Abstract
Correlative microscopy incorporates the specificity of fluorescent protein labeling into high-resolution electron micrographs. Several approaches exist for correlative microscopy, most of which have used the green fluorescent protein (GFP) as the label for light microscopy. Here we use chemical tagging and synthetic fluorophores instead, in order to achieve protein-specific labeling, and to perform multicolor imaging. We show that synthetic fluorophores preserve their post-embedding fluorescence in the presence of uranyl acetate. Post-embedding fluorescence is of such quality that the specimen can be prepared with identical protocols for scanning electron microscopy (SEM) and transmission electron microscopy (TEM); this is particularly valuable when singular or otherwise difficult samples are examined. We show that synthetic fluorophores give bright, well-resolved signals in super-resolution light microscopy, enabling us to superimpose light microscopic images with a precision of up to 25 nm in the x–y plane on electron micrographs. To exemplify the preservation quality of our new method we visualize the molecular arrangement of cadherins in adherens junctions of mouse epithelial cells.
Keywords: Correlative electron and light microscopy, Electron tomography, Adherens junctions
1. Introduction
Correlative Light- and Electron Microscopy (CLEM) is an approach that complements the information from two microscopic contrasts. Imaging of fluorescently labeled proteins localized by light microscopy (LM) is combined with the visualization of subcellular structures by electron microscopy (EM). To date, super-resolution light microscopy techniques can achieve a protein localization in the range of ~20 nm, whereas biological EM of thin-sectioned, resin-embedded samples yields a resolution of ~2–6 nm. With CLEM, superimposed images allow the protein of interest to be localized in its cellular context within a few tens of nanometers, on specimen preserved at close-to-native conditions. If optimized CLEM can yield images with such an unprecedented richness of information that is expected to surpass traditional immuno-gold labeling techniques in terms of precision, sample preservation, versatility and interpretation capabilities.
CLEM can be applied as a pre- or post-embedding technique, depending on whether the LM is performed before or after the embedding for EM. Technically, when LM is performed prior to fixation, there is a potentially small time delay (Verkade, 2008), in which the specimen can still change. Furthermore, samples might change during the EM embedding procedure, which might also obstruct CLEM. In general, the correlation of LM with EM data is challenging, since the z-resolution of the LM is much worse than the required thickness for TEM. Thus CLEM is preferably performed directly on sections on the TEM grid. For the imaging of thick samples without section preparation, block-face scanning electron microscopy can be performed (Denk and Horstmann, 2004; Heymann et al., 2006; Knott et al., 2008). This allows the generation of large volume data sets for 3D analysis of the sample in an automated fashion as well as CLEM (Murphy et al., 2011; Narayan et al., 2013).
Traditionally, protein-localization in TEM is performed by immuno-gold labeling. The gold could also be replaced by fluorophores for super-resolution microscopy. Both methods depend strongly on the quality of the available antibodies and accessibility of the epitopes on the surface (Nanguneri et al., 2012). Most closely related to the immuno-based techniques are quantum dots, which are both fluorescent and have a dense metal core that can be visualized in the EM. For increased specificity, protocols employing genetically-encoded tags applicable to photo-oxidation that utilize Tetracysteine/ReAsH (Gaietta et al., 2002), GFP (Grabenbauer et al., 2005), and miniSOG/FMN (Shu et al., 2011) have been developed, these tags are suitable for photo-oxidation yielding an enrichment of electron-dense osmium tetroxide after illumination (reviewed in Ellisman et al. (2012)).
Electron microscopy of vitrified samples offers conditions closest to the native state (Sartori et al., 2007; Schwartz et al., 2007). However, vitrified samples can only be imaged at liquid nitrogen temperatures, which puts serious constraints on the light microscopy, prohibiting the use of super-resolution techniques as well as the use of high numerical aperture objectives, thus limiting the localization precision to a few hundred nanometers (van Driel et al., 2009). Notably, cryo-fixation techniques are generally considered to be the best methods for preserving biological samples in their native state and high-pressure freezing (HPF) is often implemented as a method in CLEM (Muller-Reichert et al., 2007). Most recently, HPF and freeze substitution (FS) EM-preparation protocols have managed to preserve the post-embedding fluorescence of GFP and other fluorescent proteins (Kukulski et al., 2011; Watanabe et al., 2011). Thereby, excellent protein localization has been achieved either by employing fiducials or by using super-resolution LM techniques. While these post-embedding fluorescence-retaining methods currently represent the state-of-the-art in CLEM, they have a particular disadvantage. These approaches require very low concentrations of heavy metals during sample preparation to avoid quenching of GFP fluorescence. However, heavy metals (uranyl acetate in particular) are important during the embedding procedure for structure preservation in EM. Thus, to date, a compromise between localization precision, fluorescence preservation, and EM quality has had to be made.
Here we use genetically-encoded chemical tags to label different cellular proteins with synthetic fluorophores, in order to circumvent the fluorescence quenching of GFP. We use SNAP (Keppler et al., 2003) and Halo (Los et al., 2008) tags expressed in living cells, that are coupled with the synthetic fluorophores Alexa Fluor 647, tetramethylrhodamine (TMR) and silicon-containing rhodamine derivative SiR-carboxyl (SiR). We show that these synthetic fluorophores conserve their post-embedding fluorescence independently of the uranyl acetate (UA) concentration, thus allowing CLEM for HPF/FS EM-preparation protocols. We demonstrate that our CLEM approach is suitable for simultaneously identifying the localization of up to two intracellular proteins using conventional fluorescence microscopy (FM) correlated with both TEM and SEM, thus opening up a new range of applications. Furthermore, it is easily possible to localize intra- and extracellular proteins of interest, with a few tens of nanometers precision, by combining super-resolution light microscopy and TEM.
2. Results
2.1. Preparation
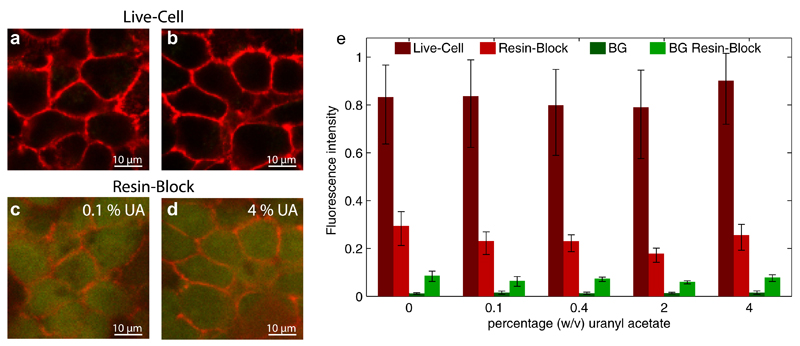
CLEM allows the functional assignment of proteins prior to their EM analysis. Here we use a pre- and post-embedding CLEM approach. During HPF and FS we varied the concentrations of UA. We found that the post-embedding fluorescence of the synthetic fluorophores was well preserved at all UA concentrations (Fig. 1a–d). This is in contrast to GFP-based fluorescence retaining methods (Kukulski et al., 2011; Watanabe et al., 2011), which either avoid the use of UA, or keep it at an extremely low concentration in order to avoid fluorescence quenching. We find that 2% UA provides the best subcellular structural preservation, which is consistent with a previous report (Hawes et al., 2007). UA concentrations even higher than 2% might be advantageous for SEM imaging, since the higher heavy metal concentration increases the amount of detectable backscattered electrons. We quantified the quenching of the fluorescence of embedded L-cells stably expressing SNAPf-NCadherin labeled with Alexa Fluor 647. We measured the intensity of several individual cell contours in live-cell confocal imaging and compared the fluorescence signal after embedding using exactly the same optical settings. The specific fluorescence in the embedded samples is decreased to ~25%, and is not decreased further at higher UA concentrations. We find that the auto-fluorescence increases in the green channel only in the area of the embedded cells by ~7-fold (Fig. 1e and Supplementary Fig. 2). Interestingly this was consistent on all investigated specimen, thus we further explored this for automated image superposition of post-embedding LM images and EM micrographs.
Fig.1.
Live-cell imaging and post-embedding fluorescence at different UA levels. (a, b) Live-cell imaging of L-cells stably expressing SNAPf-NCadherin labeled with Alexa Fluor 647. The cell–cell junctions are visualized in the red channel. The auto-fluorescence is shown in the green channel. (c, d) The corresponding fluorescence within the resin-block after high-pressure freezing and freeze-substitution with 0.1% and 4% UA. (e) Fluorescence intensity measured along the cell junctions in live-cell imaging (dark red), postembedding in the resin-block (light red), auto-fluorescence measured in the green channel in live-cell imaging (dark green), and the auto-fluorescence postembedding in the resin block (light green). At least three regions per uranyl acetate concentration were evaluated with 200 sample points each. Error bars represent the standard deviation of the intensity along the junction. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.2. Computational image superposition
The precision of superposition of the FM images on the EM micrographs is important for accurately localizing the labeled proteins within the EM micrographs. Both for embedded single cells and for cell layers the cell boundaries were well resolved by their auto-fluorescence (Fig. 1c, d and Supplementary Fig. 2) providing sufficient signal for positional identification, when compared to the cell membranes visible in the EM micrographs (Supplementary Fig. 1). The affine transformations were necessary to compensate for distortions due to the different orientation of the sample in the optical path as well as sample shrinkage due to electron irradiation.
2.3. SEM experiments
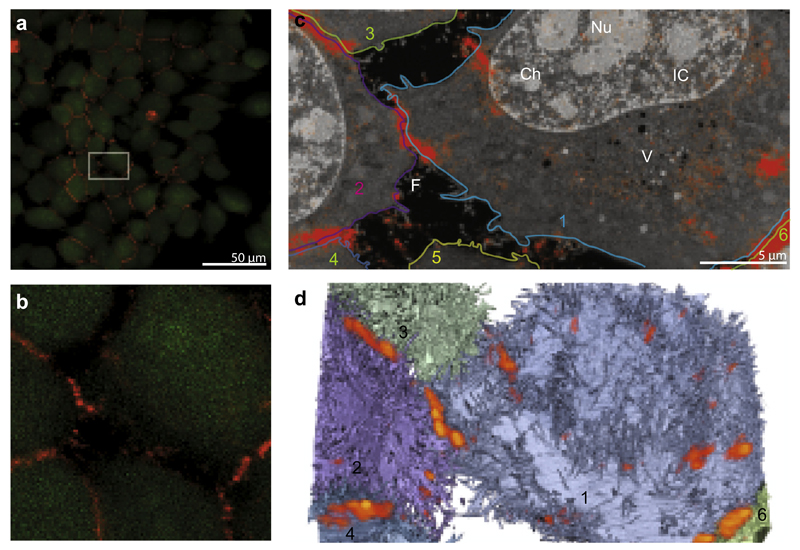
We visualized the embedded L-cells stably expressing SNAPf-NCadherin labeled with Alexa Fluor 647 using confocal fluorescence microscopy (Fig. 2a); the sample was subsequently trimmed to a block face for ion-abrasion SEM. Then, we selected a region containing cells with several cell–cell junctions for ion abrasion SEM (Fig. 2a box). In the SEM, we used backscattered electrons to record an image stack of 33 × 28 × 17 μm3 with a pixel size of ~8 nm perpendicular to the beam direction, and ~20 nm along the beam direction. The computational slices from the image stack show a good structural preservation with sufficient contrast to recognize different cellular organelles (Fig. 2c). The N-cadherin fluorescence signal (red) overlapped with the positions of the junctions seen in the SEM, and in certain locations with the positions of filopodia on the cell surface, which should contain cadherins (Fig. 2c and d).
Fig.2.
ia-SEM CLEM of NCadherin in L-cells. (a) Fluorescence image of L-cells stably expressing SNAPf-NCadherin labeled with Alexa Fluor 647 after high-pressure freezing and freeze substitution with 2% UA. The box outlines the 32 μm × 17 μm area enlarged in (b) and shows the interface of six neighboring cells. (c) Overlay of the confocal fluorescence image of SNAPf-NCadherin from (b) on a tomographic slice from the ia-SEM image stack showing the six neighboring cells outlined with different colors and numbered. The specific fluorescence of Alexa Fluor 647 shown in red is localized mainly at cell–cell contacts, and occasionally on the cell surface at densely clustered filopodia. The contrast in the SEM images allows the identification of sub-nuclear compartments: Nucleolus (Nu; medium grey), chromatin (Ch; light grey) and interchromatin compartment (IC; dark grey) as well as diverse cytoplasmic vesicles (V) and filopodia (F). (d) Volume representation of the same SEM dataset (32 μm × 17 μm × 10 μm), overlaid with the confocal fluorescence signal. The fluorescence is visualized by volume rendering in an orange heat color. The different cells are represented as transparent isosurfaces in corresponding colors to (c); blue (cell 1), purple (cell 2), grey-green (cell 3 and 6), and grey-blue (cell 4) with opaque nuclei shown in cells 1 and 2, corresponding to the contours in (c). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.4. CLEM with various fluorophores
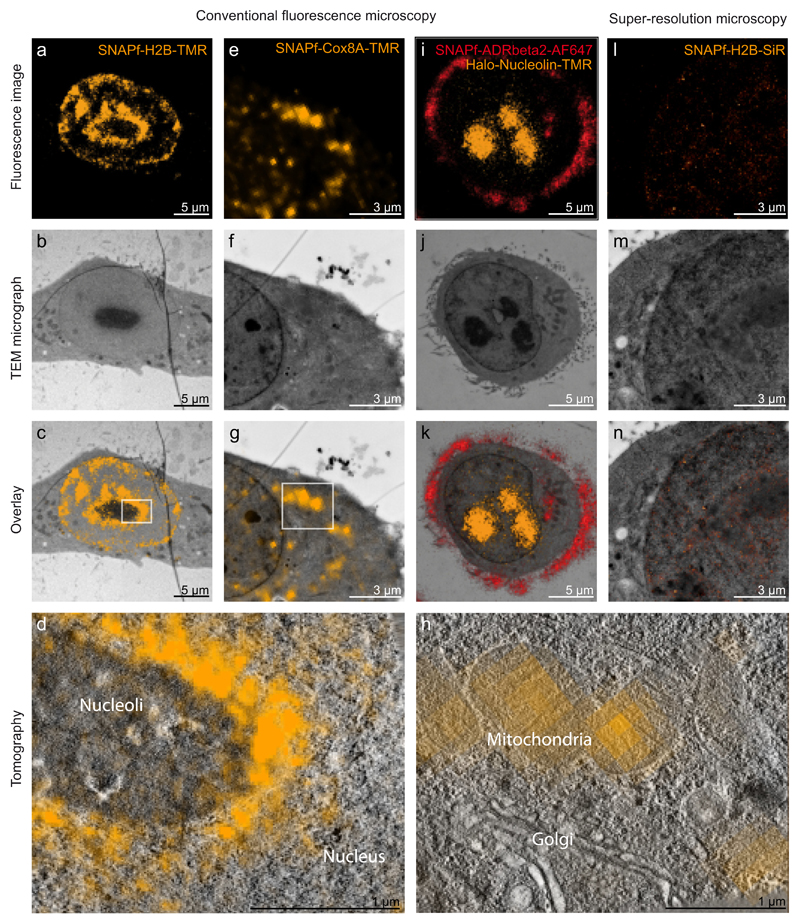
The wide range of genetically encoded chemical tags in combination with an even larger range of synthetic fluorophores offers various options for CLEM. Here we tested several tag-fluorophore combinations. In the first experiment we used HeLa cells stably expressing SNAPf-Histone 2B, which were labeled with TMR to show the feasibility of intra-nuclear labeling (Fig. 3a–d). In the overlay images of labeled Histone 2B (Fig. 3c), we observed that the fluorescence (orange) is excluded from the electron dense nucleolus that is separated from the electron lucent area of the nucleus showing areas of chromatin, in which the Histone 2B fluorescence signal is seen. To further verify the Histone 2B localization we recorded intranuclear tomograms (Fig. 3c box). In the tomographic slices the individual nucleosomes cannot be discerned, however the individual nuclear compartments can be clearly seen, and FM images superimpose nicely (Fig. 3d). In the second experiment we used HeLa cells stably expressing SNAPf-Cytochrome c oxidase subunit 8A labeled with TMR (Fig. 3e–h) to demonstrate cytoplasmic labeling. We find that the fluorescence of mitochondrial protein Cytochrome c oxidase (orange) is localized inside the mitochondria in the overlay image (Fig. 3g), and some spurious fluorescence puncta (4 out of 14) appearing within the nucleus, which were not created by the embedding process, and can be attributed to the excess of staining used to demonstrate the process. The EM structural preservation is evidenced in the tomographic slices of this data. Overall the HeLa cells show high contrast membranes, individual cytoskeletal elements like microtubules can be seen as well as the Golgi apparatus (Fig. 3h). In the third experiment we used HeLa cells stably expressing Halo-Nucleolin and SNAPf-Adrenergic receptor beta 2, which were labeled with TMR and Alexa Fluor 647 to demonstrate the possibilities of dual labeling (Fig. 3i–k). The dual labeling shows an accurate localization of Adrenergic receptor beta 2 (red) on the plasma membrane of the abundant filopodia and Nucleolin (orange) on the three electron dense intranuclear areas depicting the nucleoli (Fig. 3k). In the fourth experiment we used HeLa cells stably expressing SNAPf-Histone 2B labeled with SiR, to show the interchangeability of the different fluorophores as well as the possibility of super-resolution microscopy on the sections prepared for TEM (Fig. 3l–n). The super-resolution images show the specific overlay of Histone 2B (orange) with the nucleus (Fig. 3n). While SNAPf-Histone 2B could also be labeled with TMR enabling conventional LM, super-resolution imaging was best with SiR. We find that among the different synthetic fluorophores tested, SiR exhibited ideal photoswitching properties under the specific buffer, pH and laser intensities.
Fig.3.
CLEM of various intracellular and intranuclear proteins. (a) FM image of SNAPf-H2B-TMR. (e) FM image of SNAPf-Cox8A-TMR. (i) Dual color FM of SNAPf-ADRbeta2-AF647 (red) and Halo-Nucleolin-TMR (orange). (l) Super-resolution microscopy of SNAPf-H2B-SiR (orange). (b, f, j, m) TEM micrographs after FM imaging. (c, g, k, n) overlay of the FM images with the corresponding TEM micrographs. (c) FM signal overlaying with the nucleus sparing the darker stained nucleolus. (g) FM signal overlaying with intracellular mitochondria. (k) FM signal overlaying with cellular filipodia (red) as well as with nucleoli (orange). (n) Super-resolution signal overlaying with the nucleus. (d) tomogram acquired on the boxed area in (c). FM signal (orange) is sparing the dark nucleolus. (h) tomogram acquired on the boxed area in (g). FM signal (orange) is localized on the mitochondria. Mitochondrial cristae as well as cytoskeletal elements and parts of the Golgi apparatus are visible. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.5. Precision of the protein localization in the EM tomogram
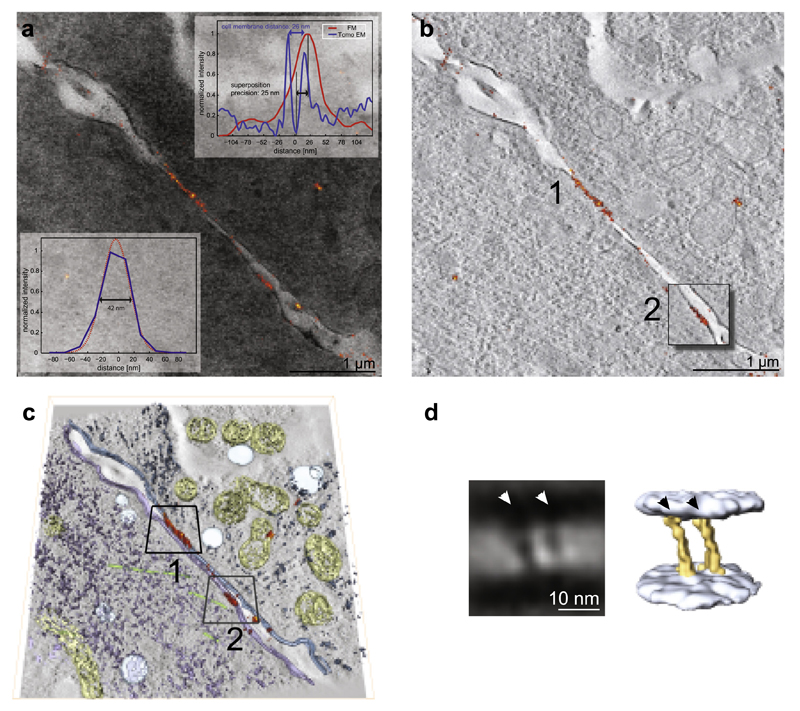
The precision of the protein localization in the EM images depends on the precision of the localization in the super-resolution images and the precision of the superposition of the LM on the EM images. Generally, conventional fluorescence microscopy indicates a continuous N-cadherin fluorescence signal across the length of intercellular boundaries, which implies that the cadherin arrangement forming cell-to-cell junctions is equally distributed across the intercellular space. FM imaging of the 300 nm TEM sections already revealed discrete clusters of cadherin molecules in the intercellular space (Fig. 4a). Those were further investigated by correlative super-resolution LM and electron tomography (ET) in order to estimate the precision with which the fluorescently labeled proteins can be localized on the EM images. Cell–cell junctions are well suited for this purpose since the cell membranes are clearly visible in EM. We estimated the precision of the protein localization of SNAPf-Ncadherin (~17 nm see Fig. 4a lower insert, see Section 4: Fluorescence Microscopy), by measuring the average deviation of the fluorescence intensity peak from the center of the two neighboring cell membranes on a 300 nm TEM section. Based on the evaluation of the line-plot data from the super-resolution signal (red) and the cell membranes from TEM (blue), the precision of the superposition of the super-resolution image on the EM tomogram was estimated to be ~25 nm in the direction perpendicular to the cell membranes (Fig. 4a upper insert).
Fig.4.
Localization-based microscopy combined with ET, and sub-tomogram averaging of NCadherin expressing L-cells. (a) Superimposed images of the super-resolution signal of SNAPf-NCadherin (orange) and the corresponding TEM micrograph. Lower insert is showing the precision of the protein localization with super-resolution microscopy. The blue line depicts the measured profile across the cellular junction in the super-resolution image, whereas the red line is the Gaussian fit to it. The full width at half maximum of the Gaussian is 42 nm, which is in good agreement with calculated precision of the localization precision of the super-resolution microscopy with sigma equal to 17 nm (2.35 * sigma). Upper insert is showing the precision of proteins localization within the tomograms, on the example of the cadherin molecules. The blue plot shows the normalized intensity profile across the junction measured on the EM images, which shows a membrane spacing of ~26 nm (blue double arrow). The red plot shows the normalized intensity profile across the super-resolution signal. The shift between the red and the blue curve indicates the precision of protein localization within the EM tomogram. The shift (black double arrow) is measured between the peak of the super-resolution signal (red) and the center of the neighboring cell membranes (blue). The precision of the protein localization within the EM tomogram was estimated at ~25 nm. (b) Overlay of a 2 nm thick tomographic slice with the super-resolution signal (orange). The first fluorescent cluster is localized between two cell membranes with a spacing of ~26 nm. The second fluorescent cluster can be associated with the cell membrane of the left cell ~80 nm higher than the first and shows a gathering of cadherins, without a connection to the juxtaposed cell (Box 2). (c) Volume representation of intracellular organelles recognizable in the tomogram; mitochondria (yellow), vesicles (light blue), microtubules (green), cytoplasmic compounds (violet). (d) Sub-tomogram averaging of the first fluorescence cluster reveals two cell membranes at a distance of ~26 nm together with two densities spanning the distance between the membranes (white arrowheads). Volume representation of the membrane (light blue) and the obliquely oriented densities (yellow) with a thickness of 3 nm (black arrowheads). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.6. Preservation of adherens junctions
In order to evaluate the cellular EM preservation as well as the preservation of fine molecular assemblies, we selected a region on the section with two distinct N-Cadherin clusters (Fig. 4a), and performed ET (He et al., 2003). We see in the overlay with the tomogram that the first longer N-cadherin cluster (orange) (Fig. 4b1) corresponds to a region with a regular spacing of ~26 nm between the two juxtaposed membranes, which is consistent with the spacing of N-cadherin estimated from crystal structures (Harrison et al., 2011). The second N-cadherin cluster (Fig. 4b2) seen in the overlay image can be associated with a membrane region ~80 nm in z-height above the previous cluster with no apparent membrane on the juxtaposed position, indicating that cadherins might first cluster within one membrane (“cis-dimer”), and then find their juxtaposed neighbor. Cellular structural preservation is demonstrated by visualization of mitochondria (yellow) microtubules (green) cytoplasmic components (blue, purple) as well as vesicles (light blue) in the tomogram (Fig. 4c). For the analysis of the preservation of fine molecular assemblies we performed sub-tomogram averaging on the first N-cadherin cluster. We find two cell-membranes spaced ~26 nm apart, and obliquely oriented densities with a periodicity of 7 nm (Fig. 4d), which resemble the X-ray model of cadherins (Boggon et al., 2002; Harrison et al., 2011).
3. Discussion
The functional assignment of ultrastructural data with fluorescence microscopy has the capacity to transform our interpretations of biological structure and function. However, the functional assignment is challenging because the harsh EM preparation methods quench the fluorescence. During the EM preparation, the proteins are denatured due to the acidic environment of the embedding material (Watanabe et al., 2011), which has direct implications for the GFP based methods, such as the quenching of the GFP fluorescence, and an increase in the overall protein background fluorescent signal. Our experiments show that synthetic fluorophores do not degrade as severely, and thus the fluorescence signal withstands the increasing background signal for a broad range of EM preparation conditions. This higher stability allows the choice of the most appropriate EM preparation protocols, and, as demonstrated here, the heavy metal concentration can be increased to such an extent that the same sample can be used for TEM and SEM. Even more importantly, the higher stability increases the reproducibility of the experiments.
Synthetic fluorophores offer a much wider palette than fluorescent proteins and cover a broader range of the wavelength spectrum usable for FM. Here we have used two different dyes, which diffuse through the cell membrane without destroying it, and without compromising the EM quality. Other synthetic fluorophores that are not membrane permeable when conjugated with substrates for chemical tagging could be used intracellularly (ATTO655 as HaloTag substrate is permeable as opposed to the SNAP substrate) (Wilmes et al., 2012). For the deposition of specific synthetic fluorophores in the cytoplasm techniques like microinjection might be a valuable tool to further exploit the entire range of synthetic fluorophores for CLEM. For the post-embedding super-resolution microscopy we tested a number of synthetic fluorophores. SiR and Alexa Fluor 647 have revealed high suitability for super-resolution imaging due to high photon yield per switching event and a large number of switching cycles.
For the thickness of the TEM section, the super-resolution images provide exclusively 2-dimensional information. Thus, in the 3D tomographic images, a z-localization without additional constraints – as in the case of the adherens junctions – is not possible, However, in any case, the 3D localization precision is better than in conventional immuno-gold labeling where epitopes must be available on the surface of the section in order to be detected. For an even better z-localization, correlative array tomography could be employed in which, a series of ultrathin sections (40–100 nm) is examined by CLEM (Wacker and Schroeder, 2013).
Generally, evaluation of the quality of preservation of the morphology in embedded samples is challenging, since, apart from specific caveats due to freezing damage, the quality cannot be assessed by objective criteria. Nevertheless, sample preservation remains the most crucial requirement in any CLEM approach, since it has direct impact on the biological interpretation. By exemplifying that the macromolecular organization of adherens junctions can be preserved to a large extent, it can be demonstrated that the molecular organization is retained. Fortunately, the broad range of preparation capabilities for EM using synthetic fluorophores allows for dedicated sample preparation. This is especially valuable for rare specimens, which cannot be processed multiple times and thus opens up new areas of biological exploration.
4. Materials and methods
4.1. Molecular cloning
The coding sequence of SNAPf was amplified by PCR (sense primer: 5′-GCGGCGCGCCATGGACAAAGACTGCGAAATGAAGCG-3′, antisense primer: 5′-GCGGCGCGCCCACCCAGCCCAGGCTTGCCCAG-3′) using the pSNAPf-vector (New England BioLabs) as a template and introducing AscI sites at both ends. The amplified region was inserted via AscI into the coding region of the extracellular domain of N-cadherin at a site (aa position 151 of the mature protein) previously reported to tolerate insertion of GFP (Kim et al., 2011).
4.2. Tissue culture, transfections and generation of stable cell lines
L cells (mouse fibroblasts, ATCC # CRL-2648) were grown in complete medium (DMEM GlutaMAX supplemented with 10% FCS and 1 mM sodium pyruvate (Life Technologies, Darmstadt, Germany)). Transfection was performed with the pNcad-SNAPf using X-tremeGENE9 (Roche, Mannheim, Germany) and CombiMag beads (OZ Bioscience, Marseille, France). Cells were trypsinized 24 h post transfection, transferred to a 6 well plate and cultivated in selection medium (complete medium + 600 μg/ml Geneticin (Life Technologies)). For generation of monoclonal cell lines, cells were trypsinized, diluted to a concentration of 2 cells/ml and plated in 96 well plates (100 μl/well). After 11 days, cells were further expanded in 24 well plates. Expression of Ncad-SNAPf was analyzed by immunoblotting with an anti-N-cadherin antibody (mouse, BD Biosciences). Cell clones, which were positive in Western blot, were stained with SNAP-Surface Alexa Fluor 647 (New England BioLabs) and analyzed by confocal laser scanning microscopy using a LSM780 (Plan-Apochromat 40×/1.4 oil objective, Carl Zeiss, Jena, Germany). Cell lines displaying high-levels of Ncad-SNAPf at the plasma membrane were chosen for further experiments.
HeLa cells were grown in complete medium (DMEM, 10% FBS, 0.29 mg/ml L-glutamine, and 100 U/ml penicillin/streptomycin). For transfections X-tremeGENE HP (Roche Applied Science) and plasmids pSNAPf-H2B, pSNAPf-ADRbeta2, pSNAPf-Cox8A (New England BioLabs) and pFN21AB5231 (HaloTag-Nucleolin; Promega, Mannheim, Germany/Kazusa DNA Research Institute) were used. To generate stable cell lines, cells transfected with pSNAPf-H2B and pSNAPf-ADRbeta2 were transferred from 24 well plates into 6 cm dishes 24 h post transfection and cultivated in selection medium (complete medium +400 μg/ml Geneticin) for 3 weeks. For dual color imaging, HeLa cells stably expressing SNAPf-ADRbeta2 were transfected with pFN21AB5231.
4.3. Cellular labeling
L cells stably expressing Ncad-SNAPf were grown to a confluency of 60–80%, stained in 2 μM SNAP-Surface Alexa Fluor 647 solution (dilution in phenolred free complete medium) at 37 °C for 5 min and subsequently washed three times with phenolred free complete medium.
For staining of HeLa cells, 3 μM SNAP-Cell TMR Star, 2 μM SNAP-Surface Alexa Fluor 647 (New England BioLabs), 3 μM SiR-SNAP (Lukinavicius et al., 2013) and 50 μM HaloTag TMR Ligand (Promega) diluted in OptiMEM (Life Technologies) with 10% FBS were used. After 30 min of staining, samples were rinsed three times and washed for at least 30 min with OptiMEM with 10% FBS.
4.4. Cryo fixation and freeze substitution
Sapphire discs with a diameter of 3 mm and thickness of 0.05 mm (Engineering Office M. Wohlwend GmbH, Sennwald, Switzerland) coated with carbon were used as support for the cells. The deposition of carbon was performed on a Cressington 208 carbon coater (Cressington Scientific Instruments) in two steps. In order to divide the working area into sub-areas identifiable by light microscope, the carbon was first evaporated over the sapphire disc covered with a finder grid. After removal of the finder grid a second layer of carbon was deposited and the coated sapphire discs were then baked at 120 °C overnight. Prior to seeding of the cells, discs were attached to the bottom of 35 mm cell culture dishes with a small drop of BD Matrigel™ (BD Biosciences, Heidelberg, Germany).
Cryo-fixation of cells grown on carbon coated sapphire discs and labeled as described above was performed by high-pressure freezing using an HPM 010 (ABRA Fluid AG) by placing the sapphire discs between two aluminum planchettes so that the cells were protected in a 50-μm cavity in one planchette. As a cryo protectant, the OptiMEM with 10% FBS was used. For freeze substitution, a temperature-controlling device (Leica EM AFS2; Leica Microsystems) was used. High pressure frozen samples were transferred in liquid nitrogen onto the substitution medium (2% (w/v) uranyl acetate, 2% water, 10% methanol, 88% acetone) precooled to −140 °C and the temperature was raised by 20 °C/h to −90 °C. For the titration uranyl acetate concentration of 0%, 0.1%, 0.4%, 2% and 4% were used. At −90 °C the substitution medium is liquid, thus the discs were separated from planchettes, kept at −90 °C for 1 h, warmed by 20 °C/h to −45 °C, transferred into glass distilled acetone and washed 3 times for 10 min each. Afterwards samples were infiltrated with increasing concentrations (10%, 25%, 50%, and 75%) of Lowicryl HM20 in acetone while warmed at 1.2 °C/h to −25 °C. Three exchanges with pure Lowicryl HM20 were done in 10 h steps. For resin polymerization the specimens were exposed to UV light at −25 °C for 48 h, warmed at 5 °C/h to 20 °C and further UV polymerized for 48 h at 20 °C.
Sections with a nominal thickness of 300 nm were cut with a microtome (Leica EM UC7) using a diamond knife (Diatome, Biel, Switzerland) and placed on formvar coated nickel (diameter 3.05 mm, parallel bars, 50 bars per inch) or copper (diameter 3.05 mm, slot 2 × 0.5 mm) TEM grids (Plano GmbH, Wetzlar, Germany). Some sections were post stained with 2% (w/v) uranyl acetate (in 70% methanol) and Reynolds lead citrate.
4.5. Fluorescence microscopy
Laser scanning fluorescence microscopy was performed on an upright confocal microscope (Carl Zeiss, LSM700) with a W Plan-Apochromat 63×/1.0 water objective or Plan-Apochromat 63×/1.4 oil objective and Immersol 518F (Carl Zeiss, Jena, Germany) using laser lines 488 nm, 555 nm and 639 nm. For confocal microscopy of resin embedded cells, the resin block was fixed onto a custom-made device that allows for translational and rotational movement and the pinhole was set to 1 airy unit. Prior to imaging of sections, TEM grids were dipped into glycerol and placed onto glass slides and covered with a cover slip (thickness 170 μm; Carl Zeiss, Jena, Germany). For imaging of sections with the LSM700, the pinhole was completely open.
Direct Stochastic Optical Reconstruction Microscopy (dSTORM) was performed on an inverted microscope (IX-71; Olympus, Hamburg, Germany or a Eclipse Ti; Nikon, Japan) using an oil-immersion objective (PlanApo 60×, NA 1.45, Olympus or a CFI Apo TIRF 100×, NA 1.49, Nikon) (Heilemann et al., 2008). Nickel TEM grids were placed into a LabTek II chamber (Nunc, Langenselbold, Germany) such that the surface of the section was on top and accessible to imaging buffer (0.1 M bicarbonate–carbonate, 100 mM, 0.5 mg/ml glucose oxidase, 40 μg/ml catalase, 10% w/v glucose, pH 7.5–8). The 647 nm and 488 nm laser beams of an argon–krypton laser (Innova 70C; Coherent, Santa Clara, USA) were selected by an acousto-optic tunable filter (AOTF) and an additional clean-up filter (Z488/568/647 RPC, AHF Analysentechnik) and were used in continuous mode for readout and activation. The laser beam was coupled into the microscope objective by a dichroic filter (HC-Dual 560/659, AHF Analysentechnik, Tuebingen, Germany). Fluorescence light was spectrally filtered with two filters (700/75 BP and 647 LP RazorEdge, AHF Analysentechnik) and detected with an EMCCD camera (Andor Ixon DV897). Typical laser powers used for dSTORM imaging were 0.5–5 mW (488 nm) and 30–80 mW (647 nm), ensuring a sufficiently low fraction of single activated fluorophores at any given time. 3000 Frames with a pixelsize of 108 nm or 5000 frames with a pixelsize of 158 nm at a frame rate of 20 Hz were recorded. Fluorescent spots identified in each image frame were analyzed with the rapidSTORM software (Wolter et al., 2012) (parameters: photon count threshold 400, reconstructed pixel size 10 nm). Experimental precision of the protein localization in the superresolution images after drift correction was estimated at 17 nm for N-Cadherin and 16 nm for H2B (Endesfelder et al., 2014). This is in good agreement to the full width at half maximum measured across the junction (Fig. 4a lower insert).
4.6. Transmission electron microscopy
15-nm Colloidal gold particles (Aurion, The Netherlands) were adsorbed on both sides of the grids (and section) to serve as fiducial markers for the alignment of the tilt-series. Grids were placed into a Model 2040 high-tilt holder (Fischione Instruments, Pittsburgh, USA) or Single Tilt Holder for Ultra Twin (#FP 6596/05, FEI, Eindhoven, The Netherlands) and imaged using a 300 kV Tecnai F30 transmission electron microscope (FEI, Eindhoven, The Netherlands). Digital micrographs and tilt-series were recorded on a US4000 CCD camera (Gatan, USA) with a 1° increment over various tilt ranges at a pixel size of 0.9–1.32 nm. For the reconstruction of tomograms, the IMOD software package (version 4.3.1; (Kremer et al., 1996)) was used. Sub-tomogram averaging was performed as previously described (Al-Amoudi et al., 2007).
4.7. Ion-abrasion scanning electron microscopy (IA-SEM)
Lowicryl embedded cell monolayers were loaded into a scanning electron microscope (Helios 600i, FEI company, Eindhoven, The Netherlands). The front surface of the block was aligned parallel to the scanning direction of the electron beam. Subsequently, the stage was brought to eucentric height and tilted to 52°. After the initial alignment, the area of interest was identified using backscattered electron (BSE) imaging with an In-lens detector. In order to reduce re-deposition and to improve imaging conditions, surplus block material around the area of interest was removed with a high ion beam current (30 keV, 2.5 nA). Afterwards a sacrificial platinum layer (500 nm) was deposited on the top surface of the sample using the gas injection system. As a final preparation step, the front surface was polished with a decreased ion current (30 keV, 2.8 nA). Serial milling and imaging of a stack of 33 × 28 × 17 μm3 was performed using the Auto Slice and View software with a pixel size of ~8 nm perpendicular to the beam direction, and ~20 nm along the beam direction (FEI company, Eindhoven, The Netherlands) with 30 keV and 0.7 nA for the milling steps and 2 keV and 1.4 nA for the imaging steps. Overlay of LSM and IA-SEM stacks and manual segmentation was performed using the Amira software package (Amira 3.0; TGS).
4.8. Automated image superposition
For the superposition of light and electron microscopic images, the auto-fluorescence, which occurs in the area of the embedded cells was exploited (Supplementary Fig. 1a). The auto-fluorescence signal provides strong and pronounced contours, in which the EM image can be matched in a template-based approach. This is comparable to template matching in electron microscopy as well as the centroid fit in super-resolution microscopy. For achieving a subpixel precise superposition of the LM image on the EM images the matching procedure had to be performed between the LM image and the lowest magnification EM image, and repeated for all EM images recorded in a magnification series of the region of interest. The FM image (99 nm pixel size) and the EM image of lowest magnification (13 nm pixel size) were low-pass filtered followed by Canny edge detection (Supplementary Fig. 1b and c) (Canny, 1986). In a brute force manner, affine transformations consisting of shift, rotations, magnifications, and stretching independently in x and y were globally applied to one image which was then compared to the other matching image, until the values with the highest cross-correlation score were found Supplementary Fig. 1). The same procedure was applied to the next pair of subsequent images in the magnification series, until the global transformation parameters between the LM image and EM image with the highest magnification was found. Afterwards the FM image could be overlaid with all the EM images by combining all transformations from the magnification series. For the implementation, custom scripts were written for Matlab (The MathWorks, Inc., Natick, USA).
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jsb.2014.03.018.
Acknowledgments
We are grateful to E.H.K. Stelzer, W. Kuehlbrandt and M. Vabulas for critical reading and discussion of the manuscript. M.H., S.M. and U.E. acknowledge funding by the BMBF (research Grant 0315262) and the CRC 902. This work was also supported by the CRC 902, ERC, and CEFII to A.S.F.
Footnotes
Contributions
A.S.F., E.S., M.H. designed the experiments. M.P., U.E., S.B., C.W., V.V.H., S.M. performed experiments. M.P., M.K., Z.Y., U.E., C.W., V.V.H., M.S., A.S. analyzed data. A.S.F., M.P., C.W., M.H., E.S., M.S., A.S. wrote the paper.
References
- Al-Amoudi A, Diez DC, Betts MJ, Frangakis AS. The molecular architecture of cadherins in native epidermal desmosomes. Nature. 2007;450:832–837. doi: 10.1038/nature05994. [DOI] [PubMed] [Google Scholar]
- Boggon TJ, Murray J, Chappuis-Flament S, Wong E, Gumbiner BM, Shapiro L. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science. 2002;296:1308–1313. doi: 10.1126/science.1071559. [DOI] [PubMed] [Google Scholar]
- Canny J. A computational approach to edge-detection. IEEE Trans Pattern Anal Mach Intell. 1986;8:679–698. [PubMed] [Google Scholar]
- Denk W, Horstmann H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2004;2:e329. doi: 10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellisman MH, Deerinck TJ, Shu X, Sosinsky GE. Picking faces out of a crowd: genetic labels for identification of proteins in correlated light and electron microscopy imaging. Methods Cell Biol. 2012;111:139–155. doi: 10.1016/B978-0-12-416026-2.00008-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endesfelder U, Malkusch S, Fricke F, Heilemann M. A simple method to estimate the average localization precision of a single-molecule localization microscopy experiment. Histochem Cell Biol. 2014 doi: 10.1007/s00418-014-1192-3. [DOI] [PubMed] [Google Scholar]
- Gaietta G, Deerinck TJ, Adams SR, Bouwer J, Tour O, Laird DW, Sosinsky GE, Tsien RY, Ellisman MH. Multicolor and electron microscopic imaging of connexin trafficking. Science. 2002;296:503–507. doi: 10.1126/science.1068793. [DOI] [PubMed] [Google Scholar]
- Grabenbauer M, Geerts WJ, Fernadez-Rodriguez J, Hoenger A, Koster AJ, Nilsson T. Correlative microscopy and electron tomography of GFP through photooxidation. Nat Methods. 2005;2:857–862. doi: 10.1038/nmeth806. [DOI] [PubMed] [Google Scholar]
- Harrison OJ, Jin X, Hong S, Bahna F, Ahlsen G, Brasch J, Wu Y, Vendome J, Felsovalyi K, Hampton CM, Troyanovsky RB, et al. The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins. Structure. 2011;19:244–256. doi: 10.1016/j.str.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawes P, Netherton CL, Mueller M, Wileman T, Monaghan P. Rapid freeze-substitution preserves membranes in high-pressure frozen tissue culture cells. J Microsc. 2007;226:182–189. doi: 10.1111/j.1365-2818.2007.01767.x. [DOI] [PubMed] [Google Scholar]
- He W, Cowin P, Stokes DL. Untangling desmosomal knots with electron tomography. Science. 2003;302:109–113. doi: 10.1126/science.1086957. [DOI] [PubMed] [Google Scholar]
- Heilemann M, van de Linde S, Schuttpelz M, Kasper R, Seefeldt B, Mukherjee A, Tinnefeld P, Sauer M. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew Chem Int Ed Engl. 2008;47:6172–6176. doi: 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]
- Heymann JA, Hayles M, Gestmann I, Giannuzzi LA, Lich B, Subramaniam S. Site-specific 3D imaging of cells and tissues with a dual beam microscope. J Struct Biol. 2006;155:63–73. doi: 10.1016/j.jsb.2006.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- Kim SA, Tai CY, Mok LP, Mosser EA, Schuman EM. Calcium-dependent dynamics of cadherin interactions at cell–cell junctions. Proc Natl Acad Sci USA. 2011;108:9857–9862. doi: 10.1073/pnas.1019003108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott G, Marchman H, Wall D, Lich B. Serial section scanning electron microscopy of adult brain tissue using focused ion beam milling. J Neurosci. 2008;28:2959–2964. doi: 10.1523/JNEUROSCI.3189-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Kukulski W, Schorb M, Welsch S, Picco A, Kaksonen M, Briggs JA. Correlated fluorescence and 3D electron microscopy with high sensitivity and spatial precision. J Cell Biol. 2011;192:111–119. doi: 10.1083/jcb.201009037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, Simpson D, et al. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- Lukinavicius G, Umezawa K, Olivier N, Honigmann A, Yang G, Plass T, Mueller V, Reymond L, Correa IR, Jr, Luo ZG, Schultz C, et al. A near-infrared fluorophore for live-cell super-resolution microscopy of cellular proteins. Nat Chem. 2013;5:132–139. doi: 10.1038/nchem.1546. [DOI] [PubMed] [Google Scholar]
- Muller-Reichert T, Srayko M, Hyman A, O’Toole ET, McDonald K. Correlative light and electron microscopy of early Caenorhabditis elegans embryos in mitosis. Methods Cell Biol. 2007;79:101–119. doi: 10.1016/S0091-679X(06)79004-5. [DOI] [PubMed] [Google Scholar]
- Murphy GE, Narayan K, Lowekamp BC, Hartnell LM, Heymann JA, Fu J, Subramaniam S. Correlative 3D imaging of whole mammalian cells with light and electron microscopy. J Struct Biol. 2011;176:268–278. doi: 10.1016/j.jsb.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanguneri S, Flottmann B, Horstmann H, Heilemann M, Kuner T. Three-dimensional, tomographic super-resolution fluorescence imaging of serially sectioned thick samples. PLoS One. 2012;7:e38098. doi: 10.1371/journal.pone.0038098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan K, Danielson CM, Lagarec K, Lowekamp BC, Coffman P, Laquerre A, Phaneuf MW, Hope TJ, Subramaniam S. Multi-resolution correlative focused ion beam scanning electron microscopy: applications to cell biology. J Struct Biol. 2013;185(3):278–284. doi: 10.1016/j.jsb.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori A, Gatz R, Beck F, Rigort A, Baumeister W, Plitzko JM. Correlative microscopy: bridging the gap between fluorescence light microscopy and cryo-electron tomography. J Struct Biol. 2007;160:135–145. doi: 10.1016/j.jsb.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Schwartz CL, Sarbash VI, Ataullakhanov FI, McIntosh JR, Nicastro D. Cryo-fluorescence microscopy facilitates correlations between light and cryo-electron microscopy and reduces the rate of photobleaching. J Microsc. 2007;227:98–109. doi: 10.1111/j.1365-2818.2007.01794.x. [DOI] [PubMed] [Google Scholar]
- Shu X, Lev-Ram V, Deerinck TJ, Qi Y, Ramko EB, Davidson MW, Jin Y, Ellisman MH, Tsien RY. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 2011;9:e1001041. doi: 10.1371/journal.pbio.1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Driel LF, Valentijn JA, Valentijn KM, Koning RI, Koster AJ. Tools for correlative cryo-fluorescence microscopy and cryo-electron tomography applied to whole mitochondria in human endothelial cells. Eur J Cell Biol. 2009;88:669–684. doi: 10.1016/j.ejcb.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Verkade P. Moving EM: the rapid transfer system as a new tool for correlative light and electron microscopy and high throughput for high-pressure freezing. J Microsc. 2008;230:317–328. doi: 10.1111/j.1365-2818.2008.01989.x. [DOI] [PubMed] [Google Scholar]
- Wacker I, Schroeder RR. Array tomography. J Microsc. 2013;252:93–99. doi: 10.1111/jmi.12087. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Punge A, Hollopeter G, Willig KI, Hobson RJ, Davis MW, Hell SW, Jorgensen EM. Protein localization in electron micrographs using fluorescence nanoscopy. Nat Methods. 2011;8:80–84. doi: 10.1038/nmeth.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes S, Staufenbiel M, Lisse D, Richter CP, Beutel O, Busch KB, Hess ST, Piehler J. Triple-color super-resolution imaging of live cells: resolving submicroscopic receptor organization in the plasma membrane. Angew Chem Int Ed Engl. 2012;51:4868–4871. doi: 10.1002/anie.201200853. [DOI] [PubMed] [Google Scholar]
- Wolter S, Loschberger A, Holm T, Aufmkolk S, Dabauvalle MC, van de Linde S, Sauer M. RapidSTORM: accurate, fast open-source software for localization microscopy. Nat Methods. 2012;9:1040–1041. doi: 10.1038/nmeth.2224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.