Abstract
To identify novel risk variants for chronic lymphocytic leukemia (CLL) we conducted a genome-wide association study of 299,983 tagging SNPs, with validation in four additional series totaling 2,503 cases and 5,789 controls. We identified four risk loci for CLL at 2q37.3 (rs757978, FARP2; odds ratio [OR] = 1.39; P = 2.11 x 10-9), 8q24.21 (rs2456449; OR = 1.26; P = 7.84 x 10-10), 15q21.3 (rs7169431; OR = 1.36; P = 4.74 x 10-7) and 16q24.1 (rs305061; OR = 1.22; P = 3.60 x 10-7). There was also evidence for risk loci at 15q25.2 (rs783540, CPEB1; OR = 1.18; P = 3.67 x 10-6) and 18q21.1 (rs1036935; OR = 1.22; P = 2.28 x 10-6). These data provide further evidence for genetic susceptibility to this B-cell hematological malignancy.
While B-cell chronic lymphocytic leukemia (CLL) shows a strong familial risk1, the genetic basis of predisposition is largely unknown. We have previously reported the results of a genome-wide association (GWA) study of CLL based on the analysis of 299,983 tagging SNPs in 505 cases and 1,438 controls (henceforth referred to as Stage 1)2 and through fast tracking analysis of the smallest P-values identified risk loci at 2q13, 2q37.1, 6p25.3, 11q24.1, 15q23, and 19q13.322. We have conducted further follow-up analyses making use of existing GWA data and report four novel susceptibility loci.
In Stage 2, we genotyped 180 SNPs in 540 UK cases (UK-replication series 1). The SNPs were chosen by a hypothesis-free strategy on the basis of P-values, excluding those correlated with an r2>0.8 with previously identified association signals. After quality control 162 SNP genotypes were recovered for 519 of the cases. We used publicly accessible data on 2,695 UK-blood donor controls to compare genotype frequencies. In Stage 3, we genotyped the 19 SNPs that showed the strongest association from combined analysis of Stages 1 and 2 in two case-control series: UK-replication series 2 (660 cases,809 controls), Spanish-replication series (424 cases,450 controls). In Stage 4 we genotyped the 10 SNPs with the strongest association from a combined analysis of Stages 1-3 in the Swedish-replication series (395 cases, 397 controls) (Supplementary Figure 1).
The combined analysis of these data provided conclusive evidence of an association between seven SNPs and CLL (i.e. P<5.0x10-7; Supplementary Table 1) mapping to five independent genomic regions. rs11668878 maps to 19q13.32, a region we have previously reported to be a risk locus for CLL (defined by rs11083846)2. Linkage disequilibrium (LD) exists between rs11668878 and rs11083846 (r2=0.27,D’=1.0) and conditional analysis did not provide evidence for a second disease locus (P>0.05). Mouthwash DNA was available from 89 of the cases in Stage 1 and Stage 2; SNP genotypes were 99% concordant with genotypes obtained from typing blood DNA. Together with the fact that these associations identified do not map to any of the regions of the genome commonly associated with copy number variation in CLL3–6 mitigates against bias from differential genotyping as a consequence of allelic imbalance influencing findings.
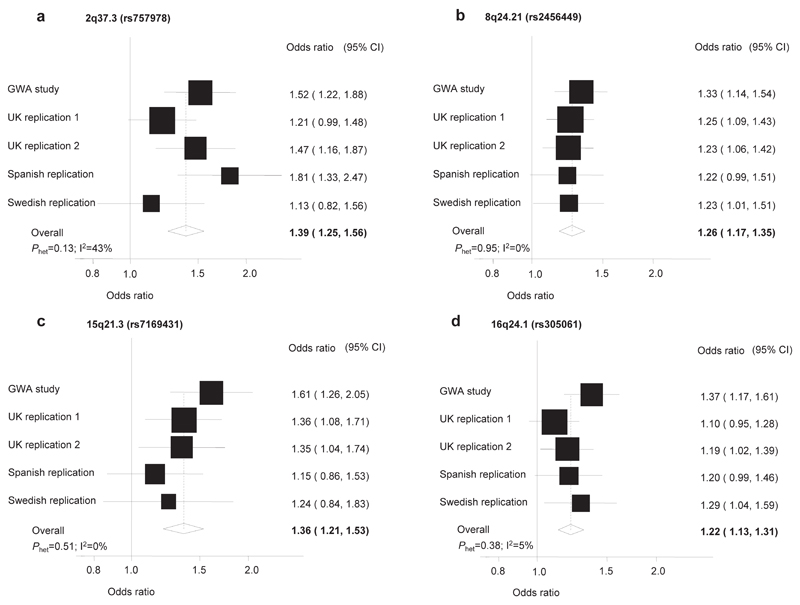
Under a fixed-effects model the strongest evidence for a novel association was attained with rs2456449 which maps to 8q24.21 at 128,262,163bp (OR=1.26; 95%CI:1.17-1.35;P=7.84x10-10; Phet=0.95, I2=0%; Figure 1, Supplementary Table 1). rs2466024, localizing to 128,257,201bp, also provided significant evidence for the 8q24.21 association (P=4.61x10-7; Supplementary Table 1). rs2456449 and rs2466024 are in strong LD (r2=0.75,D’=1.0) and map to a 40kb region of LD (128,241,868-128,282,415bp; Figure 2; Supplementary Figure 2). Conditional analysis provided no evidence for an independent role of rs2466024.
Figure 1. Forest plots of effect size and direction for the SNPs associated with CLL risk. (a) 2q37.3 (rs757978), (b) 8q24.21 (rs2456449), (c) 15q21.3 (rs7169431), (d) 16q24.1 (rs305061).
Boxes denote OR point estimates, their areas being proportional to the inverse variance weight of the estimate. Horizontal lines represent 95% confidence intervals. The diamond (and broken line) represents the summary OR computed under a fixed effects model, with 95% confidence interval given by the width of the diamond. The unbroken vertical line is at the null value (OR = 1.0).
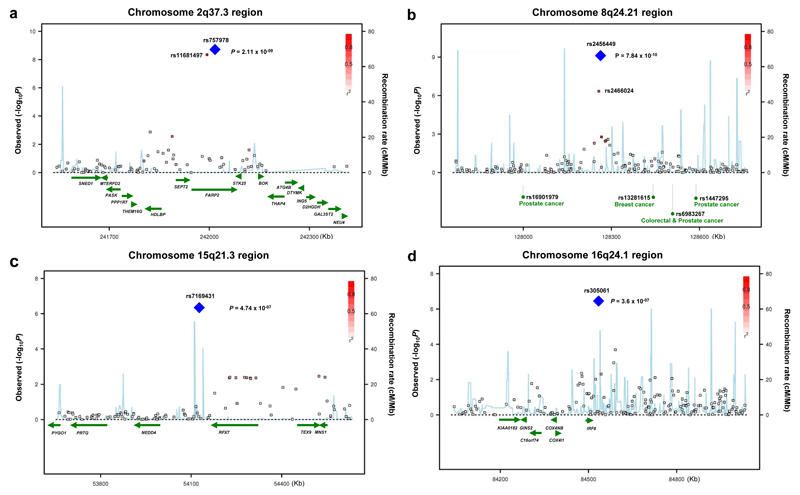
Figure 2. Four previously unidentified loci, 2q37.3, 8q24.21, 15q21.3, and 16q24.1 showing genome-wide level of evidence of association to CLL.
(a) Illustration of the 2q37.3 locus, with the local recombination rate plotted in light blue over this 600-kb chromosomal segment centered on rs757978. Each square represents a SNP found in this locus and the most associated SNP in the combined analysis, rs757978, is marked by a blue diamond. The color intensity of each square reflects the extent of LD with rs757978 - red (r2 > 0.8) through to white (r2 < 0.3). Physical positions are based on build 36 of the human genome. rs757978 is located in exon 9 of FARP2.
(b) Illustration of the 8q24.21 locus, with the most associated SNP in this locus, rs2456449, highlighted by a blue diamond. Here, we also present all SNPs found within a 600-kb window centered on rs2456449 and define SNP colors based on LD with rs2456449.
(c) Illustration of the 15q21.3 locus, with the most associated SNP in this locus, rs7169431, highlighted by a blue diamond. Here, we also present all SNPs found within a 600-kb window centered on rs7169431 and define SNP colors based on LD with rs7169431. NEDD4 and RFXDC2 map centromeric and telomeric to rs7169431.
(d) Illustration of the 16q24.1 locus, with the most associated SNP in this locus, rs305061, highlighted by a blue diamond. Here, we also present all SNPs found within a 600-kb window centered on rs305061 and define SNP colors based on LD with rs305061. In this case, IRF8 is the only gene found in the vicinity to the association signal.
Linkage disequilibrium maps are presented for all four loci in Supplementary Figure 1a–d online.
Genome-wide association studies have shown that the 128-130Mb genomic interval at 8q24.21 harbors multiple independent loci with different tumor specificities: prostate (rs16901979;128,194,098bp)7, breast (rs13281615;128,424,800bp)8, colorectal-prostate (rs6983267;128,482,487bp)9,10, prostate (rs1447295;128,554,220bp)11 and bladder (rs9642880;128,787,250bp)12 cancer. The LD blocks defining these loci are distinct from the 8q24.21 CLL association signal. This is reflected in the LD metrics between rs2456449, and rs6983267 (r2=0.00,D’=0.13), rs13281615 (r2=0.00,D’=0.01), rs16901979 (r2=0.01,D’=1.0), rs1447295 (r2=0.04,D’=1.0), rs9642880 (r2=0.02,D’=0.19). The 8q24.21 region to which the cancer associations map is bereft of genes and predicted transcripts. rs6983267 has recently been shown to affect TCF4 binding to an enhancer for the MYC promoter, providing a mechanistic basis for this 8q24.21 association13,14. It is possible that the effect of the other 8q24.21 cancer risk loci is via MYC through similar long range cis-acting mechanisms. We have shown that variation in IRF4 influences CLL risk2. If the 8q24.21 locus influences risk through differential MYC expression then the association is intriguing as MYC is a direct target of IRF4 in activated B-cells15. While no relationship between rs2456449 genotype and MYC expression was shown in EBV-transformed lymphocytes (Supplementary Figure 3) this does not preclude a subtle effect on expression.
The second strongest statistical evidence for an association signal was provided by rs757978, mapping to exon 9 of FARP2 at 2q37.3 (242,019,774bp; OR=1.39, 95%CI:1.25–1.56; P=2.11x10-9; Phet=0.13, I2=43%; Figure 1, Supplementary Figure 2, Supplementary Table 1). rs11681497, which maps to intron 4 of FARP2 (241,993,006bp) and is in LD with rs757978 (r2=1,D’=1), also provided strong evidence for the 2q37.3 association (P=4.53x10-9;Figure 2), however conditional analysis provided no evidence for an independent role. FARP2 is involved in signaling downstream of G-protein-coupled receptors16. We examined if there was a relationship between genotype and FARP2 expression in EBV-transformed lymphocytes. Data was unavailable for rs757978 but no association between rs11681497 genotype and mRNA expression level was shown (Supplementary Figure 3). rs757978 leads to the substitution of threonine for isoleucine at amino acid 260 (T260I) in the expressed protein. As in silico analysis predicts T260I to be functionally deleterious it is possible that the association signal at 2q37.3 is a consequence of T260I.
The third strongest association was provided by rs305061 (OR=1.22;95% CI:1.12-1.32; P=3.60x10-7; Phet=0.38, I2=5%; Figure 1, Supplementary Table 1), which maps within a 30kb region of LD at 16q24.1 (84,533,160bp;Figure 2). rs305061 localizes 19kb telomeric to IRF8. Variation in IRF8 represents a strong candidate for the association as IRF8 regulates α and β interferon response. Moreover, IRF8 is involved in B-cell lineage specification, immunoglobulin rearrangement, and regulation of germinal center reaction17. Variation at 16q24.1, defined by rs17445836 which maps 61kb telomeric to IRF8 (84,575,164bp), has recently been shown to be a risk locus for multiple sclerosis (MS)18. MS has been reported to be associated with CLL risk19,20. While rs17445836 and rs305061 are not strongly correlated (r2=0.13,D’=0.82), it is possible that a common genetic basis to both diseases is mediated through the same causal variant at 16q24.1.
The fourth strongest association was identified with rs7169431 which maps to 15q21.3 (54,128,188bp; OR=1.36; 95%CI:1.21-1.53; P=4.74x10-7; Phet=0.51, I2=0%; Figure 1, Supplementary Table 1). rs7169431 localizes to a 25kb region of LD flanked by NEDD4 and RFX7 (Figure 2, Supplementary Figure 2). While there is no evidence for a direct role of NEDD4 in CLL, it represents a credible candidate gene because of its role in regulating viral latency and pathogenesis of EBV. Specifically, NEDD4 regulates EBV-LMP2A which mimics signaling induced by the B-cell receptor altering B-cell development21.
In addition to these four loci there was evidence for two disease loci at 15q25.2 (rs783540) and 18q21.1 (rs1036935), although associations did not attain genome-wide significance (OR=1.18; 95%CI:1.10-1.27; P=3.67x10-6; Phet=0.07, I2=53% and OR=1.22; 95%CI:1.12-1.32; P=2.28x10-6, Phet=0.39,I2=3%, respectively; Supplementary Table 1, Supplementary Figure 2). rs783540 localizes to intron 2 of CPEB1 which has an established role in the regulation of cyclin B1 during embryonic cell division-differentiation. Two genes mapping centromeric to rs1036935, CXXC1 and MBD1 are involved in gene regulation. MBD1 expression in EBV-transformed lymphocytes correlated with risk genotype (Supplementary Figure 3). Although MBD1 has no documented role in CLL, it has potential to affect CLL development through translational control of MYC via MDBP binding22.
CLL shows male predominance and can be classified on the basis of the presence or absence of somatic hypermutations of immunoglobulin heavy-chain variable (IGVH) genes23,24, with mutated-CLL having a better prognosis. 17p-deletion is also associated with inferior survival25. We assessed the relationship between age, gender, family history of B-cell malignancy, IGVH-mutation and 17p-deletion status, and SNP-genotypes by case-only logistic regression (Supplementary Table 2). Furthermore, we examined the influence of genotype on overall survival (OS; Stages 1 and 4) and progression free survival (PFS; Stage 1; Supplementary Table 3). None of the SNPs displayed evidence of a relationship with age or gender (based on cases from all Stages). Having a family history of B-cell malignancy (Stages 1, 2) was associated with a higher frequency of FARP2 rs11681497 and rs757978 risk genotypes (P=0.031, 0.037 respectively; Supplementary Table 2), compatible with familial cases being enriched for genetic susceptibility. No relationship between 17p deletion status and genotype was shown (Stage 1). Although there was evidence rs305061 risk genotype was associated with worse OS (Supplementary Table 3), IGVH mutational status (Stages 1,2,4) was highly correlated with rs305061, with risk genotype correlating with unmutated-CLL (P=0.0002; Supplementary Table 2). Since rs305061 maps to IRF8 this relationship is compatible with dysfunctional B-cell activation signaling being associated with possession of risk genotype.
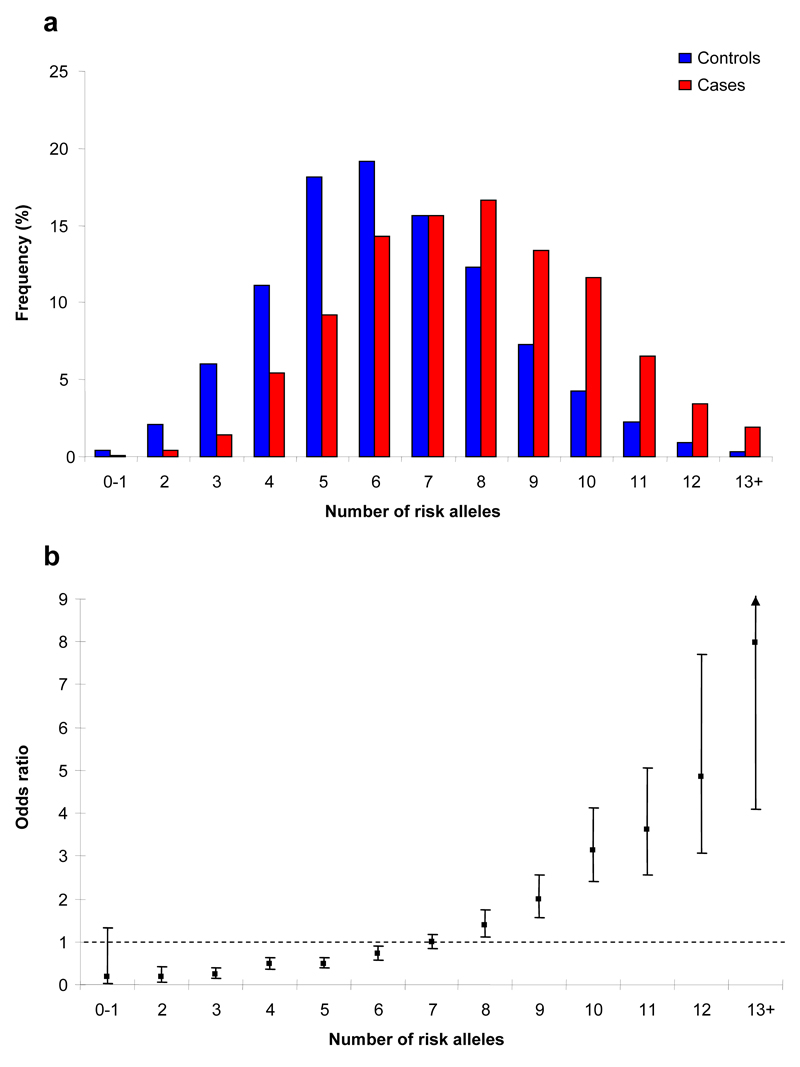
To gain insight into the allelic architecture of predisposition to CLL we examined for interactive effects between 2q37.3, 8q24.21, 15q21.3, and 16q24.1 variants and the previously identified loci at 2q13(rs17483466), 2q37.1(rs13397985), 6p25.3(rs872071), 11q24.1(rs735665), 15q23 (rs7176508) and 19q13.32(rs11083846). The only evidence for an interaction between loci was provided by rs305061 and rs7176508 (P=0.045), albeit non-significant after adjusting for multiple testing (Supplementary Table 4). The proportion of case and control subjects grouped according to the number of risk alleles that they carry is detailed in Figure 3. The distribution of risk alleles follows a normal distribution in both cases and controls, but with a shift toward a higher number of risk alleles in the cases.
Figure 3. Cumulative impact of the 10 variants on CLL risk.
(a) Distribution of risk alleles in controls (blue bars) and CLL cases (red bars) for the 10 loci (rs757978, rs2456449, rs7169431 and rs305061 and the six previously identified loci2 - rs17483466, rs13397985, rs872071, rs735665, rs7176508, and rs11083846);
(b) Plot of the increasing ORs for CLL with increasing number of risk alleles. The ORs are relative to the median number of 7 risk alleles; Vertical bars correspond to 95% confidence intervals. The distribution of risk alleles follows a normal distribution in both case and controls, with a shift towards a higher number of risk alleles in cases. Analysis is based on data from Stages 1, 2 and UK-replication series 2. Horizontal line denotes the null value (OR=1.0).
The carrier frequencies of risk alleles are high in the European population and hence the 10 loci collectively make a major contribution to the development of CLL with the population attributable fraction ascribable to these loci being ~87%. Moreover, the risk of CLL increases with increasing numbers of variant alleles for the 10 loci (ORper-allele=1.39, 95%CI:1.35-1.44; P=2.73x10-88; Figure 3, Supplementary Table 5). Individuals with 13+ risk alleles have a >7-fold increase in CLL risk compared to those with a median number of risk alleles. We estimate that the 10 loci we have identified account for ~10% of the excess familial risk of CLL, assuming a polygenic model. While the estimated effect of each of the 10 risk variants we have identified may be upwardly biased due to the phenomenon referred to as the “winners curse”, ORs may be underestimates because the additive model on which analyses are based assumes equal weighting across the SNPs. The effect on susceptibility attributable to variation at the loci and the effect of the actual causal variant responsible for the association is however, likely to be greater. Furthermore, many of the loci may carry additional risk variants, potentially including low-frequency variants with larger influence on CLL risk. Genome-wide linkage scans have provided evidence, albeit non-significant, for moderate-high penetrance susceptibility loci for CLL26–28. As none of the variants we have identified map to these regions of linkage it is unlikely that any other allele at the loci we have identified is responsible for these linkage signals.
The power of our study to detect the major common loci conferring risks of ≥1.4 (e.g. rs872071) was high. Hence, we consider that there are unlikely to be many additional SNPs with similar effects for alleles with frequencies >0.4 in populations of European ancestry. In contrast, we had low power to detect alleles with smaller effects and/or MAFs<0.1 (e.g. rs7169431). By implication, variants with such profiles are likely to represent a much larger class of susceptibility loci for CLL, whether because of truly small effect sizes or sub-maximal LD with tagging SNPs. Furthermore, GWA-based strategies are not optimally configured to identify low frequency variants with potentially stronger effects. In addition, these arrays are not ideally formatted to capture copy number variants which may also impact on CLL risk. Thus it is likely that a large number of low penetrance variants remain to be discovered. This assertion is supported by the continued excess of associations observed over those expected, in addition to the regions studied herein. Further efforts to expand the scale of GWA meta-analyses, in terms of both sample size and SNP coverage, and to increase the number of SNPs taken forward to large-scale replication may identify additional risk variants.
These results provide evidence for low risk variants predisposing to CLL and insight into the development of this hematological malignancy. Ethnic differences in the risk of CLL are recognized and it is notable that the MAF of 8q24.21 and 15q21.3 SNPs provided evidence of differences in the Spanish and UK/Swedish populations (Supplementary Table 1). Hence it will be interesting to explore how our findings translate to non-Western countries with substantially low incidence rates29. Furthermore, the reciprocal familial risks between CLL and other B-cell malignancies1 raises the possibility that these variants may also influence the risk of related B-cell tumors.
Methods
Initial genome-wide scan
This study describes the follow-up of a previously reported GWA of CLL2. Briefly 505 cases were genotyped using HumanCNV370-Duo BeadChips (Illumina, San Diego, USA). For controls we made use of publicly accessible HumanHap550K BeadChip genotype data generated on 1,438 individuals from the WTCCC British 1958 Birth Cohort. Quality control metrics included removal of samples with call rates <90% and SNP assays with call rates <95%. Subjects with evidence of cryptic relatedness and non-European background were excluded from the analysis. We considered only the 299,983 autosomal SNPs with MAF>1% in cases and controls, and with no extreme evidence of departure from Hardy-Weinberg equilibrium (HWE; P>10-5) in cases or controls. Comparison of observed and expected distributions showed little evidence for an inflation of test statistics (inflation factor λ=1.053).
Replication samples
In Stage 2 (UK-replication series 1) we genotyped SNPs in 540 CLL cases (353 male; mean age at diagnosis 60.3years; SD±12.2) ascertained through the Royal Marsden NHS Hospitals Trust, 1998-2006. All cases were UK residents and self-reported to be of European ancestry. Genotyping was conducted using the Sequenom MassARRAY system (http://www.sequenom.com/). For controls we made use of publicly accessible genotype data generated on 2,736 UK-blood donor controls. We excluded SNPs that deviated from HWE at P<1.0 x 10-5 in cases or controls. We also removed SNPs with MAF<0.05. To identify and exclude individuals with non-Western European ancestry, case and control data was merged with individuals of different ethnicities from HapMap, genome-wide IBS distances for markers shared between HapMap and our SNP panel were determined, and dissimilarity measures used to perform principal component analysis. After imposing these stringent quality control measures 162 SNP genotypes were available on 519 cases and 2,695 controls and this dataset formed the basis of our analysis.
In Stage 3 we genotyped 19 SNPs showing the strongest evidence of an association from joint analysis of Stages 1 and 2 in two independent case-control studies. UK-replication series 2: Cases were ascertained through the Newcastle CLL Consortium - 660 UK Caucasian patients with CLL diagnosed between 1970 and 2006 who attended hematology units in the UK (Newcastle Royal Victoria Infirmary, Gateshead Queen Elizabeth Hospital, Birmingham Heartlands Hospital, Hull Royal Infirmary, Liverpool Royal University Hospital, Royal Bournemouth Hospital, University of Wales College of Medicine Hospital). Peripheral blood for DNA extraction was taken between 1998 and 2006. Controls comprised 809 UK Caucasian healthy individuals (509 male) aged 16-69 (mean age 48.2 years, SD ±14.3) recruited to a study of acute leukemia conducted 1991-1996, previously described30. Spanish-replication series: 445 cases and 450 controls. Cases were Spanish residents and were of self-reported European ancestry. Controls were obtained from the MedXen Control population cohort (Xunta de Galicia) and were matched to cases by geographic origin, age, and sex.
In Stage 4 we genotyped 10 SNPs, showing the strongest evidence of an association from joint analysis of Stages 1-3 in the Swedish-replication series: 304 cases samples from SCALE (Scandinavian Lymphoma Etiology)31 and 91 samples from the biobank at Uppsala University Hospital, Uppsala, Sweden. For the SCALE samples, bloods were collected from 275 cases during 1999-2001, within a median of 4 months from diagnosis (0-29 months) while in 29 cases, follow-up samples ascertained in 2007-2008 were used. The samples received from the biobank at Uppsala University Hospital were collected between 1982 and 2005. The Swedish replication series totaled 148 females and 247 males (mean age at diagnosis 62.6). Swedish controls were collected at Karolinska Institutet, Stockholm, Sweden and included 397 healthy individuals (251 male) aged 33-71 years (mean 61.3, SD± 6.8).
Data on both progression-free survival (PFS) and overall survival (OS) was available on 356 Stage 1 cases33 who participated in the UK CLL-4 trial, a randomized phase III trial established to compare the efficacy of fludarabine, chlorambucil, and the combination of fludarabine plus cyclophosphamide as treatment for Binet stages B, C and A-progressive CLL34. Progression free survival was defined as the survival time from the date of randomisation to relapse needing further therapy, progression or death from any cause. Data on OS was also available for 395 of the Swedish replication series (Stage 4).
In all studies the diagnosis of CLL has been confirmed in accordance with the current WHO classification guidelines35.
Ethics
Collection of blood samples and clinico-pathologiocal information from patients and controls was undertaken with informed consent and relevant institutional ethical review board approval in accordance with the tenets of the Declaration of Helsinki.
Replication series genotyping
DNA was extracted from EDTA venous blood samples using standard methodologies and Picogreen quantified (Invitrogen, Paisley, United Kingdom). To ensure quality control of genotyping, a series of duplicate samples were genotyped in the same batches. For all SNP assays >99% concordant results were obtained. Genotyping in UK-replication 1 was performed by Sequenom MassARRAY system (http://www.sequenom.com/). We attempted to type 180 SNPs, although it was not possible to design functional assays for 18 SNPs with this technology. Details of the methodology are available on request. To exclude technical artifacts of genotyping, we included a random series of 24 samples previously genotyped using Illumina 550K HapMap arrays (Illumina, San Diego, USA). We successfully genotyped 519 unique subjects passing quality control metrics, excluding validation samples, and study duplicates. Genotyping of UK-replication 2 and the Swedish-replication series were conducted by competitive allele-specific PCR KASPar chemistry (KBiosciences Ltd, Hertfordshire, UK; http://www.kbioscience.co.uk/); primer sequences and conditions available on request. Excluding validation samples and study duplicates, passing quality control metrics we genotyped 1,469 unique subjects in UK-replication series 2 and 792 unique subjects in the Swedish replication series. Genotyping of Spanish samples was conducted using the Sequenom MassARRAY system (http://www.sequenom.com/) in two i-Plex assays. Excluding validation samples and study duplicates, passing quality control metrics we genotyped 874 unique subjects.
IGVH mutational and 17p deletion status
IGVH mutational status was determined according to BIOMED-2 protocols as described previously36,37 or commercial reagents (InVivoScribe Technologies, San Diego, USA). Clonality was assessed by size discrimination of PCR products using semi-automated ABI3730xl/ABI3700 sequencers in conjunction with Genescan software (Applied Biosystems, Foster City, USA). Sequences obtained were submitted to online database IMGT/V-QUEST38. In accordance with published criteria, sequences with a germline identity of ≥98% were classified as unmutated, and those displaying identity <98% as mutated. Deletion of 17p was defined as >10% loss in cells.
Statistical analysis
Statistical analyses were undertaken using R (v2.3.1) and STATA (v10.0) Software. Deviation of the genotype frequencies in the controls from those expected under HWE was assessed by the χ2-test (1d.f.). The association between each SNP and risk of CLL was assessed by the Cochran-Armitage trend test. Odds ratios (ORs) and associated 95% confidence intervals (CIs) were calculated by unconditional logistic regression. Relationships between multiple SNPs showing association with CLL risk in the same region were investigated using logistic regression analysis and the impact of additional SNPs from the same region was assessed by a likelihood-ratio test.
Associations by gender, age and mutation status were examined by logistic regression in case-only analyses. The combined effect of pairs of loci identified with risk was investigated by logistic regression modeling; evidence for interactive effects between SNPs assessed by a likelihood ratio test. The OR and trend test for increasing numbers of deleterious alleles was estimated by counting two for a homozygote and one for a heterozygote assuming equal weights.
Meta-analysis was conducted using standard methods39. Cochran’s Q-statistic to test for heterogeneity39 and the I2 statistic40 to quantify the proportion of the total variation due to heterogeneity were calculated. Large heterogeneity is typically defined as I2≥75%. The population attributable fraction was estimated from where xi = (1-p)2 + 2p(1-p)OR1 + p2OR2, p is the population allele frequency, and OR1 and OR2 are the ORs associated with hetero- and homozygosity respectively. The sibling relative risk attributable to a given SNP was calculated using the formula41:
where p is the population frequency of the minor allele, q=1-p, and r1 and r2 are the relative risks (estimated as OR) for heterozygotes and rare homozygotes, relative to common homozygotes. Assuming a multiplicative interaction the proportion of the familial risk attributable to a SNP was calculated as log(λ*)/log(λ0), where λ0 is the overall familial relative risk estimated from epidemiological studies, assumed to be 7.251.
Bioinformatics
We used Haploview software (v3.2) to infer the LD structure of the genome in the regions containing loci associated with disease risk. We applied two in silico algorithms, PolyPhen and SIFT, to predict the impact of non-synonymous SNPs on protein function.
Relationship between SNP genotype and expression levels
To examine for a relationship between SNP genotype and expression levels of MYC, FARP2, NEDD4, MBD1, and CPEB1 in lymphocytes we made use of publicly available expression data generated from analysis of 90 Caucasian derived Epstein-Barr virus–transformed lymphoblastoid cell lines using Sentrix Human-6 Expression BeadChips (Illumina, San Diego, USA)42,43. Online recovery of data was performed using WGAViewer Version 1.25 Software. Differences in the distribution of levels of mRNA expression between SNP genotypes were compared using a Wilcoxon-type test for trend44.
Supplementary Material
Acknowledgements
Leukaemia Research Fund (LRF) provided principal funding for the study (LRF05001 and 06002). Additional funding was provided by CLL Global Research Foundation, Cancer Research UK (C1298/A8362 supported by the Bobby Moore Fund), and the Arbib Fund. Dalemari Crowther-Swanepoel (DC-S) was in receipt of a PhD studentship from the Institute of Cancer Research. This study makes use of data generated by the Wellcome Trust Case-Control Consortium 1 and 2. A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Funding for the project was provided by the Wellcome Trust under award 076113 and 085475. In the UK NHS funding for the Royal Marsden Biomedical Research Centre is acknowledged. EC and AE acknowledge Fundacion Genoma España and CICYT funding (SAF 08/3630) and the RTICC from the Insituto de Salud Carlos III. AC and MT acknowledge Xunta de Galicia funding for Spanish control population (PGIDIT 08CSA005208PR) and the support of the Spanish National Genotyping Center (USC Node). We would like to thank Dr Leonid Padyukov, Karolinska Institutet, Stockholm, Sweden, and the EIRA group for providing control samples from the Swedish population for the Swedish replication study. We also acknowledge the work of Sue Richards and Rachel Wade (CTSU, Oxford) in furbishing survival data from CLL-4. We are grateful to all clinicians for their involvement in patient ascertainment. Finally we are grateful to all the patients and other individuals for their participation.
Footnotes
URLs
Online Inheritance in Man: http://www.ncbi.nlm.nih.gov/sites/entrez
The R suite can be found at http://www.r-project.org/
Detailed information on the tag SNP panel can be found at http://www.illumina.com/
dbSNP: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=snp
HAPMAP: http://www.hapmap.org/
KBioscience: http://kbioscience.co.uk/
WGAViewer: http://www.genome.duke.edu/centers/pg2/downloads/wgaviewer.php
International immunogenetics information system: http://imgt.cines.fr
The European Genome-phenome Archive (Wellcome Trust Case-Control Consortium [WTCCC]): http://www.ebi.ac.uk/ega/page.php
POLYPHEN: http://www.bork.embl-heidelberg.de/PolyPhen/
SIFT: http://blocks.fhcrc.org/sift//SIFT.html
SNAP (SNP Annotation and Proxy Search): http://www.broadinstitute.org/mpg/snap/
Author Contributions
RSH designed, obtained funding, directed the study, and oversaw analyses. RSH drafted the manuscript, with help from DC-S, PB, MCDB and SED. MCDB performed statistical analyses. DC-S, MCDB, SED performed bioinformatics analyses. RSH and DC established the parent study. RSH, DC and DC-S developed patient recruitment, sample acquisition and performed sample collection of cases. For the GWA and UK-replication series 1, DC-S and PB supervised laboratory management and oversaw genotyping of cases. DC-S conducted sequencing. JMA and DJA conceived of the Newcastle-based CLL study. JMA established the study, supervised laboratory management and oversaw genotyping of cases and controls. NJS performed sample management of cases and controls. AGH developed the Newcastle Haematology Biobank, incorporating the Newcastle-based CLL study. TM-F, GHJ, GS, RJH, ARP, DO, DJA, JRB, GP, CP, and CF developed patient recruitment, sample acquisition and performed sample collection of cases. Coordinated of the Spain replication series was conducted by EC and AE. EC and AE provided CLL samples and AC controls and compiled detailed phenotypic information from cases and controls respectively. Genotyping was performed by MT and AC and MT supervised laboratory management and quality control. For the Swedish case-control study MM performed sample collection and prepared DNA. RR performed sample collection for all cases, while JJ, GJ and KES performed sample collection of cases in the SCALE study. PB and DC-S performed genotyping of cases and controls.
All authors contributed to the final paper.
Competing Interests Statement
The authors declare no competing financial interests.
References
- 1.Goldin LR, Pfeiffer RM, Li X, Hemminki K. Familial risk of lymphoproliferative tumors in families of patients with chronic lymphocytic leukemia: results from the Swedish Family-Cancer Database. Blood. 2004;104:1850–4. doi: 10.1182/blood-2004-01-0341. [DOI] [PubMed] [Google Scholar]
- 2.Di Bernardo MC, et al. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat Genet. 2008;40:1204–10. doi: 10.1038/ng.219. [DOI] [PubMed] [Google Scholar]
- 3.Gunnarsson R, et al. Screening for copy-number alterations and loss of heterozygosity in chronic lymphocytic leukemia--a comparative study of four differently designed, high resolution microarray platforms. Genes Chromosomes Cancer. 2008;47:697–711. doi: 10.1002/gcc.20575. [DOI] [PubMed] [Google Scholar]
- 4.Ferreira BI, et al. Comparative genome profiling across subtypes of low-grade B-cell lymphoma identifies type-specific and common aberrations that target genes with a role in B-cell neoplasia. Haematologica. 2008;93:670–9. doi: 10.3324/haematol.12221. [DOI] [PubMed] [Google Scholar]
- 5.Grubor V, et al. Novel genomic alterations and clonal evolution in chronic lymphocytic leukemia revealed by representational oligonucleotide microarray analysis (ROMA) Blood. 2009;113:1294–303. doi: 10.1182/blood-2008-05-158865. [DOI] [PubMed] [Google Scholar]
- 6.Pfeifer D, et al. Genome-wide analysis of DNA copy number changes and LOH in CLL using high-density SNP arrays. Blood. 2007;109:1202–10. doi: 10.1182/blood-2006-07-034256. [DOI] [PubMed] [Google Scholar]
- 7.Gudmundsson J, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–7. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 8.Easton DF, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomlinson I, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39:984–8. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 10.Yeager M, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 11.Amundadottir LT, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–8. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 12.Kiemeney LA, et al. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat Genet. 2008;40:1307–12. doi: 10.1038/ng.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pomerantz MM, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41:882–4. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tuupanen S, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009;41:885–90. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- 15.Shaffer AL, et al. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–31. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyamoto Y, Yamauchi J, Itoh H. Src kinase regulates the activation of a novel FGD-1-related Cdc42 guanine nucleotide exchange factor in the signaling pathway from the endothelin A receptor to JNK. J Biol Chem. 2003;278:29890–900. doi: 10.1074/jbc.M301559200. [DOI] [PubMed] [Google Scholar]
- 17.Wang H, Morse HC., 3rd IRF8 regulates myeloid and B lymphoid lineage diversification. Immunol Res. 2009;43:109–17. doi: 10.1007/s12026-008-8055-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Jager PL, et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet. 2009;41:776–82. doi: 10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soderberg KC, Jonsson F, Winqvist O, Hagmar L, Feychting M. Autoimmune diseases, asthma and risk of haematological malignancies: a nationwide case-control study in Sweden. Eur J Cancer. 2006;42:3028–33. doi: 10.1016/j.ejca.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 20.Cartwright RA, et al. Chronic lymphocytic leukaemia: case control epidemiological study in Yorkshire. Br J Cancer. 1987;56:79–82. doi: 10.1038/bjc.1987.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikeda M, Longnecker R. The c-Cbl proto-oncoprotein downregulates EBV LMP2A signaling. Virology. 2009;385:183–91. doi: 10.1016/j.virol.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang XY, Supakar PC, Wu KZ, Ehrlich KC, Ehrlich M. An MDBP site in the first intron of the human c-myc gene. Cancer Res. 1990;50:6865–9. [PubMed] [Google Scholar]
- 23.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94:1848–54. [PubMed] [Google Scholar]
- 24.Damle RN, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–7. [PubMed] [Google Scholar]
- 25.Zenz T, Dohner H, Stilgenbauer S. Genetics and risk-stratified approach to therapy in chronic lymphocytic leukemia. Best Pract Res Clin Haematol. 2007;20:439–53. doi: 10.1016/j.beha.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Goldin LR, et al. A genome scan of 18 families with chronic lymphocytic leukaemia. Br J Haematol. 2003;121:866–73. doi: 10.1046/j.1365-2141.2003.04372.x. [DOI] [PubMed] [Google Scholar]
- 27.Sellick GS, et al. A high-density SNP genome-wide linkage search of 206 families identifies susceptibility loci for chronic lymphocytic leukemia. Blood. 2007;110:3326–33. doi: 10.1182/blood-2007-05-091561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sellick GS, et al. A high-density SNP genomewide linkage scan for chronic lymphocytic leukemia-susceptibility loci. Am J Hum Genet. 2005;77:420–9. doi: 10.1086/444472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weiss NS. Geographical variation in the incidence of the leukemias and lymphomas. Natl Cancer Inst Monogr. 1979:139–42. [PubMed] [Google Scholar]
- 30.Allan JM, et al. Polymorphism in glutathione S-transferase P1 is associated with susceptibility to chemotherapy-induced leukemia. Proc Natl Acad Sci U S A. 2001;98:11592–7. doi: 10.1073/pnas.191211198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smedby KE, et al. Ultraviolet radiation exposure and risk of malignant lymphomas. J Natl Cancer Inst. 2005;97:199–209. doi: 10.1093/jnci/dji022. [DOI] [PubMed] [Google Scholar]
- 32.Hallek M, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111:5446–56. doi: 10.1182/blood-2007-06-093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wade R, et al. Genome-wide scan of SNPs in CLL4 trial patients identifies genetic variants influencing prognosis. Submitted for publication. doi: 10.1182/blood-2007-08-110130. [DOI] [PubMed] [Google Scholar]
- 34.Catovsky D, et al. Assessment of fludarabine plus cyclophosphamide for patients with chronic lymphocytic leukaemia (the LRF CLL4 Trial): a randomised controlled trial. Lancet. 2007;370:230–9. doi: 10.1016/S0140-6736(07)61125-8. [DOI] [PubMed] [Google Scholar]
- 35.Müller-Hermelink H, M E, Catovsky D, Harris N. Chronic lymphocytic leukaemia/small lymphocytic lymphoma. In: Jaffe ES, Harris NL, Stein H, Vardiman JW, editors. World Health Organization Classification of Tumours: Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: IARC Press; 2001. pp. 127–130. [Google Scholar]
- 36.van Dongen JJ, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17:2257–317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 37.van Krieken JH, et al. Improved reliability of lymphoma diagnostics via PCR-based clonality testing: report of the BIOMED-2 Concerted Action BHM4-CT98-3936. Leukemia. 2007;21:201–6. doi: 10.1038/sj.leu.2404467. [DOI] [PubMed] [Google Scholar]
- 38.Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503–8. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petitti D. Meta-analysis Decision Analysis and Cost-Effectiveness Analysis. Oxford, New York: Oxford; 1994. [Google Scholar]
- 40.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 41.Houlston RS, Ford D. Genetics of coeliac disease. QJM. 1996;89:737–43. doi: 10.1093/qjmed/89.10.737. [DOI] [PubMed] [Google Scholar]
- 42.Stranger BE, et al. Genome-wide associations of gene expression variation in humans. PLoS Genet. 2005;1:e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stranger BE, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–53. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cuzick J. A Wilcoxon-type test for trend. Stat Med. 1985;4:87–90. doi: 10.1002/sim.4780040112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.