Abstract
Lineage-selective expression of developmental genes is dependent on the interplay between activating and repressive mechanisms. Gene activation is dependent on cell-specific transcription factors that recognize transcriptional enhancer sequences. Gene repression often depends on the recruitment of Polycomb group (PcG) proteins, although the sequences that underlie the recruitment of PcG proteins, also known as Polycomb response elements (PREs), remain poorly understood in vertebrates. While distal PREs have been identified in mammals, a role for positive-acting enhancers in PcG-mediated repression has not been described. Here we have used a highly efficient procedure based on lentiviral-mediated transgenesis to carry out in vivo fine-mapping of, cis-regulatory sequences that control lineage-specific activation of Neurog3, a master regulator of pancreatic endocrine differentiation. Our findings reveal an enhancer region that is sufficient to drive correct spacio-temporal expression of Neurog3 and demonstrate that this same region serves as a PRE in alternative lineages where Neurog3 is inactive.
Introduction
Embryonic development involves the establishment of progressively divergent transcriptional programs. This process relies on the dynamic activation of developmental regulatory genes, which is largely determined through the interaction of lineage-specific transcriptional activators with cis-acting sequences located in enhancers [1,2].
Strict spatiotemporal regulation is further determined by mechanisms that repress lineage-specific regulatory genes. Polycomb group (PcG) proteins are thought to play a major role in the repression of such genes [3–5]. PcG-dependent repression is mediated by two major complexes known as PRC1 and PRC2 that act in an interdependent manner. PRC2 contains Ezh2 and Ezh1 in its non-canonical form, both of which catalyzes the trimethylation of Lysine 27 on histone 3 (H3K27me3). This histone mark contributes to the recruitment of the PRC1 complex, which promotes H2A ubiquitylation, interference with transcriptional machinery, and chromatin compaction [4–8].
In contrast to the extensive knowledge of how DNA binding factors are recruited to specific genomic regions to promote lineage-specific transcription, the recruitment of PcG complexes remains poorly understood. In Drosophila, Polycomb response elements (PREs) are enriched in specific DNA binding factor motifs, and can therefore often be predicted [9]. To demonstrate an element functions as a PRE, it must be shown that it can autonomously recruit polycomb proteins outside of its normal genomic environment, which has been done for several Drosophila PREs [9,10]. In vertebrates, our understanding of PREs is more limited, and so far no combination of DNA-binding motifs has been able to predict their existence. Recent studies, however, have shown that PcG proteins primarily target GC-rich promoter regions, and suggest that a high density of unmethylated CpGs may be sufficient to promote PcG recruitment and repression in the absence of activating signals [11–14]. Other work however suggests that CpG islands are not required for the recruitment of PcG- proteins in the HoxD gene cluster in vivo [15]. In addition to promoter associated PREs, distal PREs have been identified in the mammalian genome, adding to the challenge of understanding what characterizes a PRE and how they contribute to gene regulation [13,15–17]. Thus, current evidence implicates both CpG-rich promoter-proximal regions and distal regions in the recruitment of PcG repressive complexes. However, enhancer regions exerting a dual regulatory function of promoting transcription in one lineage and ensuring PcG-mediated repression in another lineage, have not been shown.
In the current study we have examined the cis-regulatory mechanisms of Neurog3, which encodes a transcription factor that is essential for the formation of all pancreatic endocrine lineages, including insulin-producing β-cells [18–20]. Neurog3 is transiently activated in a subpopulation of cells of the ductal epithelium of the embryonic pancreas, and orchestrates a lineage-committed progenitor program that leads to the differentiation of hormone-producing cells [19–22]. Understanding the mechanisms that control Neurog3 activation is therefore critical for efforts to artificially program functional β-cells.
Previous work demonstrated that the 5.7 kb upstream sequence of human NEUROG3 can drive a correct spatio-temporal expression pattern in transgenic mice [23], yet critical cis-elements within this sequence remain to be established. Several studies have revealed transcription factors that interact with specific sequences in the Neurog3 5’ region [23–26], although the in vivo function of such regulatory elements has not been examined.
So far, studies of the transcriptional mechanisms that underlie pancreas development have largely focused on activating transcription factors and cis-regulatory elements [23–29]. Pancreatic cell differentiation, however, is also linked to dynamic changes in PcG-repressed chromatin [30–32], although nothing is known about the sequence elements that direct the repressive programs that are relevant for pancreatic endocrine differentiation.
Here we used an efficient lentiviral transgenesis strategy to dissect the function of Neurog3 cis-regulatory elements in vivo. Using this approach, we identify an enhancer region that is sufficient to direct correct expression of the Neurog3 gene in embryonic pancreatic progenitors, and in this enhancers we map discrete cis-elements that are recuired for its function. In addition, our study shows that the same enhancer region acts as a PRE in alternate cellular lineages, thereby illustrating how one genomic region can exert a dual function as an enhancer and as a PRE.
Results
An enhancer region that activates Neurog3 in endocrine progenitors
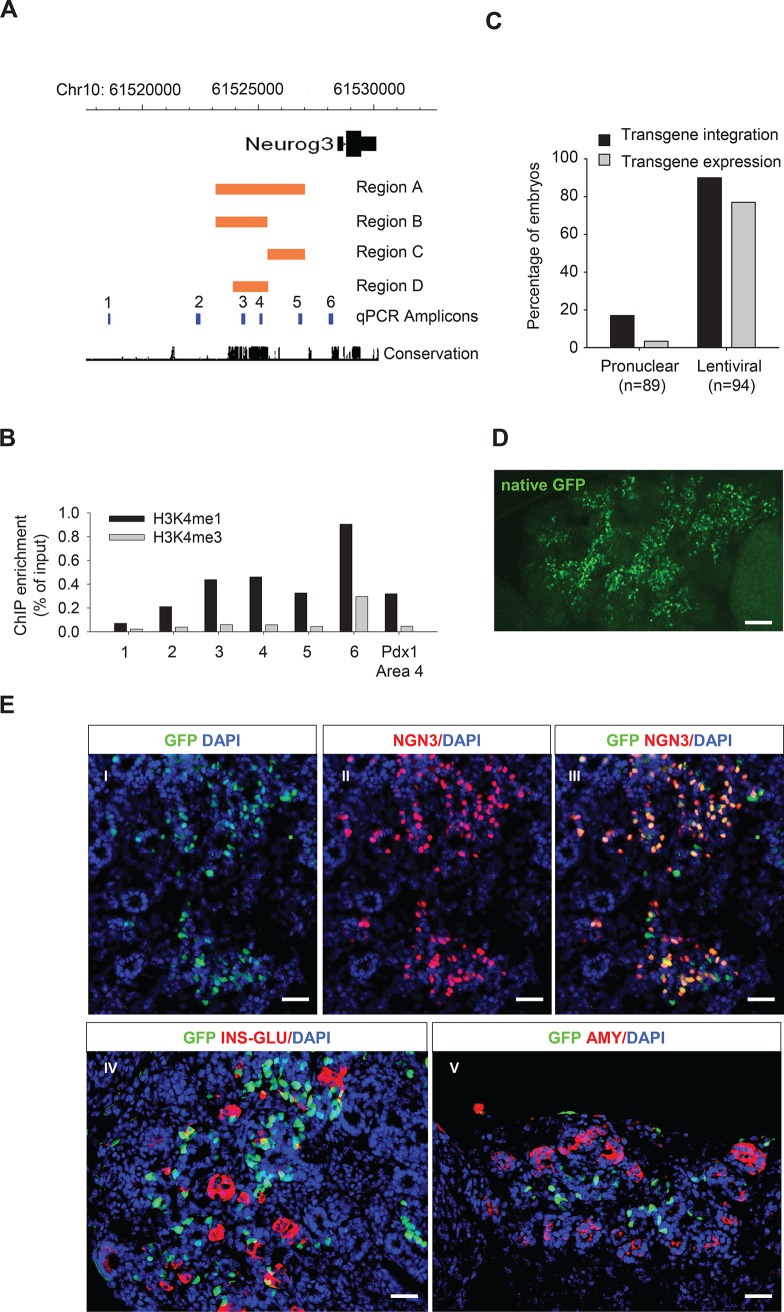
Sequence conservation and monomethylated Histone 3 Lysine 4 (H3K4me1) in the absence of trimethylated H3K4 (H3K4me3) are known hallmarks of transcriptional enhancers [33]. To identify sequences that regulate the expression of Neurog3 during development, we screened the Neurog3 5’ region for evolutionary sequence conservation and H3K4me1-enrichment in mouse pancreatic bud from embryonic day 13.5, when Neurog3 expression peaks. Conservation was particularly high in the region stretching from approximately -5kb to -3kb relative to the transcription start site (TSS) (Fig 1A). This same region also showed H3K4me1 enrichment in the absence of H3K4me3, to a similar extent as a well-known pancreatic enhancer of the Pdx1 gene (Fig 1B) [34].
Fig 1. Mapping a Neurog3 enhancer region through highly efficient lentiviral transgenesis.
A. Schematic representation of the screened Neurog3 locus. Orange bars depict regions A-D that were selected for characterization through standard transgenesis. PCR amplicons that were used for ChIP analysis (in B) are shown in blue. The conservation track represents conservation over 17 vertebrate species. B. ChIP revealed H3K4me1 without H3K4me3 enrichment in E13.5 pancreatic buds in putative Neurog3 enhancer regions (n = 1; note that the H3K4me3 signal results from Neurog3+ cells which represent <5% of the pancreatic bud). Pdx1 Area 4 is shown as a positive control as it represents a known enhancer. C. Lentiviral integration increases the efficiency of transgenesis and transgene expression compared with standard pronuclear injections. D, E. Expression of GFP under the control of the Neurog3 enhancer region B in lentiviral transgenics. Native GFP in a whole mount follows the trunk of E14.5 pancreas (D) and coincides with endogenous Neurog3 in (E, panel I-III), whereas no overlap is seen with Amylase (red; panel V) and only rare overlap is seen with Insulin and Glucagon (panel IV). Occasional GFP-positive Neurog3-negative cells were found at E14.5, most of which belonged to the endocrine lineage (panel III, IV). Scale bars are 75 μm (D) and 50μm (E).
Based on these findings we first tested the ability of a fragment stretching from approximately -5.5 kb to -1.5 kb of the Neurog3 gene to drive expression of a reporter gene selectively in Neurog3+ cells using standard transgenesis (S1 Fig). This 3.9kb element (construct A; Fig 1A) was able to reproduce selective expression of the LacZ reporter gene in all Neurog3+ cells, namely the ventral hypothalamus, the ventral spinal cord, the duodenum and the pancreas, with only minimal ectopic expression (S1A Fig). To assess if a similar result could be obtained by a smaller fragment, we generated construct B, C and D (Fig 1A) and demonstrated that only construct B (representing the 5 prime 2.2kb of construct A) consistently reproduced the expression pattern of Neurog3 with the exception of expression in the ventral spinal cord (S1B Fig). These results demonstrate that enhancer region B, hereafter referred to as the Neurog3 enhancer region, contains all the regulatory sequences to drive correct spatio-temporal expression of Neurog3 in the pancreas.
Recapitulation of Neurog3 enhancer activity using highly efficient lentiviral-mediated transgenesis
Next we aimed to further dissect the Neurog3 enhancer region by generating deletions of putative regulatory elements. However, the efficiency of standard transgenesis is prohibitively low to allow for a comprehensive screen of this nature. For example, in the abovementioned analysis, 263 embryos were collected, of which only 38 (14.5%) showed transgene integration and only 3 of them expressed LacZ in the pancreas at embryonic day 14.5. We thus tested lentiviral transduction in fertilized eggs followed by embryo implantation in pseudopregnant mice as an alternative strategy to produce transgenic animals [35]. After initial transduction with a lentiviral vector expressing GFP under the control of the Neurog3 enhancer region, we confirmed transgene integration in 85 out of 94 (90.4%) embryos and transgene expression in 72 (76.6%) embryos. In comparison, after pronuclear injection of the same Neurog3 enhancer region, only 3 out of 89 (3.4%) collected embryos expressed the transgene (Fig 1C). Taken together, the data indicate that in our hands lentiviral transgenesis is at least 20-fold more efficient than standard transgenesis (Fig 1C). In lentiviral transgenic embryos, selectivity of GFP expression in Neurog3+ cells was similar to the results obtained through standard transgenesis (Fig 1D and 1E, S1 and S2 Figs). Having thus validated lentiviral transgenesis for the assessment of enhancer function, we set out to identify critical activating elements within the Neurog3 enhancer region.
A 50 bp cis-element is essential for Neurog3 enhancer activation in pancreatic progenitors
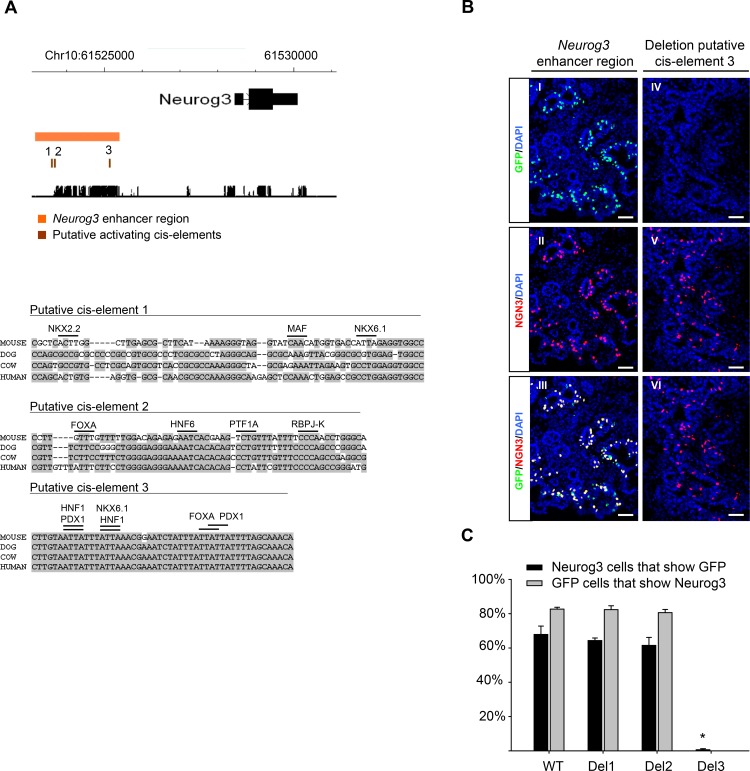
As a first step to identify critical activating sequence elements within the Neurog3 enhancer region, we searched for ~50 bp elements enriched in binding site sequence motifs for pancreatic transcription factors. We identified 3 motif clusters of 50–65 bp length located ~ -4.9 to -3.3kb upstream of the Neurog3 TSS, which we refer to as candidate cis-elements 1, 2 and 3 (Fig 2A). We assessed if these elements were essential for Neurog3 expression by generating lentiviral-mediated transgenic animals with the Neurog3 enhancer region carrying deletions for each of these 3 cis-elements. While loss of element 1 or 2 did not change the expression pattern in any of the transgenic embryos (16 and 17 transgenics studied, respectively), loss of element 3 led to a full abrogation of expression in 28 out of 30 transgenic embryos (Fig 2B and 2C). A critical role for this same element (previously described as ‘Cluster 1’ in human [23]) had been suggested previously based on interactions with key pancreatic transcription factors, although its in vivo function was not tested [23,26]. Taken together, these results uncover a distal 50 bp cis-element that is essential for activation of the Neurog3 enhancer in embryonic pancreatic endocrine progenitors.
Fig 2. An essential activating cis-element within the Neurog3 enhancer region.
A. Three elements in the Neurog3 enhancer displayed clustered binding motifs for known pancreatic transcription factors. Black lines above sequence indicate 4 bp core of transcription factor motif. B. Deletion of cis-element 3 (Del3; but not 1 or 2) from the Neurog3 enhancer region leads to a near complete loss of expression. The panels show GFP (green) and Neurog3 (red) in an E14.5 transgenic for the wild-type Neurog3 enhancer region (panel I-III) and a transgenic for the Neurog3 enhancer region with activating cis-element 3 deleted (panel IV-VI). C. Quantification of percentage of Neurog3 cells that are GFP positive (black bars) and the percentage of GFP cells that are Neurog3 positive (grey bars; not present for Del3 because in 7 out of 8 embryo’s no GFP cells were detected. Quantification was done on 8 embryos for each genotype in which slides amounting to at least 1000 Neurog3 cells were counted ([*] P = 5.3 x 10−9, Student’s t-test with Bonferroni correction). Scale bars: I-VI = 50 μm.
Pdx1, Foxa2 and Hnf1b act on cis-element 3 to activate Neurog3
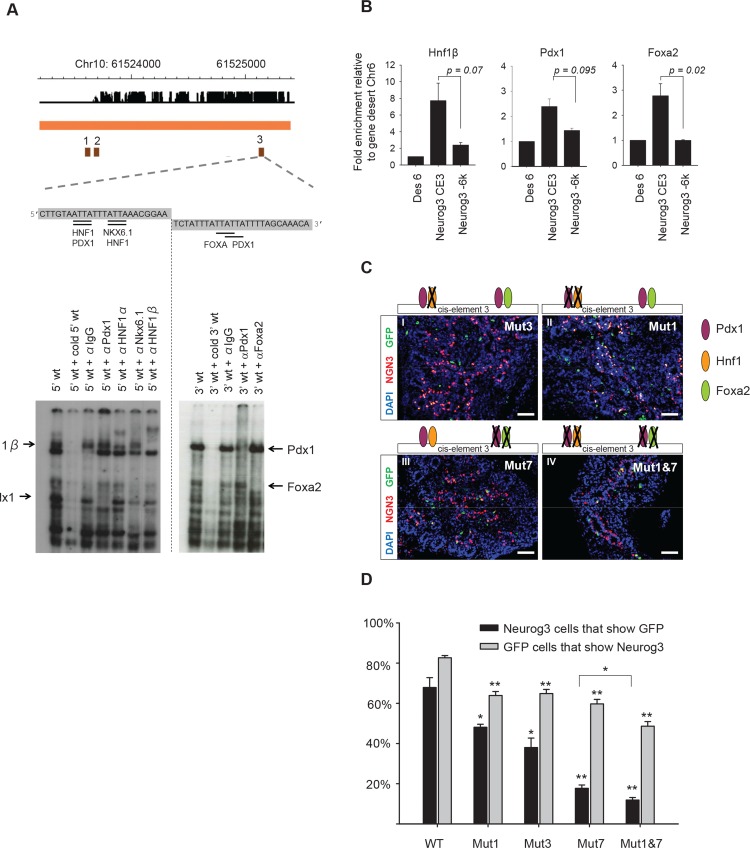
The analysis of transcription factor binding motifs together with previous work suggested that the Neurog3 enhancer region exerts its activating effect through the binding of Pdx1, Foxa2 and Hnf1b to cis-element 3 [23,26] (Fig 2A). To address this hypothesis we first tested binding of these factors to cis-element 3 using EMSA. Using nuclear extracts from pancreatic buds isolated at E13.5 and specific antibodies we confirmed binding of Pdx1, Foxa2, and Hnf1b (Fig 3A). Next, we performed ChIP on E13.5 pancreatic bud chromatin. This experiment, while technically challenging as <5% of the cells in pancreatic buds express Neurog3, suggested that these three factors also bind to the cis-element 3 of the endogenous enhancer region in vivo (Fig 3B).
Fig 3. Identification of cis-regulatory mutations that disrupt activation of Neurog3 in pancreatic progenitors.
A. Binding of Hnf1b, Pdx1 and Foxa2 to cis-element 3 of the Neurog3 enhancer using EMSAs with nuclear extracts from E13.5 pancreatic buds. The 5’ part of the cis-element 3 sequence binds Hnf1b and Pdx1 while the 3’ part binds Pdx1 and Foxa2. B. ChIP for Hnf1b (n = 3), Pdx1 (n = 2) and Foxa2 (n = 3) in E13.5 pancreatic buds. Binding to the cis-element 3 (Neurog3 CE3) was compared to control regions Des6 (locus in gene-desert) and Neurog3 -6K (Neurog3 5’ upstream region). Indicated P-values were calculated with Students t-test. C. Lentiviral transgenesis using enhancers with indicated mutations, all of which disrupt specific transcription factor binding sites (S3 Fig). Note how the strongest reduction of expression is found upon disruption of all identified binding sites within cis-element 3. D. Quantification of percentage of Neurog3 cells that are GFP positive (black bars) and the percentage of GFP cells that are Neurog3 positive (grey bars). Quantification was done on 8 embryos for each genotype in which at least 1000 Neurog3 cells were counted ([**] P<1.0 x 10−6, [*] P<0.02, Student’s t-test with Bonferroni correction). Scale bars: C I-VI = 50 μm.
To further address the activating role of this regulatory element we designed mutations that selectively disrupt binding of Hnf1 (Mut3), or both Pdx1 and Hnf1 (Mut1) to the 5’ side of cis-element 3, or else binding of both Pdx1 and Foxa2 to the 3’ side of cis-element 3 (Mut7) (S3 Fig, S1 Table and S2 Table). In lentiviral-mediated transgenic animals, all three mutations, and most clearly Mut7, caused a reduction in the expression penetrance (number of Neurog3 cells that show GFP) and cell-type specificity of expression (number of GFP cells that express Neurog3, Fig 3C and 3D). Next, we combined Mut1 and Mut7 and found a further 33% reduction in the combined mutant as compared to Mut7 alone for the number of Neurog3 cells that express GFP (Fig 3C and 3D). These results suggest that Pdx1, Foxa2 and Hnf1b activate Neurog3 through cis-element 3 in a partially redundant manner. Importantly, the data identifies discrete nucleotides that are essential to for the ability of the Neurog3 enhancer to drive expression in the embryonic pancreas in vivo.
The Neurog3 enhancer represses transcription in alternate lineages
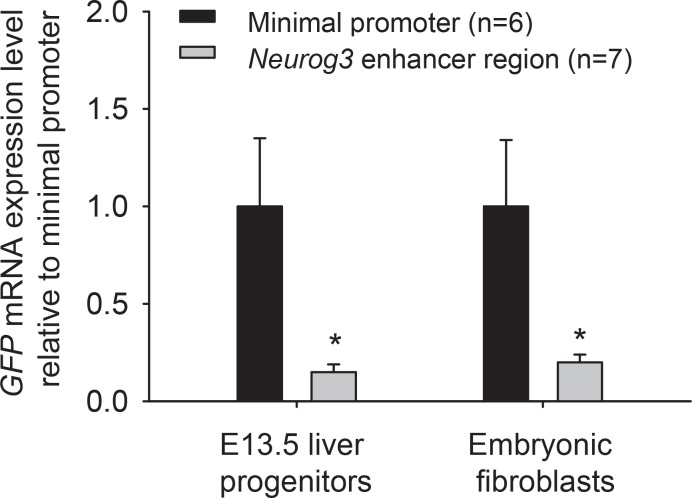
Next, we assessed whether the Neurog3 enhancer region, in addition to its ability to selectively drive expression in endocrine progenitors, could also contribute to transcriptional repression in non-expressing lineages. We therefore isolated transgenic mouse embryonic liver progenitors from E13.5 transgenic embryos that carried GFP under the control of the minimal promoter with or without the Neurog3 enhancer region and studied GFP expression. As expected, GFP was not detectable by fluorescence microscopy in both groups. However, low levels of transcript were detectable by qPCR, and they were 5-fold lower in transgenics that carried the Neurog3 enhancer region than in trangenics that integrated the backbone vector containing only the minimal promoter and GFP (Fig 4, see Fig 5B for a schematic representation of the two constructs). Similar results were obtained in transgenic mouse embryonic fibroblasts in the presence of the Neurog3 enhancer region (Fig 4). This effect did not result from differences in the number of integration events of the two constructs (S4 Fig). These results fit the notion that in addition to its ability to drive expression in endocrine progenitors, the Neurog3 enhancer region can exert transcriptional repression in alternative developmental lineages.
Fig 4. The Neurog3 enhancer region contributes to transcriptional repression in alternative lineages.
qRT-PCR analysis showed that in E13.5 liver progenitors and embryonic fibroblasts from transgenics carrying the Neurog3 enhancer, GFP mRNA was significantly reduced. GFP transcript levels were normalized for Tbp and compared to the construct that lacked the Neurog3 enhancer region ([*] (P<0.05, Student’s t-test).
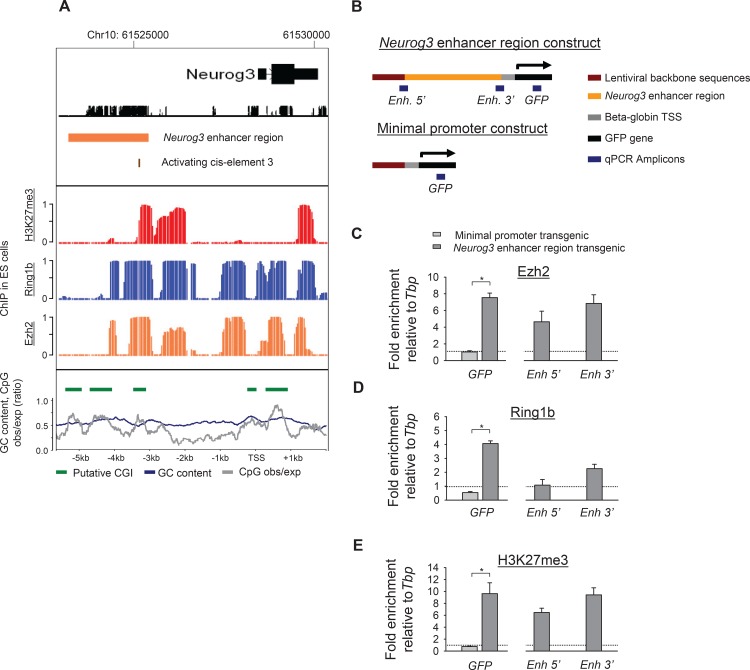
Fig 5. The minimal Neurog3 enhancer functions as a PRE in vivo.
A. ChIP-oligonucleotide array analysis of the endogenous mouse Neurog3 locus in ES cells. The graph shows significance of enrichments for H3K27me3 (red), Ring1b (blue), and Ezh2 (orange) in posterior probability ranging from 0 to 1. The bottom panel shows a CpG content analysis for the same genomic coordinates. The average GC content in 400 bp sliding windows is shown in blue and the CpG fold over expected is shown in grey. Green bars depict putative CpG islands. B. Schematic representation of amplicons used in ChIP-qPCR to selectively study the integrated exogenous Neurog3 enhancer region or the control minimal promoter (note that only GFP can be used on both). C-E. ChIP-qPCR analysis for Ezh2, Ring1b and H3K27me3 on the exogenous Neurog3 enhancer region in transgenic E13.5 liver (n = 2) ([*] P<0.01, Student’s t-test).
The Neurog3 enhancer region functions as a PRE in vivo
Next, we investigated whether the Neurog3 enhancer region induces transcriptional repression in alternative cellular lineages by functioning as a PRE. To address this, we first analyzed H3K27me3 enrichment in cell types that do not express Neurog3 including Pdx1 positive purified E10.5 pancreatic progenitor and adult islets that represent developmental stages that precede and follow Neurog3 expression, respectively. We observed H3K27me3 within and around the endogenous enhancer region in virtually all cell types analyzed [30] (S5 Fig). Moreover, we performed ChIP for the PcG subunits Ezh2 and Ring1b in ES cells and found these to be enriched in and around the Neurog3 enhancer region (Fig 5A). It is known that PcG-mediated repression preferentially occurs among CpG-island containing genes, and recently it was proposed that CpG islands themselves play a role in recruiting PcG proteins [12,13]. Consistent with this notion, the Neurog3 enhancer region contains several putative CpG-rich areas (Fig 5A). These findings suggested that this enhancer region might act as a PRE, although the same results could arise if other sequences that are adjacent to the Neurog3 enhancer region are responsible for recruiting PcG subunits to the enhancer region. We thus tested if transgenes that carried the Neurog3 enhancer region are capable of recruiting PcG subunits Ezh2 and Ring1b and can establish H3K27me3-enriched chromatin in liver from E13.5 embryos. We first examined embryonic liver cells from lentiviral transgenics that lacked the enhancer region, and found that they did not exhibit H3K27me3 enrichment and were not bound by PcG proteins at the transgenic locus (Fig 5C–5E). By contrast, in transgenic animals that integrated the Neurog3 enhancer region, the exogenous transgene locus was bound by Ezh2 and Ring1b and displayed H3K27me3 enrichment that stretched into the GFP coding region (Fig 5C–5E). To assess the role of cis-element 3 in the establishment of PcG-mediated repression we performed ChIP for H3K27me3 in animals that were transgenic for the Neurog3 enhancer region with a deletion of cis-element 3. The results demonstrated that the 50 bp cis-element was dispensable for the establishment of H3K27me3 enrichment (S6A Fig). Finally, we tested whether the Neurog3 enhancer promotes PcG-mediated repression that extends into adulthood, and found that indeed H3K27me3 enrichment was present in pancreatic exocrine tissue and skeletal muscle from adult transgenic mice (S6B and S6C Fig). Taken together, these data demonstrate that the minimal Neurog3 enhancer region functions as a PRE in vivo.
Discussion
The exact manner in which discrete genomic sequences provide either activating or repressive instructions to ensure lineage-specific gene transcription is still incompletely understood. Here we applied an efficient lentiviral transgenic strategy to understand the cis-regulatory sequences that underlie the initiation of the pancreatic endocrine differentiation program. Our data revealed a distal Neurog3 enhancer region that serves as a platform through which activating transcription factors direct Neurog3 expression in the pancreatic epithelium. This same enhancer region simultaneously acts as a PRE in alternate developmental lineages. Thus, we have identified a genomic region that functions as a lineage-specific enhancer and as a PRE.
A highly efficient transgenic approach to dissect cis-regulatory function
Understanding the cis-regulatory mechanisms that underlie organogenesis and differentiation is a fundamental challenge for biomedical research. Current epigenomic assays theoretically allow for high-throughput prediction of genomic regulatory elements. For example, active enhancers can be predicted by the combinatorial presence of specific histone modifications, nucleosome depletion, or transcription factor occupancy patterns [36–38]. Understanding the function of such predicted elements requires experimental analysis in meaningful developmental contexts. Existing technologies for this purpose, however, remain inefficient. Zebrafish provide an efficient model system to test putative regulatory elements, but it remains unproven that most mammalian cis-regulatory elements are appropriately recognized in this species. A significant example of a divergent regulatory mechanism in zebrafish is that although they develop an endocrine pancreas, the putative orthologue of Neurog3 is not expressed in endocrine progenitors [39]. Given their evolutionary proximity and extensive validation, mouse transgenics remain a desirable model system to study mammalian cis-regulatory mechanisms [37].
We have here used lentiviral mediated transgenesis to increase the throughput of mouse transgenesis for the discovery and fine mapping of Neurog3 regulatory sequences. This approach enhanced the efficiency of transgene expression by approximately 20-fold relative to conventional transgenesis, without compromising the specificity of expression. The results suggest that mouse lentiviral transgenesis is a robust tool for discovery, fine-mapping, and characterization of mammalian regulatory sequences, and can thus prove useful to elucidate the role of cis-regulatory elements in development and disease.
Dissection of an enhancer that recapitulates pancreatic Neurog3 expression
We have identified a Neurog3 enhancer region that is sufficient to selectively drive expression in Neurog3+ multipotent ductal epithelial cells, and have fine-mapped a critical cis-element in this enhancer. Although our studies are the first to define this enhancer region and demonstrate the critical role of a ~50bp cis-element in vivo, a critical role for this cis-element had been proposed previously based on the fact that it was bound by key pancreatic regulators [23,24,26]. Within this activating sequence we performed a mutation screen to further finemap the critical bases. Notably, two of the mutations that inhibit Neurog3 activation were high-affinity binding sites for Pdx1, a transcription factor previously shown to be a critical regulator of Neurog3 activation and of the specification of pancreatic endocrine progenitors [26]. We also identified a functionally important binding site for Hnf1b, which plausibly mediates the role of this transcription factor in pancreatic endocrine specification [40]. Other cis-elements and transcription factor binding sites were found to be functionally redundant, underscoring the importance of functional assessment of such regulatory elements in vivo. These findings can be exploited to address how Neurog3 becomes activated in the multipotent pancreatic epithelium, and to efficiently target prospective pancreatic endocrine progenitors.
The Neurog3 enhancer region functions as a PRE in vivo
The demarcation of sharp lineage boundaries of gene expression is directed by cell specific DNA-binding activators as well as by repressive mechanisms. Distal regulatory regions play a pivotal role by providing binding sites for cell-specific transcriptional activators, whereas the recruitment of repressive complexes to specific genomic sites is less understood.
Recently it was proposed that in mammals, unlike current knowledge based on studies in Drosophila, the main factor determining the recruitment of PcG complexes is the presence of non-methylated GC-rich elements [12,13]. This has supported a model in which GC-rich promoter regions are repressed by default and become activated upon clearing of PcG repression through local or distal binding of activating transcription factors [12,13,41,42]. Our experiments have now revealed an enhancer region that not only activates its target gene in a lineage-selective manner, but also recruits PcG-mediated repression in non-expressing developmental lineages. While previous studies in mammals had shown PREs that do not overlap promoters [13,15,17] and even presence of H3K27me3 at poised enhancers [43,44], the findings described here are of interest in that they reveal that an enhancer can direct both lineage-specific transcriptional activation and PcG-mediated repression. Deletion experiments, nevertheless, demonstrated that the sequences required for activation were distinct from the sequences required for PcG-mediated repression. Further studies should explore the prevalence of this developmental cis-regulatory mechanism in metazoan genomes.
Methods
Mouse models
All experiments were approved by the Institutional Animal Care Committee of the University of Barcelona or by the Direction Départementale de Protection des Populations (DDPP) de Paris, under accreditation number A75-13-19.
Animals were sacrificed by cervical dislocation according to the recommended practices of the French legislation.
DNA constructs for conventional transgenesis
A HindIII–HindIII fragment of the 5’flanking region of Neurog3 located between nucleotide -5284 and -1428 relative to transcription start site was amplified by PCR using Phusion DNA polymerase (Finnzyme) a high fidelity thermostable DNA polymerase and the following primers: -5284 sense AAGCTTTGTGTGGAAGGA and -1428 antisense AAGCTTCTAGTACGTTCTAACT. The amplification was performed on a lambda clone containing a 20kb fragment of the Neurog3 mouse locus. The resulting PCR product was digested by HindIII and cloned into the HindIII site of the pBGZ40 reporter plasmid upstream a human beta globin minimal promoter linked to lacZ and a SV40 poly adenylation signal [45]. The resulting construct-A was entirely sequenced to rule out undesired mutations and further digested by XhoI and XbaI prior to pronuclear injection. A similar approach was used to generate constructs-B, -C and–D using the following primers. Construct-B: -5284 sense and -3061 antisense AAGCTTGCTAGCATTGCCTGGGGG. Construct-C: -3057 sense TTTACCCCCTCCCAACAG and -1428 antisense. Construct-D -4916 AAGCTTCCAGCCATAAGGTTTATT and -3061 antisense.
DNA constructs for lentiviral transgenesis
The construct-B PCR fragment was cloned into the pENTR/D-TOPO vector (Invitrogen) according to the manufacturer’s instructions to generate the 2.2 kb wild type enhancer entry clone. A destination vector containing a Gateway recombination cassette rfa followed by the pBGZ40 beta globin minimal promoter linked to eGFP was constructed in the pTRIP lentiviral backbone using a similar strategy as described [46]. The 2.2 kb enhancer was inserted into the lentiviral destination vector by in vitro gateway recombination using LR clonase II following manufacturer’s instructions. Note that all lentiviral vectors containing cis element deletions or specific point mutations were all cloned into the lentiviral destination vector using the same gateway recombination procedure.
Cis-element deletions and site directed mutagenesis
Deletions and point mutations were all performed on the pENTR 2.2 kb enhancer plasmid using a PCR based mutagenesis system. Briefly, sense and antisense complementary primers overlapping either deletion area or containing the point mutations were designed. Such mutation containing primers were used for a first round of PCR with two external 5’ and 3’ primers to generate two PCR products. The left product was obtained using the mutated primer antisense along with the 5’ external primer and the right PCR product with the sense mutated primer and the 3’ external primer. Both PCR products were generated using Phusion DNA polymerase (Finnzyme) and no more than 25 PCR cycles. Both DNA fragments were purified on Wyzard PCR cleanup columns (Promega) and 1ng of each resulting purification were used as matrix for a second round of PCR using the 2 external primers. The resulting PCR products contained the desired deletions or point mutations and were further digested with BstEII and PstI for deletions of cis-element 1 and 2 or by SpeI and NcoI for all other deletions or point mutations. The digested fragments were cloned into the pENTR 2.2 kb enhancer plasmid digested with BstEII and PstI or SpeI and NcoI to replace a wild type cassette by a mutation containing one. Note that the double mutation 1–7 was generated using mutation 1 as original matrix with the PCR primers of mutation 7. The complete list of mutation primers and external primer is presented in S2 Table. For constructs with deletions of cis-element 1, 2 or 3 the deleted fragments are located from nucleotide -4877 to -4812 (relative to transcription start site), -4877 to -4812pb and -3336 to– 3287 respectively.
Conventional transgenesis
Transgenic embryos were generated by pronuclear injection into fertilized eggs from FVB/NRj mouse strain (Elevage Janvier). The injected eggs were next re-implanted into F1 hybrids (DBA/2 × C57Bl6) pseudo pregnant females. Most constructs were assayed in founder (F0) transgenic embryos with the exception of construct-A for which 2 different mouse transgenic lines have being generated.
Production of lentiviral vectors
Lentiviral vector stocks were produced as previously described [47]. The amount of p24 capsid protein was quantified by the HIV-1 p24 ELISA antigen assay (Helvetica Health Care). All transductions of fertilized eggs were performed using normalized titers relative to p24 capsid protein quantification ranging between 50 to 60 ng of p24 per microliter of viral stock.
Lentiviral transgenesis
Transgenic embryos were generated by injection of lentiviral vectors into the perivitelline space of fertilized eggs from RjOrl:SWISS mouse strain as described previously [48]. The injected eggs were next re-implanted into F1 hybrids (DBA/2 × C57Bl6) pseudo pregnant females. All constructs were assayed in founder (F0) transgenic embryos.
Genotyping of transgenic animals
Genomic DNA was extracted from either tail of adult mice or yolk sac of embryos using proteinase K digestion followed by PCR amplification to detect LacZ or GFP transgenes with the following primers. LacZ1: 5’ ACCCTGGCGTTACCCAACTTAATCG 3’; LacZ2: 5’ ACAAACGGCGGATTGACCGTAATGG 3’; GFP sense: 5’ GACCACATGAAGCAGCACGACTTCT 3’; GFP antisense: 5’ TTCTGCTGGTAGTGGTCGGCGAGCT 3’.
Derivation of embryonic fibroblast culture from transgenic embryos
E14.5 transgenic embryos containing either the Neurog3 enhancer region or the minimal promoter GFP construct were collected. For each collected transgenic embryo a separate fibroblast culture was derived. Briefly, a small part of the skin of the embryo was collected under sterile condition and placed in a 24 well culture plate under cover glass and culture at 37°C under 5% CO2 for 2 days in culture medium containing 10% FCS 1% essential amino acids (Invitrogen), 1% Penicillin and Streptomycin (Invitrogen) in DMEM 4.5 g/l D-glucose (Invotrogen). After 2 days, the skin explant was removed and the fibroblasts were passaged by Trypsin / EDTA treatment in a 6 well plate. When confluence was observed cells were passaged into a 6 cm diameter plate and next into a 10 cm diameter plate. Cells were amplified subsequently by passages at confluence with a 1 to 5 dilution until frozen or used.
RNA extraction
Total RNA was extracted using RNeasy micro kit (Qiagen) followed by DNAase I treatment. RNA integrity was verified with a 2100 Bioanalyzer (Agilent) and reverse transcribed using Superscript 2 (invitrogen).
Quantitative PCR analysis
Quantitative PCR of reverse-transcribed RNA, ChIP or gDNA samples was performed on a 7300 Realtime PCR System (Applied Biosystems) using the Power SYBR Green reagent (Applied Biosystems). Quantities were determined using 2(-Delta Delta C(T)). A full list of primers used is provided as S3 Table.
Tissue preparation for histological analysis
Transgenic embryos were collected at different stages. The date of reimplantation of the one cell stage embryos in pseudo-pregnant females was considered as embryonic day 0.5 (E0.5). Embryos were dissected and a tissue block containing the stomach; pancreas and duodenum was removed and fixed by immersion in 4% PFA for 24 h. Tissues were cryoprotected by incubation for 48 h in 15% sucrose prepared in PBS and next embedded in 15% sucrose, 7% gelatin prepared in PBS then frozen in isopentane at -50°C. 10 μm sections were performed using a cryostat (Leica). Whole embryos were fixed in 4% PFA for 20 min and stained for lacZ reporter activity as described and embedded in BSA-gelatin [49]. 300-μm sections were cut with a Vibratome (Leica VT1000S) as described [50].
Immunostaining
10μm cryosections were used for immunofluorescence staining as described previously [51]. The following primary antibodies were used: rabbit anti-insulin (1/1500 Euromedex Ref. 20056), rabbit anti-glucagon (1/1500 Euromedex Ref. 20076), rabbit anti-amylase (1/350 Sigma Ref. A8273) mouse anti-Neurog3 (1/1000 Beta Cell Biology Consortium Ref. AB2013) and chicken anti-GFP (1/1000 Antibodies-Online GMBH Germany, Ref. ABIN 147441). The corresponding secondary antibodies were used: Alexa 488 Goat anti Chicken (1/2000 Life Technologies Ref. A11039) and Alexa 555 Goat anti Rabbit (1/1000 Life Technologies Ref. A21428). Neurog3 detection was amplified as described by Zahn et al. 2004 [52]. TSA amplification (Perkin Elmer TSA-Direct NEL702) was preformed prior to GFP staining. Beta-galactosidase was detected by immuno-staining using rabbit anti-βGal (1/1000 Cappel Inc.) and stained with the Vectastain DAB kit (Vector Inc.) as described [53].
ISH
Riboprobes were synthesized from various cDNAs inserted into pGEM-T Easy vector (Promega). After linearization of the plasmids, non-radioactively labeled riboprobes were synthesized in the presence of 3.5 nmol of either digoxigenin (Dig)-11-UTP or fluorescein (Fluo)-12-UTP (Roche) by using the riboprobe combination system Sp6/T7 (Promega). The LacZ riboprobe spans the whole coding sequence. The Neurog3 and Insulin riboprobes were described previously [50,54]. Hybridization experiments were performed essentially as described previously on 10μm cryosections [55]. For double in situ staining probes were detected with alkaline phosphatase-conjugated anti-Dig and anti-Fluo antibodies (Roche). Colorimetric detection was achieved with 5-bromo-4-chloro-3-indolylphosphate (BCIP) and either nitroblue tetrazolium (NBT; Promega) or 2-(4-iodophenyl)-3-(4-nitrophenyl)-5-phenyltetrazolium chloride (INT; Roche). Dig- and Fluo-labeled riboprobes were applied together to the slides. After detection of the Dig-labeled probe with NBT-BCIP, the anti-Dig antibody was removed by washes with 0.1 M glycine (pH 2.2)-0.1% Tween before incubation with the anti-Fluo antibody. No background signal was observed on slides hybridized with the sense control probes and incubated with NBT-BCIP for at least as long as the corresponding antisense probes.
Quantification of Neurog3 and GFP expression in transgenic embryos
Transgenic embryos at E14.5 generated with all lentiviral constructs (wild type enhancer, deletions of cis-element 1, 2 and 3 and point mutations 1, 3, 7 and 1–7) were analyzed. GFP + cells, Neurog3 + cells and GFP/Neurog3 double + cells were counted on successive 10μm sections located every 50 μm. A total number of at least 1000 Neurog3 positive cells were counted per embryo. 8 transgenic embryos were thus analyzed for each transgenic construct.
Chromatin Immunoprecipitation (ChIP)
ChIPs were performed as described [30]. In short, mouse tissues or mouse ES cells (CGR8; [56]) were fixed in 1% formaldehyde for 10 min after which nuclei were purified and sonicated using Bioruptor (Diagenode) to a length of 200 to 1000 bp. Samples were precleared with protein A+G-Sepharose (1:1) and immunoprecipitated with rabbit anti-Ring1b [57], rabbit anti-H3K27me3 (Upstate, 07–449), mouse anti-Ezh2 [58] overnight at 4°C. Immune complexes were collected by adsorption to protein A+G-Sepharose for 2 hr at 4°C. Beads were washed and immunocomplexes eluted prior to DNA purification with Qiaquick columns (Qiagen). Precipitations on pancreatic buds were performed with at least 20 pancreatic buds per IP and in the presence of 2.5 mg/mL BSA and 25 mg/mL tRNA. For tiling array experiments, ChIP and input DNA were amplified as described previously using the Sigma GenomePlex WGA2 kit while adding dUTPs to a final concentration of 0.4 mM during the amplification reaction to enable subsequent fragmentation [30]. We fragmented 6–7.5 μg DNA, labeled it using the Affymetrix GeneChip WT Double-Stranded DNA terminal Labeling Kit, and hybridized to GeneChip® Mouse Promoter 1.0R Arrays.
EMSA
Single-stranded oligonucleotides (S1 Table) were annealed and labeled with [γ-32P]ATP (GE healthcare Rediprime labeling kit). The labeled oligonucleotides were column-purified (GE microspin G-50 columns). 32P-labeled oligonucleotides were incubated for 20 min at room temperature with nuclear extracts in 20mM Hepes (pH 7.9), 90 mM KCl, 5 mM MgCl2, and 0.05% Nonidet P-40, 1mg/ml BSA, 5% Glycerol, 1mM DTT, 1mg/ml poly(dI-dC).poly(dI-dC) and 0.1mM ZnCl. For supershifts, antibodies or preimmune serum was added to the reaction and incubated for 20 min on ice before incubation with the labeled probe. Samples were electrophoresed on a 5% acrylamide gel in 0.5 X TBE buffer and autoradiographed. For competition assays, the ratio of labelled probe to cold competitor was 1:100. The antibodies were Rabbit anti-mouse IgG (Abcam; ab6709), goat anti-Pdx1 (abcam; ab47383), goat anti-Foxa2 (santa-cruz; SC-6554), goat anti-Hnf1b (santa-cruz; SC-7411) and rabbit anti-Hnf1a [59].
Statistical and integrated data analysis
For GeneChip ChIP experiments enrichment relative to input DNA was determined using Cisgenome [60]. We applied a Hidden Markov Model as described [61].
CpG island analysis was done using CpGPlot in EMBOSS [62]. We scanned the Neurog3 region with a 400 bp sliding window in steps of one base to calculate GC content and the ratio of observed CpGs and expected CpGs (CpG (obs/exp)) for each centred position. The cut-off for CpG (obs/exp) and GC content was set to an average of 0.6 and 0.5 respectively in a set of 10 windows. Furthermore we required an island to be at least 100 bp in length.
Motif search was performed on the complete Neurog3 enhancer region using free access version of Genomatix Matinspector with the default settings [63]. The obtained output was manually searched for known pancreatic factors. For the selected sites the lowest Core similarity was 0.767 and the lowest matrix similarity was 0.744. These results were manually curated based on more recent ChIP-seq based motifs where available.
All barplots are shown as means with error bars representing the SEM. Comparisons were done as indicated with Student’s two-sample, two-sided t-test and Bonferroni corrections were done for the number of comparisons.
Data access
Microarray data for ChIP experiments of Ezh2 in ES cells will be made publicly available through ArrayExpress under accession number E-MTAB-1460 (reviewer password: nxgEEa62). Other data sets have been described before [30,31].
Supporting information
Regions A and B recapitulate the Neurog3 expression pattern in conventional trangenics. A. LacZ expression under the control of enhancer region A (see Fig 1 for representation of enhancer segments). In E11.5 embryos (panel I-IV), region A recapitulated the previously reported pattern of expression of Neurog3 in the spinal cord (SC) that displays 2 stripes, the medial stripe (MS) and the ventral stripe (VS) (panel I), the ventral hypothalamus (vHyp; panels II, III) and the pancreatic bud (PB; panel IV) with ectopic expression only in the cephalic mesenchyme (CM; panel III) and the notochord (NC; panel III). In panel IV the stomach (St) and the liver (L) are indicated. At E14.5 LacZ was observed in the pancreas (P; panel V) and the duodenum (D; panel VI) B. LacZ expression in the E15.5 pancreas from transgenics carrying enhancer region B. LacZ immunostaining show that LacZ (panel II) follows the expected distribution detected by double in situ hybridization of Neurog3 in the pancreatic epithelial tree (blue; panel I, adjacent section to II), whereas insulin (brown; panels I and III) shows the expected distribution in more peripheral regions. Panel III shows and enlargement of the inset in panel I. Panel IV shows an in situ hybridization co-staining for LacZ (brown) and Neurog3 (blue) with an enlargement of the inset shown as panel V. Scale bars: A I, II = 200μm, III = 400μm, IV-VI = 200μm, B I, II = 100μm, III- V = 25 μm
(TIF)
Expression of GFP was analyzed in the pancreases of E18.5 transgenic embryos. GFP expression (green) was compared to Neurog3 expression (red upper panel) and also compared to Insulin (red left lower panel) and Glucagon (red right lower panel). GFP-positive Neurog3-negative cells were in the endocrine lineage. This became more prevalent at E18.5 compared to E14.5, presumably because of the different stability of GFP and Neurog3. Scale bars: 50 μm.
(TIF)
The 5’ part of cis-element 3 (5’WT) was screened with overlapping substitutions of 3 wild-type bases by G nucleotides. The effect of these mutations on DNA binding affinities was assessed by incubating the unlabeled mutant probes at 100-fold excess relative to the wild-type labeled sequence in Min6 nuclear extracts. Note that mutation 1 was found to disrupt both Pdx1 and Hnf1b binding, while Mutation 3 selectively disrupted Hnf1b binding. For the 3’ part of cis-element 3 (3’WT) two mutations were designed and tested in E13.5 pancreatic bud extracts. Both mutations (mut7, mut8) were found to disrupt Pdx1 binding, and mut7 moderately affected Foxa2 binding.
(TIF)
qPCR was performed on genomic DNA isolated from embryonic tails using the GFP 3’UTR primer pair, and Cdx2 was used to normalize the data. Each cross represents an animal. The horizontal lines represent the means. N.S., not significant, Student’s t-test.
(TIF)
The graph shows the results of an oligonucleotide tiling array analysis of ChIPs for H3K4me3 and H3K27me3 as reported [30]. Enrichment values for H3K4me3 (green) and H3K27me3 (red) are expressed as posterior probability values ranging from 0 to 1.
(TIF)
ChIP analysis of H3K27me3 in the exogenous Neurog3 enhancer region with or without cis-element 3 in E13.5 liver (A), adult acinar tissue (B), or adult skeletal muscle (C).
(TIF)
(DOCX)
(DOC)
(DOCX)
Acknowledgments
We thank Finn Cilius Nielsen, Rehannah Borup and Susanne Smed (RH Microarray Center, Rigshospitalet, Copenhagen) for array hybridizations, Anouchka L. Skoudy for ES cells, Mark Kalisz, Jose Luis Gómez-Skarmeta and Jacques Mallet for insightful comments and funding from Ministerio de Economía y Competitividad (SAF2011-27086), (BFU2014-54284-R) and a Wellcome Trust Senior Investigator Award (101033) to J.F. P.R. received funding for the Association de Langue Française pour l'Etude du Diabète et des Maladies Métaboliques (ALFEDIAM) and a joint JDRF / INSERM grant.
Data availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Funding from Ministerio de Economía y Competitividad (SAF2011-27086), (BFU2014-54284-R) and a Wellcome Trust Senior Investigator Award (101033) to J.F. P.R. received funding for the Association de Langue Française pour l'Etude du Diabète et des Maladies Métaboliques (ALFEDIAM) and a joint JDRF / INSERM grant.
References
- 1.Heintzman ND, Ren B (2009) Finding distal regulatory elements in the human genome. Curr Opin Genet Dev 19: 541–549. 10.1016/j.gde.2009.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ong CT, Corces VG (2012) Enhancers: emerging roles in cell fate specification. EMBO Rep 13: 423–430. 10.1038/embor.2012.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Margueron R, Reinberg D (2011) The Polycomb complex PRC2 and its mark in life. Nature 469: 343–349. 10.1038/nature09784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morey L, Helin K (2010) Polycomb group protein-mediated repression of transcription. Trends Biochem Sci 35: 323–332. 10.1016/j.tibs.2010.02.009 [DOI] [PubMed] [Google Scholar]
- 5.Schuettengruber B, Cavalli G (2009) Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 136: 3531–3542. 10.1242/dev.033902 [DOI] [PubMed] [Google Scholar]
- 6.de Napoles M., Mermoud JE, Wakao R, Tang YA, Endoh M et al. (2004) Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell 7: 663–676. 10.1016/j.devcel.2004.10.005 [DOI] [PubMed] [Google Scholar]
- 7.Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S et al. (2010) Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell 38: 452–464. 10.1016/j.molcel.2010.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P et al. (2004) Role of histone H2A ubiquitination in Polycomb silencing. Nature 431: 873–878. 10.1038/nature02985 [DOI] [PubMed] [Google Scholar]
- 9.Ringrose L, Paro R (2007) Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development 134: 223–232. 10.1242/dev.02723 [DOI] [PubMed] [Google Scholar]
- 10.Dejardin J, Rappailles A, Cuvier O, Grimaud C, Decoville M et al. (2005) Recruitment of Drosophila Polycomb group proteins to chromatin by DSP1. Nature 434: 533–538. 10.1038/nature03386 [DOI] [PubMed] [Google Scholar]
- 11.Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M et al. (2008) Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4: e1000242 10.1371/journal.pgen.1000242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lynch MD, Smith AJ, De GM, Flenley M, Hughes JR et al. (2012) An interspecies analysis reveals a key role for unmethylated CpG dinucleotides in vertebrate Polycomb complex recruitment. EMBO J 31: 317–329. 10.1038/emboj.2011.399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mendenhall EM, Koche RP, Truong T, Zhou VW, Issac B et al. (2010) GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet 6: e1001244 10.1371/journal.pgen.1001244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E et al. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560. 10.1038/nature06008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schorderet P, Lonfat N, Darbellay F, Tschopp P, Gitto S et al. (2013) A Genetic Approach to the Recruitment of PRC2 at the HoxD Locus. PLoS Genet 9: e1003951 10.1371/journal.pgen.1003951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sing A, Pannell D, Karaiskakis A, Sturgeon K, Djabali M et al. (2009) A vertebrate Polycomb response element governs segmentation of the posterior hindbrain. Cell 138: 885–897. 10.1016/j.cell.2009.08.020 [DOI] [PubMed] [Google Scholar]
- 17.Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE (2010) A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell 140: 99–110. 10.1016/j.cell.2009.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Apelqvist A, Li H, Sommer L, Beatus P, Anderson DJ et al. (1999) Notch signalling controls pancreatic cell differentiation. Nature 400: 877–881. 10.1038/23716 [DOI] [PubMed] [Google Scholar]
- 19.Gradwohl G, Dierich A, LeMeur M, Guillemot F (2000) neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci U S A 97: 1607–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwitzgebel VM, Scheel DW, Conners JR, Kalamaras J, Lee JE et al. (2000) Expression of neurogenin3 reveals an islet cell precursor population in the pancreas. Development 127: 3533–3542. [DOI] [PubMed] [Google Scholar]
- 21.Beucher A, Martin M, Spenle C, Poulet M, Collin C et al. (2012) Competence of failed endocrine progenitors to give rise to acinar but not ductal cells is restricted to early pancreas development. Dev Biol 361: 277–285. 10.1016/j.ydbio.2011.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Solar M, Cardalda C, Houbracken I, Martin M, Maestro MA et al. (2009) Pancreatic exocrine duct cells give rise to insulin-producing beta cells during embryogenesis but not after birth. Dev Cell 17: 849–860. 10.1016/j.devcel.2009.11.003 [DOI] [PubMed] [Google Scholar]
- 23.Lee JC, Smith SB, Watada H, Lin J, Scheel D et al. (2001) Regulation of the pancreatic pro-endocrine gene neurogenin3. Diabetes 50: 928–936. [DOI] [PubMed] [Google Scholar]
- 24.Jacquemin P, Durviaux SM, Jensen J, Godfraind C, Gradwohl G et al. (2000) Transcription factor hepatocyte nuclear factor 6 regulates pancreatic endocrine cell differentiation and controls expression of the proendocrine gene ngn3. Mol Cell Biol 20: 4445–4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lynn FC, Smith SB, Wilson ME, Yang KY, Nekrep N et al. (2007) Sox9 coordinates a transcriptional network in pancreatic progenitor cells. Proc Natl Acad Sci U S A 104: 10500–10505. 10.1073/pnas.0704054104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliver-Krasinski JM, Kasner MT, Yang J, Crutchlow MF, Rustgi AK et al. (2009) The diabetes gene Pdx1 regulates the transcriptional network of pancreatic endocrine progenitor cells in mice. J Clin Invest 119: 1888–1898. 10.1172/JCI37028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arda HE, Benitez CM, Kim SK (2013) Gene regulatory networks governing pancreas development. Dev Cell 25: 5–13. 10.1016/j.devcel.2013.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujitani Y, Fujitani S, Boyer DF, Gannon M, Kawaguchi Y et al. (2006) Targeted deletion of a cis-regulatory region reveals differential gene dosage requirements for Pdx1 in foregut organ differentiation and pancreas formation. Genes Dev 20: 253–266. 10.1101/gad.1360106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raum JC, Hunter CS, Artner I, Henderson E, Guo M et al. (2010) Islet beta-cell-specific MafA transcription requires the 5'-flanking conserved region 3 control domain. Mol Cell Biol 30: 4234–4244. 10.1128/MCB.01396-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Arensbergen J, Garcia-Hurtado J, Moran I, Maestro MA, Xu X et al. (2010) Derepression of Polycomb targets during pancreatic organogenesis allows insulin-producing beta-cells to adopt a neural gene activity program. Genome Res 20: 722–732. 10.1101/gr.101709.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Arensbergen J, Garcia-Hurtado J, Maestro MA, Correa-Tapia M, Rutter GA et al. (2012) Ring1b bookmarks genes in pancreatic embryonic progenitors for repression in adult beta cells. Genes Dev [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie R, Everett LJ, Lim HW, Patel NA, Schug J et al. (2013) Dynamic Chromatin Remodeling Mediated by Polycomb Proteins Orchestrates Pancreatic Differentiation of Human Embryonic Stem Cells. Cell Stem Cell [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A et al. (2009) Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112. 10.1038/nature07829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerrish K, Van Velkinburgh JC, Stein R (2004) Conserved transcriptional regulatory domains of the pdx-1 gene. Mol Endocrinol 18: 533–548. 10.1210/me.2003-0371 [DOI] [PubMed] [Google Scholar]
- 35.Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D (2002) Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295: 868–872. 10.1126/science.1067081 [DOI] [PubMed] [Google Scholar]
- 36.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW et al. (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318. 10.1038/ng1966 [DOI] [PubMed] [Google Scholar]
- 37.May D, Blow MJ, Kaplan T, McCulley DJ, Jensen BC et al. (2012) Large-scale discovery of enhancers from human heart tissue. Nat Genet 44: 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xi H, Shulha HP, Lin JM, Vales TR, Fu Y et al. (2007) Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet 3: e136 10.1371/journal.pgen.0030136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Chu LT, He J, Emelyanov A, Korzh V et al. (2001) A novel zebrafish bHLH gene, neurogenin3, is expressed in the hypothalamus. Gene 275: 47–55. [DOI] [PubMed] [Google Scholar]
- 40.Haumaitre C, Barbacci E, Jenny M, Ott MO, Gradwohl G et al. (2005) Lack of TCF2/vHNF1 in mice leads to pancreas agenesis. Proc Natl Acad Sci U S A 102: 1490–1495. 10.1073/pnas.0405776102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klose RJ, Cooper S, Farcas AM, Blackledge NP, Brockdorff N (2013) Chromatin sampling-an emerging perspective on targeting polycomb repressor proteins. PLoS Genet 9: e1003717 10.1371/journal.pgen.1003717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vernimmen D, Lynch MD, De GM, Garrick D, Sharpe JA et al. (2011) Polycomb eviction as a new distant enhancer function. Genes Dev 25: 1583–1588. 10.1101/gad.16985411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA et al. (2011) A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283. 10.1038/nature09692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zentner GE, Tesar PJ, Scacheri PC (2011) Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res 21: 1273–1283. 10.1101/gr.122382.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manzanares M, Nardelli J, Gilardi-Hebenstreit P, Marshall H, Giudicelli F et al. (2002) Krox20 and kreisler co-operate in the transcriptional control of segmental expression of Hoxb3 in the developing hindbrain. EMBO J 21: 365–376. 10.1093/emboj/21.3.365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Norrman K, Fischer Y, Bonnamy B, Wolfhagen SF, Ravassard P et al. (2010) Quantitative comparison of constitutive promoters in human ES cells. PLoS One 5: e12413 10.1371/journal.pone.0012413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ravassard P, Emilie B, Hazhouz Y, Pechberty S, Mallet J et al. (2009) A new strategy to generate functional insulin-producing cell lines by somatic gene transfer into pancreatic progenitors. PLoS One 4: e4731 10.1371/journal.pone.0004731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Collombat P, Xu X, Ravassard P, Sosa-Pineda B, Dussaud S et al. (2009) The ectopic expression of Pax4 in the mouse pancreas converts progenitor cells into alpha and subsequently beta cells. Cell 138: 449–462. 10.1016/j.cell.2009.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whiting J, Marshall H, Cook M, Krumlauf R, Rigby PW et al. (1991) Multiple spatially specific enhancers are required to reconstruct the pattern of Hox-2.6 gene expression. Genes Dev 5: 2048–2059. [DOI] [PubMed] [Google Scholar]
- 50.Ravassard P, Chatail F, Mallet J, Icard-Liepkalns C (1997) Relax, a novel rat bHLH transcriptional regulator transiently expressed in the ventricular proliferating zone of the developing central nervous system. J Neurosci Res 48: 146–158. [DOI] [PubMed] [Google Scholar]
- 51.Khalfallah O, Ravassard P, Lagache CS, Fligny C, Serre A et al. (2009) Zinc finger protein 191 (ZNF191/Zfp191) is necessary to maintain neural cells as cycling progenitors. Stem Cells 27: 1643–1653. 10.1002/stem.88 [DOI] [PubMed] [Google Scholar]
- 52.Zahn S, Hecksher-Sorensen J, Pedersen IL, Serup P, Madsen O (2004) Generation of monoclonal antibodies against mouse neurogenin 3: a new immunocytochemical tool to study the pancreatic endocrine progenitor cell. Hybrid Hybridomics 23: 385–388. 10.1089/hyb.2004.23.385 [DOI] [PubMed] [Google Scholar]
- 53.Trocme C, Sarkis C, Hermel JM, Duchateau R, Harrison S et al. (1998) CRE and TRE sequences of the rat tyrosine hydroxylase promoter are required for TH basal expression in adult mice but not in the embryo. Eur J Neurosci 10: 508–521. [DOI] [PubMed] [Google Scholar]
- 54.Castaing M, Guerci A, Mallet J, Czernichow P, Ravassard P et al. (2005) Efficient restricted gene expression in beta cells by lentivirus-mediated gene transfer into pancreatic stem/progenitor cells. Diabetologia 48: 709–719. 10.1007/s00125-005-1694-6 [DOI] [PubMed] [Google Scholar]
- 55.Kiefer H, Chatail-Hermitte F, Ravassard P, Bayard E, Brunet I et al. (2005) ZENON, a novel POZ Kruppel-like DNA binding protein associated with differentiation and/or survival of late postmitotic neurons. Mol Cell Biol 25: 1713–1729. 10.1128/MCB.25.5.1713-1729.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Skoudy A, Rovira M, Savatier P, Martin F, Leon-Quinto T et al. (2004) Transforming growth factor (TGF)beta, fibroblast growth factor (FGF) and retinoid signalling pathways promote pancreatic exocrine gene expression in mouse embryonic stem cells. Biochem J 379: 749–756. 10.1042/BJ20031784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Garcia E, Marcos-Gutierrez C, del Mar LM, Moreno JC, Vidal M (1999) RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex, and with the transcription factor YY1. EMBO J 18: 3404–3418. 10.1093/emboj/18.12.3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K (2006) Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boj SF, Parrizas M, Maestro MA, Ferrer J (2001) A transcription factor regulatory circuit in differentiated pancreatic cells. Proc Natl Acad Sci U S A 98: 14481–14486. 10.1073/pnas.241349398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ji H, Jiang H, Ma W, Johnson DS, Myers RM et al. (2008) An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol 26: 1293–1300. 10.1038/nbt.1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ji H, Wong WH (2005) TileMap: create chromosomal map of tiling array hybridizations. Bioinformatics 21: 3629–3636. 10.1093/bioinformatics/bti593 [DOI] [PubMed] [Google Scholar]
- 62.Rice P, Longden I, Bleasby A (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16: 276–277. [DOI] [PubMed] [Google Scholar]
- 63.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M et al. (2005) MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21: 2933–2942. 10.1093/bioinformatics/bti473 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Regions A and B recapitulate the Neurog3 expression pattern in conventional trangenics. A. LacZ expression under the control of enhancer region A (see Fig 1 for representation of enhancer segments). In E11.5 embryos (panel I-IV), region A recapitulated the previously reported pattern of expression of Neurog3 in the spinal cord (SC) that displays 2 stripes, the medial stripe (MS) and the ventral stripe (VS) (panel I), the ventral hypothalamus (vHyp; panels II, III) and the pancreatic bud (PB; panel IV) with ectopic expression only in the cephalic mesenchyme (CM; panel III) and the notochord (NC; panel III). In panel IV the stomach (St) and the liver (L) are indicated. At E14.5 LacZ was observed in the pancreas (P; panel V) and the duodenum (D; panel VI) B. LacZ expression in the E15.5 pancreas from transgenics carrying enhancer region B. LacZ immunostaining show that LacZ (panel II) follows the expected distribution detected by double in situ hybridization of Neurog3 in the pancreatic epithelial tree (blue; panel I, adjacent section to II), whereas insulin (brown; panels I and III) shows the expected distribution in more peripheral regions. Panel III shows and enlargement of the inset in panel I. Panel IV shows an in situ hybridization co-staining for LacZ (brown) and Neurog3 (blue) with an enlargement of the inset shown as panel V. Scale bars: A I, II = 200μm, III = 400μm, IV-VI = 200μm, B I, II = 100μm, III- V = 25 μm
(TIF)
Expression of GFP was analyzed in the pancreases of E18.5 transgenic embryos. GFP expression (green) was compared to Neurog3 expression (red upper panel) and also compared to Insulin (red left lower panel) and Glucagon (red right lower panel). GFP-positive Neurog3-negative cells were in the endocrine lineage. This became more prevalent at E18.5 compared to E14.5, presumably because of the different stability of GFP and Neurog3. Scale bars: 50 μm.
(TIF)
The 5’ part of cis-element 3 (5’WT) was screened with overlapping substitutions of 3 wild-type bases by G nucleotides. The effect of these mutations on DNA binding affinities was assessed by incubating the unlabeled mutant probes at 100-fold excess relative to the wild-type labeled sequence in Min6 nuclear extracts. Note that mutation 1 was found to disrupt both Pdx1 and Hnf1b binding, while Mutation 3 selectively disrupted Hnf1b binding. For the 3’ part of cis-element 3 (3’WT) two mutations were designed and tested in E13.5 pancreatic bud extracts. Both mutations (mut7, mut8) were found to disrupt Pdx1 binding, and mut7 moderately affected Foxa2 binding.
(TIF)
qPCR was performed on genomic DNA isolated from embryonic tails using the GFP 3’UTR primer pair, and Cdx2 was used to normalize the data. Each cross represents an animal. The horizontal lines represent the means. N.S., not significant, Student’s t-test.
(TIF)
The graph shows the results of an oligonucleotide tiling array analysis of ChIPs for H3K4me3 and H3K27me3 as reported [30]. Enrichment values for H3K4me3 (green) and H3K27me3 (red) are expressed as posterior probability values ranging from 0 to 1.
(TIF)
ChIP analysis of H3K27me3 in the exogenous Neurog3 enhancer region with or without cis-element 3 in E13.5 liver (A), adult acinar tissue (B), or adult skeletal muscle (C).
(TIF)
(DOCX)
(DOC)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.