Abstract
Cluster of differentiation 36 (CD36) variants influence fasting lipids and risk of metabolic syndrome, but their impact on postprandial lipids, an independent risk factor for cardiovascular disease, is unclear. We determined the effects of SNPs within a ∼410 kb region encompassing CD36 and its proximal and distal promoters on chylomicron (CM) remnants and LDL particles at fasting and at 3.5 and 6 h following a high-fat meal (Genetics of Lipid Lowering Drugs and Diet Network study, n = 1,117). Five promoter variants associated with CMs, four with delayed TG clearance and five with LDL particle number. To assess mechanisms underlying the associations, we queried expression quantitative trait loci, DNA methylation, and ChIP-seq datasets for adipose and heart tissues that function in postprandial lipid clearance. Several SNPs that associated with higher serum lipids correlated with lower adipose and heart CD36 mRNA and aligned to active motifs for PPARγ, a major CD36 regulator. The SNPs also associated with DNA methylation sites that related to reduced CD36 mRNA and higher serum lipids, but mixed-model analyses indicated that the SNPs and methylation independently influence CD36 mRNA. The findings support contributions of CD36 SNPs that reduce adipose and heart CD36 RNA expression to inter-individual variability of postprandial lipid metabolism and document changes in CD36 DNA methylation that influence both CD36 expression and lipids.
Keywords: dietary lipids, lipoproteins, dyslipidemia, genetics, cholesterol/metabolism, cluster of differentiation 36, low density lipoprotein, single nucleotide polymorphism, deoxyribonucleic acid
Elevated fasting lipid levels have traditionally been associated with increased risk of heart disease, diabetes, and stroke. More recently, similar and independent associations were found with levels of nonfasting or postprandial lipids (1, 2). Nonfasting measurements incorporate those of chylomicrons (CMs) produced in response to dietary fat ingestion. The TGs in CM particles are hydrolyzed by LPL in adipose, skeletal muscle, and heart vascular beds to deliver FFAs for cellular uptake facilitated by membrane cluster of differentiation 36 (CD36) (3). These tissues quantitatively contribute to CM clearance in humans (4–6) and express high levels of CD36 at the level of both endothelial and parenchymal cells (7, 8). Elevated concentrations of postprandial CM remnants after an oral fat load were reported in four subjects with complete CD36 deficiency (9), suggesting that low CD36 might impair CM clearance and result in sustained high levels of circulating CM remnants (10, 11). Studies support a heritable component in determining postprandial lipid metabolism (12, 13). A number of candidate genes were implicated, including apolipoproteins and LPL, but together they account for a modest fraction of postprandial lipid variability (14).
CD36 is a highly conserved membrane protein expressed on metabolic and immune cells that functions in cellular recognition and uptake of many lipid-related molecules (15, 16), including dietary long chain fatty acids and native or oxidized lipoproteins (17). In addition, CD36 functions in transducing cell type-specific intracellular signaling that serves to regulate lipid utilization (17–19) or the immune response (20). Variants in the CD36 gene have been shown to influence fasting lipids (21, 22) and risk of metabolic syndrome (MetS) (22). Evidence from CD36-null mouse models and CD36-deficient humans suggests that the protein regulates postprandial TGs and cholesterol metabolism. In mice, CD36 deficiency increases cholesterol levels, impairs CM secretion into the lymph, and delays postprandial TG clearance (9, 23–25). A similar phenotype is found in CD36-deficient subjects who were reported to have increased CM remnants and cholesterol levels (9, 26, 27). Despite the established role of CD36 in lipid absorption and CM metabolism, there is no information on whether polymorphisms in the CD36 gene influence variability in postprandial lipid metabolism in humans. Such information is important considering the documented link between CM remnants and cardiovascular disease.
In this study, we determined the effects of common CD36 variants [minor allele frequency (MAF) 5–45%] on lipoprotein levels before and after ingestion of a high-fat shake in the family-based study of 1,117 Caucasian participants, the Genetics of Lipid Lowering Drugs and Diet Network (GOLDN) (28). We identified several SNPs that influence CM remnants, TG response, and LDL. The most significant SNPs overlap those previously associated with MetS. Examination of the potential mechanisms underlying the observed effects highlighted transcriptional and epigenetic regulatory regions of the gene.
MATERIALS AND METHODS
Study population
GOLDN participants of European ancestry were recruited from the National Heart, Lung, and Blood Institute Family Heart Study. The data shown are from 1,117 individuals (52% females): age (48.4 ± 16.3 years); BMI (28.3 ± 5.6 kg/m2); and blood pressure (systolic 115.3 ± 16.6 mmHg, diastolic 68.1 ± 9.3 mmHg) (supplemental Table S1). Individuals ≥19 years old with fasting TG ≤1,500 mg/dl and normal liver and kidney function were included, while individuals with a history of pancreas, gallbladder, or malabsorption diseases and use of lipid lowering drugs, insulin, and warfarin were excluded, as previously reported (29). Fasted (8 h) participants ingested (within 15 minutes) a high-fat milkshake (700 calories/m2 body surface area, with 3% from protein, 14% from carbohydrate, and 83% from fat). Blood was collected at fasting and after the meal at 3.5 and 6 h for measuring lipid levels and lipoprotein profiling using NMR (28). Blood collection at 3.5 and 6 h was based on TG kinetics and designed to assess differences in lipid absorption and clearance, respectively (30–33).
SNP selection and analysis
Genotype data for 81 SNPs within ∼410 kb (chr7: 79,900,000–80,310,331) encompassing the CD36 gene region met quality control procedures and were used to impute 337 SNPs based on phased genotype data for HapMap CEU (release 22, Human Genome build 36) with MAFs ≥5% (28). SNP genotyping was performed using the Affymetrix Human 6.0 platform (34). Linkage disequilibrium (LD) block structure was determined using the default method of Haploview for the Gabriel protocol (D′ confidence interval 0.70–0.98) (35). Conventional and NMR-derived lipid variables were transformed to render a normal distribution and adjusted for age, age2, and sex within a stepwise regression. Lead SNPs were tested against lipid traits using the same linear mixed-effects model, accounting for familial relationship (36). TG uptake and clearance post meal were calculated from normalized growth curve models where uptake was based on time points 1 (fasting) and 2 (3.5 h post meal), and clearance on time point 3 (6 h post meal) (28). Bonferroni corrections for multiple tests (p ≤ 0.006) were based on the major variables, LDL, TG, and seven LD blocks, across the CD36 region (HapMap CEU, version 4.2).
Metabolic phenotype, transcript level, DNA methylation, and genotypes in the MuTHER cohort
The Multiple Tissue Human Expression Resource (MuTHER) cohort includes subcutaneous adipose tissue collected from 856 females of European descent recruited from the TwinsUK adult registry (37, 38). Adipose RNA was used for expression profiling (Illumina Human HT-12 V3 BeadChips) (38), while DNA was bisulfite converted (EZ-96 DNA methylation kit; Zymo Research) and profiled on the Illumina HumanMethylation450 array (Illumina Inc.) (37). Genotypes were generated using a combination of Illumina arrays (HumanHap300, HumanHap610Q, 1M Duo, and 1.2MDuo 1M) and imputed with IMPUTE (version 2) using two reference panels, P0 (HapMap2, rel 22, combined CEU+YRI+ASN panels) and P1 (610k+, including the combined HumanHap610k and 1M array). The SNPs were filtered at MAF >5% and IMPUTE INFO value of >0.8.
The MuTHER study analyzed DNA methylation and transcript levels for 210,984 methylation and 18,818 expression probes situated on or 1,500 bp upstream of 13,532 genes. For this study, only sites localized to the CD36 promoter region (chr7: 80,060,000–80,095,000) were examined. Association of DNA methylation and transcript levels with probabilities of imputed genotypes (MAF >5%, INFO >0.8) were tested in samples of related individuals using a two-step statistical approach implemented in the GenABEL/ProbABEL packages. Age, beadchip, BS conversion efficiency, and BS-treated DNA input were included as cofactors in the DNA methylation analysis (metQTL, Ntotal = 603) (37) and age and experimental batch in the expression analysis [expression quantitative trait loci (eQTL), Ntotal = 776] (38), respectively. The cis-metQTL analysis was limited to SNPs within 100 kb on either side of the probe location, whereas the cis-eQTL analysis considered SNPs located within 1 MB on either side of the transcription start or end site or within the gene body. False discovery rate was calculated using the qvalue package 30 implemented in R2.11 26 (37, 38) where 1% false discovery rate in the cis-metQTL, cis-eQTL, and methylation versus expression analysis corresponded to p < 8.6E-4, p < 5.0E-5, and p < 2.6E-4, respectively.
Associations between phenotypes or CD36 transcript levels with DNA methylation were tested with a linear mixed-effects model in R (39) using the lmer() function in the lme4 package (40) fitted by maximum-likelihood. The linear mixed-effects model was adjusted for both fixed (age, beadchip, BS conversion efficiency, and BS-treated DNA input) and random effects (family relationship and zygosity). A likelihood ratio test was used to assess significance of the phenotype effect. The p value of the phenotype effect in each model was calculated from the Chi-square distribution with 1° of freedom using −2log (likelihood ratio) as the test statistic.
RESULTS
Common CD36 SNPs influence CM remnants and TG levels
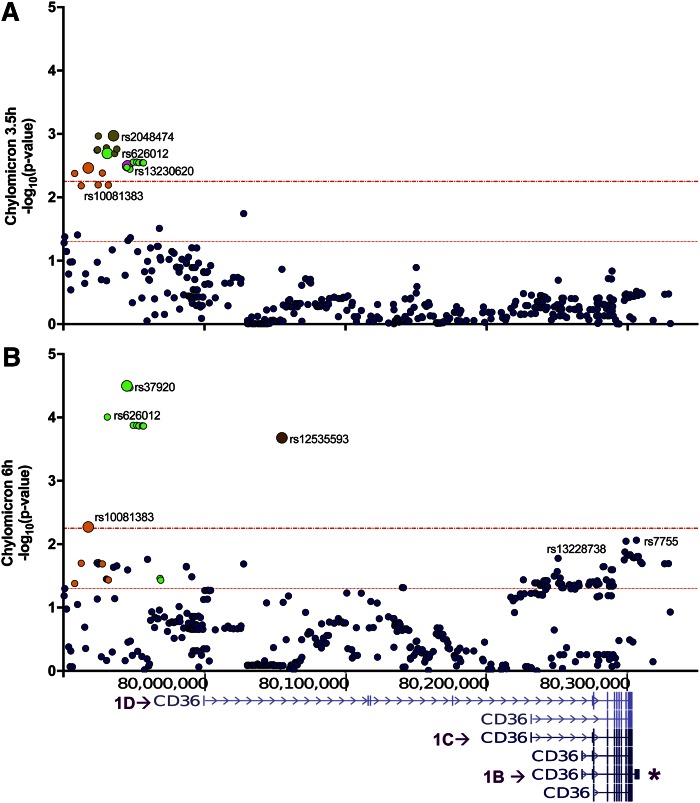
The GOLDN cohort includes relatively healthy overweight to obese individuals (as indicated in the Materials and Methods). The plasma lipid profile for this cohort before and after the meal is in supplemental Table S1. To comprehensively examine the effects of common CD36 variants on postprandial lipids, we tested SNPs across the CD36 locus encompassing all six alternative first exons (1A–1F) (41, 42) and the two 3′UTRs (short and long) (Fig. 1). SNP associations with CM levels were identified (Table 1, Fig. 1) at 3.5 and 6 h post meal. At 3.5 h, independent common SNPs near the distal promoter (1D in Fig. 1A), tagged by rs10081383, rs2048474, and rs13230620, associated with lower CM levels, while CM levels were higher for the less frequent rs37920 (7%) (Table 1). At 6 h post meal, the same relationships were maintained for SNPs rs10081383 and rs37920 (Fig. 1B, Table 1). The effect of rs37920 (C allele) was much more significant at 6 h, suggesting that the major impact of the allele is to reduce clearance, a more substantial contributor to CM levels at 6 h post meal. The rs10081383 (A-allele) and rs12535593 (intronic ∼60 kb downstream of 1D) SNPs associated with lower CM at 6 h, implying either reduced CM input or enhanced CM clearance. In addition, there was an effect in the 3′UTR at rs7755 (G-allele, 46%) on increasing CM remnants (Fig. 1B), but this association was not significant after Bonferroni correction.
Fig. 1.
SNPs near distal first exon 1D of CD36 associate with CM remnants. Regional plot of CD36 SNPs that associate with CM remnant concentration at 3.5 h (A) and 6 h (B) after a the fat meal. p values (−log10) are plotted based on chromosomal position (human genome GRCh37/hg19). Colors differentiate LD blocks (r2 ≥ 0.8). The most significant SNP (representative tag SNP) is identified by a larger symbol and the rs identification number. The overlay of the CD36 gene from the UCSC genome browser illustrates the SNP position relative to alternative CD36 promoters and transcripts (referred to as 1B, 1C, 1D) and the long 3′UTR (asterisk). Dotted red lines indicate p ≤ 0.05 and the Bonferroni-corrected threshold, p < 0.006.
TABLE 1.
CD36 tag SNPs influence CM levels
| SNP | Effect Allele | MAF | β (SE) | a |
| 3.5 h post meal | ||||
| rs10081383 | G/A | 0.24 | −0.18 (0.06) | 0.0035 |
| rs37920 | T/C | 0.07 | 0.40 (0.14) | 0.0034 |
| rs2048474 | A/G | 0.32 | −0.19 (0.06) | 0.0011 |
| rs13230620 | G/C | 0.28 | −0.18 (0.06) | 0.0032 |
| 6.0 h post meal | ||||
| rs10081383 | G/A | 0.24 | −0.18 (0.06) | 0.0005 |
| rs37920 | T/C | 0.07 | 0.57 (0.14) | 0.00003 |
| rs12535593 | T/C | 0.27 | −0.26 (0.07) | 0.0002 |
Shown are β-estimates (β) ± SEs and raw p values for the most significant CD36 tag SNP (within respective LD blocks) that associate with CM levels. SNPs are listed based on genomic positions (see Fig. 1). The effect allele is bold and underlined.
Bonferroni correction threshold p < 0.006.
We then tested SNP associations with changes in TG levels post meal using slope of growth curve modeling (43), which distinguishes between TG appearance (0–3.5 h) and clearance (3.5–6.0 h) phases. As shown in Table 2, three common SNPs near the distal first exon 1D (rs304763, rs304798, rs304802) associated with slower TG appearance (or negative β estimate) and three other SNPs with delayed TG removal (rs37920, rs13228738, rs3211842). Of interest, rs37920 associated with higher CM remnants (Table 1), which is consistent with slow TG removal (Table 2). These findings support a role for the distal 1D region of CD36 in the regulation of postprandial lipid metabolism.
TABLE 2.
Effect of CD36 SNP on growth curve slopes for TG appearance (0–3.5 h) and clearance (3.5–6.0 h) after the meal
| SNP | Effect Allele | MAF | β (SE) | a |
| 0–3.5 h | ||||
| rs304763 | G/A | 0.38 | −0.15 (0.05) | 0.0049 |
| rs304798 | A/G | 0.21 | −0.21 (0.07) | 0.0019 |
| rs304802 | T/C | 0.32 | −0.20 (0.06) | 0.0009 |
| 3.5–6.0 h | ||||
| rs37920 | T/C | 0.07 | 0.44 (0.13) | 0.0008 |
| rs13228738 | G/A | 0.38 | 0.16 (0.05) | 0.0032 |
| rs1931694 | G/C | 0.47 | 0.15 (0.05) | 0.0048 |
| rs3211842 | G/A | 0.42 | 0.17 (0.05) | 0.0013 |
Shown are β-estimates (β) ± SEs and raw p-values for the most significant CD36 tag SNP (within respective LD blocks). The effect allele is bold and underlined. For TG appearance, negative β-estimates indicate that the allele reduces absorption. For clearance, negative β indicates enhanced TG removal and positive β indicates reduced TG removal.
Bonferroni correction threshold p < 0.006.
LDL
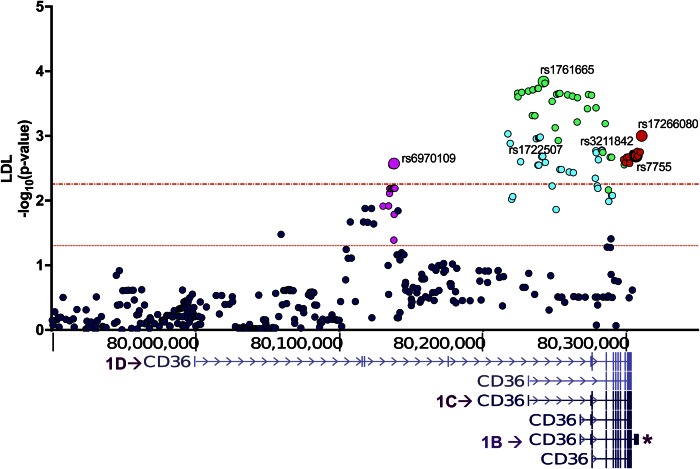
Associations with higher LDL particle numbers at fasting and at the post meal times (Fig. 2, Table 3, supplemental Table 2) were identified for SNPs in the central (near first exon 1C) and proximal (near 1B) promoters and in the longer 3′UTR. The most significant observations were with tag rs1761665 (C-allele, 44%) and rs6970109 (A-allele, 10%). Two additional SNPs sharing modest LD with rs1761665 showed similar effects. In the 3′UTR, rs7755 (A-allele, 46%) (Table 3), previously linked to risk of MetS and stroke (21, 44), associated with higher LDL. As noted earlier, it also associated with higher CM remnants at 6 h post meal, but this did not survive the Bonferroni correction. Of note, the associations shown for fasting LDL remained significant at 3.5 and 6 h post meal (supplemental Table S2) as would be expected.
Fig. 2.
CD36 SNPs in the proximal and central promoter regions associate with fasting LDL. p-values (−log10) are plotted based on chromosomal position (human genome GRCh37/hg19). Colors differentiate LD blocks (r2 ≥ 0.8) with the most significant SNP (representative tag) identified by a larger symbol and rs identification number. Dotted red lines indicate p ≤ 0.05 and Bonferroni threshold, p < 0.006.
TABLE 3.
CD36 tag SNPs associate with higher LDL
| SNP | Effect Allele | MAF | β (SE) | a |
| rs6970109 | C/A | 0.10 | 0.26 (0.09) | 0.0027 |
| rs1722507c | A/G | 0.39 | 0.17 (0.05) | 0.0011 |
| rs1761665b,c | T/C | 0.44 | 0.20 (0.05) | 0.0001 |
| rs3211842b | G/A | 0.42 | 0.02 (0.05) | 0.0017 |
| rs7755d | G/A | 0.46 | 0.16 (0.05) | 0.0020 |
SNPs are listed by genomic location. The effect allele is bold and underlined. Shown are β-estimates (β) ± SEs and raw p-values for most significant CD36 tag SNP (within respective LD blocks). Data shown are for fasting LDL. Supplemental Table S2 shows data for 3.5 and 6 h post meal.
Bonferroni correction threshold p < 0.006.
LD between SNPs: r2 > 0.80, D′ = 0.93.
LD between SNPs: r2 = 0.68, D′ = 1.0.
LD between SNPs: r2 > 0.80, D′ = 0.96.
Lipid-associated SNPs influence CD36 mRNA levels in adipose tissue and heart
To determine the potential functional relevance of the lipid-associated SNPs, we examined their influence on CD36 level in adipose and heart, tissues that are highly active in lipid metabolism and where CD36 is abundant. eQTL were available for adipose tissue from 776 healthy individuals of the MuTHER project (38). As shown in Table 4, analysis of the adipose tissue dataset documented strong influence of the SNPs on the most abundantly expressed CD36 transcript (1B long 3′UTR, NM_001001548) (41, 42). Alleles that related to increased CM remnants or higher LDL associated with lower adipose tissue CD36 mRNA (Table 4). Another independent genome-wide eQTL analysis performed with nondiseased human heart samples from 129 Western Europeans (45) showed similar negative relationships between the lipid-SNPs and CD36 mRNA (Table 4). These findings document strong effects of the SNPs on CD36 transcripts in heart and adipose tissue and suggest that expression in these tissues inversely relates to higher lipids, consistent with the role of CD36 in enhancing lipid clearance and reducing circulating lipid levels.
TABLE 4.
Lipid-associated SNPs influence CD36 transcript level
| SNP | Effect Allele | Adipose Tissue | Hearta | ||
| β (SE) | p | β (SE) | p | ||
| rs1931694 (TGc) | G/C | −0.17 (0.03) | 1.24 × 10−09 | — | — |
| rs1722507c (TGc) | A/G | −0.25 (0.03) | 5.58 × 10−17 | — | — |
| rs1761665b (LDL) | T/C | −0.27 (0.03) | 3.31 × 10−22 | −0.40 (0.08) | 7.25 × 10−7 |
| rs13228738c (TGc) | G/A | −0.24 (0.03) | 1.03 × 10−16 | — | — |
| rs3211842b (TGc) | G/A | −0.28 (0.03) | 2.27 × 10−22 | — | — |
| rs7755 (LDL) | G/A | −0.44 (0.03) | 1.12 × 10−50 | −0.53 (0.08) | 4.32 × 10−11 |
Correlations between alleles and CD36 mRNA were determined using simple regression. SNPs are listed by effect size. Frequency of the effect allele is ≥40% for all listed SNPs. The effect allele is bold and underlined. The lipid trait affected by the SNP is in parentheses: SNPs are listed based on genomic location. CD36 mRNA levels (ILMN_1784863; Illumina HT-12 v3) in abdominal subcutaneous adipose tissue in the MuTHER were retrieved from GENEVAR database. TGc, TG clearance.
SNP effects on heart CD36 expression (ILMN_1784863; Illumina HumanHT-12 v4) from a genome wide eQTL dataset in nondiseased tissue of 129 Western Europeans (45).
SNPs in LD: r2 > 0.80, D′ > 0.8.
SNPs in LD: r2 = 0.93, D′ > 0.8.
SNPs align near sites for PPARγ and DNA methylation in adipose tissue CD36
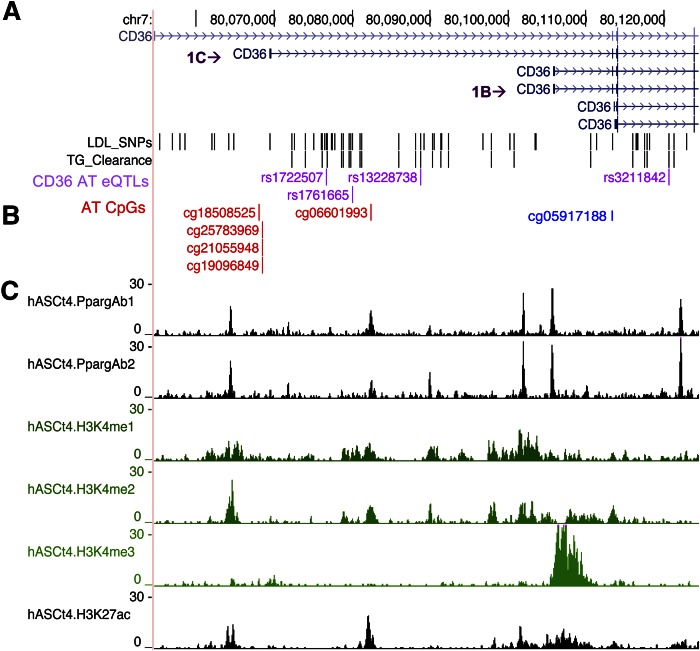
Human adipocyte ChiP-seq data (46) showed that many of the associated promoter SNPs lie within close proximity of previously validated binding sites for PPARγ, a major CD36 transcriptional regulator (Fig. 3). In addition, this region is characterized by active histone H3 lysine methylation and acetylation marks (46).
Fig. 3.
TG clearance and LDL-associated CD36 SNPs align in proximity to PPARγ motifs and to CpG sites in the CD36 promoter. The central CD36 promoter region (chr7: 80,060,000–80,095,000) is tagged by rs1761665, which associates with methylation levels at CpG sites in adipose tissue near CD36 alternative exon 1C that is specific to adipocytes (absent from immune cells and endothelial cells). A: Purple text indicates SNPs that associate with both lipid levels and adipose tissue CD36 expression. B: SNP-associated methylation sites (red text). The location of a previously published TG-associated CpG site is shown in blue text (47). C: Chromatin state maps of the CD36 locus from mature human adipocytes. Histograms of ChIP-seq fragments aligned to the human reference genome (hg18) highlighting active PPARγ binding sites (peaks) and epigenetic signatures (H3K4me1, H3K4me2, H3K4me3, H3K27ac).
Previously, an adipose tissue epigenome study in MuTHER showed the most significant association with fasting TG levels at a cytosine phosphate guanine (CpG) site (cg05917188) in the CD36 promoter (47). In the same dataset (37), we tested to determine whether a significant relationship exists between the lipid-associated SNPs we identified and methylation using a multivariate linear regression model with age and BMI as covariates. Interestingly, the lipid-SNPs that negatively related to adipose CD36 level associated with DNA methylation at several CpG sites (cg25783969, cg21055948, cg19096849 and cg18508525, cg26138637, cg06601993) (Table 5) within the central promoter region. Hypomethylation of four of these sites strongly related to CD36 mRNA levels (Table 6). To gain a better perspective of the contribution of altered methylation to the SNP effects on CD36 mRNA levels, a mixed-linear model for effects of the two most significant eQTLs, rs1761665 (promoter) or rs7755 (3′UTR), was adjusted for methylation (supplemental Table S3). Comparison of the adjusted β estimates shown in supplemental Table S3 to those shown in Table 4, where associations are unadjusted, demonstrates that adjustment for methylation does not significantly attenuate the associations between the SNPs and the most abundant CD36 transcript (1B with long 3′UTR). This indicated that the effect of the SNPs on CD36 is independent of methylation at the above sites and is more likely to involve transcriptional regulation of the gene by PPARγ or posttranslational regulatory sites in the 3′UTR (rs7755). We then examined whether methylation also relates to higher lipid levels. As shown in Table 7, DNA methylation correlated with higher TG and LDL, suggesting that it independently impacts lipid metabolism through its influence on CD36 level (Table 6).
TABLE 5.
Lipid-SNPs associate with DNA methylation in adipose tissue
| CpG ID | rs13228738 | rs3211842 | rs1722507 | rs1761665 | rs7755 |
| cg18508525a | 9.65 × 10−06 | 3.83 × 10−04 | 2.76 × 10−07 | 3.20 × 10−05 | 3.04 × 10−04 |
| cg25783969a | 1.24 × 10−08 | — | 7.23 × 10−11 | 6.72 × 10−05 | 9.26 × 10−05 |
| cg21055948a | 1.57 × 10−11 | 5.01 × 10−06 | 2.37 × 10−13 | 3.08 × 10−07 | 6.14 × 10−06 |
| cg19096849 | 3.36 × 10−09 | 5.87 × 10−06 | 3.64 × 10−11 | 1.40 × 10−07 | 7.97 × 10−07 |
| cg26138637a | — | — | 8.38 × 10−05 | — | — |
| cg06601993 | — | — | 6.75 × 10−04 | 4.00 × 10−04 | — |
Adipose tissue CpG methylation levels determined in the MuTHER study using the Infinium HumanMethylation450 platform. CpGs listed based on chromosomal position. p-values are for CpG-SNP associations.
CpG associates with adipose CD36 mRNA (Table 6).
TABLE 6.
Hypomethylation of promoter CpG sites associates with adipose tissue CD36 expression levels
| CpG Identification | p |
| cg18508525 | 2.40 × 10−05 |
| cg25783969 | 2.03 × 10−05 |
| cg21055948 | 3.06 × 10−05 |
| cg26138637 | 6.66 × 10−05 |
CD36 gene expression (ILMN_1784863) was quantified in adipocyte nuclei from 776 individuals using Illumina whole genome expression array (Human HT-12 v3).
TABLE 7.
Methylation at CD36 expression-associated CpG sites correlates with TG and LDL concentrations
| CpG Identification | TG | LDL | ||
| β | P | β | P | |
| cg18508525a | 0.24 | 1.36 × 10−06 | 0.17 | 3.05 × 10−05 |
| cg25783969a | 0.21 | 5.17 × 10−04 | 0.11 | 1.49 × 10−02 |
| cg21055948a | 0.22 | 3.99 × 10−04 | 0.12 | 7.23 × 10−03 |
| cg26138637a | 0.20 | 1.12 × 10−05 | 0.13 | 2.82 × 10−03 |
Adipose tissue CpG methylation levels were determined in the MuTHER study using the Infinium HumanMethylation450 platform. CpGs are listed based on chromosomal position.
CpG associates with adipose CD36 mRNA (Table 6).
DISCUSSION
Complex interactions between diet, lifestyle, and genetic factors can lead to sustained elevation of postprandial lipids, which independently increases the risk of cardiovascular disease (1, 2). Earlier genetic studies linked CD36 to fasting lipids (21, 48), MetS (22, 49–51), and stroke risk (44). However, information related to the role of the gene in postprandial lipid metabolism, which plays an important role in etiology of MetS and cardiovascular disease, remains limited. This study is the first comprehensive examination of the impact of CD36 variants on postprandial lipids. We present findings implicating common promoter SNPs in inter-individual variability of the CM remnant response after a fatty meal. We also identify SNPs that associate with LDL particle number at both fasting and two postprandial time points. We document that the identified lipid SNPs, which align closely to validated PPARγ sites, associate with reduced adipose and heart CD36 and higher lipid traits, supporting the role of CD36 in enhancing lipid clearance. In addition, we provide evidence that higher serum lipids associate with altered DNA methylation and reduced CD36 mRNA.
The most significant promoter SNPs identified to influence CM levels and TG clearance (rs1931694, rs13228738, rs3211842) are in strong LD with variants previously associated with MetS or coronary artery disease (22, 48, 49). Similarly, the 3′UTR variant, rs7755, identified to associate with LDL and to trend with higher CM remnants has been linked to diabetes-associated coronary artery disease (48) and stroke (44), as well as MetS (22). Overall, our findings support an important role of CD36 in the handling of dietary lipids in humans and are consistent with data in CD36-null mice showing defective CM metabolism and persistently high levels of CM remnants (9, 23–25). Elevated CM remnant levels were reported in small cohorts of CD36-deficient subjects (9, 26, 27). Our study indicates that common CD36 alleles (5–45% frequency) contribute to variability of the postprandial response and possibly to risk of diet-induced metabolic abnormalities. GOLDN is the largest cohort to-date in which postprandial lipid measures are available and, ideally, our findings will need replication in other large studies as they become available.
We identified five CD36 SNPs to associate with higher LDL, while previous GWASs did not link LDL to the CD36 locus. Several considerations might explain the discrepancy. The GOLDN family-based study consists of relatively healthy overweight to obese Caucasians who underwent a washout period to neutralize effects of lipid-lowering medications. Earlier GWASs included cohorts with a wide range of ages, metabolic disease traits, medications, and BMI (52). We queried available Global Lipid Genetics Consortium datasets using LocusZoom and found that most cohorts did not include sufficient SNP coverage of the CD36 promoter region comparable to this study. In one of the Global Lipid Genetics studies (53) with similar coverage and that included SNPs in LD with those we genotyped, we found modest raw associations (p < 0.05) with LDL cholesterol. Several of the identified variants, rs799979, rs646722, rs6949840, rs2030711, and rs17154155, are in strong LD with rs1722507 and rs1761665 in this study. In addition, our study measured LDL particle number as opposed to LDL cholesterol obtained by the Friedewald calculation. While both measures predict cardiovascular risk, they do not always correlate (1, 54). In support of our findings are the reports of higher cholesterol levels in partially (55) or completely CD36-deficient individuals (9) and the higher LDL levels and abnormal cholesterol absorption of CD36-null mice (24, 56). In addition, a recent linkage analysis of LDL particle response to fenofibrate treatment in GOLDN reported a peak encompassing the CD36 locus (57). Thus, the LDL associations we identified and their potential relationship to disease risk warrant further investigation.
Our findings highlight regulatory regions in the CD36 gene: SNPs near the distal promoter (designated 1D), which is almost exclusive to endothelial cells (42), associated with TG uptake/clearance and CM remnants. The central promoter region had signals for LDL and TG clearance, and contained the validated PPARγ sites and the lipid-associated DNA methylation sites. Finally, the long 3′UTR had signals for LDL and contained predicted miRNA and RNA editing sites.
We showed that SNPs that associated with higher CM remnants and LDL reduced adipose and heart CD36. The rs1761665 SNP, in particular, is in strong LD with promoter SNPs previously shown to influence monocyte and platelet CD36 (58, 59). Several of the promoter SNPs we identified here lie in close proximity to validated binding sites for PPARγ and to methylation sites that associate with CD36 mRNA levels (Fig. 3). Further analyses involving the two most significant eQTLs (rs1761665 and rs7755) for CD36 indicated that the effect of these SNPs is independent of methylation and is more likely to involve altered transcriptional regulation by PPARγ (rs1761665) or posttranscriptional effects involving the 3′UTR (rs7755). Although changes in methylation do not account for the SNP effect on CD36 level, our data suggest that they influence lipid levels. Methylation might contribute to the metabolic and dietary regulation of the CD36 gene. For example, fatty acids were reported to alter global DNA methylation and to participate in shaping metabolic disease-related methylomes (60).
In summary, our findings link common CD36 SNPs that reduce adipose and heart CD36 levels to higher CM remnants and LDL in humans. These SNPs would contribute to individual differences in the handling of dietary lipid and in susceptibility to diet-induced metabolic abnormalities. Finally, factors that alter methylation of the CD36 gene impact lipid levels and potentially disease risk.
Supplementary Material
Acknowledgments
The authors thank Drs. Tarjei Mikkelsen (Broad Institute) and Evan Rosen (Harvard University) for sharing the adipocyte chromatin state alignment data.
Footnotes
Abbreviations:
- CD36
- cluster of differentiation 36
- CM
- chylomicron
- CpG
- cytosine phosphate guanine
- eQTL
- expression quantitative trait loci
- GOLDN
- Genetics of Lipid Lowering Drugs and Diet Network
- LD
- linkage disequilibrium
- MAF
- minor allele frequency
- MetS
- metabolic syndrome
- MuTHER
- Multiple Tissue Human Expression Resource
This study was supported by grants from the Longer Life Foundation, the American Heart Association Scientist Development Grant, Washington University Nutrition and Obesity Research Center P&F DK56341 and CTSA, National Center for Advancing Translational Sciences Grant UL1TR000448 (L.L-G.), National Institute of Diabetes and Digestive and Kidney Diseases Grants R01 DK33301 and R01 DK060022 (N.A.A.), and Cooperative State Research, Education, and Extension Service Grant 58-1950-4-003 (J.M.O.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare that they have no financial conflicts of interest.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Mora S., Rifai N., Buring J. E., and Ridker P. M.. 2008. Fasting compared with nonfasting lipids and apolipoproteins for predicting incident cardiovascular events. Circulation. 118: 993–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nordestgaard B. G., and Varbo A.. 2014. Triglycerides and cardiovascular disease. Lancet. 384: 626–635. [DOI] [PubMed] [Google Scholar]
- 3.Goldberg I. J., Eckel R. H., and Abumrad N. A.. 2009. Regulation of fatty acid uptake into tissues: lipoprotein lipase- and CD36-mediated pathways. J. Lipid Res. 50(Suppl): S86–S90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bickerton A. S., Roberts R., Fielding B. A., Hodson L., Blaak E. E., Wagenmakers A. J., Gilbert M., Karpe F., and Frayn K. N.. 2007. Preferential uptake of dietary Fatty acids in adipose tissue and muscle in the postprandial period. Diabetes. 56: 168–176. [DOI] [PubMed] [Google Scholar]
- 5.Karpe F., Bickerton A. S., Hodson L., Fielding B. A., Tan G. D., and Frayn K. N.. 2007. Removal of triacylglycerols from chylomicrons and VLDL by capillary beds: the basis of lipoprotein remnant formation. Biochem. Soc. Trans. 35: 472–476. [DOI] [PubMed] [Google Scholar]
- 6.Potts J. L., Coppack S. W., Fisher R. M., Humphreys S. M., Gibbons G. F., and Frayn K. N.. 1995. Impaired postprandial clearance of triacylglycerol-rich lipoproteins in adipose tissue in obese subjects. Am. J. Physiol. 268: E588–E594. [DOI] [PubMed] [Google Scholar]
- 7.Greenwalt D. E., Scheck S. H., and Rhinehart-Jones T.. 1995. Heart CD36 expression is increased in murine models of diabetes and in mice fed a high fat diet. J. Clin. Invest. 96: 1382–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abumrad N. A., and Goldberg I. J.. 2016. CD36 actions in the heart: lipids, calcium, inflammation, repair and more? Biochim. Biophys. Acta. 1860: 1442–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masuda D., Hirano K., Oku H., Sandoval J. C., Kawase R., Yuasa-Kawase M., Yamashita Y., Takada M., Tsubakio-Yamamoto K., Tochino Y., et al. . 2009. Chylomicron remnants are increased in the postprandial state in CD36 deficiency. J. Lipid Res. 50: 999–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adiels M., Matikainen N., Westerbacka J., Soderlund S., Larsson T., Olofsson S. O., Boren J., and Taskinen M. R.. 2012. Postprandial accumulation of chylomicrons and chylomicron remnants is determined by the clearance capacity. Atherosclerosis. 222: 222–228. [DOI] [PubMed] [Google Scholar]
- 11.Frayn K., Bernard S., Spalding K., and Arner P.. 2012. Adipocyte triglyceride turnover is independently associated with atherogenic dyslipidemia. J. Am. Heart Assoc. 1: e003467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Irvin M. R., Zhi D., Aslibekyan S., Claas S. A., Absher D. M., Ordovas J. M., Tiwari H. K., Watkins S., and Arnett D. K.. 2014. Genomics of post-prandial lipidomic phenotypes in the Genetics of Lipid lowering Drugs and Diet Network (GOLDN) study. PLoS One. 9: e99509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uiterwaal C. S., Grobbee D. E., Witteman J. C., van Stiphout W. A., Krauss X. H., Havekes L. M., de Bruijn A. M., van Tol A., and Hofman A.. 1994. Postprandial triglyceride response in young adult men and familial risk for coronary atherosclerosis. Ann. Intern. Med. 121: 576–583. [DOI] [PubMed] [Google Scholar]
- 14.Perez-Martinez P., Lopez-Miranda J., Perez-Jimenez F., and Ordovas J. M.. 2008. Influence of genetic factors in the modulation of postprandial lipemia. Atheroscler. Suppl. 9: 49–55. [DOI] [PubMed] [Google Scholar]
- 15.Hoebe K., Georgel P., Rutschmann S., Du X., Mudd S., Crozat K., Sovath S., Shamel L., Hartung T., Zahringer U., et al. . 2005. CD36 is a sensor of diacylglycerides. Nature. 433: 523–527. [DOI] [PubMed] [Google Scholar]
- 16.Abumrad N. A., and Davidson N. O.. 2012. Role of the gut in lipid homeostasis. Physiol. Rev. 92: 1061–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Endemann G., Stanton L. W., Madden K. S., Bryant C. M., White R. T., and Protter A. A.. 1993. CD36 is a receptor for oxidized low density lipoprotein. J. Biol. Chem. 268: 11811–11816. [PubMed] [Google Scholar]
- 18.Hames K. C., Vella A., Kemp B. J., and Jensen M. D.. 2014. Free fatty acid uptake in humans with CD36 deficiency. Diabetes. 63: 3606–3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pepino M. Y., Kuda O., Samovski D., and Abumrad N. A.. 2014. Structure-function of CD36 and importance of fatty acid signal transduction in fat metabolism. Annu. Rev. Nutr. 34: 281–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stewart C. R., Stuart L. M., Wilkinson K., van Gils J. M., Deng J., Halle A., Rayner K. J., Boyer L., Zhong R., Frazier W. A., et al. . 2010. CD36 ligands promote sterile inflammation through assembly of a Toll-like receptor 4 and 6 heterodimer. Nat. Immunol. 11: 155–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Love-Gregory L., and Abumrad N. A.. 2011. CD36 genetics and the metabolic complications of obesity. Curr. Opin. Clin. Nutr. Metab. Care. 14: 527–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Love-Gregory L., Sherva R., Sun L., Wasson J., Schappe T., Doria A., Rao D. C., Hunt S. C., Klein S., Neuman R. J., et al. . 2008. Variants in the CD36 gene associate with the metabolic syndrome and high-density lipoprotein cholesterol. Hum. Mol. Genet. 17: 1695–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tran T. T., Poirier H., Clement L., Nassir F., Pelsers M. M., Petit V., Degrace P., Monnot M. C., Glatz J. F., Abumrad N. A., et al. . 2011. Luminal lipid regulates CD36 levels and downstream signaling to stimulate chylomicron synthesis. J. Biol. Chem. 286: 25201–25210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nauli A. M., Nassir F., Zheng S., Yang Q., Lo C. M., Vonlehmden S. B., Lee D., Jandacek R. J., Abumrad N. A., and Tso P.. 2006. CD36 is important for chylomicron formation and secretion and may mediate cholesterol uptake in the proximal intestine. Gastroenterology. 131: 1197–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drover V. A., Ajmal M., Nassir F., Davidson N. O., Nauli A. M., Sahoo D., Tso P., and Abumrad N. A.. 2005. CD36 deficiency impairs intestinal lipid secretion and clearance of chylomicrons from the blood. J. Clin. Invest. 115: 1290–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamashita S., Hirano K., Kuwasako T., Janabi M., Toyama Y., Ishigami M., and Sakai N.. 2007. Physiological and pathological roles of a multi-ligand receptor CD36 in atherogenesis; insights from CD36-deficient patients. Mol. Cell. Biochem. 299: 19–22. [DOI] [PubMed] [Google Scholar]
- 27.Kuwasako T., Hirano K., Sakai N., Ishigami M., Hiraoka H., Yakub M. J., Yamauchi-Takihara K., Yamashita S., and Matsuzawa Y.. 2003. Lipoprotein abnormalities in human genetic CD36 deficiency associated with insulin resistance and abnormal fatty acid metabolism. Diabetes Care. 26: 1647–1648. [DOI] [PubMed] [Google Scholar]
- 28.Wojczynski M. K., Glasser S. P., Oberman A., Kabagambe E. K., Hopkins P. N., Tsai M. Y., Straka R. J., Ordovas J. M., and Arnett D. K.. 2011. High-fat meal effect on LDL, HDL, and VLDL particle size and number in the Genetics of Lipid-Lowering Drugs and Diet Network (GOLDN): an interventional study. Lipids Health Dis. 10: 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corella D., Arnett D. K., Tsai M. Y., Kabagambe E. K., Peacock J. M., Hixson J. E., Straka R. J., Province M., Lai C. Q., Parnell L. D., et al. . 2007. The -256T>C polymorphism in the apolipoprotein A-II gene promoter is associated with body mass index and food intake in the genetics of lipid lowering drugs and diet network study. Clin. Chem. 53: 1144–1152. [DOI] [PubMed] [Google Scholar]
- 30.Patsch J. R., Miesenbock G., Hopferwieser T., Muhlberger V., Knapp E., Dunn J. K., Gotto A. M. Jr., and Patsch W.. 1992. Relation of triglyceride metabolism and coronary artery disease. Studies in the postprandial state. Arterioscler. Thromb. 12: 1336–1345. [DOI] [PubMed] [Google Scholar]
- 31.Dubois C., Armand M., Azais-Braesco V., Portugal H., Pauli A. M., Bernard P. M., Latge C., Lafont H., Borel P., and Lairon D.. 1994. Effects of moderate amounts of emulsified dietary fat on postprandial lipemia and lipoproteins in normolipidemic adults. Am. J. Clin. Nutr. 60: 374–382. [DOI] [PubMed] [Google Scholar]
- 32.López-Miranda J., Cruz G., Gómez P., Marín C., Paz E., Pérez-Martínez P., Fuentes F. J., Ordovas J. M., and Pérez-Jiménez F.. 2004. The influence of lipoprotein lipase gene variation on postprandial lipoprotein metabolism. J. Clin. Endocrinol. Metab. 89: 4721–4728. [DOI] [PubMed] [Google Scholar]
- 33.Williams C. M. 1997. Postprandial lipid metabolism: effects of dietary fatty acids. Proc. Nutr. Soc. 56: 679–692. [DOI] [PubMed] [Google Scholar]
- 34.Irvin M. R., Kabagambe E. K., Tiwari H. K., Parnell L. D., Straka R. J., Tsai M., Ordovas J. M., and Arnett D. K.. 2010. Apolipoprotein E polymorphisms and postprandial triglyceridemia before and after fenofibrate treatment in the Genetics of Lipid Lowering and Diet Network (GOLDN) study. Circ Cardiovasc Genet. 3: 462–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gabriel S. B., Schaffner S. F., Nguyen H., Moore J. M., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., et al. . 2002. The structure of haplotype blocks in the human genome. Science. 296: 2225–2229. [DOI] [PubMed] [Google Scholar]
- 36.Kenward M. G., and Roger J. H.. 1997. Small sample inference for fixed effects from restricted maximum likelihood. Biometrics. 53: 983–997. [PubMed] [Google Scholar]
- 37.Grundberg E., Meduri E., Sandling J. K., Hedman A. K., Keildson S., Buil A., Busche S., Yuan W., Nisbet J., Sekowska M., et al. . 2013. Global analysis of DNA methylation variation in adipose tissue from twins reveals links to disease-associated variants in distal regulatory elements. Am. J. Hum. Genet. 93: 876–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grundberg E., Small K. S., Hedman A. K., Nica A. C., Buil A., Keildson S., Bell J. T., Yang T. P., Meduri E., Barrett A., et al. . 2012. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet. 44: 1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.R Development Core Team. 2011. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 40.Bates D., Maechler M. ,, and Bolker B.. 2011. lme4: linear mixed-effects models using S4 classes. Accessed October 28, 2016, at http://CRAN.R-project.org/package=lme4. [Google Scholar]
- 41.Andersen M., Lenhard B., Whatling C., Eriksson P., and Odeberg J.. 2006. Alternative promoter usage of the membrane glycoprotein CD36. BMC Mol. Biol. 7: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pietka T. A., Schappe T., Conte C., Fabbrini E., Patterson B. W., Klein S., Abumrad N. A., and Love-Gregory L.. 2014. Adipose and muscle tissue profile of CD36 transcripts in obese subjects highlights the role of CD36 in fatty acid homeostasis and insulin resistance. Diabetes Care. 37: 1990–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wojczynski M. K., Gao G., Borecki I., Hopkins P. N., Parnell L., Lai C. Q., Ordovas J. M., Chung B. H., and Arnett D. K.. 2010. Apolipoprotein B genetic variants modify the response to fenofibrate: a GOLDN study. J. Lipid Res. 51: 3316–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ikram M. A., Seshadri S., Bis J. C., Fornage M., DeStefano A. L., Aulchenko Y. S., Debette S., Lumley T., Folsom A. R., van den Herik E. G., et al. . 2009. Genomewide association studies of stroke. N. Engl. J. Med. 360: 1718–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koopmann T. T., Adriaens M. E., Moerland P. D., Marsman R. F., Westerveld M. L., Lal S., Zhang T., Simmons C. Q., Baczko I., dos Remedios C., et al. . 2014. Genome-wide identification of expression quantitative trait loci (eQTLs) in human heart. PLoS One. 9: e97380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mikkelsen T. S., Xu Z., Zhang X., Wang L., Gimble J. M., Lander E. S., and Rosen E. D.. 2010. Comparative epigenomic analysis of murine and human adipogenesis. Cell. 143: 156–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allum F., Shao X., Guenard F., Simon M. M., Busche S., Caron M., Lambourne J., Lessard J., Tandre K., Hedman A. K., et al. . 2015. Characterization of functional methylomes by next-generation capture sequencing identifies novel disease-associated variants. Nat. Commun. 6: 7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma X., Bacci S., Mlynarski W., Gottardo L., Soccio T., Menzaghi C., Iori E., Lager R. A., Shroff A. R., Gervino E. V., et al. . 2004. A common haplotype at the CD36 locus is associated with high free fatty acid levels and increased cardiovascular risk in Caucasians. Hum. Mol. Genet. 13: 2197–2205. [DOI] [PubMed] [Google Scholar]
- 49.Farook V. S., Puppala S., Schneider J., Fowler S. P., Chittoor G., Dyer T. D., Allayee H., Cole S. A., Arya R., Black M. H., et al. . 2012. Metabolic syndrome is linked to chromosome 7q21 and associated with genetic variants in CD36 and GNAT3 in Mexican Americans. Obesity (Silver Spring). 20: 2083–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coram M. A., Duan Q., Hoffmann T. J., Thornton T., Knowles J. W., Johnson N. A., Ochs-Balcom H. M., Donlon T. A., Martin L. W., Eaton C. B., et al. . 2013. Genome-wide characterization of shared and distinct genetic components that influence blood lipid levels in ethnically diverse human populations. Am. J. Hum. Genet. 92: 904–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elbers C. C., Guo Y., Tragante V., van Iperen E. P., Lanktree M. B., Castillo B. A., Chen F., Yanek L. R., Wojczynski M. K., Li Y. R., et al. . 2012. Gene-centric meta-analysis of lipid traits in African, East Asian and Hispanic populations. PLoS One. 7: e50198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Global Lipids Genetics Consortium, Willer C. J., Schmidt E. M., Sengupta S., Peloso G. M., Gustafsson S., Kanoni S., Ganna A., Chen J., Buchkovich M. L., et al. . 2013. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 45: 1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kathiresan S., Willer C. J., Peloso G. M., Demissie S., Musunuru K., Schadt E. E., Kaplan L., Bennett D., Li Y., Tanaka T., et al. . 2009. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 41: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Otvos J. D., Mora S., Shalaurova I., Greenland P., Mackey R. H., and Goff D. C. Jr. 2011. Clinical implications of discordance between low-density lipoprotein cholesterol and particle number. J. Clin. Lipidol. 5: 105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yanai H., Chiba H., Morimoto M., Abe K., Fujiwara H., Fuda H., Hui S. P., Takahashi Y., Akita H., Jamieson G. A., et al. . 2000. Human CD36 deficiency is associated with elevation in low-density lipoprotein-cholesterol. Am. J. Med. Genet. 93: 299–304. [DOI] [PubMed] [Google Scholar]
- 56.Febbraio M., Abumrad N. A., Hajjar D. P., Sharma K., Cheng W., Pearce S. F., and Silverstein R. L.. 1999. A null mutation in murine CD36 reveals an important role in fatty acid and lipoprotein metabolism. J. Biol. Chem. 274: 19055–19062. [DOI] [PubMed] [Google Scholar]
- 57.Hidalgo B., Aslibekyan S., Wiener H. W., Irvin M. R., Straka R. J., Borecki I. B., Tiwari H. K., Tsai M. Y., Hopkins P. N., Ordovas J. M., et al. . 2015. A family-specific linkage analysis of blood lipid response to fenofibrate in the Genetics of Lipid Lowering Drug and Diet Network. Pharmacogenet. Genomics. 25: 511–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghosh A., Murugesan G., Chen K., Zhang L., Wang Q., Febbraio M., Anselmo R. M., Marchant K., Barnard J., and Silverstein R. L.. 2011. Platelet CD36 surface expression levels affect functional responses to oxidized LDL and are associated with inheritance of specific genetic polymorphisms. Blood. 117: 6355–6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Love-Gregory L., Sherva R., Schappe T., Qi J. S., McCrea J., Klein S., Connelly M. A., and Abumrad N. A.. 2011. Common CD36 SNPs reduce protein expression and may contribute to a protective atherogenic profile. Hum. Mol. Genet. 20: 193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Silva-Martínez G. A., Rodríguez-Ríos D., Alvarado-Caudillo Y., Vaquero A., Esteller M., Carmona F. J., Moran S., Nielsen F. C., Wickström-Lindholm M., Wrobel K., et al. . 2016. Arachidonic and oleic acid exert distinct effects on the DNA methylome. Epigenetics. 11: 321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.