by translation elongation inhibition were required for ribosome-associated QC. To test this hypothesis, we generated cell lines with stable expression of FLAG-hemagglutinin (HA)-tagged wild-type or mutant RPS2, RPS20, or RPS10 proteins. We converted lysine residues identified as ubiquitylated in response to translation elongation inhibition (RPS2-K58, RPS2-K275, RPS20-K4, RPS20-K8, RPS10-K138, and RPS10-K139) to arginine individually or in combination. Immunoblotting confirmed stable expression of the wild-type or mutant proteins, and, for RPS2 and RPS20, exogenous expression resulted in a corresponding decrease in endogenous protein production, resulting in more than 90% of cellular RPS2 or RPS20 being comprised of the exogenous protein (Figure S3B; Higgins et al., 2015). Although stable expression of RPS10 did not effectively replace endogenous RPS10, the exogenous RPS10 contributes ~50% of total cellular RPS10 (Figure S3B). We expressed our K20-containing reporter in the wild-type and RRub mutant cell lines and observed that cells expressing mutant RPS20 or RPS10, but not RPS2, displayed enhanced readthrough of the poly(A)-stall sequence (Figure 2A). This result supports a role for specific RRub events in mediating the proper resolution of stalled ribosomes.
Figure 2. Regulatory Ubiquitylation of RPS20 and RPS10 Is Mediated by ZNF598 and Facilitates Translation Stall Resolution.
(A) K20 reporter ChFP:GFP ratios in 293T cells with stable expression of FLAG-HA (FH)-tagged wild-type (WT) RPS20, RPS2, or RPS10 or versions containing the indicated lysine-to-arginine mutations. Error bars represent SEM for three separate transfections and flow cytometry measurements. *p < 0.01 using Student’s t test, comparing cells expressing mutant ribosomal proteins with cells expressing wild-type ribosomal proteins.
(B) K20 reporter ChFP:GFP ratios in parental HCT116 cells, ZNF598-KO cells, or ZNF598-KO cells with stable ectopic expression of FLAG-HA-tagged wild-type or C29A mut ZNF598. *p < 0.01 using Student’s t test, comparing parental cells with ZNF598-engineered cell lines. Inset: whole-cell extracts from parental HCT116 cells, ZNF598-KO cells, or ZNF598-KO cells with exogenous FLAG-HA-tagged wild-type or mutant ZNF598 expression were immunoblotted with the indicated antibodies. #, non-specific band.
(C) Schematic of SILAC-based proteomic experiments.
(D) Scatterplot of the Log2 SILAC ratio (L:H) for all quantified ubiquitin-modified peptides from ZNF598-KO cells and ZNF598-KO cells with exogenous wild-type ZNF598 expression (light) compared with unlabeled (heavy) parental HCT116 cells. Selected ubiquitin-modified peptides from RPS10 are shown in red.
(E) Log2 SILAC ratio (L:H) for selected ubiquitin-modified peptides from 40S ribosomal proteins in ZNF598-KO cells (black bars), ZNF598-KO cells with exogenous wild-type ZNF598 expression (red bars), or ZNF598-KO cells with exogenous mutant ZNF598 expression (blue bars) compared with parental cells. The site of ubiquitin modification is indicated. Error bars represent SEM from multiple peptide mass spectrometry (MS) quantifications.
(F) Parental HCT116 cells, ZNF598-KO cells, or ZNF598-KO cells rescued with exogenous wild-type or mutant ZNF598 expression were untreated or treated with HTN or DTT for 4 hr. Whole-cell extracts were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies. s and l denote short or long exposures, respectively.
See also Figure S3.
ZNF598 Mediates Regulatory Ubiquitylation of RPS10 and RPS20 during Ribosome-Associated QC Events
Because both knockdown of ZNF598 and expression of specific 40S ribosomal proteins containing RRub-blocking mutations resulted in readthrough of poly(A)-stall sequences, we reasoned that ZNF598 may be an RRub ubiquitin ligase. To test this idea, we generated somatic ZNF598 knockout (KO) HCT116 cells using clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 genome engineering (Ran et al., 2013). We validated the loss of ZNF598 protein expression in several isolated clones by immunoblotting and showed that ZNF598-KO cell lines display elevated ChFP expression relative to parental HCT116 cells upon introduction of the ribosome stall reporter (Figure 2B; Figure S3C). We selected one ZNF598-KO clone and rescued ZNF598 expression by exogenous stable expression of FLAG-HA-tagged wild-type or C29A mutant (mut) ZNF598. The C29A mutation occurs in the really interesting new gene (RING) domain of ZNF598, and we validated that this mutation blocks in vitro ZNF598 ubiquitylation activity (Figure S3D). Exogenous overexpression of wild-type ZNF598 in ZNF598-KO cells restored and even enhanced poly(A)-induced ribosome stall resolution, resulting in lower levels of ChFP expression compared with parental HCT116 cells (Figure 2B). This rescue of stall resolution was partially dependent on the ubiquitin ligase activity of ZNF598 (Figure 2B). These results validate our previous results using transient ZNF598 knockdown and establish the importance of ZNF598’s ubiquitin ligase activity for its ribosome-associated QC function.
We took an unbiased proteomics approach to identify critical ZNF598 substrates and to evaluate whether ZNF598 is an RRub ligase. We utilized our ZNF598-KO and rescue cells to perform quantitative proteomic profiling of alterations to both the total and ubiquitin-modified proteome upon loss or gain of ZNF598 function (Figure 2C; Gendron et al., 2016; Kim et al., 2011). We reasoned that ZNF598 substrates would show decreased ubiquitylation in ZNF598-KO and that the decrease would be reversed upon expression of wild-type but not mutant ZNF598. Cells without ZNF598 expression or those overexpressing ZNF598 displayed little to no alteration to the total proteome (Figure S3E). However, examination of the ubiquitin-modified proteome identified RPS10 ubiquitylation events at K138 and K139 as being dramatically altered upon loss of ZNF598 function (Figure 2D). The doubly ubiquitin-modified RPS10 peptide displayed the largest change upon loss or reintroduction of ZNF598 (Figure 2E; Table S1). Lysine 107 in RPS10 as well as K214 in RPS3 and K8 in RPS20 were also affected in the expected manner upon loss or reintroduction of wild-type or mutant ZNF598, although the magnitude of the effect was smaller than that observed for RPS10 K138 and K139 ubiquitylation (Figure 2E). This result indicates that ZNF598 catalyzes RRub events on RPS10 at K138 and K139 and, to a lesser extent, on RPS20 at K8 and on RPS3 at K214.
We validated our proteomic results by immunoblotting lysates from parental, ZNF598-KO, and rescue cell lines before and after treatment with the unfolded protein response (UPR) agonist DTT or the translation inhibitor harringtonine (HTN). We used DTT and HTN treatment to stimulate RPS2 and RPS3 regulatory ubiquitylation to better examine the site specificity of ZNF598 ubiquitin ligase activity. ZNF598-KO cells contain less ubiquitylated RPS10 and RPS20 in untreated cells, which confirms our findings using ubiquitin-specific proteomics (Figure 2F). Loss of ZNF598 did not prevent HTN or DTT-induced RPS2 or RPS3 ubiquitylation, although the extent of RPS3 ubiquitylation upon DTT and HTN treatment was reduced in ZNF598 KO cells (Figure 2F). RPS10 and RPS20 ubiquitylation increased robustly upon overexpression of wild-type but not mutant ZNF598, with a smaller effect on HTN- or DTT-induced RPS3 ubiquitylation and no effect on RPS2 ubiquitylation. Taken together, these results suggest that loss of ZNF598 results in an inability to catalyze ubiquitylation events on RPS10 and RPS20 that are needed for the proper resolution of stalled ribosomes.
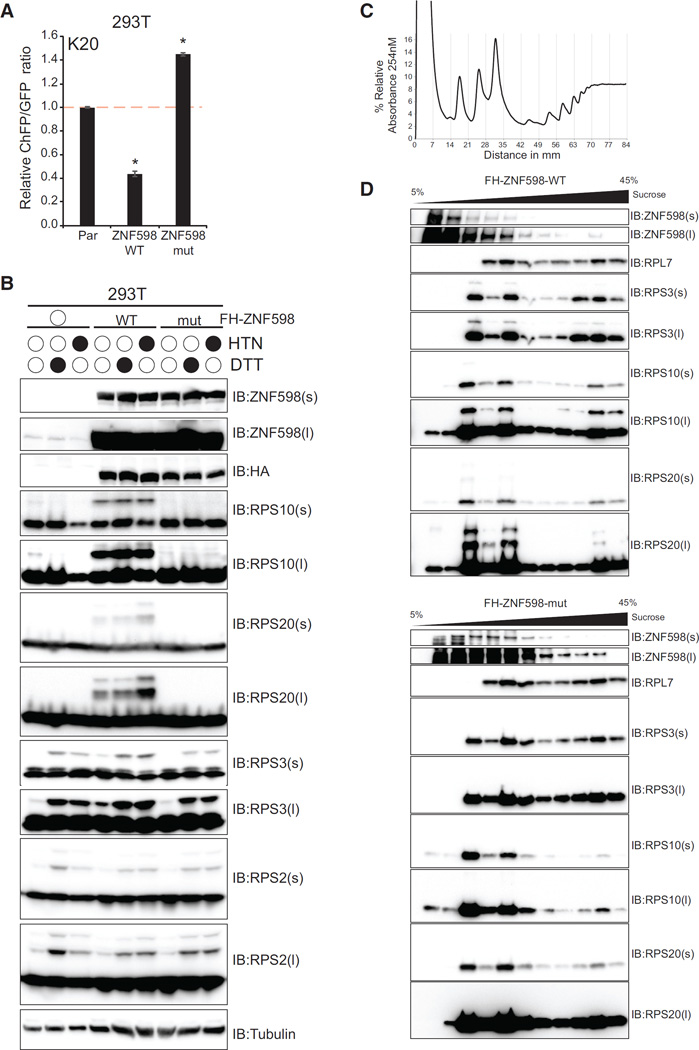
We generated ZNF598 overexpression cell lines in 293T cells with endogenous ZNF598 expression to further validate ZNF598 as a RPS10 and RPS20 RRub ligase and to examine whether overexpression of catalytically inactive ZNF598 acts in a dominant-negative fashion. Expression of wild-type ZNF598 resulted in reduced ChFP expression from the poly(A) reporter compared with parental 293T cells (Figure 3A). This effect was dependent on the ubiquitin ligase activity of ZNF598 because overexpression of mutant ZNF598 not only failed to reduce ChFP expression but also resulted in enhanced readthrough of poly(A)-induced stall events (Figure 3A). These results suggest that ZNF598 may be a limiting factor during ribosome-associated QC events and that catalytically inactive ZNF598 can act as a dominant negative. As observed in the ZNF598-KO HCT116 cells, overexpression of wild-type but not mutant ZNF598 in 293T cells strongly stimulated RPS10 and RPS20 ubiquitylation as well as HTN- and DTT-induced RPS3 ubiquitylation without altering RPS2 ubiquitylation (Figure 3B). Over-expression of mutant ZNF598 blocked RPS10 ubiquitylation in untreated cells without altering RPS2 or RPS3 steady-state ubiquitylation.
Figure 3. Overexpression of ZNF598 Enhances PolyA-Mediated Stall Resolution and RPS10, RPS20, and RPS3 Ubiquitylation in a Ligase-Dependent Manner.
(A) K20 reporter ChFP:GFP ratios in parental 293T cells or 293T cells with stable expression of wild-type or mutant ZNF598. *p < 0.01 using Student’s t test compared with parental 293T cells.
(B) Parental 293T cells or 293T cells with stable expression of wild-type or mutant ZNF598 were untreated or treated with HTN or DTT for 4 hr. Whole-cell extracts were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies.
(C) Native whole-cell lysates from 293T cells with stable expression of wild-type or mutant ZNF598 were separated on a linear 5%–45% sucrose density gradient. The percent relative absorbance at 254 nm is depicted as fractions collected, with vertical gray lines signifying fraction boundaries. 0-mm distance indicates the top of the tube.
(D) Fractions from the linear sucrose density gradient from cells overexpressing wild-type (top) or mutant (bottom) ZNF598 were immunoblotted with the indicated antibodies.
To investigate the ribosomal subpopulation in which ZNF598 catalyzes RRub events, we performed sucrose density gradient analysis on cell lysates from 293T cells overexpressing wild-type or mutant ZNF598. Both wild-type and mutant ZNF598 can be found in fractions throughout the gradient, suggesting that ZNF598 may associate with actively translating ribosomes and that this association is not dependent upon ZNF598 ligase activity (Figures 3C and 3D). Ubiquitylated RPS3, RPS20, and RPS10 can be readily observed in 40S-, 60S-, and polysome-containing fractions from cells expressing wild-type but not mutant ZNF598, which suggests that ZNF598 can catalyze RPS20, RPS10, and RPS3 ubiquitylation on ribosomes that are actively translating (Figures 3C and 3D). Overall, these results support the hypothesis that ZNF598 affects ribosome-associated QC function primarily through catalyzing regulatory ubiquitylation of RPS10 and RPS20. Our results also suggest that ZNF598 regulates RPS3 ubiquitylation in response to UPR activation and translation elongation inhibition, but whether RPS3 ubiquitylation regulates ribosome-associated QC function remains unknown because of our inability to stably express wild-type or mutant RPS3 to functionally relevant levels.
Ribosome-Associated RACK1 Assists in Ribosome-Associated QC and Facilitates Regulatory Ubiquitylation of RPS2 and RPS3
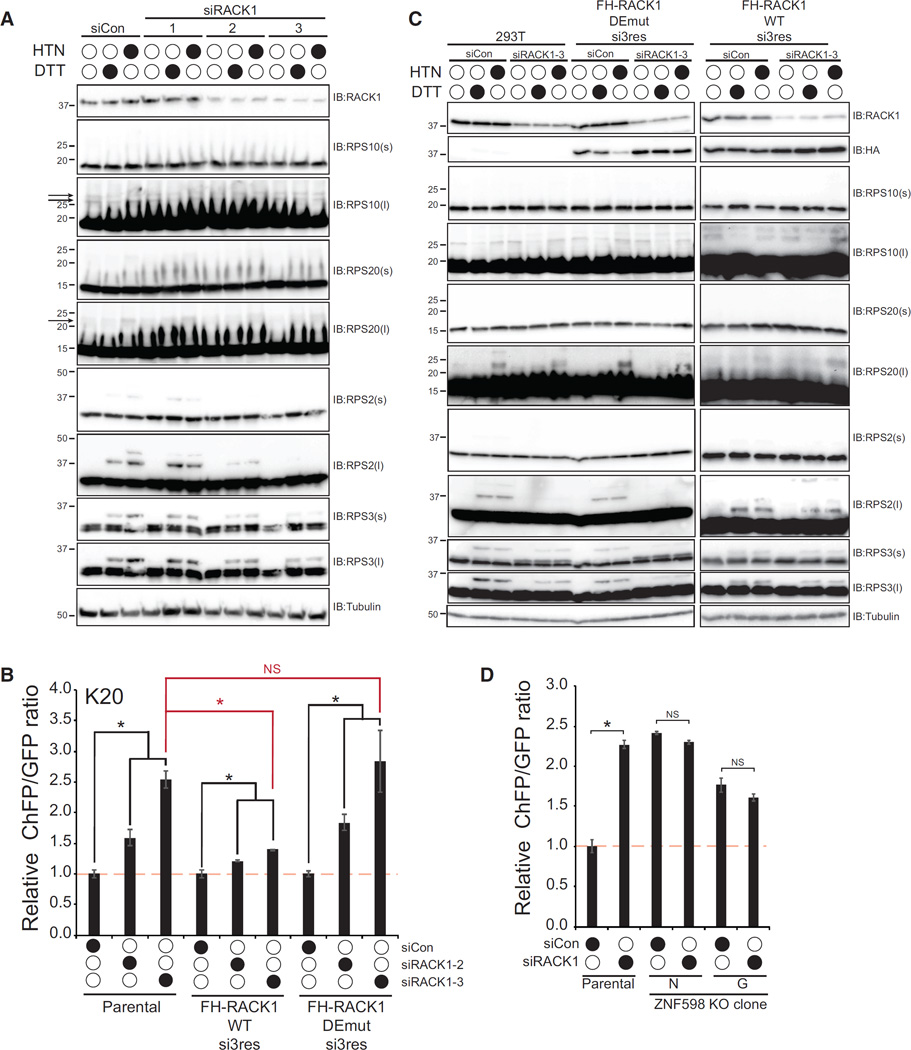
Having demonstrated that, like ZNF598, loss of RACK1 resulted in readthrough of a poly(A) stall-inducing sequence, we wanted to examine whether loss of RACK1 also affected similar ZNF598-mediated RRub events. Transient knockdown of RACK1 resulted in a near ablation of HTN- and DTT-stimulated RPS2 ubiquitylation and reduced HTN- and DTT-stimulated RPS3 ubiquitylation (Figure 4A). Steady-state RPS20 ubiquitylation was difficult to detect in 293T cells, but HTN-stimulated RPS20 ubiquitylation was reduced by RACK1 knockdown. Distinct to what was observed with ZNF598, loss of RACK had little to no effect on steady-state ubiquitylation of RPS10 (Figure 4A). This result revealed that the presence of RACK1 is required for a distinct subset of RRub events compared with ZNF598. RPS3 ubiquitylation in response to UPR activation or translation elongation inhibition is similarly repressed by loss of either ZNF598 or RACK1, whose contribution to ribosome-associated QC events is unknown.
Figure 4. Ribosome-Associated RACK1 Facilitates RRub and Translation Stall Resolution.
(A) 293T cells were transfected with control siRNA or three separate siRNA oligos targeting RACK1. Cells were then left untreated or treated with HTN or DTT for 4 hr. Whole-cell extracts were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies. Arrows indicate the position of ubiquitylated RP20 or RPS10.
(B) Parental 293T cells or cells expressing siRNA-resistant versions (resistant to siRNA #3) of FLAG-HA-tagged wild-type RACK1 or DEmut RACK1 (R36D;K38E) were transfected with control siRNA or two separate RACK1-targeting siRNAs. K20 reporter ChFP:GFP ratios are depicted. Error bars represent SEM from triplicate experiments. *p < 0.01 using Student’s t test, comparing control siRNA transfected cells with RACK1 knockdown cells. The red asterisk denotes the comparison between the parental 293T cell line transfected with si#3 targeting RACK1 and the cell lines expressing si-resistant wild-type or DEmut RACK1 transfected with si#3. NS, not significant.
(C) Parental 293T cells or cells expressing siRNA-resistant versions of wild-type RACK1 or DEmut RACK1 were transfected with control siRNA or siRNA#3 targeting the endogenous RACK1 and then left untreated or treated with HTN or DTT for 4 hr. Whole-cell extracts were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies.
(D) K20 reporter ChFP:GFP ratios from parental HCT116 cells or two separate ZNF598-KO clones (G,N) transfected with control siRNA or siRNA targeting RACK1 (oligo#3). Error bars represent SEM from triplicate experiments. *p < 0.01 using Student’s t test, comparing control siRNA-transfected cells with RACK1 knockdown cells.
See also Figure S4.
Because RACK1 has functions in cellular pathways separate from the ribosome, we wanted to interrogate whether ribosome association was necessary for RACK1 to participate in ribosome-associated QC events (Gandin et al., 2013). We first made mutations in RACK1 (R36D;K38E, DEmut) that block its association with the ribosome (Coyle et al., 2009; Sengupta et al., 2004). We combined these mutations with silent mutations to render exogenous RACK1 resistant to small interfering RNA (siRNA) knockdown (oligo #3) and generated stable expression cell lines in 293T cells. We confirmed that DEmut RACK1 failed to co-migrate with ribosomes in sucrose density gradients compared with wild-type RACK1 (Figure S4A). We then performed transient knockdown of RACK1 in parental 293T cells and our siRNA-resistant wild-type and DEmut RACK1 expression lines, followed by introduction of our stall reporter plasmid. Knockdown of RACK1 again resulted in poly(A) readthrough and elevated ChFP levels, which was rescued by ectopic expression of wild-type but not DEmut RACK1 (Figure 4B; Figure S4B). We used the same knockdown rescue approach to examine whether ribosome association of RACK1 was required for its effect on RPS2, RPS3, and RPS20 ubiquitylation. As observed previously, RACK1 knockdown in parental cells completely blocked HTN- and DTT-induced RPS2 ubiquitylation with a lesser but still strong reduction in RPS3 ubiquitylation (Figure 4C). HTN-induced RPS20 ubiquitylation was also reduced by RACK1 knockdown, although to a smaller magnitude than observed for RPS2 or RPS3 ubiquitylation. These effects on RPS2 and RPS3 ubiquitylation were rescued by the presence of wild-type but not DEmut RACK1 (Figure 4C). Taken together, these results demonstrate that ribosome-associated RACK1 primarily facilitates RPS2 and RPS3 ubiquitylation, with lesser effects on RPS20 ubiquitylation. Because loss of both RACK1 and ZNF598 did not have an additive effect on poly(A)-induced stall readthrough (Figure 1D), we wondered whether ZNF598 was required for RACK1 to participate in ribosome-associated QC events. Knockdown of RACK1 in parental HCT116 cells resulted in the expected increase in ChFP expression (Figure 4D). However, RACK1 knockdown in two separate ZNF598-KO cell lines did not result in a further increase in ChFP expression (Figure 4D; Figure S4C). This result indicates that loss of ZNF598 function eliminates the role for RACK1 in ribosome-associated QC.
Summing up our observations, we establish that ZNF598 and RACK1 participate in mammalian ribosome-associated QC. Further, we demonstrate that ZNF598 participates in regulatory ubiquitylation of RPS10 and RSP20 and that an inability to catalyze these RRub events results in defective resolution of stalled ribosomes. Regulatory ribosomal ubiquitylation now appears to be an early and critical ribosome-associated QC event.
DISCUSSION
Errors during mRNA transcription, processing, or decoding can lead to the production of defective translation products that require ubiquitin-mediated destruction (Brandman and Hegde, 2016; Lykke-Andersen and Bennett, 2014; Rodrigo-Brenni and Hegde, 2012). Indeed, it has been estimated that ~12%–15% of nascent chains are ubiquitylated and that nascent chains make up the majority of ubiquitylated proteins in mammalian cells (Kim et al., 2011; Wang et al., 2013). Together, these observations argue that targeting of defective nascent chains occupies a substantial fraction of ubiquitin-proteasome activity and that ribosome-associated QC pathways play a key role in regulating protein homeostasis. Much of the pioneering work on ribosome-associated QC function has been done in the yeast Saccharomyces cerevisiae. Whether similar or distinct pathways and mechanisms regulate mammalian ribosome-associated QC remains largely unknown. Here we demonstrate that human ZNF598 and RACK1 are key factors that regulate mammalian ribosome-associated QC through regulatory ubiquitylation of 40S ribosomal proteins.
Numerous studies have demonstrated that reporter mRNAs encoding consecutive polybasic amino acids are sufficient to elicit mRNA decay and destruction of the nascent chain (Bengtson and Joazeiro, 2010; Brandman et al., 2012; Dimitrova et al., 2009; Ito-Harashima et al., 2007; Kuroha et al., 2010). Upstream frameshifting of the polybasic encoding region resulted in production of stable mRNA and protein products, indicating that the polybasic region within the nascent chain was critical to trigger ribosome-associated QC activity in S. cerevisiae (Kuroha et al., 2010). Our results, as well as previous studies, suggest that long poly(A) sequences, and not polybasic amino acids, are the key feature that results in stalled ribosomes in mammalian cells (Arthur et al., 2015). Decoding poly(A) sequences results in ribosome stalling and frameshifting in vitro using E. coli ribosomes (Koutmou et al., 2015; Yan et al., 2015). Consistent with these observations, loss of ZNF598 function allows readthrough of poly(A)-stimulated stall events in multiple reading frames. Although it is clear that 60 consecutive adenosines can trigger ribosomal stalling that requires ZNF598 and RACK1 activity for proper resolution, the physiological relevance of such sequences is uncertain. Few human proteins contain consecutive poly-lysine tracts larger than seven within their coding region (Arthur et al., 2015). However, the usage of alternative mRNA cleavage and polyadenylation as well as errors in the processing of the more complex mammalian transcriptome likely contribute to an elevated requirement to sense poly(A)-induced ribosome stalls in higher eukaryotes (Tian and Manley, 2013). This reasoning may explain why the elements that trigger ribosome stalls during translation may differ between yeast and mammals.
Our results clearly demonstrate a role for RRub in regulating mammalian ribosome-associated QC events, but the exact mechanism governing that regulation remains unclear. One possibility is that RRub triggers the nucleation of downstream factors that prevent frameshifting and progression of ribosomes through stall-inducing sequences. ZN598 likely directly contributes to RPS10 and RPS20 ubiquitylation, making its role during ribosome stalling more clear. ZNF598 has also been shown to bind to a translation repression complex comprising eIF4E2 (4EHP) and GIGYF2, indicating that ZNF598 may participate in multiple levels of translational control (Morita et al., 2012). How RACK1 contributes to RRub events is less clear. It is possible that RACK1 contributes a scaffolding role to assist with structural remodeling of the mRNA contacts with the ribosome during stalling events (Coyle et al., 2009; Rabl et al., 2011). It appears that ZNF598 and RACK1 function prior to recruitment of the ribosome splitting factors and the Ltn1 RQC ubiquitin ligase (Figure 1C; Figure S2A). Recent work demonstrated that yeast Asc1 was required for endonucleolytic mRNA cleavage of a nonstop decay reporter in cells lacking Dom34 (Pelota) (Ikeuchi and Inada, 2016). Taken together, these results suggest that RACK1- and ZNF598-mediated RRub events may be required to recruit unidentified RNA endonucleases to stalled ribosomes to facilitate downstream ribosome splitting, mRNA decay, and nascent chain destruction. More precise biochemical and structural work is needed to fully examine the mechanism by which RRub regulates ribosome-associated QC.
STAR*METHODS
Detailed methods are provided in the online version of this paper and include the following:
CONTACT FOR REAGENT AND RESOURCE SHARING
-
METHOD DETAILS
Plasmids
Cell lines, siRNA and transfections
Dual fluorescence translation stall assay
SILAC-based quantitative ubiquitin proteomics
LC-MS-MS analysis
Peptide Identification and Quantification
Purification of ZNF598
In vitro ubiquitylation assays
Sucrose gradient separation
Immunoblotting
QUANTIFICATION AND STATISTICAL ANALYSIS
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-RPS10 | Abcam | Cat#ab151550 |
| Rabbit monoclonal anti-RPS20 | Abcam | Cat#ab133776 |
| Rabbit polyclonal anti-ZNF598 | Sigma-Aldrich | Cat#HPA041760; RRID:AB_10792490 |
| Rabbit polyclonal anti-RACK1 | Bethyl Laboratories | Cat# A302-545A; RRID:AB_1999012 |
| Rabbit polyclonal anti-RPS2 | Bethyl Laboratories | Cat# A303-794A; RRID:AB_11218192 |
| Rabbit polyclonal anti-RPS3 | Bethyl Laboratories | Cat# A303-840A; RRID:AB_2615588 |
| Mouse monoclonal anti-Ubiquitin | EMD Millipore/Chemicon | Cat# MAB1510; RRID:AB_2180556 |
| Mouse monoclonal anti-HA | Biolegend | Cat# MMS-101P; RRID:AB_2314672 |
| Mouse monoclonal anti-tubulin | Cell Signaling Technology | Cat# 3873S; RRID:AB_1904178 |
| Mouse monoclonal anti-FLAG | Sigma | Cat# F3165 RRID:AB_259529 |
| Rabbit polyclonal anti-RPL7 | Bethyl Laboratories | Cat# A300-741A RRID:AB_2301241 |
| Mouse monoclonal Anti-GFP | Roche | Cat#11814460001 RRID:AB_390913 |
| Anti-Rabbit IgG (H+L), HRP Conjugate antibody | Promega | Cat# W4011, RRID:AB_430833 |
| Anti-Mouse IgG (H+L), HRP Conjugate antibody |
Promega | Cat# W4021, RRID:AB_430834 |
| Lipofectamine RNAiMax | Thermo-Fisher | Cat# 13778030 |
| Clarity Western ECL Substrate | Biorad | Cat# 1705061 |
| Lipofectamine 2000 | Thermo fisher | Cat# 11668019 |
| Immun-Blot® PVDF Membrane | Biorad | Cat# 1620177 |
| Mirus TransIT 293 | Mirus Bio llc | Cat# MIR 2700 |
| Protease inhibitor cocktail tablet | Roche | Cat# 11836170001 |
| Pierce Protein A Plus UltraLink Resin | Thermo-Fisher | Cat# 53142 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 13C6, 15N2 L-Lysine-hydrochloride | Cambridge Isotopes Labratories | Cat# CNLM-291-H-0.05 |
| L-Arginine hydrochloride | Sigma Aldrich | Cat# A5131 |
| L-Lysine hydrochloride | Sigma Aldrich | Cat# 62929 |
| N-Ethylmaleimide | Sigma Aldrich | Cat# E3876-5G |
| 3X FLAG peptide | Sigma Aldrich | Cat# F4799 |
| anti-FLAG M2 agarose | Sigma Aldrich | Cat# A4596 |
| DL-Dithiothreitol | ACROS organics | Cat# 165680050 |
| Harringtonine | LKT labs | Cat# H0169 |
| Lys-C | Wako | Cat# 125-05061 |
| FBS | Mediatech (Corning) | N/A |
| Puromycin | Mediatech (Corning) | Cat# 61-385-RA |
| Trypsin | Sigma Aldrich | Cat# T1426 |
| Human recombinant ubiquitin | Boston Biochem | Cat# U-100H |
| Hexadimethrine bromide (Polybrene) | Sigma Aldrich | Cat# H9268 |
| Critical Commercial Assays | ||
| BCA Protein Assay | Thermo Scientific (Pierce) | Cat# 23225 |
| Deposited Data | ||
| Mass spectrometry | http://massive.ucsd.edu/ProteoSAFe/static/massive.jsp | MSV000080390 |
| Experimental Models: Cell Lines | ||
| 293T | ATCC | Cat# CRL-3216 |
| HCT116 | ATCC | Cat# CCL-247 |
| Recombinant DNA | ||
| pHAGE-FH-RPS10-WT | This paper | N/A |
| pHAGE-FH-RPS10-K138R,K139R | This paper | N/A |
| pHAGE-FH-RPS20-WT | This paper | N/A |
| pHAGE-FH-RPS20-K4R | This paper | N/A |
| pHAGE-FH-RPS20-K8R | This paper | N/A |
| pHAGE-FH-RPS20-K4R,K8R | This paper | N/A |
| pHAGE-FH-RPS2-WT | Higgins et.al. (2015) | N/A |
| pHAGE-FH-RPS2-K58R | Higgins et.al. (2015) | N/A |
| pHAGE-FH-RPS2-K275R | Higgins et.al. (2015) | N/A |
| pHAGE-FH-RPS2-K58R,K275R | Higgins et.al. (2015) | N/A |
| pHAGE-FH-ZNF598-WT | This paper | N/A |
| pHAGE-FH-ZNF598-mut (C29A) | This paper | N/A |
| pHAGE-FH-RACK1-WT | This paper | N/A |
| pHAGE-FH-RACK1-DEmut (R36D;K38E) (si3res) |
This paper | N/A |
| pCMV-GFP-2A–VHP-2A–CHFP (linker control) |
Juszkiewicz and Hegde (2017), this issue of Molecular Cell |
N/A |
| pCMV-GFP-2A–VHP-K20-2A–CHFP (K20 stall reporter) |
This paper | N/A |
| pSpCas9(BB)-2A–GFP | Addgene | Cat# 48138 |
| Sequence-Based Reagents | ||
| See Table S2 for siRNA reagents and primers | ||
| Software and Algorithms | ||
| FlowJo (v9.1) | BD biosciences |
STAR*METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Eric J Bennett (e1bennett@ucsd.edu)
METHOD DETAILS
Plasmids
All dual-fluorescence translation stall reporter plasmids were a generous gift from Manu Hegde and Susan Shao (MRC, Cambridge, UK). RPS2, RPS10, RPS20, RACK1, and ZNF598 coding regions were cloned into lentiviral-expression vectors using Gateway cloning (Invitrogen). Mutations were introduced using standard PCR-based strategies and were confirmed by sequencing.
Cell lines, siRNA and transfections
HEK293 and HCT116 cells were grown in DMEM (high glucose, pyruvate and L-Glutamine) containing 10% fetal bovine serum (FBS) in a humidified incubator maintained in a 5% CO2 atmosphere. For SILAC labeling, HCT116 cells were grown in DMEM medium free of lysine and arginine, supplemented with 10% dialyzed FBS, light lysine and arginine (light media) or heavy lysine (K8) (heavy media) (Cambridge Isotope laboratories) and light arginine at a concentration of 50 µg/mL and 85 µg/mL, respectively. Where indicated, cells were treated with 5mM DTT or 2 µg/mL Haringtonine for 4 hr before harvesting.
ZNF598 knockout cells were generated using CRISPR/Cas9 approaches (Ran et al., 2013). The guide sequence (listed in Table S2) was used to target the ZNF598 locus, and was cloned into the BbsI restriction sites in the pSpCas9(BB)-2A-GFP plasmid, which was used for transfecting HCT116. Transfected cells were single cell sorted, based on GFP fluorescence intensity into 96 well plates and screened for ZNF598 expression by immunoblotting. siRNA (listed in Table S2) transfection was performed using Lipofectamine RNAiMAX reagent according to manufacturer’s instructions. Stable cells lines expressing Flag-HA-tagged proteins were generated using lentiviral transduction and subsequent puromycin selection as follows. 293T cells were transfected with the following helper plasmids pHAGE - GAG-POL; pHAGE - VSVG; pHAGE - tat1b; pHAGE - rev and pHAGE (gene of interest (listed in key resources table)) using Mirus TransIT 293 transfection reagent. After 24 hr fresh media was added to the cells. The supernatant containing the infectious virions was filtered using a 0.45 mm sterile syringe filter and mixed with 2 µL of 6mg/ml polybrene for (1.5ml media). The mixture was added to cells seeded at 30% confluency and infected for 24hours, prior to selection with 1 µg/ml Puromycin to obtain stable expression clones.
Dual fluorescence translation stall assay
Dual fluorescence reporter plasmids were transfected into cells using Lipofectamine 2000 (Thermo Fisher) according to manufacturer guidelines. Two days after transfection cellular GFP and ChFP fluorescence was measured on a LSRFortessa X-20 (BD Biosciences). Subsequent analysis of FACS data was done using FlowJo (v9.1).
SILAC-based quantitative ubiquitin proteomics
HCT116 cells were grown in media containing either light (K0) lysine or 13C615N2-labeled (K8) lysine were processed for diGly peptide immuno purification and mass spectrometry as follows. A 1:1 mixture of heavy and light cells were lysed in 4 mL of denaturing lysis buffer (8 M urea, 75 mM NaCl, 50 mM Tris-Cl pH 8.2, Roche complete protease inhibitor, 1 mM NaF, 1 mM β-glycerophosphate, 1 mM sodium orthovanadate, 1 mM PMSF, 5 mM NEM). Lysates were sonicated twice for 5 s with 30 s rest on ice between cycles at a 10% power cycle before clarified by centrifuging at 21,000 g. The clarified supernatant was digested with Lys-C (Wako, Richmond, VA) for 4 hr at 37° Cat a final concentration of 10 ng/µl. Trypsin (Sigma) digestion at 37°C (1:100 enzyme substrate ratio) was performed after lysates were diluted to 2 M urea with 50 mM Tris-Cl pH 7.2 and was stopped by addition of Trifluoroacetic acid (TFA, Sigma) to a final concentration of 0.4%. The digested samples were centrifuged at 2500 × g for 15 min in a room temperature centrifuge to obtain clear supernatant and were desalted using C18 solid-phase extraction cartridges (Waters) the desalted peptides were flashfrozen and were dried in a lyophilizer. To enrich for diGLY-modified peptide, dried peptides were resuspended in 1.3 mL of 2 × IAP buffer (50 mM MOPS-NaOH pH 7.5, 10 mM Na2HPO4, 50 mM NaCl) and were incubated with α-diGly antibody (Cell Signaling Technologies) preconjugated to Protein-A (Thermo) beads. The incubation was performed for 2 hr at 4°C and the peptides with non-specific binding were removed by four washes with 1 mL IAP buffer with 10 min room temperature incubation between washes. Peptides were eluted with 5% formic acid and were desalted with in-house prepared C18 stage-tips and dried in a vacuum centrifuge. Samples were resuspended in 10 µL 5% formic acid, 5% ACN and transferred to autosampler vials.
LC-MS-MS analysis
All the samples were analyzed in triplicate by LC-MS/MS using a Q-Exactive mass spectrometer (Thermo Scientific, San Jose, CA) with the following conditions. A fused silica microcapillary column (100 µm ID, 20 cm) packed with C18 reverse-phase resin (XSELECT CSH 130 C18 2.5 µm, Waters Co., Wilford, MA) using an in-line nano-flow EASY-nLC 1000 UHPLC (Thermo Scientific) was used to resolve the peptides. Peptides were eluted over a 2 min 0%–5% ACN gradient, followed by a 158 min 5%–30% ACN gradient, a 15 min 30%–45% ACN gradient, a 1 min 45%–98% gradient, with a final 14 min isocratic step at 98% ACN for a total run time of 190 min at a flow rate of 250 nl/min. All gradient mobile phases contained 0.1% formic acid. MS/MS data were collected in a data-dependent fashion using a top 10 method with a full MS mass range from 300–1750 m/z, 70,000 resolution, and an AGC target of 3e6. MS2 scans were triggered when an ion intensity threshold of 1e5 was reached with a maximum injection time of 250 ms. Peptides were fragmented using a normalized collision energy setting of 22. A dynamic exclusion time of 40 s was used and the peptide match setting was disabled. Singly charged ions, charge states above 8 and unassigned charge states were excluded. All mass spectrometry data files are available through the MassIVE archive (http://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) ID: MSV000080390.
Peptide Identification and Quantification
The spectral analysis was performed as detailed in Gendron et al. (2016).
Purification of ZNF598
Wild-type and RING mutant (C29A) versions of Flag-HA-ZNF598 were purified from stable 293T cell lines. Cells were grown to 80% confluence, harvested in ice-cold phosphate-buffered saline, and stored at −80°C. Cell pellets were lysed in non-denaturing lysis buffer (1% (v/v) Triton X-100, 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 5 mM MgCl2, 50 mM NEM, 1 mM NaF, 1 mM sodium orthovanadate, 1 mM β-glycerophosphate, 10% (v/v) glycerol, EDTA-free protease inhibitor cocktail (Roche)) on ice. The subsequent steps were carried out at 4°C. The lysate was clarified at 21,000 × g for 15 min and applied to anti-FLAG M2 agarose (Sigma) with end-over-end rotation for 1 hr. The agarose was washed twice with lysis buffer and twice with wash buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 5 mM MgCl2, 10% (v/v) glycerol) with a duration of 15 min per wash step. Bound proteins were eluted in wash buffer supplemented with 0.1 mg/ml 3X FLAG peptide (Sigma) for 1 hr and stored at −80°C. The purity and concentration of the protein eluate were estimated visually by SDS-PAGE and silver staining (Invitrogen).
In vitro ubiquitylation assays
In vitro ubiquitylation reactions were carried out at room temperature for 1 hr. Each reaction (20 µl) contained 1 µM Uba1, 1 µM UbcH5c, 20 mM Tris-Cl pH 7.6, 150 mM NaCl, 2 mM ATP, 5 mM MgCl2, 60 µM human recombinant ubiquitin (Boston Biochem), 5 ng FLAG-HA-ZNF598, and 5 µg of human ribosomal protein complexes isolated by 35% sucrose cushion ultracentrifugation. Reactions were inactivated by the addition of 4 × Laemmli buffer and a 5 min incubation step at 95°C. Proteins were separated by 4%-15% SDS-PAGE and visualized by immunoblotting.
Sucrose gradient separation
Cells were collected using trypsin-EDTA (GIBCO) and lysed in Polysome Extraction Buffer (10 mM Tris pH 7.4, 0.5% Triton X-100, 15 mM MgCl2, 50 µg/mL cycloheximide, 0.3 M NaCl, 0.2 µg/mL Heparin). 3 mg of protein were run through 10%-50% linear sucrose gradients using a Beckmann Coulter SW 41 Ti rotor at 41,000 rpm, at 4°C for 2 hr. Gradients were formed using a Biocomp Gradient Master and fractions were collected using a Biocomp Gradient Station. Protein was precipitated from the solution using a final concentration of 20% Trichloroacetic acid (TCA). The resulting pellets were washed with 10% TCA, followed by 100% acetone, and dried using a Concentrator Plus (Eppendorf). Samples were resuspended in Laemmli sample buffer containing β-mercaptoethanol and boiled at 95°C for 5 min.
Immunoblotting
For Immunoblot analysis, cells were resuspended in denaturing lysis buffer (8 M urea, 50 mM Tris-Cl, pH 8.0, 75 mM NaCl, 1 mM NaF, 1 mM NaV, 1 mM β-glycerophosphate, 5 mM NEM) in the presence of EDTA-free Protease Inhibitor Cocktail (Roche Diagnostics, 5056489001). Cells/lysates were kept on ice during preparation. Lysis buffer was added to cells, and cells were then sonicated briefly (10 s at output of 3 W on a membrane dismembrator model 100 (Fisher Scientific) with a microtip probe and centrifuged for 10 min at 21,000 × g at 4°C. Supernatants were transferred to clean 1.5 mL tubes, and protein levels were determined by BCA Protein Assay (Thermo Scientific Pierce, 23225). Laemmli sample buffer with β-mercaptoethanol was then added to cell lysates. Cell lysates were heated at 95°C for 5 min, then cooled to room temperature and centrifuged briefly. Cell lysates were resolved by Tris-glycine SDS-polyacrylamide gel electrophoresis (SDS -PAGE). Proteins were transferred using Bjerrum semi-dry transfer buffer (48 mM Tris Base, 39 mM Glycine-free acid, 0.0375% SDS, 20% MeOH, pH 9.2) to PVDF membranes (Bio-Rad Immun-Blot, 1620177) using a semi-dry transfer apparatus (Bio-Rad Turbo Transfer) for 30 min at 25V. Immunoblots were developed using Clarity Western ECL Substrate (Bio-Rad, 1705061), and imaged on a Bio-Rad Chemi-DocX RS+ system. Blots images were processed using Imagelab (Biorad) software and were subsequently handled in Adobe Photoshop/Illustrator for preparing final images.
QUANTIFICATION AND STATISTICAL ANALYSIS
For all FACS-based reporter assays, siRNA transfections and reporter plasmid transfections were done in triplicate (N = 3). The mean and SEM for the CHFP/GFP ratio was calculated and compared to control siRNA knockdown or parental cell type and significance (p value) was determined using a two-tailed Student’s t test using Microsoft Excel.
Supplementary Material
In Brief.
Ribosomes stall on poly(A) tracts. Sundaramoorthy et. al identify regulatory ubiquitylation of RPS10 and RPS20 as a key regulator of poly(A)-induced ribosome stalling. ZNF598 catalyzes these ubiquitylation events and is needed for efficient stall resolution. Ribosome-associated RACK1 also facilitates stall resolution by mediating regulatory ubiquitylation of distinct but overlapping 40S proteins.
Highlights.
Poly(A) sequences stall ribosomes and repress downstream translation in mammals
Ribosome stall resolution requires regulatory ubiquitylation of RPS10 and RPS20
ZNF598 and RACK1 are needed for initiation of ribosome stall resolution in mammals
ZNF598 catalyzes regulatory ribosomal ubiquitylation of RPS20, RPS10, and RPS3
Acknowledgments
We thank R. Hegde and S. Shao (MRC) for providing the stall reporter plasmids and for insightful conversations; A. Goldrath, A.T. Phan, and L. Shaw (USCD) for providing guidance for all FACS-based experiments; and Gary Kleiger (UNLV) for purified Uba1 and UbcH5c enzymes. Brian Tsu (UCSD) provided the initial characterization of the stall reporter plasmids. We thank Joshua Gendron (Yale) for critical analysis of the manuscript. This work was supported by a New Scholar award (grant number AG-NS-0902-12) from the Ellison Medical Foundation (to E.J.B.), a Hellman Fellowship (to E.J.B.), and the NIH (DP2-GM119132 and PGM085764) (to E.J.B). E.S. is a fellow in the San Diego Center for Systems Biology (PGM085764).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes four figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2016.12.026.
AUTHOR CONTRIBUTIONS
Conceptualization, E.S. and E.J.B; Methodology, E.S. and E.J.B; Investigation, E.S., M.L., R.M., A.F., and J.L; Writing – Original Draft, E.S. and E.J.B.; Writing – Review & Editing, E.S., R.M., M.L., and E.J.B; Visualization, E.S., R.M., and E.J.B.; Funding Acquisition, E.J.B; Supervision, E.J.B.
REFERENCES
- Arthur L, Pavlovic-Djuranovic S, Smith-Koutmou K, Green R, Szczesny P, Djuranovic S. Translational control by lysine-encoding A-rich sequences. Sci. Adv. 2015;1:1. doi: 10.1126/sciadv.1500154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtson MH, Joazeiro CA. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature. 2010;467:470–473. doi: 10.1038/nature09371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, Hegde RS. Ribosome-associated protein quality control. Nat. Struct. Mol. Biol. 2016;23:7–15. doi: 10.1038/nsmb.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li GW, Zhou S, King D, Shen PS, Weibezahn J, et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012;151:1042–1054. doi: 10.1016/j.cell.2012.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle SM, Gilbert WV, Doudna JA. Direct link between RACK1 function and localization at the ribosome in vivo. Mol. Cell. Biol. 2009;29:1626–1634. doi: 10.1128/MCB.01718-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova LN, Kuroha K, Tatematsu T, Inada T. Nascent pep-tide-dependent translation arrest leads to Not4p–mediated protein degradation by the proteasome. J. Biol. Chem. 2009;284:10343–10352. doi: 10.1074/jbc.M808840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandin V, Senft D, Topisirovic I, Ronai ZA. RACK1 Function in Cell Motility and Protein Synthesis. Genes Cancer. 2013;4:369–377. doi: 10.1177/1947601913486348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron JM, Webb K, Yang B, Rising L, Zuzow N, Bennett EJ. Using the Ubiquitin-modified Proteome to Monitor Distinct and Spatially Restricted Protein Homeostasis Dysfunction. Mol. Cell. Proteomics. 2016;15:2576–2593. doi: 10.1074/mcp.M116.058420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins R, Gendron JM, Rising L, Mak R, Webb K, Kaiser SE, Zuzow N, Riviere P, Yang B, Fenech E, et al. The unfolded protein response triggers site-specific regulatory ubiquitylation of 40S ribosomal proteins. Mol. Cell. 2015;59:35–49. doi: 10.1016/j.molcel.2015.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi K, Inada T. Ribosome-associated Asc1/RACK1 is required for endonucleolytic cleavage induced by stalled ribosome at the 3′ end of nonstop mRNA. Sci. Rep. 2016;6:28234. doi: 10.1038/srep28234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito-Harashima S, Kuroha K, Tatematsu T, Inada T. Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev. 2007;21:519–524. doi: 10.1101/gad.1490207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszkiewicz S, Hegde RS. Initiation of quality control during poly(A) translation requires site-specific ribosome ubiquitination. Mol. Cell. 2017;65 doi: 10.1016/j.molcel.2016.11.039. Published online January 5, 2017. http://dx.doi.org/10.1016/j.molcel.2016.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutmou KS, Schuller AP, Brunelle JL, Radhakrishnan A, Djuranovic S, Green R. Ribosomes slide on lysine-encoding homopolymeric A stretches. eLife. 2015;4:4. doi: 10.7554/eLife.05534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroha K, Akamatsu M, Dimitrova L, Ito T, Kato Y, Shirahige K, Inada T. Receptor for activated C kinase 1 stimulates nascent poly-peptide-dependent translation arrest. EMBO Rep. 2010;11:956–961. doi: 10.1038/embor.2010.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letzring DP, Dean KM, Grayhack EJ. Control of translation efficiency in yeast by codon-anticodon interactions. RNA. 2010;16:2516–2528. doi: 10.1261/rna.2411710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letzring DP, Wolf AS, Brule CE, Grayhack EJ. Translation of CGA codon repeats in yeast involves quality control components and ribosomal protein L1. RNA. 2013;19:1208–1217. doi: 10.1261/rna.039446.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J, Bennett EJ. Protecting the proteome: Eukaryotic cotranslational quality control pathways. J. Cell Biol. 2014;204:467–476. doi: 10.1083/jcb.201311103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Ler LW, Fabian MR, Siddiqui N, Mullin M, Henderson VC, Alain T, Fonseca BD, Karashchuk G, Bennett CF, et al. A novel 4EHP-GIGYF2 translational repressor complex is essential for mammalian development. Mol. Cell. Biol. 2012;32:3585–3593. doi: 10.1128/MCB.00455-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo-Brenni MC, Hegde RS. Design principles of protein biosynthesis-coupled quality control. Dev. Cell. 2012;23:896–907. doi: 10.1016/j.devcel.2012.10.012. [DOI] [PubMed] [Google Scholar]
- Sengupta J, Nilsson J, Gursky R, Spahn CM, Nissen P, Frank J. Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM. Nat. Struct. Mol. Biol. 2004;11:957–962. doi: 10.1038/nsmb822. [DOI] [PubMed] [Google Scholar]
- Shao S, Hegde RS. Reconstitution of a minimal ribosome-associated ubiquitination pathway with purified factors. Mol. Cell. 2014;55:880–890. doi: 10.1016/j.molcel.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S, von der Malsburg K, Hegde RS. Listerin-dependent nascent protein ubiquitination relies on ribosome subunit dissociation. Mol. Cell. 2013;50:637–648. doi: 10.1016/j.molcel.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R. Translation drives mRNA quality control. Nat. Struct. Mol. Biol. 2012;19:594–601. doi: 10.1038/nsmb.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Manley JL. Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem. Sci. 2013;38:312–320. doi: 10.1016/j.tibs.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Durfee LA, Huibregtse JM. A cotranslational ubiquitination pathway for quality control of misfolded proteins. Mol. Cell. 2013;50:368–378. doi: 10.1016/j.molcel.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Wen JD, Bustamante C, Tinoco I., Jr Ribosome excursions during mRNA translocation mediate broad branching of frameshift pathways. Cell. 2015;160:870–881. doi: 10.1016/j.cell.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.