Abstract
Objective
Dual vector AAV systems are being utilised to enable gene therapy for disorders in which the disease gene is too large to fit into a single capsid. Fragmented adeno-associated viral (fAAV) vectors containing single inverted terminal repeat truncated transgenes have been considered as one such gene replacement strategy. Here we aim to add to the current understanding of the molecular mechanisms employed by fAAV dual vector systems.
Methods
Oversized (>8kb) transgene constructs containing ABCA4 coding sequence were packaged as truncated fragments <5kb in size into various AAV serotypes. In vitro transductions with these fAAV vector preparations were conducted with mRNA and protein expression products assessed by way of RT-PCR, qPCR and western blot techniques.
Results
Transductions with fAAV vector preparations yielded ABCA4 mRNA, but did not generate detectable levels of protein. Sequencing of the transcript population revealed the presence of full length ABCA4 CDS with additional hybrid ABCA4 variants, indicating truncated transgenes without regions of overlap were joining and forming stable hybrid transgenes. In contrast, an ABCA4 overlapping dual vector system (OV) with a defined complementary region generated only full length mRNA transcripts plus detectable ABCA4 protein.
Conclusion
Despite previous success shown with the fAAV approach, the lack of repeatability and identification of stable hybrid transcripts capable of protein production suggests there is more refinement required before considering this approach in a clinical setting.
Introduction
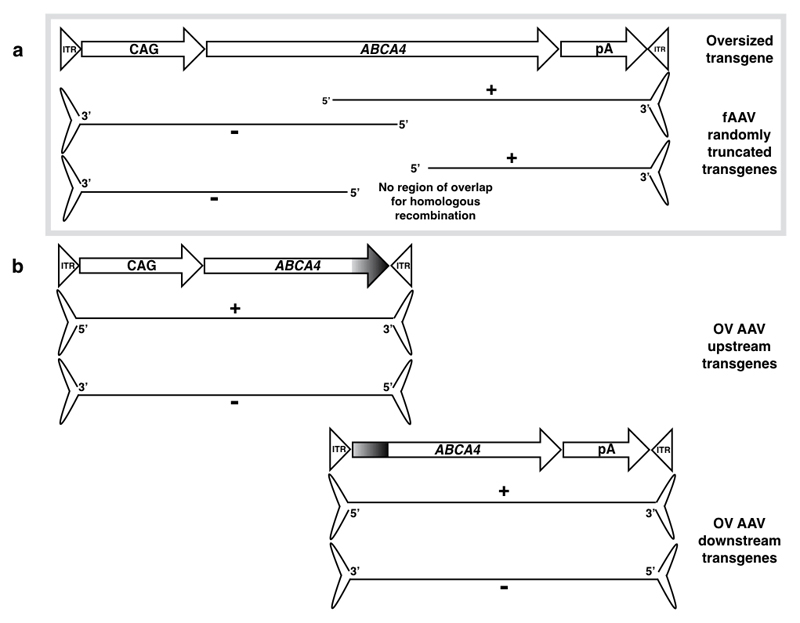
Adeno-associated virus (AAV) vectors are known to have an optimal packaging capacity of ~4.7kb (the size of their native genome) but can be encouraged to package slightly larger transgenes1. With AAV showing encouraging safety and efficacy in human clinical trials, there is a desire to utilise this vector for the treatment of diseases for which the causative gene is larger than the packaging capacity. For example, Stargardt disease is the most common cause of autosomal recessive childhood blindness and is the result of mutations in the ATP-binding cassette transporter protein, family member 4 (ABCA4), the gene for which is 6.8kb and therefore far exceeds the size of the native AAV genome. In 2008, Allocca et al. attempted to package much larger genomes of up to 8.9kb and given the positive outcomes of their strategy, assumed the packaging to have been successful. However, since then reports appear consistent in showing that AAV capsids do indeed fail to package complete transgenes larger than 5.2kb3–5. The success from Allocca et al. is now acknowledged to be due to AAV capsids truncating the oversized transgenes during the packaging process5. When creating a recombinant AAV vector, a double-stranded DNA (dsDNA) template is provided from which single-stranded transgenes are generated from the 3’ inverted terminal repeat sequence (ITR) of both the plus and minus strand. For oversized constructs, the replicated transgenes are truncated at the 5’ end at an undefined point (Figure 1a). This method provides no designated limit of packaging at the 5’ end of either the plus or minus strands of the transgene and results in a collection of AAV particles carrying transgenes of different length containing only a single ITR, referred to herein as fragmented AAV (fAAV)5,6. The success of the approach relies on the majority of AAV particles containing large enough fragments of the oversized transgene such that complementary overlaps between truncated transgenes exist. Within the host nucleus these have been predicted to regenerate the original oversized transgene through homologous recombination7 or annealing of plus and minus strand complementary regions prior to second-strand synthesis8 (Figure 6). Trapani et al. have shown success with the fAAV approach in HEK293 cells and Hirsch et al. suggested fAAV vectors are a better gene therapy approach for delivering large genes to the retina and skeletal muscle than an alternative trans-splicing dual vector approach. The mechanisms by which transgenes recombine is still being elucidated but both non-homologous end joining (NHEJ)11 and homologous recombination10 have been implicated. The latter study provided evidence that fAAV transgenes recombine in a RAD51-dependent manner, which indicates recombination occurs following second-strand synthesis at which stage truncated transgenes would mimic a double-stranded break. Homologous recombination enables error-free joining of transgenes whereas NHEJ mechanisms are error prone and joining by these routes would create incorrect coding sequence within joined transgenes. Thus far evidence of fAAV joining by HR has been shown but in this report we reveal evidence of fAAV transgenes joining by NHEJ mechanisms. Given the success of fAAV vectors exhibited by other research groups and the potential for this approach to move beyond pre-clinical testing, we have attempted to investigate further the molecular outcomes of transducing cells with an AAV vector population carrying truncated transgenes. Until now, reports for fAAV have focused only on the successful expression of the target product generated following the desired formation of the large transgene via regions of sequence overlap between truncated transgenes.
Figure 1.
(a) Depiction of randomly truncated transgenes generated in fAAV vectors and (b) OV AAV vector transgenes. ITR = inverted terminal repeat; CAG = ubiquitous promoter; ABCA4 CDS = ATP-binding cassette transporter protein member 4 coding sequence; pA = polyA signal; fAAV = fragmented AAV; OV = overlapping.
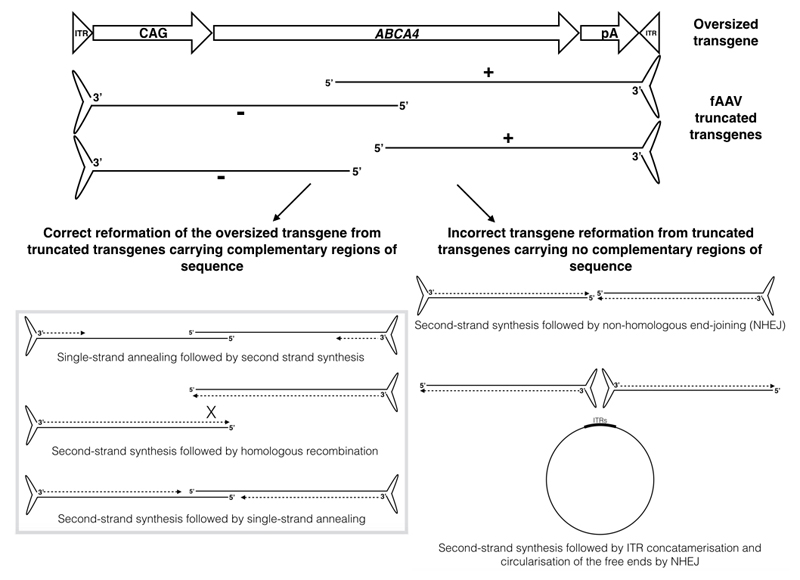
Figure 6.
Prediction of the methods by which truncated transgenes from fAAV vectors may join to form correct and incorrect transgenes for gene expression. ITR = inverted terminal repeat; CAG = ubiquitous promoter; ABCA4 CDS = ATP-binding cassette transporter protein member 4 coding sequence; pA = polyA signal; fAAV = fragmented AAV; NHEJ = non-homologous end-joining.
In this report we demonstrate that truncated transgenes that carry no regions of sequence overlap are able to combine and form stable transgenes. Transcript analysis of fAAV transduced cells revealed that whilst larger truncated transgenes did successfully recombine to form complete ABCA4 CDS within a large transgene, the resulting expression levels were low and any subsequent translation was not detectable by western blot. A significant finding was that a proportion of truncated ABCA4 transgenes that did not carry any sequence overlap, an area thought to be critical for reformation of the complete large construct, joined and formed hybrid constructs from which stable transcripts were detected. Our results suggest that despite the success seen by other research groups employing this dual vector strategy, the lack of control in generating fAAV dual vectors leads to unwanted hybrid expression products.
Materials and Methods
Vector production
ABCA4 CDS was inserted into pFastBac1 (ThermoFisher Scientific) and further cloned into a vector carrying AAV2 ITRs to generate a pTransgene of: pCAG.ABCA4.WPRE.bGHpA. This was packaged in AAV5 (fAAV5-WPRE) by GeneDetect® (Auckland, New Zealand). A second transgene (pCAG.ABCA4.SV40pA) was also packaged in AAV5 (fAAV5) and pCAG.ABCA4.bGHpA was further generated and packaged in AAV2 Y444F (fAAV2Y444F) and AAV8 Y733F capsids (fAAV8Y733F). For overlapping dual vector (OV) preparations, pCMV.CBA.ABCA4 and pABCA4.WPRE.pA were generated with each carrying a fragment of the ABCA4 CDS that included an overlapping region of 506 nucleotides. Both dual vector transgenes were 4.8kb in size and packaged in AAV8 Y733F capsids (OV AAV). Packaging for fAAV5 was achieved in HEK293 cells with a co-transfection of pTransgene with pRep2Cap5Helper using CaPO4. For all other preparations, HEK293T cells were transfected using a typical PEI protocol to deliver pRepCap, pHelper (pDeltaAdF6) and pTransgene. Cells were harvested three days post-transfection, lysed and the AAV population isolated by ultracentrifugation with a double cesium chloride gradient for fAAV5 and an iodixanol gradient followed by purification in Amicon Ultra-15 100K filter units (MerckMillipore, UK) for all other variants. The final preparations were collected in PBS. SDS-PAGE analysis confirmed good purification of each preparation and qPCR titres were determined using primers targeting upstream and downstream portions of the ABCA4 CDS. Upstream FW GCACCTTGGCCGTATTTGGACAG; Upstream RC TGAGTCAGACAGGCCGATGT; Downstream FW AGGGTGGCTTCCGGAGTA; Downstream RC CGTCAGGTCTTGTAGAAT. MCPA FW GCAGACAATGCGAGAGAATG and MCPA RC CAGTCATCCAAATCCACATTGAC were used to detect the MCPA intron in AAV preparations.
Transduction protocol
HEK293T cells were plated at 1E+05 cells per well in 24 well plates. 24 hours after plating, one well of cells was lifted and counted. Each AAV was applied at an MOI of 10,000. Transductions were conducted in triplicate with a media change 48 and 96 hours post-transduction. At five days post-transduction (unless otherwise stated) media was removed, cells were rinsed and lifted in cold PBS then washed three times in cold PBS using a gentle spin protocol. Cells from replicate wells were combined then split into aliquots for protein or mRNA processing and washed cell pellets frozen at -80°C.
mRNA extraction and cDNA synthesis
HEK293T frozen cell pellets were thawed on ice and mRNA extracted using mRNA DIRECT Dynabeads-oligodT (Life Technologies, UK) with 500ng of mRNA then used in a SuperScript III cDNA synthesis reaction with oligodT primer as per the manufacturers guidelines. The cDNA was cleaned in QIAGEN spin columns and eluted in 50μl DEPC-treated water.
PCR/qPCR detection of ABCA4 CDS
Transcripts containing ABCA4 CDS were amplified for sequencing using Primer F7 GGCTCCAGTTGGAATCTACC and Primer RCviii TCTGAAACCCGATGAAGAT. Bands were gel extracted and sequenced. QPCR detection of ABCA4 transcripts utilised the Upstream FW and Upstream RC listed above and Middle FW TCCAGAGCCAAAGGAAAG with Middle RC AGGAAGATAAGTTCTTGA. All qPCR reactions contained 2μl of the prepared cDNA and were run under the same cycling conditions of: 95°C 5 minutes, 40x[95°C 30 seconds, 55°C 20 seconds], + melt curve. The melt curve consisted of an increase from 55°C to 95°C at increments of 0.5°C for 5 seconds per temperature. The melt curve analyses for each primer set generated a single peak confirming specificity of the primers and amplification of a single target. ABCA4 transcript levels were normalised to βACTIN levels and presented as the fold change relative to untreated cells.
Southern blot
AAV samples containing 5E+5 genome copies were DNase treated, heated at 90°C for 10 minutes and run on a 0.7% agarose gel containing no ethidium bromide. The ssDNA fragments were transferred in 0.4M NaOH to a Hybond-N+ membrane (GE Healthcare) and incubated with DIGEasyHyb then DIG-labelled probes overnight. Membranes were washed in Low and High Stringency buffers before blocking with bovine serum albumin (BSA) and incubating with anti-DIG-AP. Following final washes, the membranes were treated with Detection Buffer and CDP-Star before being exposed. All DIG-associated buffers and blotting reagents were obtained from Roche. Plasmid control samples pCMV.CBA.ABCA4 and pABCA4.WPRE.pA (plasmids generated for production of upstream and downstream OV vectors) were digested with SwaI as per the manufacturers guidelines. This digestion removed the genetic elements of the transgenes within the ITRs, leaving a backbone fragment of ~2kb (undetected by either probe) and a detectable fragment of ~4.8kb for each. The fragment from pCMV.CBA.ABCA4 contained a portion of ABCA4 CDS detectable by the upstream probe and the fragment from pABCA4.WPRE.pA contained a portion of ABCA4 CDS detectable by the downstream probe.
Western blot
HEK293T frozen cell pellets were thawed on ice then lysed in RIPA buffer (MerckMillipore, UK) plus proteasome inhibitor (Roche, UK). Samples were left to rotate at 4°C for one hour prior to spinning at 24,000xg for 30 minutes. The supernatant was removed with 20μl added to 5μl protein loading buffer (GeneFlow, UK). This was left for 15 minutes at room temperature prior to loading on a 7.5% TGX gel (BioRad, UK). Proteins were transferred to a PVDF membrane using a TransBlotTurbo with subsequent ABCA4 detection (ab72955, Abcam, UK) conducted using a SNAPiD system (MerckMillipore, UK).
Statistical analysis
Data in Figure 4 are represented as the mean of n=3 with error bars depicting standard error of the mean (SEM). Differences between fAAV Upstream and Middle qPCRs were determined using the Student’s t-test. Treatment effect differences between fAAV and OV AAV were determined by two-way ANOVA.
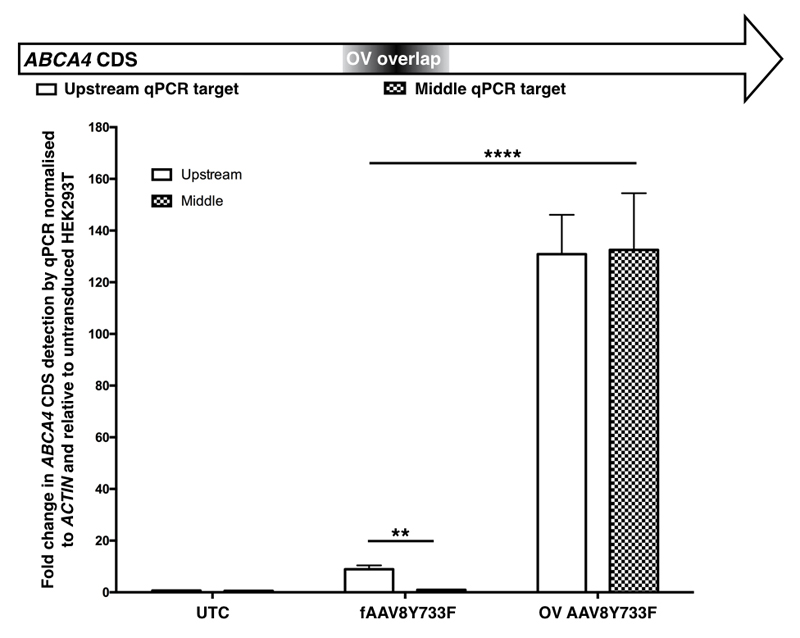
Figure 4.
qPCR detection of ABCA4 transcripts in cDNA generated from HEK293T cells transduced with fAAVY733F or OV AAV8Y733F. Targeting of ABCA4 transcripts at two different sites in the coding sequence revealed equivalent detection levels for OV treated samples but a significant difference (**p=0.0056) in fAAV treated samples between the two primer sets. There was also a significant difference between the two treatment groups with the OV treated samples having higher levels of ABCA4 transcripts than fAAV treated samples (****p=<0.0001). Error bars represent mean + SEM, n=3 per treatment group. - = untransduced HEK293T; fAAV = fragmented AAV2/8Y733F treated HEK293T; OV = overlapping dual vector AAV2/8Y733F treated HEK293T.
Results
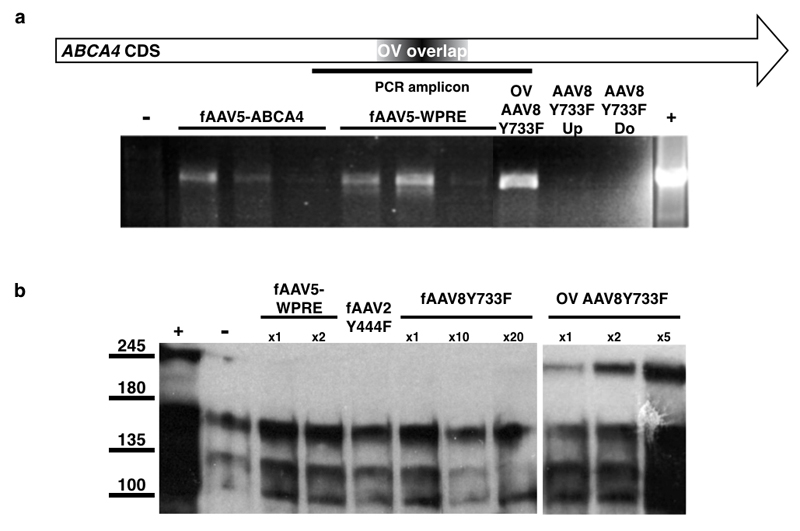
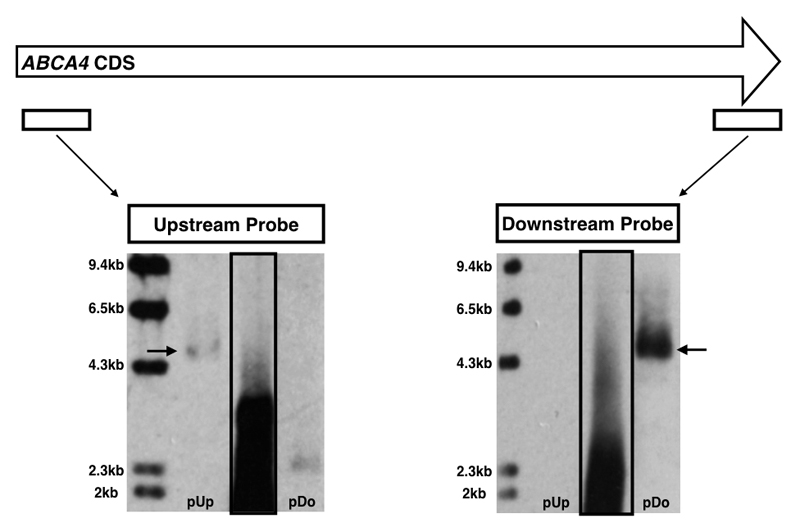
ABCA4 transgene constructs exceeding the AAV packaging limit were generated with (9.1kb) and without (8.5kb) a Woodchuck hepatitis virus post-transcriptional regulatory element (WPRE) and packaged into AAV5 capsids, fAAV5-WPRE and fAAV5, respectively. Numerous experiments were conducted using these preparations with cDNA analyses providing evidence of ABCA4 transcription in treated cells that was clearly detectable up to 5 days post-transduction but much reduced at seven days (Figure 2a). Despite consistent detection of ABCA4 transcripts, ABCA4 protein was repeatedly not evident by western blot in fAAV treated samples (Figure 2b). This indicated the fAAV approach was working but not to the extent required for protein detection by western blot. In case this was due to use of the AAV5 serotype, the large ABCA4 transgene without WPRE was then packaged into AAV8 Y733F capsids (fAAV8Y733F) for comparison to an overlapping dual vector (OV) system containing ABCA4 transgenes of 4.8kb in length with a defined 506 base pair complementary region (Figure 1b). The fAAV8Y733F transgenes were assessed by Southern blot analysis (Figure 3). Probes were designed to bind upstream and downstream portions of ABCA4 CDS and the assessments indicated an expected heterogeneous population of fAAV8Y733F transgenes existed, with the majority <4.8kb in length. Two control samples were prepared from plasmids containing known fragments of ABCA4 CDS (used for generating the OV AAV8Y733F dual vectors), these were digested with SwaI to isolate 4.8kb fragments containing either an upstream portion of ABCA4 CDS appropriate for the upstream probe detection (pUp) or a downstream portion of ABCA4 CDS suitable for detection by the downtream probe (pDo). Detection of the control samples indicated the probes used were specific to their targets.
Figure 2.
(a) Representative agarose gel showing amplifications of ABCA4 transcripts isolated from fAAV (5, 6 and 7 days post-transduction left to right) and OV AAV (5 days post-transduction) treated HEK293T cells. (b) Representative western blot detection of ABCA4 (ab72955, Abcam) from HEK293T cells transduced with fAAV and OV AAV variants. Cells were transduced with a MOI of 10,000: x1, x2, x5, x10 & x20 indicate multiples of the MOI. ABCA4 CDS = ATP-binding cassette transporter protein member 4 coding sequence; WPRE = Woodchuck hepatitis virus post-transcriptional regulatory element; - = untransduced HEK293T; + = pABCA4 transfected HEK293T; fAAV = fragmented AAV in various serotypes; OV = overlapping dual vector AAV2/8Y733F; OV overlap = region of overlap contained in upstream and downstream transgenes that form the OV system.
Figure 3.
Southern blot detection of ABCA4 CDS in transgenes of fAAV8Y733F vectors using upstream and downstream specific probes. Solid boxed lanes identify fAAV sample. Arrows indicate 4.8kb ABCA4 released from digested plasmid control samples containing either an upstream portion of ABCA4 or a downstream portion (for probe binding controls). pUp = digested ABCA4 plasmid for upstream probe positive control; pDo = digested ABCA4 plasmid for downstream probe positive control.
Given that the fAAV approach will generate a heterogeneous transgene population3–5, we performed further in vitro transduction experiments to discover what these truncated transgenes would initiate once inside cells. Transductions were conducted with both fAAV8Y733F and OV AAV8Y733F preparations with mRNA extracted and cDNA prepared five days post-transduction. QPCR analysis was conducted targeting two regions of the ABCA4 CDS (Figure 4). cDNA was generated from the polyA tails of transcripts and therefore a primer set was deisgned at an upstream portion of the ABCA4 CDS in order to prioritise targeting successfully generated full length transcripts. As the Southern blot assessments indicated that the majority of packaged transgenes of the fAAV8Y733F preparation were less than 4.8kb in size, we extrapolated from the original large transgene design that a proprotion of our plus and minus strand transgenes would be lacking a middle section of the ABCA4 CDS that would be necessary for successful homologous recombination between transgenes (Figure 1a). A primer set was designed to this middle section that was predicted to be absent from a proportion of our fAAV8Y733F transgenes to determine whether our ABCA4 transcript population existed with or without this section of CDS. This middle primer set was also designed to be within the overlap sequence contained in both OV AAV8Y733F transgenes. It was anticipated that ABCA4 transcripts isolated from the OV approach would be generated from successful homologous recombination at the known overlap sequence and that a homogenous population of ABCA4 transcripts would exist. Under this scenario, targeting two regions of such transcripts would provide equivalent detection of ABCA4 CDS from both the upstream and middle primer set.
For both treatment groups, stable ABCA4 transcripts were detected by qPCR whilst untreated samples had an absence of ABCA4 transcripts, indicating transgenes from both fAAV8Y744F and OV AAV8Y733F dual vector systems initiated ABCA4 expression. Overall, there was a significant difference in ABCA4 transcript levels between the two treatment groups (p=<0.0001), indicating the OV AAV8Y733F was a more efficient dual vector strategy in vitro than fAAV8Y733F. For the OV treated samples, equivalent high levels of ABCA4 expression were observed from both primer sets whereas a significant difference in levels of ABCA4 transcripts were detected from fAAV8Y733F treated samples between the two primer sets (p=0.0056). This indicated the ABCA4 CDS transcript structures were not consistent in the transcript population of fAAV8Y733F treated cells whereas they were in the OV AAV8Y733F treated samples.
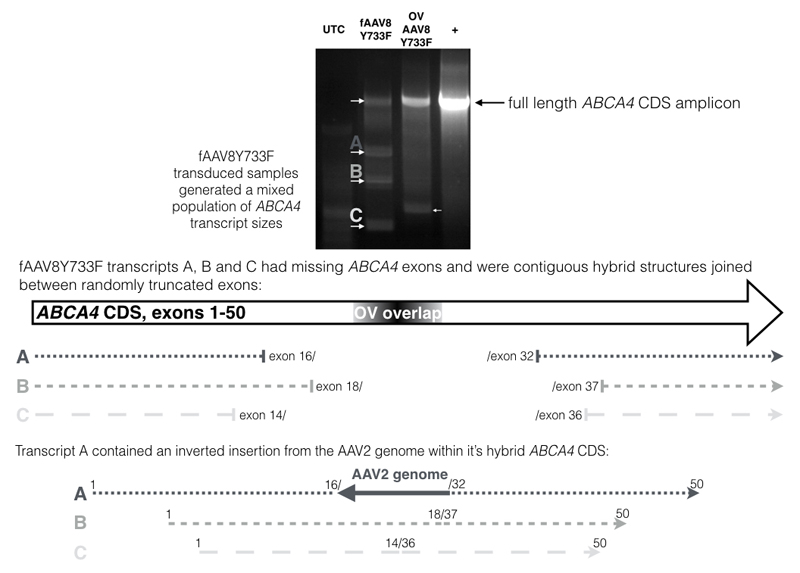
The cDNA samples were further assessed to determine what form of ABCA4 transcripts were being detected. Primers were designed to amplify across the ABCA4 CDS with the forward primer designed to bind only to CDS originating from upstream transgenes and the reverse primer only to CDS originating from downstream transgenes. The PCR results (Figure 5) revealed positive amplification across the full length of the ABCA4 CDS for both fAAV8Y733F and OV AAV8Y733F treated samples. This indicated there had been successful formation of the oversized ABCA4 transgene via complementary regions contained in opposing transgenes with both approaches, which led to subsequent ABCA4 expression. These transcripts were sequenced, which confirmed no errors in the ABCA4 CDS, suggesting reformation by homologous recombination had occurred in both treatment groups. From the strength of the amplicons it was apparent that more full-length transcripts were amplified for the OV AAV8Y733F treated samples than the fAAV8Y733F treated samples, which corresponded with the qPCR data and successful western blot detection of ABCA4 from OV AAV8Y733F treated samples but not fAAV8Y733F treated samples (Figure 2b).
Figure 5.
PCR amplifications of ABCA4 transcripts in cDNA from transduced HEK293T cells with depictions of the transcripts identified following sequencing. Both fAAV8Y733F and OV AAV8Y733F treated samples contained correctly formed full length ABCA4 transcripts. For fAAV8Y733F treated samples, hybrid transcript sequences were identified for which upstream and downstream portions of ABCA4 CDS were fused within exons. The exons within which the hybrids were joined are shown for transcript variants A, B and C. UTC = untransduced HEK293T; fAAV = fragmented AAV2/8Y733F treated HEK293T; OV = overlapping dual vector AAV2/8Y733F treated HEK293T; + = ABCA4 transfected HEK293T; AAV2 genome = genetic material from the AAV2 genome containing major coat protein A intron.
For a given AAV treatment, we would aim to identify only a single transcript product from the target gene following transduction. However, in addition to the full length ABCA4 fragment, both the fAAV8Y733F and OV AAV8Y733F samples consistently generated extra PCR products. For the OV AAV8Y733F treated samples, a single extra band was noted of ~0.6kb in size. Sequencing revealed this to be a mixed product resulting from appropriate binding of the reverse primer with non-specific binding of the forward primer to a second gene (GRINA) leading to a heterogeneous product. This was concluded to be a non-specific PCR product.
For the fAAV8Y733F treated samples, multiple additional bands were evident of discrete sizes, which occurred in repeat PCRs. Sequencing of the additional bands unveiled a population of multiple hybrid ABCA4 transcripts, Figure 5. A further finding was that one of the hybrid transcript forms contained an insertion between the two segments of ABCA4 CDS. This insertion was identified to be a fragment of the AAV2/2 major intron (MCPA) in the reverse orientation. The AAV Rep2Cap8Y733F genome (without ITRs) was provided in the vector production phase. The MCPA intron is 322 nucleotides in length and lies between Rep40/68 CDS and VP1 CDS in the pRep2Cap8Y733F construct. We designed qPCR primers to amplify the MCPA intron in our fAAV8Y733F and OV AAV8Y733F vector preparations. We obtained consistent detection of the MCPA intron in all three AAV preparations. For fAAV8Y733F, MCPA was detected at 3.1E+09 gc/ml (compared to an average transgene titre of 5.75E+11 gc/ml), for the upstream OV AAV8Y733F preparation the MCPA titre was 2.7E+09 gc/ml (transgene 9.7E+11 gc/ml) and for the downstream OV AAV8Y733F preparation the MCPA titre was 3.3E+09 gc/ml (transgene 3.4E+12 gc/ml). This revealed that in our AAV production process, any given AAV preparation packaged the pRepCap genetic element at a consistent level yet detection of the MCPA intron in subsequent ABCA4 transcripts was unique to the fAAV8Y733F treated samples.
Discussion
Packaging fragmented single-ITR AAV genomes from over-sized transgenes has been shown by several research groups to successfully lead to expression of the target protein in transduced cells in vitro and in vivo with proof of principle demonstrated in muscle, lung and retina2,7,9–10,12. In contrast to the successful data reported by other research groups, we have been unable to replicate the in vitro success with fAAV vectors. We have tested different fAAV preparations made from ABCA4 large transgene variants in multiple serotypes with numerous independent experiments conducted per vector generated. We utilised HEK293T cells for which there is a master cell bank stock that is well characterised and validated for use in pre-clinical testing of gene therapy vectors. Expression of ABCA4 was consistently detected by transcript anlaysis in transduced samples but not in untransduced samples, providing evidence that these fAAV systems do work. However, the absence of detectable protein indicated they do not work to the extent we had hoped (or as shown by other research groups). A similar lack of success with fAAV vectors has previously been reported4 and whilst refinement of the AAV preparation technique by enriching the population containing larger transgenes may improve the success of the approach12, as a dual vector system it appears to be prone to variability and less efficient than other dual vector methods.
fAAV preparations are known to generate a heterogeneous vector population3–5 and here we have shown that these randomly truncated transgenes lead to a heterogeneous mRNA population following fAAV transduction, which does not occur following treatment with a defined overlapping dual vector system. It has previously been shown that fAAV transgenes undergo homologous recombination to enable formation of complete and correct large coding sequences that then express the desired product2,9,10,12. We did identify expression of error-free transcripts that were therefore presumed to be formed following HR as previously described. However, we have also shown that truncated transgenes with no region of overlap are capable of joining and forming hybrid transgenes from which stable mRNA transcripts are generated. Sequencing of the transcript population indicated there had been joining of truncated opposing transgenes prior to transcription as the transcripts identified were not truncated but rather were stable hybrid forms of ABCA4 CDS. Given the region of ABCA4 CDS missing in these hybrid transcripts, the transgenes from which they originated are unlikely to have contained a complementary region of overlap for homologous recombination or complementary single-strand annealing. We hypothesise that their method of joining is likely to have occurred following second-strand synthesis of the truncated transgenes, which then underwent non-homologous end joining (NHEJ) to form hybrid ABCA4 transgenes or by ITR joining with subsequent circularisation of the free ends (Figure 6). Either way, the resulting formations would resemble the normal structure of a transgene with 5’ITR, promoter, hybrid ABCA4 CDS, polyA signal and 3’ITR and as such would be capable of generating stable transcripts, which were subsequently detected. The ABCA4 CDS detected in the hybrid transcripts was consistently truncated within exons, not at exon boundaries. There were no apparent genetic features consistent at the regions at which the transgenes appeared to have been truncated and ligated (such as GC content, purine composition or repeat regions). The hybrid transcript forms were predicted to lead to stop codons in the fused downstream nucleotide sequence, which if translated would generate truncated proteins of 80-110kDa with 90+% homology to ABCA4.
Importantly, one of the hybrid transcript forms contained an insertion of the AAV2/2 major coat protein (MCPA) intron derived from the native AAV2 genome. AAV packaging is known to be imperfect and whilst the majority of a vector population will carry the transgene, preparations also contain rep and cap sequences13.. The presence of the MCPA intron in the AAV preparation was not unique to the fAAV mixture and nor was it present in excess compared to other AAV preparations used. However, the presence of the MCPA intron in subsequent ABCA4 transcripts was unique to the fAAV treated samples, indicating that single ITR transgenes are able to incorporate additional DNA elements. This insertional capability may be unique to fAAV ABCA4 transgenes but there are no apparent sequence features at the site of insertion that would indicate it to be a hotspot for such activity.
The inconsistency of success between research groups attempting the fAAV approach should be taken into account when considering the future of multi-vector AAV treatments for disorders requiring delivery of large genes, such as ABCA4. The data presented here highlight the need for further investigations into the methods and mechanisms of delivering large genes using a dual AAV vector approach.
Table 1.
QPCR titres of AAV preparations.
| AAV | Upstream titre (gc/ml) | Downstream titre (gc/ml) |
|---|---|---|
| fAAV5 | 3.5E+10 | 3.4E+10 |
| fAAV5-WPRE | 5.6E+11 | 3.6E+11 |
| fAAV2Y444F | 6.5E+09 | 6.6E+09 |
| fAAV8Y733F | 5.0E+10 | 1.1E+12 |
| Upstream OV AAV8Y733F | 9.7E+11 | - |
| Downstream OV AAV8Y733F | - | 3.4E+12 |
Acknowledgements
MRC DPFS scheme, Fight for Sight (UK), Royal College of Surgeons of Edinburgh, Knoop Fellowship, Marie Curie Foundation
Footnotes
All authors declare no conflicts of interest.
References
- 1.Grieger JC, Samulski RJ. Packaging capacity of adeno-associated virus serotypes: impact of larger genomes on infectivity and postentry steps. J Virol. 2005;79:9933–9944. doi: 10.1128/JVI.79.15.9933-9944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allocca M, Doria M, Petrillo M, et al. Serotype-dependent packaging of large genes in adeno-associated viral vectors results in effective gene delivery in mice. J Clin Invest. 2008;118:1955–1964. doi: 10.1172/JCI34316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu Z, Yang H, Colosi P. Effect of genome size on AAV vector packaging. Mol Ther. 2010;18:80–86. doi: 10.1038/mt.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lai Y, Yue Y, Duan D. Evidence for the failure of adeno-associated virus serotype 5 to package a viral genome > or = 8.2 kb. Mol Ther. 2010;18:75–79. doi: 10.1038/mt.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong B, Nakai H, Xiao W. Characterization of genome integrity for oversized recombinant AAV vector. Mol Ther. 2010;18:87–92. doi: 10.1038/mt.2009.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kapranov P, Chen L, Dederich D, et al. Native molecular state of adeno-associated viral vectors revealed by single-molecule sequencing. Hum Gene Ther. 2012;23:46–55. doi: 10.1089/hum.2011.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grose WE, Clark KR, Griffin D, et al. Homologous recombination mediates functional recovery of dysferlin deficiency following AAV5 gene transfer. PLoS ONE. 2012;7:e39233. doi: 10.1371/journal.pone.0039233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakai H, Storm TA, Kay MA. Recruitment of single-stranded recombinant adeno-associated virus vector genomes and intermolecular recombination are responsible for stable transduction of liver in vivo. J Virol. 2000;74:9451–9463. doi: 10.1128/jvi.74.20.9451-9463.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trapani I, Colella P, Sommella A, et al. Effective delivery of large genes to the retina by dual AAV vectors. EMBO Mol Med. 2013;6:n/a–n/a. doi: 10.1002/emmm.201302948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirsch ML, Li C, Bellon I, et al. Oversized AAV transduction is mediated via a DNA-PKcs-independent, Rad51C-dependent repair pathway. Mol Ther. 2013;21:2205–2216. doi: 10.1038/mt.2013.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inagaki K, Ma C, Storm TA, Kay MA, Nakai H. The role of DNA-PKcs and artemis in opening viral DNA hairpin termini in various tissues in mice. J Virol. 2007;81(20):11304–11321. doi: 10.1128/JVI.01225-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pryadkina M, Lostal W, Bourg N, et al. A comparison of AAV strategies distinguishes overlapping vectors for efficient systemic delivery of the 6.2 kb Dysferlin coding sequence. Mol Ther Methods Clin Dev. 2:15009. doi: 10.1038/mtm.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halbert CL, Metzger MJ, Lam S-L, Miller AD. Capsid-expressing DNA in AAV vectors and its elimination by use of an oversize capsid gene for vector production. Gene Ther. 2011;18:411–417. doi: 10.1038/gt.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]