Abstract
We have proposed a computer-assisted framework for machine-learning–based delineation of gross tumor volumes (GTVs) following an optimum contour selection (OCS) method. The key idea of the proposed framework was to feed image features around GTV contours (determined based on the knowledge of radiation oncologists) into a machine-learning classifier during the training step, after which the classifier produces the ‘degree of GTV’ for each voxel in the testing step. Initial GTV regions were extracted using a support vector machine (SVM) that learned the image features inside and outside each tumor region (determined by radiation oncologists). The leave-one-out-by-patient test was employed for training and testing the steps of the proposed framework. The final GTV regions were determined using the OCS method that can be used to select a global optimum object contour based on multiple active delineations with a LSM around the GTV. The efficacy of the proposed framework was evaluated in 14 lung cancer cases [solid: 6, ground-glass opacity (GGO): 4, mixed GGO: 4] using the 3D Dice similarity coefficient (DSC), which denotes the degree of region similarity between the GTVs contoured by radiation oncologists and those determined using the proposed framework. The proposed framework achieved an average DSC of 0.777 for 14 cases, whereas the OCS-based framework produced an average DSC of 0.507. The average DSCs for GGO and mixed GGO were 0.763 and 0.701, respectively, obtained by the proposed framework. The proposed framework can be employed as a tool to assist radiation oncologists in delineating various GTV regions.
Keywords: gross tumor volume (GTV), planning computed tomography, 18F-fluorodeoxyglucose (FDG)-positron emission tomography (PET), machine learning, image segmentation
INTRODUCTION
The uncertainties of gross tumor volume (GTV) regions have a great impact on the precision of entire radiation treatment courses, including treatment planning and patient positioning [1]. Target volumes such as the clinical target volume (CTV) and the planning target volume (PTV) in treatment planning are determined based on the GTV region. In particular, the uncertainty of the GTV is critical in the setting of stereotactic body radiation therapy (SBRT), because precise positioning is required for delivering a higher dose per fraction to a tumor rather than to the surrounding normal tissue. In this study, we focused on computer-assisted frameworks for automated delineating the GTV.
The first reason for the necessity of computer-assisted delineation is the intra- and interobserver variability of GTV contours determined by radiation oncologists, which indicates low reproducibility [2, 3]. Chao et al. reported that the variation in delineating CTVs from scratch among radiation oncologists is significant and that using computer-assisted methods reduces volumetric variation and improves geometric consistency [3]. The second reason is that manual contouring is tedious and time-consuming. According to Chao's study [3], the average percentage of time saved for contouring using a computer-assisted method is 26–29% for experienced physicians and 38–47% for less experienced physicians.
A number of automated contouring approaches for determining the GTV have been proposed to reduce the inter- and intraobserver variability and planning time, and to increase the segmentation accuracy of the GTV [4–13]. Conventional methods can be categorized into two major types: positron emission tomography (PET)-based and PET/computed tomography (CT)-based approaches. PET-based approaches are based on model-based methods [4], thresholding of the standardized uptake value (SUV) [5], region-growing methods using the SUV [6], Gaussian mixture model–based segmentation [7], gradient-based segmentation methods [8], the fuzzy locally adaptive Bayesian approach [9], the fuzzy c-means algorithm [10], and a total lesion glycolysis (TLG) algorithm of PET Response Criteria in Solid Tumors (PERCIST) [11]. Kerhet et al. developed a machine-learning framework to assist in the threshold-based segmentation of non–small-cell lung cancer tumors in PET/CT imaging for use in treatment planning [12]. 18F-fluorodeoxyglucose (FDG) PET directly shows biological information on higher metabolic rates of radiolabeled glucose compared with that observed in normal tissues, a phenomenon associated with malignant neoplasms. El Naqa et al. developed a multimodality segmentation method using a multivalued level set method (LSM) that provides a feasible and accurate framework for combining imaging data obtained from various modalities (PET/CT) [13]. This method is a promising tool for delineating biophysical structure volumes for radiotherapy treatment planning. However, there are no studies regarding GTV segmentation approaches using pixel-based machine-learning techniques, which have the potential to learn the contours (delineated by radiation oncologists) for assessing GTV regions based on datasets of planning CT and PET/CT images, including biological as well as morphological information.
In this study, we attempted to feed PET biological and CT morphological information of GTV contours determined by radiation oncologists into a machine-learning classifier during the training step, after which the classifier produced the ‘degree of GTV’ for each voxel in the testing step. The aim of this study was to develop a computer-assisted framework for delineation of the GTVs of lung tumors using a machine-learning classifier, i.e. a support vector machine (SVM) (in this study) that can learn biological and morphological information (PET and CT image features) regarding various contours determined by radiation oncologists, and an optimum contour selection (OCS) method that was developed in the previous study [14].
MATERIALS AND METHODS
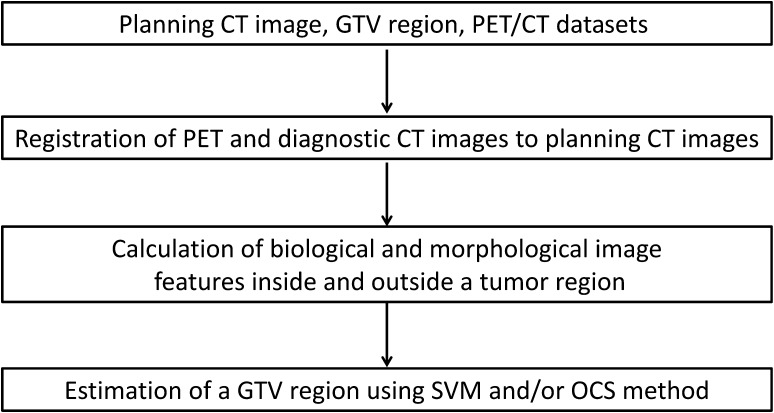
This study was performed with the approval of the institutional review board of our university hospital. Figure 1 shows the overall scheme of the proposed framework. The overall procedure of the proposed framework is described below.
The PET and diagnostic CT images were registered to the planning CT images based on the centroid of the lung regions in two CT images.
The biological and morphological image features were calculated from the planning CT, PET and diagnostic CT images, respectively.
The initial GTV regions were estimated using a SVM that learned the image features inside and outside each GTV region determined by radiation oncologists.
Final GTV regions were determined by thresholding the SVM outputs and/or the OCS method [14] that can be used to select a global optimum object contour based on multiple active delineations with a LSM around the GTVs.
Fig. 1.
An overall scheme of the proposed framework.
Clinical cases
Datasets consisting of planning CT and PET/CT images of 14 lung cancer patients (mean age: 76 years; range: 65–86 years; female: 7; male: 7; mean effective diameter of GTV: 20.4 mm; range: 13.8–29.4 mm) who had undergone SBRT were selected for this study. Table 1 shows a summary of the patient characteristics. A four-slice CT scanner (Mx 8000; Philips, Amsterdam, The Netherlands) was employed to acquire planning CT images with dimensions of 512 × 512 pixels, an in-plane pixel size of 0.977 mm and a slice thickness of 2 mm. The planning CT images with anisotropic voxels were transformed to images with matrix sizes of 512 × 512 × 266–378 and an isovoxel size of 0.977 mm, using a cubic interpolation method.
Table 1.
Summary of patient characteristics
| Case no. | Gender | Age (years) | GTV sizea (mm) | Tumor location | SUVmax b | Tumor type | Tumor CT imaging characteristics |
|---|---|---|---|---|---|---|---|
| 1 | Fc | 71 | 17.7 | RULe | 8.43 | Solid | Homogeneous Irregular |
| 2 | F | 67 | 24.2 | RUL | 12.2 | Solid | Homogeneous Irregular Vascular |
| 3 | Md | 65 | 25.3 | RUL | 6.79 | Solid | InhomogeneousIrregular |
| 4 | M | 75 | 20.2 | LULf | 8.74 | Solid | Inhomogeneous Irregular Adjacent Pleural |
| 5 | M | 86 | 29.4 | LUL | 9.68 | Solid | Cavity Irregular |
| 6 | F | 81 | 25.8 | RUL | 4.43 | Solid | Homogeneous Irregular Pleural Indentation |
| 7 | M | 76 | 17.8 | LUL | 1.73 | GGO | Irregular Pleural Indentation Vascular |
| 8 | F | 74 | 16.4 | RLLg | 1.29 | GGO | Regular |
| 9 | M | 81 | 18.5 | LUL | 2.56 | GGO | Regular |
| 10 | F | 79 | 21.2 | RUL | 1.45 | GGO | Irregular |
| 11 | M | 77 | 20.5 | LLLh | 6.5 | Mixed GGO | Inhomogeneous Irregular Cavity |
| 12 | F | 85 | 13.8 | RUL | 1.72 | Mixed GGO | Irregular |
| 13 | M | 65 | 18.3 | RUL | 1.29 | Mixed GGO | Regular Inhomogeneous |
| 14 | F | 84 | 16.3 | LLL | 1.39 | Mixed GGO | Irregular Pleural Indentation Vascular |
aEffective diameter.
bMaximum standardized uptake value.
cFemale.
dMale.
eRight upper lobe.
fLeft upper lobe.
gRight lower lobe.
hLeft lower lobe.
The 14 lung cancers were classified into the three types of lung tumor, which were solid (6), ground glass opacity (GGO) (4) and mixed GGO (4), as shown in Table 1. The lung window (window level and width were set at −600 and 1500 Hounsfield units [HU], respectively) and the mediastinal window (window level and width were set at 50 and 400 HU, respectively) were used to determine the tumor types on the planning CT image by a radiologist (H.Y.).
Each patient was scanned on an integrated PET/CT scanner (Discovery STE; General Electric Medical Systems, Milwaukee, WI, and Biograph mCT; SIEMENS, Berlin, Germany) with their arms down while free breathing 60 min after the FDG injection, because PET images could help treatment planners to delineate GTV regions with relevant biological information. The PET data were acquired in the 3D mode and reconstructed with correction for attenuation, scatter, decay, random and dead time using a 3D ordered subset–expectation maximization (OS-EM) algorithm (VUE Point Plus; GE Healthcare, and ultraHD; SIEMENS). The number of iterations, the number of subsets, and the full width at half maximum of a Gaussian filter were set as 2, 21 and 6 mm, respectively. The PET images consisted of 128 × 128 pixels with an in-plane pixel size of 5.47 mm and a slice thickness of 3.27 mm, or 256 × 256 pixels with an in-plane pixel size of 3.18 mm and a slice thickness of 3.0 mm. In addition, diagnostic CT images were acquired using a 16-slice CT scanner (512 × 512 matrix, in-plane pixel size of 0.977 mm and slice thickness of 3.27 or 3 mm) in the PET/CT system.
The PET and diagnostic CT images were converted into isotropic images with a voxel size of 0.977 mm by using a linear and cubic interpolation method, respectively. The matrix sizes with an isotropic voxel of 0.977 mm were 716 × 716 × 793–920 or 834 × 834 × 811–992 for PET images, and 512 × 512 × 811–992 for diagnostic CT images. The edge portions in the PET images were likely to be enhanced by a cubic interpolation method. To avoid this problem, the linear interpolation method was applied to the PET images. The isotropic PET images were larger than the isotropic diagnostic CT images, but the matrix size of both images should be the same size to obtain image features at the same voxels in both images. Therefore, the isotropic diagnostic CT images were put on the centers of the isotropic PET images, and then isotropic PET images were cropped by the same sizes as the isotropic diagnostic CT images.
The SUV was employed as one of the features of PET images to be input to the SVM. The SUV was calculated as the ratio of the radioactivity concentration of the tissue at one time-point to the injected dose of radioactivity concentration at that time-point divided by the body weight:
| (1) |
where C represents the radioactivity concentration in kBq/ml obtained from the pixel value in the PET image multiplied by a cross-calibration factor, D is the injected dose of 18FDG administered in MBq (decay corrected) and W is the body weight of the patient in kilograms.
Radiation treatment plans were approved by two experienced radiation oncologists using a commercially available radiation treatment planning (RTP) system (Eclipse version 6.5 and 8.1; Varian Medical Systems Inc., Palo Alto, USA). GTV contours were delineated based on a consensus between two experienced radiation oncologists by using the RTP system on the planning CT images with reference to fusion images of the PET and diagnostic CT images.
Registration of PET and diagnostic CT images to planning CT images
In order to utilize both the biological and morphological information of the PET, diagnostic CT and planning CT images in the same coordinate system, the PET and diagnostic CT images were registered to the planning CT images. Each patient body region was extracted from the planning CT and diagnostic CT images for the image registration [15].
A diagnostic CT image was aligned with a planning CT image using centroid matching of lung regions with bronchus. Lung regions with bronchus were extracted from a diagnostic CT and planning CT images. (The algorithm for extraction of the lung regions with bronchus is explained in Appendix 1.) The centroids of the lung regions in the two images were calculated. The diagnostic CT image was registered to the planning CT image using lung-region-centroid matching. Finally, the diagnostic CT and PET images were cropped by the matrix size of the planning CT image, so that the matrix sizes of the PET and diagnostic CT images in the PET/CT datasets agreed with those of the planning CT image, as described in the previous section.
After the lung-region-centroid matching, the displacement of a lung tumor was corrected by aligning an extracted tumor region in a PET image with a center of a volume of interest (VOI). A rectangular VOI that was slightly larger than a circumscribed parallelepiped of the tumor was determined on the planning CT image by manually selecting the minimum and maximum coordinates of the circumscribed parallelepiped and calculating three widths in x, y and z directions. The size of the VOI was determined by adding an isotropic margin of 10 mm to the circumscribed parallelepiped of the tumor. The tumor region was extracted by binarizing the PET image based on a threshold value of 80% of the maximum SUV (SUVmax) within the VOI. Finally, the centroid of the extracted tumor region was aligned with the center of the VOI.
Biological and morphological image features inside and outside the tumor regions
Voxel values and the magnitudes of image gradient vectors were obtained as the biological and morphological image features at each voxel on the planning CT, diagnostic CT and PET images of the PET/CT dataset. The six image features were given to the machine-learning system, i.e. the SVM in this study. The image gradient image was calculated from the following first-order polynomial within a 5 × 5 × 5 voxel region obtained by use of the least squares method [16]:
| (2) |
where x, y and z are the coordinates of one of the three types of images, f (x, y, z) is the first-order polynomial and a, b, c and d are constants. The gradient magnitude G was calculated by
| (3) |
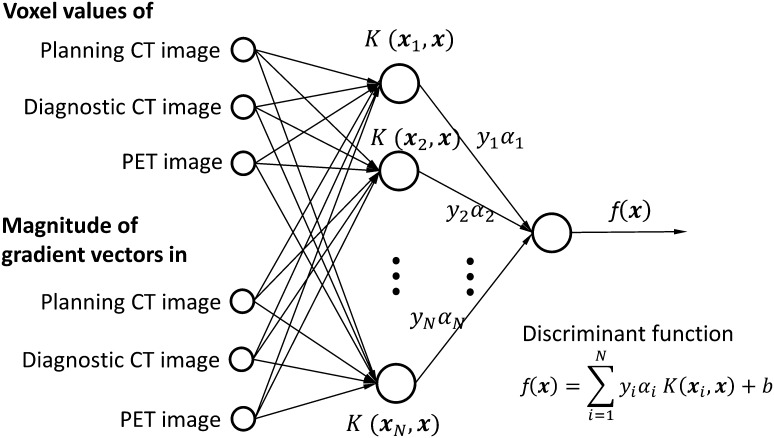
Support vector machine
The SVM [17] is one of the machine-learning techniques that can classify data into several (generally two) categories based on the output of a discriminant function. The SVM constructs a discriminant function in a linearly separable space by applying a non-linear kernel function to given training dataset. We consider a training dataset of training data and teacher signals, [, ] (), that we would like to classify. The discriminant function constructed by the SVM is expressed by [17]:
| (4) |
where ( number of support vectors) is the support vector, b and , are parameters that determines the discriminant function, is the non-linear kernel function, which can map a linearly non-separable dataset to a linearly separable dataset. The training procedure for construction of the SVM is shown as follows:
Step 1: A training dataset of image features and teacher signals [, ] is prepared, where (F: number of image features). In this study, F is 6 or 4, which depends on lung tumor type (see subsection ‘Extraction of initial GTV regions using SVM’).
Step 2: All parameters of the discriminant function are optimized by repeatedly calculating the parameters using image features of the training dataset based on a quadratic programming approach [17]. For this step, an open source software package SVMlight [18] was employed for this study.
Figure 2 illustrates the structure of an SVM constructed using six image features, which are explained in a subsection ‘Biological and morphological image features inside and outside the tumor regions’ of this report. The image voxels of unknown cases, which were not used for the training, were classified into inside GTV and outside GTV voxels, based on the output ( in Fig. 2) of the discriminant function. The output was referred to as ‘degree of GTV’ in this study.
Fig. 2.
An illustration of the structure of a support vector machine constructed using six image features. In this figure, indicates the support vector, and refers to the weight that determines the discriminant function .
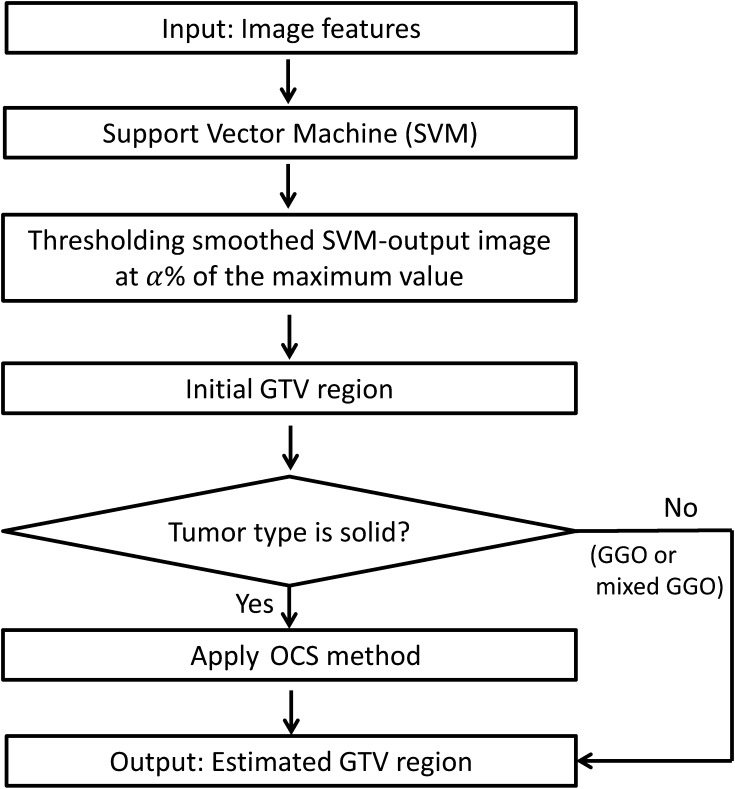
Extraction of initial GTV regions using SVM
The basic idea of this study was to feed image features around GTV contours determined based on the knowledge and experience of radiation oncologists into a machine-learning system during the training step, after which the classifier produced the ‘degree of GTV’ for each voxel in the testing step. The initial GTVs were extracted using a SVM that learned four or six image features inside and outside each true GTV region (ground truth). Six image features, i.e. the voxel value and the magnitude of the image gradient vector on the planning CT and PET/CT image datasets, were used for the solid type of lung tumor. Four image features, i.e. the voxel value and the magnitude of the image gradient vector on the planning CT and the diagnostic CT images, were used for GGO and mixed GGO types of lung tumor. The reason why the image feature sets were changed according to tumor types was that the proposed framework with the four image features provided better results than that with the six features, as mentioned in Discussion (Table 3).
Table 3.
Comparisons of DSCs for three types obtained by the proposed framework using four and six features with and without the OCS method
| Tumor type | Four featuresa | Six featuresb | ||
|---|---|---|---|---|
| Without OCSc method | With OCS method | Without OCS method | With OCS method | |
| Solid | 0.834 ± 0.034 | 0.822 ± 0.049 | 0.829 ± 0.024 | 0.836 ± 0.044 |
| GGO | 0.763 ± 0.043 | 0.674 ± 0.051 | 0.758 ± 0.043 | 0.636 ± 0.169 |
| Mixed GGO | 0.701 ± 0.145 | 0.553 ± 0.092 | 0.699 ± 0.145 | 0.591 ± 0.058 |
aThe voxel values and the magnitudes of the image gradient vector on the planning CT and the diagnostic CT images.
bThe voxel values and the magnitudes of the image gradient vector on the planning CT, PET and diagnostic CT images.
cOptimum contour selection.
The teacher signal was plus one if the voxel was inside the GTV region, and it was minus one if the voxel was outside the GTV region. The area outside the GTV region was defined as the region dilated six times by a circle kernel with a radius of 1.0 mm. The training voxels were selected at various sampling intervals, depending on the ratio of the numbers of inside and outside voxels, so that the number of inside voxels was the same as that of outside voxels. We constructed an SVM classifier with a Gaussian kernel, i.e. using the open source software package SVMlight [18]. In this study, the value γ and soft margin parameter C were set at 0.001 and 10 for solid, at 0.001 and 0.01 for GGO and at 0.00001 and 10 for mixed GGO, respectively, and the average number of support vectors was 37 070 for solid, 18 941 for GGO and 7 119 for mixed GGO. Finally, the initial GTV region was determined by successively applying the following post-processing to the rough GTV region obtained with the SVM.
Figure 3 depicts the post-processing technique applied for the original SVM-output images. First, the original SVM-output images were linearly converted into the voxel value of 0 to 2. Second, false-positive pixels outside the lung regions were removed by masking the SVM-output image with the corrected lung region. We assumed that the proposed framework could detect GTV regions that attach to chest wall even if the chest wall is included in the VOI. The algorithm for the correction of the lung region was explained in Appendix 2. Third, the SVM-output images were smoothed by applying a Gaussian filter with a standard deviation of 3 pixels. Finally, the initial GTV regions were obtained by thresholding the smoothed SVM-output images at an α value. In this study, the αvalues were 75% of the maximum smoothed SVM-output value (SVMmax) for solid type, 56% for GGO type and 25% for mixed GGO type. In the preliminary study, the suitable threshold percentages of the SVMmax for initial regions on the SVM-output images were determined for all cases by changing the percentage of SVMmax from 20 to 80%, so that the average Dice similarity coefficient (DSC), which was explained in a subsection ‘Evaluation of the proposed framework’, can achieve the maximum value.
Fig. 3.
A post-processing algorithm to apply for an original SVM-output image.
As for the solid type of lung tumor, the final GTV regions were estimated by applying the OCS method to the initial regions, which were determined on the SVM-output images, whereas the initial GTV regions for GGO and mixed GGO types of tumors were regarded as the estimated final GTV regions.
Estimation of final GTV regions using the OCS method
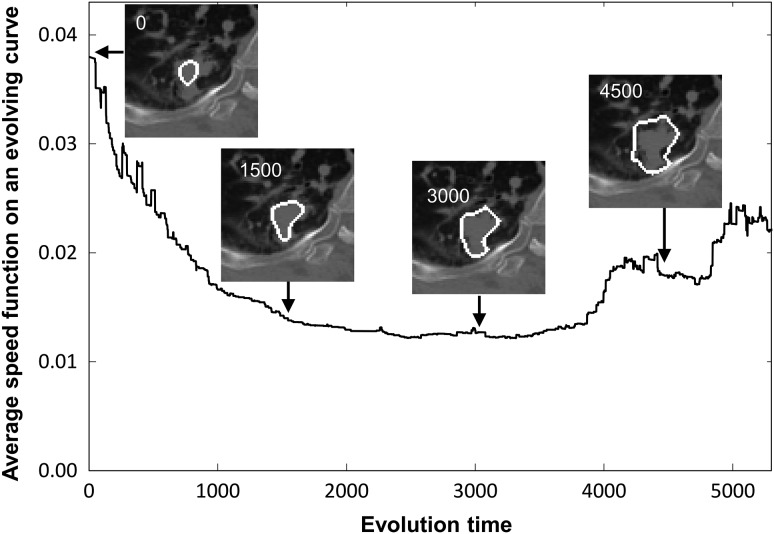
The final GTV regions for solid type tumors were estimated by applying the OCS method to initial regions determined on the SVM-output images. The basic concept of the OCS method is to retrospectively select a global optimal object contour from among multiple active delineations with a LSM around the tumor according to the relationship between the evolution time and the average speed function value on an evolving contour [14]. In the OCS method, the LSM [19] is employed to obtain object contours and the relationship mentioned above.
In the first step, the GTV contour and speed function value obtained with the LSM were recorded at each evolution time until the evolution time reached 10 000 or the evolving curve reached the edge of the VOI on the planning CT image. The level set function was iteratively updated from the initial GTV contour in accordance with the following discrete partial differential equation:
| (5) |
where n is the evolution number, t is the evolution time (iteration time), is the evolution time interval and is the speed function, which is inversely proportional to the gradient of the planning CT image to be processed. The speed function is relatively larger in homogeneous regions around the object, whereas the speed function becomes smaller when the evolving curve approaches the object edge.
In the second step, the optimal GTV contour was determined from among multiple active contours derived using the LSM by searching for the minimum point in the relationship between the average speed function value on an evolving curve and the evolution time. Figure 4 illustrates the relationship on the LSM between the evolution time and the average speed function on a moving contour.
Fig. 4.
Illustrations of the relationship on the LSM between the evolution time and the average speed function on a moving contour. The inserted images in this figure show the resulting contours on a lung tumor image at evolution times of 0, 1500, 3000 and 4500.
Evaluation of the proposed framework
The leave-one-out-by-patient cross-validation test was employed for training and testing of the SVM in this study [20]. In this test, the one case was left out for the testing, and then the parameters of a discriminant function in the SVM were determined based on the remaining cases, not including the left-out case. Finally, the left-out case was tested by using the discriminant function. This procedure was repeatedly performed for all cases. The performance of the proposed framework was evaluated using a 3D DSC [21]. The DSC denotes the degree of region similarity between the GTV ground truth region and the GTV region estimated using the proposed framework. The DSC was calculated according to the following equation:
| (6) |
where T is the GTV ground truth region determined by two radiation oncologists (Y.S., T.S.), D is the GTV region estimated using the proposed framework, n(T) is the number of pixels in the region T, n(D) is the number of pixels in the region D and n(T ∩ D) is the number of logical AND pixels between T and D. The DSC ranges from 0 (no overlap between T and D) to 1 (T and D are identical). The GTV regions were obtained from the Digital Imaging and Communications in Medicine (DICOM) for radiation therapy (DICOM-RT) files. The isotropic GTV regions were used as ground truths, which were produced using shape-based interpolation [22] to match with the isotropic planning CT images with an isovoxel size of 0.977 mm.
RESULTS
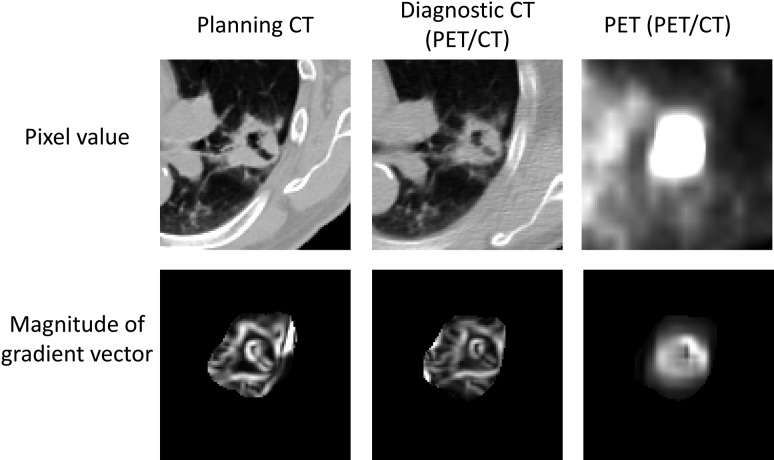
The proposed framework is referred to as the SVM-based framework because the SVM-output images were used as initial regions for the OCS method. Figure 5 shows the original images and gradient vector magnitude images obtained from the planning CT and PET/CT image datasets for a Case 5 (solid type) that were fed as image features to the SVM. Three kinds of images included the edge and surrounding information of the GTVs.
Fig. 5.
Original images and gradient vector magnitude images obtained from the planning CT and PET/CT image datasets that were fed as image features to the SVM.
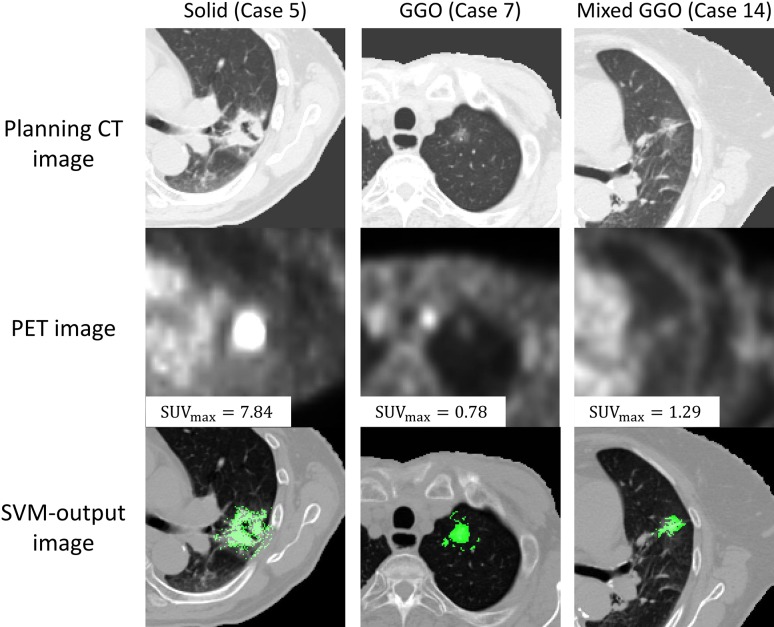
Figure 6 shows the planning CT, PET and SVM-output images for three types of lung tumors (Cases 5, 7, 14). A tumor of the solid type has high intensity with an SUVmax of 7.84, but the GGO and mixed GGO types of tumors show low intensities with an SUVmax of 0.78 and 1.29, respectively. On the other hand, the SVM enhanced not only the solid type of tumor, but also the GGO and mixed GGO types of tumors, in spite of low SUVmax.
Fig. 6.
Illustrations of the original planning CT, the PET and SVM-output images for solid, GGO and mixed GGO types of lung tumors.
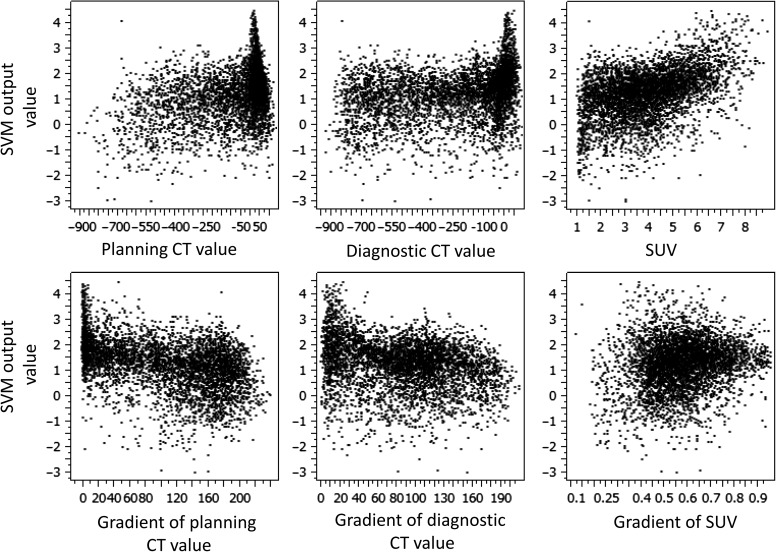
Figure 7 shows the relationship between the six image features and the SVM-output value. According to Fig. 7, the correlation between the SUV and SVM-output value was the largest, with 0.449 of the correlation coefficient.
Fig. 7.
Relationship between the six image features and SVM-output value. The gradient refers to the magnitude of a gradient vector.
Table 2 shows a comparison between the DSCs of the proposed framework (SVM-based framework) and the conventional framework (OCS-based framework) in the 14 cases. The conventional framework was the OCS-based framework developed in a previous study [14]. In this previous study, 80% of the SUV regions were employed as initial GTV regions for the OCS method. The average 3D DSC between the GTV ground truths contoured by the radiation oncologists and the GTV regions obtained using the proposed framework was 0.777 in the 14 cases, which was higher than the average DSC of 0.507 obtained using the conventional OCS-based framework.
Table 2.
Three-dimensional Dice similarity coefficients (DSCs) of the proposed framework (SVM-based framework) and conventional framework (OCSa-based framework) for 14 cases
| Case no. | Tumor type | OCSa-based framework | SVM-based frameworkb |
|---|---|---|---|
| 1 | Solid | 0.788 | 0.841 |
| 2 | Solid | 0.758 | 0.835 |
| 3 | Solid | 0.801 | 0.799 |
| 4 | Solid | 0.832 | 0.897 |
| 5 | Solid | 0.778 | 0.870 |
| 6 | Solid | 0.791 | 0.778 |
| 7 | GGO | 0.000 | 0.793 |
| 8 | GGO | 0.000 | 0.751 |
| 9 | GGO | 0.000 | 0.706 |
| 10 | GGO | 0.438 | 0.800 |
| 11 | Mixed GGO | 0.419 | 0.487 |
| 12 | Mixed GGO | 0.444 | 0.729 |
| 13 | Mixed GGO | 0.516 | 0.795 |
| 14 | Mixed GGO | 0.620 | 0.791 |
| Mean 1c | 0.645 | 0.784 | |
| Mean 2d | 0.507 | 0.777 |
aOptimum contour selection.
bSolid: SVM with six features using OCS method, and GGO and mixed GGO: SVM with four features.
cMean for 11 cases excluding 3 cases of Cases 7, 8 and 9, whose GTV regions were not segmented
dMean for 14 cases.
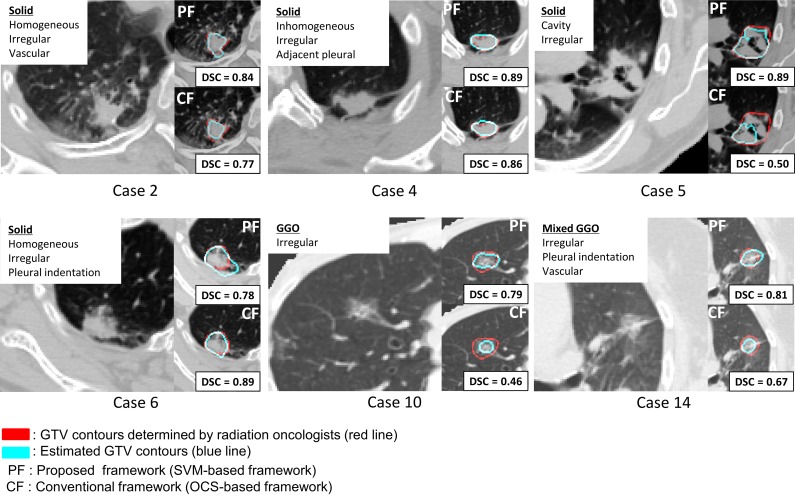
Figure 8 compares the segmentation results of the proposed (SVM-based) framework and the conventional (OCS-based) framework for six cases. For Case 2, the proposed framework provided the superior segmentation of a homogeneous and irregular tumor with vascularization to the conventional frameworks. For Case 4, the results of both the proposed framework and the conventional framework were comparable for an inhomogeneous and an irregular tumor connected to a chest wall. For Case 5, the GTV region was not segmented well by using the conventional framework at a global minimum point. Although brighter tumor regions on PET images indicate regions in which tumor cells may be active, SUV regions were not appropriate as initial regions for the OCS method. On the other hand, in the proposed framework, the initial regions were properly extracted by the SVM, which learned the ‘degree of GTV’ according to the knowledge of radiation oncologists. For Case 6, the DSC of the proposed framework was lower than that of the conventional framework, because the SVM extracted the tumor region as well as the blood vessels around the tumor. For Case 10, the proposed framework more correctly segmented even a GGO type of lung tumor than the conventional framework. For Case 14, the proposed framework produced a GTV region of the mixed GGO type of lung tumor that was irregular and connected to a chest wall.
Fig. 8.
A comparison between results of the proposed framework and the conventional framework in terms of tumor CT imaging characteristics.
DISCUSSION
An SUVmax smaller than 2.5 may indicate a lower malignancy of non–small cell lung cancer [23]. The GGO types of GTV regions were not able to be extracted by using the conventional OCS-based framework because the GGO types of lung tumors showed the SUVmax as <2.5. On the other hand, the proposed framework enabled us to extract the GGO-type GTV regions with low SUVmax.
Table 3 shows the comparisons of DSCs for the three types obtained by the proposed framework using four and six features with and without the OCS method. For solid types of tumors, the average DSC of 0.836 obtained using six features with the OCS method was larger than the average DSC of 0.829 obtained without the OCS method. For GGO types of tumors, the framework using four features without the OCS method reached the DSC of 0.763 (higher than the 0.674 obtained with the OCS method). For mixed GGO types of tumors, the framework using four features without the OCS method achieved the highest DSC of 0.701, but that with the OCS method produced a DSC of 0.553. In the proposed framework, therefore, six features and the OCS method were necessary for solid types, and four image features were applied for GGO types and mixed GGO types without using the OCS method. According to these results, we developed the post-processing algorithm in the proposed framework shown in Fig. 3.
There are two main types of errors in registration with respect to the lung tumor region, i.e. translation errors and shape variation errors. After the lung-region-centroid matching, the displacements of lung tumors were corrected to reduce the translation errors by aligning an extracted tumor region in a PET image with a center of a VOI. However, shape variation errors could remain residual errors and affect the segmentation accuracy. Therefore, deformable registration techniques should be applied to reduce the shape variation errors.
There have been several studies [13, 24, 25] on development of computerized segmentation methods for lung tumors, which have evaluated the segmentation accuracy using the DSC. El Naqa et al. (2007) developed a segmentation method based on a multivalued LSM using multimodalities, such as CT, PET and magnetic resonance (MR) imaging. The average DSC was 0.90 for a phantom study with CT/PET/MR images, although MRI was not applied for lung cancer patients. Wanet et al. (2011) validated a gradient-based segmentation method for performing GTV delineation on FDG-PET of non–small cell lung cancer patients using surgical specimens in comparison with threshold-based approaches and CT. The average DSC was 0.58 to 0.62 on the CT images and 0.62 to 0.68 on the PET images. Cui et al. (2012) developed an automated localization and segmentation method for assessing lung tumors based on PET/CT lung volumes using image feature analysis. They dealt with lung tumors adjacent to the chest wall or the mediastinum, but did not mention types of lung tumors. Their method reached an average DSC of 0.89 in 20 lung cancer patients.
Three limitations can be raised with respect to this study. The first limitation is false extraction of the blood vessels around the tumor. In Case 6 in Fig. 8, the blood vessels were extracted as false positives, because they had a similar CT value to the lung tumor due to the partial volume effect. The second limitation is that the proposed framework depends on the knowledge of radiation oncologists. Therefore, contour data based on some guidelines for delineation of GTV should be collected from experts in radiation oncology in Japan by organizing clinical trials. The third limitation is the number of cases that was used for this study. Fourteen patients with various types of tumor CT imaging characteristics were selected for this study; however, the proposed framework should be applied to various types of lung tumors in future work in order to improve the segmentation accuracy, particularly mixed GGO types of lung tumors. In addition, independent databases obtained from various institutions should be used to improve the robustness of the proposed framework.
We herein have proposed an automated framework for extracting GTVs using a machine-learning classifier that has learned from the knowledge of radiation oncologists using datasets for planning CT and FDG-PET/CT images. The proposed framework achieved an average DSC of 0.777 in 14 lung cancer patients, while the OCS-based framework was 0.507. The framework proposed in this study can therefore be employed as a tool to assist radiation oncologists in delineating tumor regions.
ACKNOWLEDGEMENTS
The authors are grateful to all members of the Arimura Laboratory (http://www.shs.kyushu-u.ac.jp/~arimura) for their valuable comments and helpful discussion.
APPENDIX 1: ALGORITHM FOR EXTRACTION OF THE LUNG REGIONS AND BRONCHUS
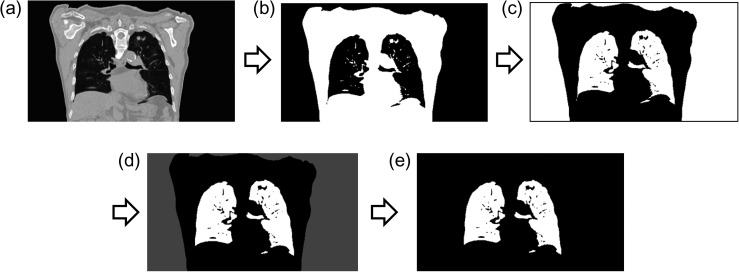
The algorithm for extraction of the lung regions and bronchus is elaborated in this Appendix 1. Figure A1 illustrates the extraction of the lung region and bronchus from the CT image. First, pixel values in an original image (Fig. A1a) were normalized from 0 to 1023. Second, the CT image was binarized by thresholding using an Otsu's method (Fig. A1b), and then pixel values were inverted (Fig. A1c). Third, the opening processing and labeling processing were performed (Fig. A1d), then the largest, the second largest, and the third largest regions in the labeled image were determined. Fourth, the ratio of the third largest to the second largest regions was calculated to identify the connection of right and left lung regions, one of which was connected with the bronchus region. In this study, the largest region in the labeled image was considered background. The second largest and the third largest regions were regarded as separated lung regions if the ratio was >0.26 and as a jointed lung region if the ratio was ≤0.26, which was determined by using 50 CT images. Finally, both lung and the bronchus regions were extracted (Fig. A1e).
Fig. A1.
Illustrations of (a) an original CT image, (b) a binary image obtained by using an Otsu's method after the normalization of pixel values, (c) an image inverted pixel values, (d) the labeled image after the opening processing and (e) an extracted lung region including the bronchi from CT image.
APPENDIX 2: CORRECTION OF LUNG REGIONS WITH CONCAVE REGIONS USING SELECTIVE DILATION FOR LUNG PLEURAL ADJACENT TUMORS
The correction of lung regions with concave regions using selective dilation for lung pleural adjacent tumors is elaborated in this Appendix 2. Concave regions on the planning CT images, which were caused by lung tumors adjacent to the lung pleura (i.e. lung pleural adjacent tumors), were filled by using a 3D selective dilation method proposed in this study (because extracted lung regions may not contain a pleural adjacent tumor region), which could result in a concave area in the lung region. First, a rectangular VOI that was slightly larger than a circumscribed parallelepiped of the tumor was determined on the planning CT image by manually selecting the minimum and maximum coordinates of the circumscribed parallelepiped and calculating its width in the x, y and z directions. Figure A2a shows an original CT image with a lung pleural adjacent tumor in the VOI (white line box). Figure A2b shows the lung region enlarged in the VOI. Second, the initial lung regions in the VOI were extracted by thresholding the CT image at a certain value that was empirically determined as a CT value of −300 on the gray level histogram of the same VOI image, as mentioned above. Figure A2c shows an initial lung region (white color region) with concave regions, in which the tumor region is not included. Therefore, the concave region was filled by using the selective dilation method in order to correct the lung region. In this method, binary dilation is repeatedly performed in a concave volume, which was identified using a selective dilation kernel until no concave areas exist in the lung region. Figure A2d shows the lung region (red color region) corrected using the selective dilation method.
Fig. A2.
Illustrations of (a) an original CT image with a lung pleural adjacent tumor in the VOI (white line box), (b) the lung region enlarged in the VOI, (c) an initial lung region (white color region) with concave regions in which the tumor region is not included and (d) the lung region (red color region) corrected according to the selective dilation method.
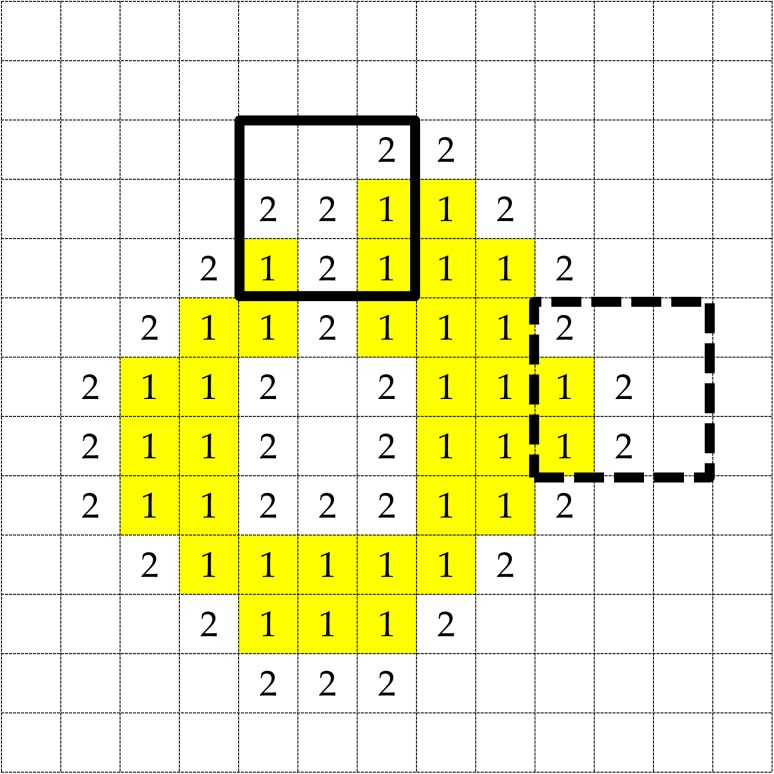
The algorithm of the selective dilation method is elaborated. Figure A3 shows an illustration of the definitions of lung region voxels and two types of background voxels (refer to Step 2). In addition, examples of concave and convex regions are shown in a selective dilation kernel of solid and dotted line boxes, respectively, as depicted in Fig. A3. The concave region was defined as the region, in which the number of objective pixels was larger than that of background pixels, as shown in a 3 × 3 solid line square. The convex region was defined as the region, in which the number of objective pixels was smaller than the number of background pixels, as shown in a 3 × 3 dotted line box.
Step 1: The initial lung region is segmented by thresholding a certain voxel value, with the lung region voxels set s 1.
Step 2: Background voxels nearest to the lung region detected using six neighborhood voxels are set s 2; the other background pixels are set s 0.
Step 3: If there are no nearest background voxels, the algorism stops. Otherwise, go to Step 4.
Step 4: If the selective dilation kernel includes a concave area, the center of the kernel is filled with a 1-value voxel as a lung voxel. Go to Step 2.
Fig. A3.
An illustration of the definitions of lung region voxels and two types of background voxels. Examples of concave and convex regions are shown in a selective dilation kernel of solid and dotted line boxes, respectively.
FUNDING
This research was partially supported by the JSPS KAKENHI Grant no. 26670301 (Grant-in-Aid for Challenging Exploratory Research).
CONFLICT OF INTEREST
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
REFERENCES
- 1.Weiss E, Hess CF. The impact of gross tumor volume (GTV) and clinical target volume (CTV) definition on the total accuracy in radiotherapy theoretical aspects and practical experiences. Strahlenther Onkol 2003;179:21–30. [DOI] [PubMed] [Google Scholar]
- 2.International Commission on Radiation Units & Measurements (ICRU) Prescribing, Recording and Reporting Photon Beam Therapy. Supplement to ICRU Report 50, ICRU Report 62 ICRU, Bethesda, MD, USA, 1999, 833–4.
- 3.Chao KS, Bhide S, Chen H, et al. Reduce in variation and improve efficiency of target volume delineation by a computer-assisted system using a deformable image registration approach. Int J Radiat Oncol Biol Phys 2007;68:1512–21. [DOI] [PubMed] [Google Scholar]
- 4.Rousson M, Khamene A, Diallo M, et al. Constrained surface evolutions for prostate and bladder segmentation in CT images. Lect Notes Comput Sci 2005;3765:251–60. [Google Scholar]
- 5.Zhang T, Tachiya Y, Sakaguchi Y, et al. Phantom study on three-dimensional target volume delineation by PET/CT-based auto-contouring. Fukuoka Igaku Zasshi 2010;101:238–46. [PubMed] [Google Scholar]
- 6.Day E, Betler J, Parda D, et al. A region growing method for tumor volume segmentation on PET images for rectal and anal cancer patients. Med Phys 2009;36:4349–58. [DOI] [PubMed] [Google Scholar]
- 7.Aristophanous M, Penney BC, Martel MK, et al. A Gaussian mixture model for definition of lung tumor volumes in positron emission tomography. Med Phys 2007;34:4223–35. [DOI] [PubMed] [Google Scholar]
- 8.Geets X, Lee JA, Bol A, et al. A gradient-based method for segmenting FDG-PET images: methodology and validation. Eur J Nucl Med Mol Imaging 2007;34:1427–38. [DOI] [PubMed] [Google Scholar]
- 9.Hatt M, Cheze Le Rest C, Albarghach N, et al. PET functional volume delineation: a robustness and repeatability study. Eur J Nucl Med Mol Imaging 2011;38:3663–72. [DOI] [PubMed] [Google Scholar]
- 10.Belhassen S, Zaidi H.. A novel fuzzy C-means algorithm for unsupervised heterogeneous tumor quantification in PET. Med Phys 2010;37:1309–24. [DOI] [PubMed] [Google Scholar]
- 11.Niyazi M, Landrock S, Elsner A, et al. Automated biological target volume delineation for radiotherapy treatment planning using FDG-PET/CT. Radiat Oncol 2013;8:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kerhet A, Small C, Quon H, et al. Application of machine learning methodology for PET-based definition of lung cancer. Curr Oncol 2010;17:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El Naqa I, Yang D, Apte A, et al. Concurrent multimodality image segmentation by active contours for radiotherapy treatment planning. Med Phys 2007;34:4738–49. [DOI] [PubMed] [Google Scholar]
- 14.Jin Z, Arimura H, Shioyama Y, et al. Computer-assisted delineation of lung tumor regions in treatment planning CT images with PET/CT image sets based on an optimum contour selection method. J Radiat Res 2014;55:1153–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arimura H, Itano W, Shioyama Y, et al. Computerized estimation of patient setup errors in portal images based on localized pelvic templates for prostate cancer radiotherapy. J Radiat Res 2012;53:961–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tokunaga C, Arimura H, Yoshiura T, et al. Automated measurement of three-dimensional cerebral cortical thickness in Alzheimer's patients using localized gradient vector trajectory in fuzzy membership maps. J Biomed Sci 2013;6:327. [Google Scholar]
- 17.Vapnik VN. The Nature of Statistical Learning Theory. 2nd edn. New York: Springer, 1999. [Google Scholar]
- 18.Joachims T. SVMlight. Version: 6.02. Cornell University, ; 2008. http://svmlight.joachims.org/ [Google Scholar]
- 19.Sethian JA. Level Set Methods and Fast Marching Methods: Evolving Interfaces in Computational Geometry, Fluid Mechanics, Computer Vision, and Materials Science. Cambridge: Cambridge University Press, 1999. [Google Scholar]
- 20.Arimura H, Magome T, Yamashita Y, et al. Computer-aided diagnosis systems for brain diseases in magnetic resonance images. Algorithms 2009;2:925–52. [Google Scholar]
- 21.Crum WR, Camara O, Hill DL.. Generalized overlap measures for evaluation and validation in medical image analysis. IEEE Trans Med Imaging 2006;25:1451–61. [DOI] [PubMed] [Google Scholar]
- 22.Herman GT, Zheng J, Bucholtz C.. Shape-based interpolation. IEEE Comput Graph Appl 1992;12:69–79. [Google Scholar]
- 23.Nakayama H, Okumura S, Daisaki H, et al. Value of integrated positron emission tomography revised using a phantom study to evaluate malignancy grade of lung adenocarcinoma: a multicenter study. Cancer 2010;116:3170. [DOI] [PubMed] [Google Scholar]
- 24.Wanet M, Lee JA, Weynand B, et al. Gradient-based delineation of the primary GTV on FDG-PET in non-small cell lung cancer: a comparison with threshold-based approaches, CT and surgical specimens. Radiother Oncol 2011;98:117–25. [DOI] [PubMed] [Google Scholar]
- 25.Cui H, Wang X, Feng D.. Automated localization and segmentation of lung tumor from PET-CT thorax volumes based on image feature analysi. Conf Proc IEEE Eng Med Biol Soc 2012:5384–7. [DOI] [PubMed] [Google Scholar]