Abstract
The non-canonical amino acid labeling techniques BONCAT (bioorthogonal non-canonical amino acid tagging) and FUNCAT (fluorescent non-canonical amino acid tagging) enable the specific identification and visualization of newly synthesized proteins. Recently, these techniques have been applied to neuronal systems to elucidate protein synthesis dynamics during plasticity, identify stimulation-induced proteomes and subproteomes and to investigate local protein synthesis in specific subcellular compartments. The next generation of tools and applications, reviewed here, includes the development of new tags, the quantitative identification of newly synthesized proteins, the application of NCAT to whole animals, and the ability to genetically restrict NCAT labeling. These techniques will enable not only improved detection but also allow new scientific questions to be tackled.
Introduction
Neuronal plasticity, the ability to change on the molecular, cellular and/or the systems level in response to chemical, electrophysiological and/or behavioral stimuli, is a key characteristic of the nervous system and the basis for learning and memory. During many forms of plasticity the neuronal proteome is regulated by protein synthesis and degradation [1]. Insight into protein synthesis dynamics, the subcellular localization of newly synthesized proteins, as well as the identity of these proteins is therefore crucial to understanding neuronal plasticity.
In all fields of biology the techniques to visualize the levels or location of newly synthesized proteins have been predominantly limited to genetic labeling of specific candidate proteins with fluorescent protein tags such as GFP [2]. Although genetically encoded fluorescent protein tags have had a tremendous impact on the fields of cellular and molecular biology, these tags are usually large and require genetic manipulation of the protein in question, both of which may affect the in vivo function and localization of the labeled protein. As only a handful of predetermined candidate proteins can be tracked at the same time, global protein synthesis dynamics are missed entirely. In turn, advances in mass spectrometry (MS) based approaches such as ‘Stable Isotope Labeling with Amino acids in Cell culture’ (SILAC) now permit the identification and comparative quantification of proteomes of differentially stimulated cell populations [3]. However, the complexity of the neuronal proteome may hinder identification of proteins of low abundance without preliminary steps to enrich newly synthesized proteins specifically. To visualize global protein synthesis dynamics in an unbiased way, as well as enable identification of newly synthesized proteins of low abundance, new experimental approaches were needed.
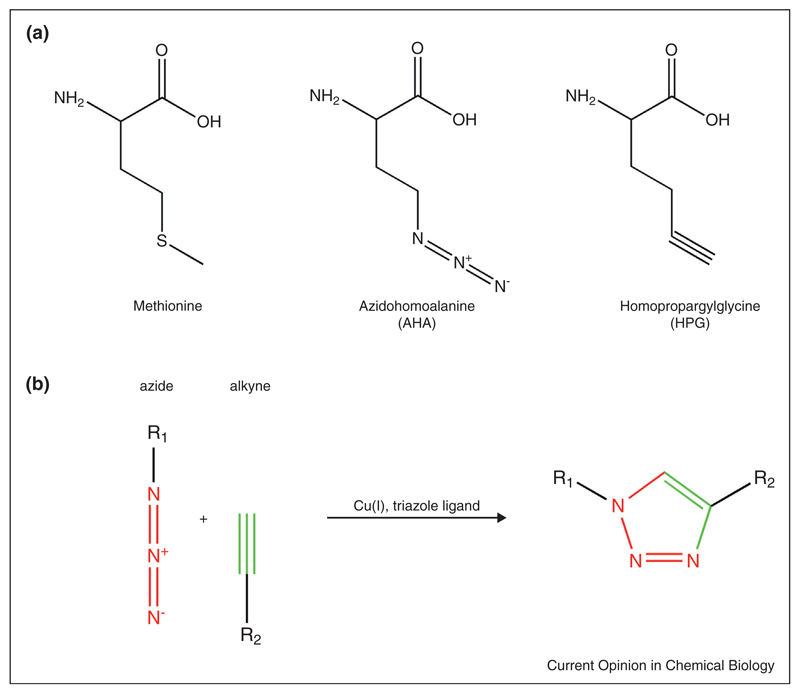
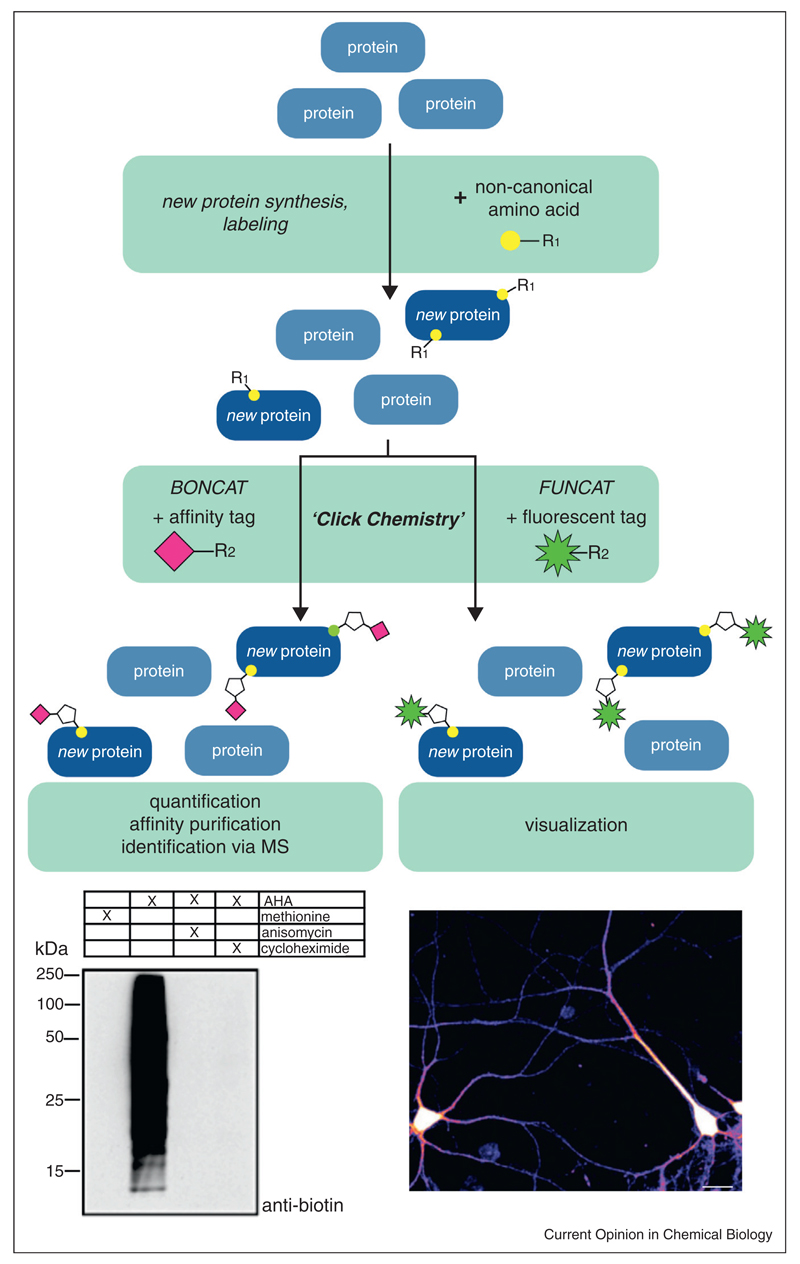
Over the last decade, Tirrell and coworkers established the use of the azide-bearing non-canonical amino acid azidohomoalanine (AHA) and the alkyne-bearing noncanonical amino acid homopropargylglycine (HPG) as surrogates for methionine in bacterial cells (Figure 1a) [4–6]. Azides and alkynes can be covalently linked via selective Cu(I)-catalyzed [3+2] azide-alkyne cycloaddition (termed ‘click chemistry’) (Figure 1b) [7,8] under biological conditions, making them ideal candidates to label proteins. Using this approach, Dieterich et al. developed the sister techniques bioorthogonal non-canonical amino acid tagging (BONCAT), and fluorescent noncanonical amino acid tagging (FUNCAT) [9–12]. During BONCAT, proteins labeled with non-canonical amino acids (bearing either azide or alkyne moieties) are tagged using affinity tags (bearing the respective alkyne or azide functional groups) to enable new protein purification, while FUNCAT utilizes fluorescent tags to enable visualization of newly synthesized proteins in mammalian cells (Figure 2). Affinity-tagged proteins can be quantified using immunoblot analysis or separated from the preexisting proteome by affinity purification and identified by tandem-MS. BONCAT has already been successfully applied to study the proteome of HEK293 cells during a two-hour time window, allowing the identification of 195 newly synthesized proteins [9]. Fluorescent tags can be used to visualize newly synthesized proteins, including those proteins of interest whose identities may not be known. In this manner, FUNCAT has been used to investigate temporally defined protein populations in Rat-1 fibroblasts [13,14] and local protein synthesis in dissociated hippocampal neurons and organotypic hippocampal slices [11].
Figure 1.
Chemical structures and ‘click chemistry’ reaction scheme. (a) Chemical structures of methionine, azidohomoalanine (AHA) and homopropargylglycine (HPG). All amino acids are L-isomers. (b) Scheme of Cu(I)-catalyzed of [3+2] azide-alkyne cycloaddition.
Figure 2.
BONCAT and FUNCAT scheme. Actively translating cells are incubated with an azide or alkyne (R1) bearing non-canonical amino acid. Newly synthesized proteins incorporate the non-canonical amino acid and can be tagged via a ‘click’ chemistry reaction with either an affinity or a fluorescent tag bearing the respective alkyne or azide moiety (R2). Newly synthesized, affinity tagged proteins can be quantified on immunoblots, affinity purified and subsequently identified via tandem-MS. Fluorescently labeled proteins, instead, enable visualization of protein synthesis dynamics in vivo and determination of subcellular regions of protein synthesis. Sample immunoblot of HEK293 cells incubated with non-canonical amino acid and ‘clicked’ to a biotin affinity tag [taken from [9]], showing that labeling is protein synthesis-dependent. Sample fluorescence image of dissociated hippocampal neurons labeled with non-canonical amino acid and ‘clicked’ to fluorescent tag [taken from [11]], showing that regions of protein synthesis can be visualized in vivo. Scale bar, 20 μm.
BONCAT and FUNCAT were developed as complementary approaches to fluorescent protein tagging and SILAC-based proteome identification. Azides and alkynes are small chemical groups, making AHA and HPG ideal metabolic tags, unlikely to cause significant perturbations of protein folding, localization [9] and therefore function of the labeled proteins in vivo. As azides and alkynes are essentially absent from biological systems, the ‘click chemistry’ tagging reaction is highly sensitive with low background levels despite the complex cellular environment. Furthermore, as AHA and HPG can act as surrogates for naturally occurring amino acids and be charged onto wild-type methionyl-tRNAs by endogenous methionyl-tRNA synthetases (MetRS), they can be metabolically incorporated into newly synthesized proteins by the cell’s own synthesis machinery without a requirement for genetic manipulation. As a result, BONCAT and FUNCAT have enabled non-candidate based investigation of proteome dynamics in both neuronal and non-neuronal cells.
In this review we will first discuss how the BONCAT and FUNCAT techniques have been applied to neurons and then explore a number of exciting new technical extensions of the non-canonical amino acid tagging approaches and how these new strategies can be used to investigate proteome dynamics in the future. This review will not discuss bio-orthogonal labeling of other biomolecules such as sugars, lipids, RNA and DNA [see [15] for a comprehensive review], nor will we discuss recent advances in site-specific labeling with non-canonical amino acids [16].
BONCAT and FUNCAT applied to neuronal systems
Investigating changes in global protein synthesis during plasticity
A number of studies have recently used non-canonical amino acid labeling to visualize changes in global protein synthesis in neurons after different stimulation protocols. Roche and co-workers used AHA labeling and a subsequent ‘click’ reaction to a fluorescent alkyne to confirm previous findings that axon guidance cues increase protein synthesis in neurons [17]. They also demonstrated that stimulation of dorsal root ganglion (DRG) neurons with nerve growth factor (NGF) or semaphorin3A (Sema3A) increased the amount of nascent protein in both cell bodies and axons. By contrast, Baez et al. show that stimulation of dissociated hippocampal cultures with the glutamate receptor agonist NMDA caused a decrease in protein synthesis in dendrites [18], replicating previous findings [19]. In a more sophisticated approach, these researchers pulse labeled cells with HPG and AHA sequentially to evaluate both baseline protein synthesis and protein synthesis after stimulation in individual dendrites. Furthermore, the cytokine interleukin 6 (IL-6), as well as NGF, both linked to nociceptive plasticity, were shown to cause ERK and mTOR-dependent increases in translation [20] in DRG neurons, while peripheral nerve injury led to increases in protein synthesis in the sciatic nerve [21].
Identifying stimulation induced proteins and subproteomes
Identifying the relatively small number of proteins translated in response to a specific pharmacological stimulation can be extremely difficult using established stable isotope labeling techniques, as there is no means to enrich for the labeled subproteome. In addition, proteins of low abundance, though potentially significant, may be overlooked against the backdrop of baseline cellular protein complexity. However, Hodas and colleagues recently demonstrated that BONCAT can be applied to neuronal systems to identify the dopaminergic subproteome in rat hippocampal neuropil [22•]. To do so, acute hippocampal slices were incubated with AHA for 2.5 h, either with or without a D1/D5 dopamine receptor agonist. Subsequently, the slices were microdissected, and homogenized, then newly synthesized proteins were ligated to a biotin alkyne tag, affinity purified and identified using tandem-MS. 891 unique proteins, involved in a number of different biological processes and molecular functions, were identified across all samples. 616 proteins were common to both the vehicle and D1/D5-agonist-treated samples, while 100 proteins were unique to the agonist-treated sample. Many of the candidate proteins identified in the agonist-treated sample belonged to gene ontology (GO) categories specific for protein synthesis and synaptic function, such as the presynaptic protein Munc13-1, the voltage-gated potassium channel subunit beta-1 (KCNAB1), as well as ribosomal proteins (e.g. 40S ribosomal protein S25 (RPS25) and 60S ribosomal protein L13a (RPL13A)). These data indicate that BONCAT can be applied to neuronal systems to identify stimulation-specific subproteomes. Moreover, Yoon and coworkers used AHA labeling to identify Engrailed-1 (En-1) as the axonal guidance cue that causes the greatest increase in protein synthesis in Xenopus retinal ganglion cell (RGC) axons [23•]. To identify which proteins are differentially synthesized upon stimulation with En-1, the researchers then combined FUNCAT with 2D difference gel electrophoresis (2D-DIGE). This allowed them to identify Lamin B2 (LB2), an intermediate filament protein normally associated with the nuclear membrane, as the most differentially translated protein in RGC axons stimulated with En-1. Interestingly, axonally translated LB2 plays a crucial role in axonal mitochondria function and, thus, axon maintenance.
Visualizing local protein synthesis
Although local synthesis of a number of candidate proteins has been described in neurons [24], FUNCAT can be applied to study global protein synthesis in specific subcellular compartments, such as dendrites and axonal growth cones. In the first demonstration of FUNCAT in neurons, Dieterich et al. showed that while perfusing the cell body of a dissociated hippocampal neuron with the protein synthesis inhibitor anisomycin, AHA was incorporated into proteins in the dendrite [11]. When anisomycin was bath-applied to the culture, perfusion of AHA at the dendrite led to detectable de novo protein synthesis. By using non-canonical amino acids to label newly synthesized proteins in specific subcellular compartments, these findings provide further evidence for local protein synthesis in the dendrite. Furthermore, Tcherkenzian and colleagues showed with FUNCAT that DCC, a transmembrane receptor that is stimulated by netrin, colocalized with sites of new protein synthesis in the filopodia of commissural neurons and hippocampal neurons [25•]. As DCC was also shown to physically associate with translation machinery, the authors hypothesized that DCC is involved in regulation of local translation at axon growth cones. Without the ability to metabolically incorporate a chemical handle into newly synthesized proteins via non-canonical amino acid tagging, these studies would be limited to candidate-based approaches. However, it is important to note that localized non-canonical amino acid labeling is limited to specific cellular subcompartments that contain the necessary amino acid transporters as well as the protein synthesis machinery. Furthermore, as non-canonical amino acids readily diffuse between cellular compartments, application of protein synthesis inhibitors to areas outside the cellular subcompartment of interest [11] may be necessary.
Visualizing global protein synthesis to investigate protein turnover in synapses
Chemical synapses are composed of proteins with finite lifetimes. Hence, for synapses to persist and maintain their individual characteristics, they need to be continuously and precisely replenished with newly synthesized proteins. The turnover dynamics of synaptic proteins has thus far only been studied for a limited number of synaptic proteins [26]. In a very recent study Cohen et al. [27] combined SILAC, MS, quantitative immunohistochemistry and bioinformatics with FUNCAT to systematically measure the metabolic half-lives of hundreds of synaptic proteins. With FUNCAT the authors were able to directly measure that the fluorescence of pulse-labeled AHA-bearing proteins in synapses was reduced to 70% and 55% after 24 and 48 h respectively, as compared to samples fixed directly after 24 h pulse labeling. This, combined with SILAC and MS-determined turnover rates, leads to the surprising conclusion that all synaptic proteins investigated in this study showed similar half-lives in the range of 2–5 days regardless of their presynaptic or postsynaptic localization or the presence of a dendritically localized mRNA. For some structurally and functionally related proteins very similar turnover rates suggest a tightly coupled synthesis and degradation regime.
New developments
Although there has been a dramatic increase in number of studies that use non-canonical amino acid tagging techniques to study neuronal plasticity, recent innovations may enable new applications and increases in sensitivity that could fundamentally change the way we investigate neuronal plasticity. We describe below a few of the most exciting recent developments.
BONCAT and FUNCAT in vivo
Although some forms of neuronal plasticity can be studied in cell culture, molecular correlates of memory and learning, areas of key interest in neuroscience, are best studied in vivo. Therefore, developing BONCAT and FUNCAT for use in an intact organism in which simple forms of learning may be investigated is an essential next step. Very recently, we described the application of these techniques to the 7-day-old larval zebrafish [28••]. BONCAT and FUNCAT can be easily applied to the larval zebrafish as non-canonical amino acids introduced in the swim water simply diffuse into the larvae. We showed that AHA is metabolically incorporated into newly synthesized proteins, in a time-dependent and concentration-dependent manner, but has no apparent toxic cellular or behavioral effects. This enables fluorescent labeling of newly synthesized proteins in whole-mount larval zebrafish. In addition, the regulation of protein synthesis can also be observed: stimulation of the whole larva with the GABA anatagonist PTZ during AHA exposure causes an increase in protein synthesis throughout the nervous system [29]. This experiment paves the way for future studies examining changes in protein synthesis levels following learning.
Several groups are currently working on adapting BONCAT and FUNCAT to a wider range of model organisms including Caenorhabditis elegans, fruit fly and mouse (unpublished data). Although incubation periods necessary for significant labeling in deep structures and tissues of the whole organism are currently still relatively long (~24 h), in the future one may be able to pair noncanonical amino acid tagging with simple associative learning paradigms to reveal the neuronal circuits underlying memory formation, as well as to identify the proteins that are newly synthesized during learning.
QuaNCAT – quantifying affinity purified proteins
SILAC labeling has allowed for the quantitative analysis of proteomic changes [3], however proteins newly synthesized and labeled at low levels may be missed when a complex proteome is analyzed via MS. An enrichment step subsequent to labeling, but before identification would therefore be highly beneficial. In three recent papers, SILAC and BONCAT techniques were combined to affinity purify and then quantitatively identify proteins that were differentially synthesized in the absence of a translational regulator [29], during a period of pharmacological stimulation [30••], or following secretion [31]. Most notably, using a combination of SILAC and BONCAT Howden et al. documented a high dynamic range of 1.6–589-fold changes in expression at 2 h and/or 4 h after stimulation of CD4-positive primary T cells [30••]. Furthermore, Howden et al. also discovered ‘new’ proteins that were not annotated or previously detected during T cell activation.
New affinity and fluorescent tags
Cleavable Biotin Probes — Currently, following labeling with a non-canonical amino acid, affinity resin bound proteins are usually released by boiling the affinity resin in denaturing buffer or by enzymatic cleavage of bound peptides. However, these elution protocols may lead to contamination by nonspecifically bound proteins, endogenously biotinylated proteins, or resin-based peptides released. To eliminate these types of contamination, Szychowski et al. designed a set of cleavable biotin probes for the efficient and easy cleavage of peptides from the affinity purification matrix [32••]. One of the most promising tags contains a dialkoxydiphenylsilane (DADPS) linker and can be cleaved efficiently when treated with 10% formic acid for only 30 min, thereby leading to higher accuracy and sensitivity in peptide identification.
Qdot and Pdot fluorescent ‘click chemistry’ tags — The most widely available and commonly used fluorescent ‘click chemistry’ tags contain Alexa fluorophores. These fluorophores are bright, insensitive to pH over a broad range and soluble in water — making them ideal for a wide range of applications. However, conventional fluorophores are not bright enough to enable single-particle tracking and visualized puncta will often contain tens to hundreds of labeled molecules. Instead, Dieterich et al. made use of quantum-dot (Qdot) conjugates, which show improved brightness and photostability over traditional fluorescent dyes, to live-image diffusion properties of single particles in neurons using FUNCAT [11]. The intrinsic toxicity of Qdots, caused by the potential leaching of heavy metal ions, may however have deleterious effects on the cells probed in long-term imaging experiments. Instead, Wu et al. developed ultrabright fluorescent ‘click chemistry’ tags using semiconducting polymer dots (Pdots), which are not cytotoxic [33•]. These Pdots are brighter than Qdots, can have up to a thousand-fold faster emission rates and do not blink, widening the door for single-particle tracking in neuronal systems. Unfortunately, because of their size Pdots are not cell membrane permeable, preventing tracking of molecules within the cytoplasm, and require potentially phototoxic UV illumination.
Live ‘Click Chemistry’ labeling — Live cell imaging of newly synthesized cytoplasmic proteins using FUNCAT has been hindered by the toxicity of the copper catalyst [34] necessary for traditional [3+2] azide-alkyne cycloaddition, as well as the size of the fluorophore conjugate. To overcome the toxicity of the copper catalyst, Soriano del Amo et al. developed the tris(triazolylmethyl)amine-based ligand BTTES, which in combination with 75 μM CuSO4, was able to facilitate the ‘click’ reaction between cell-surface sialylated glycans bearing azide or alkyne groups and their respective biotin-tags in a variety of cell lines within minutes [35]. No cell death was observed during a 17-minute reaction in CHO cells, or 24 h after a 3-minute reaction in Jurkat or HEK293T cells. Alternatively, the Bertozzi group has developed a strain-promoted [3+2] cycloaddition between cyclooctynes and azides that proceeds under physiological conditions without the need for a catalyst and is therefore not toxic in vivo [36,37,11]. Unfortunately, these fluorescent difluorinated cyclooctyne (DIFO) tags are difficult to synthesize and neither the DIFO-tags nor the standard Alexa Fluor-conjugated or biotin-conjugated azide-tags or alkyne-tags that have been tested in combination with the BTTES ligand are cell membrane-permeable. However, progress towards live labeling of cytoplasmic proteins is being made. Beatty and coworkers are currently developing a set of cyclooctyne tags coupled to small, cell membrane-permeable fluorophores such as coumarin and BODIPY [38•,39•]. A coumarin-based cyclooctyne conjugate 2 [38•] is very promising. Using this tag, Beatty and co-workers demonstrated an up to 30-fold fluorescence enhancement of AHA-labeled Rat-1 fibroblasts compared to methionine control, after a 4-hour AHA labeling and 10-minute tagging reaction [38•]. Unfortunately, the coumarin fluorophore requires excitation in the UV range (360 nm), which is both damaging to cells and has poor tissue penetration, and is therefore less common in standard biological imaging systems. By contrast, BODIPY has an excitation/emission spectrum similar to that of GFP, but the BODIPY-cyclooctyne appears to be less membrane-permeable than the coumarin-cyclooctyne. Although imaging experiments show bright labeling and little background signal in Rat-1 fibroblasts after 4-hour treatment with AHA followed by a 10 min ‘click’ reaction, cell fractionation experiments show that most of the labeled proteins are detected in cell membranes [39•]. The further development of cell membrane-permeable tags will permit live imaging of newly synthesized proteins in neurons and possibly whole organisms, thereby opening new avenues for investigating dynamic metabolic responses to chemical and perhaps even behavioral stimuli in complex systems.
Genetically restricted metabolic labeling
BONCAT and FUNCAT have exploited the somewhat promiscuous nature of MetRS that enables the charging of the structurally similar methionine analogs AHA and HPG to a methionyl-tRNA resulting in the incorporation of these non-canonical amino acids into newly synthesized proteins in wild-type cells. However, the Tirrell group has recently shown that altering the specificity of Escherichia coli MetRS by introducing specific mutations into its binding-pocket region enables the metabolic incorporation of otherwise inert non-canonical amino acids, such as the long-chain azide-bearing azidonorleucine (ANL) [40,41]. This opens the door for genetically restricted cell-specific metabolic labeling of proteins.
Building on previous work, Ngo et al. showed that both E. coli and mammalian cells expressing a mutant MetRS are able to utilize ANL as a surrogate for methionine in protein synthesis, while wild-type cells are inert to ANL and proteins made in these cells are not labeled [42,43•]. In co-culture experiments, labeling of newly synthesized proteins with affinity reagents or fluorescent dyes is restricted to cells expressing the mutant MetRS, therefore enabling cell-specific enrichment, identification and visualization, even in mixtures of different cell types. This approach was extended to show that when the mutant MetRS expression is driven by state-selective promoters proteins synthesized in predetermined physiological states, such as oxidative stress, can be identified [44••].
When applied to multicellular organisms, cell-type-specific metabolic labeling allows for the visualization and identification of proteomes in genetically defined cells and circuits. The ensuing sparse FUNCAT labeling will improve the detection of protein synthesis differences between cells within the labeled group as well as the visualization of labeled cell morphology. Subproteomes of specific cell types can thus be enriched and identified. Furthermore, genetically restricted metabolic labeling allows for the investigation of subproteomes translated under specific physiological or behavioral states, such as starvation or learning, respectively.
Conclusions
BONCAT and FUNCAT have been widely adopted in neuroscience to study the protein synthesis dynamics underlying neuronal plasticity in a variety of different cell types and organisms. New technical developments, such as quantitative identification of newly synthesized proteins, translation of these techniques into whole organisms, development of new tags and genetically restricted labeling will enable not only improved detection but also allow completely new scientific questions to be tackled. Researchers may soon be able to monitor live global protein synthesis in specific subcellular compartments in response to stimulation or quantitatively identify changes in protein synthesis underlying specific physiological and pathophysiological states. But most interestingly, application of genetically restricted metabolic labeling in whole organisms may soon lead to the elucidation of which proteins and neuronal circuits are involved in different forms of behavioral adaption — one of the great, unsolved questions of neuroscience.
Acknowledgements
Flora Hinz acknowledges support from NIH/NRSA Institutional training grant 5T32 GM07616 and the Max Planck Society. Daniela Dieterich receives funding from the DFG Emmy Noether Program, DFG SFB 772, DFG GRK 1167, DFG/BMBF DIP and the Center for Behavioural Brain Science (CBBS), Magdeburg. Erin Schuman receives funding from the Max Planck Society, European Research Council (Advanced Investigator Award), DFG CRC 902, DFG CRC 1080, and the Cluster of Excellence for Macromolecular Complexes, Goethe University. The authors thank Dr. Anke Müller, Magdeburg, for providing Figure 3a–d and Ina Bartnik, Lisa Kochen and Belquis Nassim-Assir, MPI Brain Research, for providing Figure 3e–h.
Figure 3.
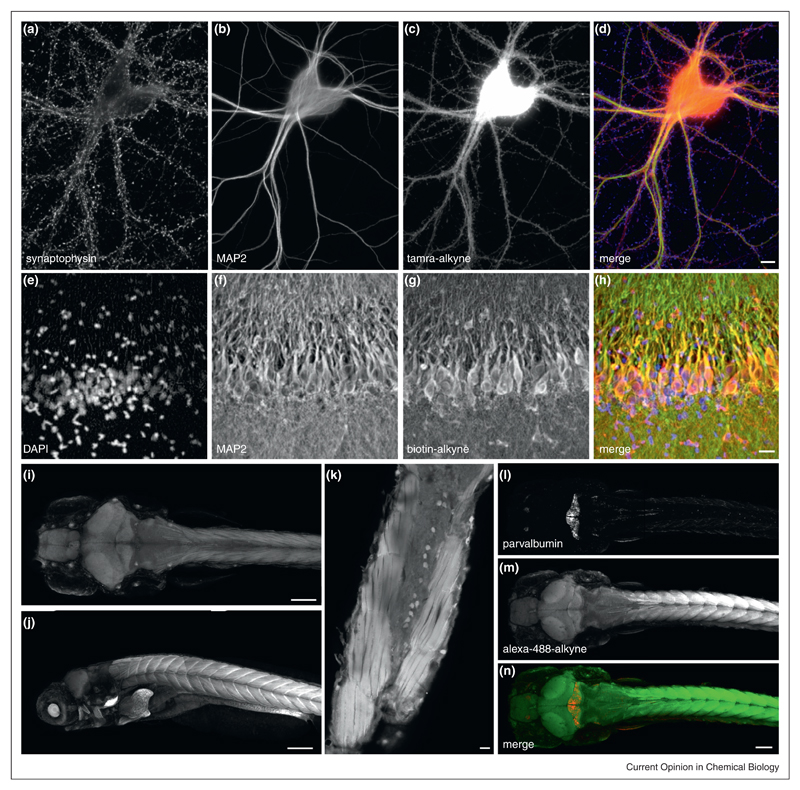
FUNCAT labeling in dissociated neurons, organotypic hippocampal slices and whole mount zebrafish larvae. (a)–(d) Antibody and FUNCAT labeling of dissociated hippocampal neurons. Scale bar = 10 μm. (e)–(h) Antibody and FUNCAT labeling of organotypic hippocampal slice. FUNCAT labeling was achieved by incubating slice with AHA, ‘clicking’ to biotin alkyne and subsequent immunostaining with mouse anti-biotin primary antibody and Alexa Fluor-594 anti-mouse secondary antibody. Scale bar = 20 μM. (i)–(k) FUNCAT labeling of whole-mount 7-day-old larval zebrafish. (i) dorsal view; (j) lateral view. Scale bar = 150 μm. (k) Dorsal view of cross-section of tail, showing muscles and spinal cord. Scale bar = 20 μm. (l)–(n) Antibody and FUNCAT labeling of whole-mount 7-day-old larval zebrafish. Scale bar = 100 μm.
Footnotes
This review comes from a themed issue on In vivo chemistry
Edited by Carolyn Bertozzi and Peng Wu
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Davis HP, Squire LR. Protein synthesis and memory: a review. Psychol Bull. 1984;96:518–559. [PubMed] [Google Scholar]
- 2.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 3.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;5:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 4.Kiick KL, Saxon E, Tirrell DA, Bertozzi CR. Incorporation of azides into recombinant proteins for chemoselective modification by the Staudinger ligation. Proc Natl Acad Sci U S A. 2002;1:19–24. doi: 10.1073/pnas.012583299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Link AJ, Vink MK, Tirrell DA. Presentation and detection of azide functionality in bacterial cell surface proteins. J Am Chem Soc. 2004;126:10598–10602. doi: 10.1021/ja047629c. [DOI] [PubMed] [Google Scholar]
- 6.Beatty KE, Xie F, Wang Q, Tirrell DA. Selective dye-labeling of newly synthesized proteins in bacterial cells. J Am Chem Soc. 2005;127:14150–14151. doi: 10.1021/ja054643w. [DOI] [PubMed] [Google Scholar]
- 7.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem. 2002;14:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 8.Tornøe CW, Christensen C, Meldal M. Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(i)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J Org Chem. 2002;9:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 9.Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT) Proc Natl Acad Sci U S A. 2006;103:9482–9487. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dieterich DC, Lee JJ, Link AJ, Graumann J, Tirrell DA, Schuman EM. Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat Protoc. 2007;2:532–540. doi: 10.1038/nprot.2007.52. [DOI] [PubMed] [Google Scholar]
- 11.Dieterich DC, Hodas JJ, Gouzer G, Shadrin IY, Ngo JT, Triller A, Tirrell DA, Schuman EM. In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat Neurosci. 2010;7:897–905. doi: 10.1038/nn.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tom Dieck S, Müller A, Nehring A, Hinz FI, Bartnik I, Schuman EM, Dieterich DC. Metabolic labeling with noncanonical amino acids and visualization by chemoselective fluorescent tagging. Curr Protoc Cell Biol. 2012;7 doi: 10.1002/0471143030.cb0711s56. Unit 7.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beatty KE, Liu JC, Xie F, Dieterich DC, Schuman EM, Wang Q, Tirrell DA. Fluorescence visualization of newly synthesized proteins in mammalian cells. Angew Chem Int Ed Engl. 2006;45:7364–7367. doi: 10.1002/anie.200602114. [DOI] [PubMed] [Google Scholar]
- 14.Beatty KE, Tirrell DA. Two-color labeling of temporally defined protein populations in mammalian cells. Bioorg Med Chem Lett. 2008;18:5995–5999. doi: 10.1016/j.bmcl.2008.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Best MD. Click chemistry and bioorthogonal reactions: unprecedented selectivity in the labeling of biological molecules. Biochemistry. 2009;28:6571–6584. doi: 10.1021/bi9007726. [DOI] [PubMed] [Google Scholar]
- 16.Ngo JT, Tirrell DA. Noncanonical amino acids in the interrogation of cellular protein synthesis. Acc Chem Res. 2011;9:677–685. doi: 10.1021/ar200144y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roche FK, Marsick BM, Letourneau PC. Protein synthesis in distal axons is not required for growth cone responses to guidance cues. J Neurosci. 2009;3:638–652. doi: 10.1523/JNEUROSCI.3845-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baez MV, Luchelli L, Maschi D, Habif M, Pascual M, Thomas MG, Boccaccio GL. Smaug1 mRNA-silencing foci respond to NMDA and modulate synapse formation. J Cell Biol. 2011;7:1141–1157. doi: 10.1083/jcb.201108159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marin P, Nastiuk KL, Daniel N, Girault JA, Czernik AJ, Glowinski J, Nairn AC, Premont J. Glutamate-dependent phosphorylation of elongation factor-2 and inhibition of protein synthesis in neurons. J Neurosci. 1997;17:3445–3454. doi: 10.1523/JNEUROSCI.17-10-03445.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melemedjian OK, Asiedu MN, Tillu DV, Peebles KA, Yan J, Ertz N, Dussor GO, Price TJ. IL-6- and NGF-induced rapid control of protein synthesis and nociceptive plasticity via convergent signaling to the eIF4F complex. J Neurosci. 2010;45:15113–15123. doi: 10.1523/JNEUROSCI.3947-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melemedjian OK, Asiedu MN, Tillu DV, Sanoja R, Yan J, Lark A, Khoutorsky A, Johnson J, Peebles KA, Lepow T, et al. Targeting adenosine monophosphate-activated protein kinase (AMPK) in preclinical models reveals a potential mechanism for the treatment of neuropathic pain. Mol Pain. 2011;7:70. doi: 10.1186/1744-8069-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hodas JJ, Nehring A, Höche N, Sweredoski MJ, Pielot R, Hess S, Tirrell DA, Dieterich DC, Schuman EM. Dopaminergic modulation of the hippocampal neuropil proteome identified by bioorthogonal noncanonical amino acid tagging (BONCAT) Proteomics. 2012;15–16:2464–2476. doi: 10.1002/pmic.201200112. [• The first study to apply BONCAT to acute hippocampal slices to identify a stimulation specific subproteome.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoon BC, Jung H, Dwivedy A, O’Hare CM, Zivraj KH, Holt CE. Local translation of extranuclear lamin B promotes axon maintenance. Cell. 2012;4:752–764. doi: 10.1016/j.cell.2011.11.064. [• This study pairs FUNCAT with 2D-DIGE to identify the most differentially translated protein in axons after stimulation with an axon guidance cue.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30:489–502. doi: 10.1016/s0896-6273(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 25.Tcherkezian J, Brittis PA, Thomas F, Roux PP, Flanagan JG. Transmembrane receptor DCC associates with protein synthesis machinery and regulates translation. Cell. 2010;4:632–644. doi: 10.1016/j.cell.2010.04.008. [• In this study researchers use FUNCAT to demonstrate that a specific transmembrane receptor co-localized with sites of protein synthesis in filopodia of commissural neurons and hippocampal neurons, thereby implicating that it is involved in regulation of local translation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ehlers MD. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat Neurosci. 2003;3:231–242. doi: 10.1038/nn1013. [DOI] [PubMed] [Google Scholar]
- 27.Cohen LD, Zuchman R, Sorokina O, Müller A, Dieterich DC. Metabolic turnover of synaptic proteins: kinetics, interdependencies and implications for synaptic maintenance. PLoS ONE. 2013;5:e63191. doi: 10.1371/journal.pone.0063191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinz FI, Dieterich DC, Tirrell DA, Schuman EM. Non-canonical amino acid labeling in vivo to visualize and affinity purify newly synthesized proteins in larval zebrafish. ACS Chem Neurosci. 2012;3:40–49. doi: 10.1021/cn2000876. [•• The first study to apply BONCAT and FUNCAT techniques to an intact organism, the larval zebrafish.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Somasekharan SP, Stoynov N, Rotblat B, Leprivier G, Galpin JD, Ahern CA, Foster LJ, Sorensen PH. Identification and quantification of newly synthesized proteins translationally regulated by YB-1 using a novel Click-SILAC approach. J Proteomics. 2012;77:1–10. doi: 10.1016/j.jprot.2012.08.019. [DOI] [PubMed] [Google Scholar]
- 30.Howden AJ, Geoghegan V, Katsch K, Efstathiou G, Bhushan B, Boutureira O, Thomas B, Trudgian DC, Kessler BM, Dieterich DC, et al. QuaNCAT: quantitating proteome dynamics in primary cells. Nat Methods. 2013;4:343–346. doi: 10.1038/nmeth.2401. [•• Here researchers develop QuaNCAT, a technique that allows for the quantitative identification of subproteomes, by dual labeling with both heavy amino acids and non-canonical amino acids.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eichelbaum K, Winter M, Diaz MB, Herzig S, Krijgsveld J. Selective enrichment of newly synthesized proteins for quantitative secretome analysis. Nat Biotechnol. 2012;10:984–990. doi: 10.1038/nbt.2356. [DOI] [PubMed] [Google Scholar]
- 32.Szychowski J, Mahdavi A, Hodas JJ, Bagert JD, Ngo JT, Landgraf P, Dieterich DC, Schuman EM, Tirrell DA. Cleavable biotin probes for labeling of biomolecules via azide-alkyne cycloaddition. J Am Chem Soc. 2010;51:18351–18360. doi: 10.1021/ja1083909. [•• In this study researchers develop a set of cleavable biotin probes to increase sensitivity while reducing peptide contamination for BONCAT.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu C, Jin Y, Schneider T, Burnham DR, Smith PB, Chiu DT. Ultrabright and bioorthogonal labeling of cellular targets using semiconducting polymer dots and click chemistry. Angew Chem Int Ed Engl. 2010;49:9436–9440. doi: 10.1002/anie.201004260. [• This paper describes ultrabright polymer dots with copper-catalyzed click reactivity for membrane protein labeling.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sletten EM, Bertozzi CR. Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew Chem Int Ed Engl. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soriano Del Amo D, Wang W, Jiang H, Besanceney C, Yan AC, Levy M, Liu Y, Marlow FL, Wu P. Biocompatible copper(I) catalysts for in vivo imaging of glycans. J Am Chem Soc. 2010;132:16893–16899. doi: 10.1021/ja106553e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agard NJ, Prescher JA, Bertozzi CR. A strain-promoted [3+2] azide-alkyne cycloaddition for covalent modification of biomolecules in living systems. J Am Chem Soc. 2004;126:15046–15047. doi: 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
- 37.Baskin JM, Prescher JA, Laughlin ST, Agard NJ, Chang PV, Miller IA, Lo A, Codelli JA, Bertozzi CR. Copper-free click chemistry for dynamic in vivo imaging. Proc Natl Acad Sci U S A. 2007;104:16793–16797. doi: 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beatty KE, Fisk JD, Smart BP, Lu YY, Szychowski J, Hangauer MJ, Baskin JM, Bertozzi CR, Tirrell DA. Live-cell imaging of cellular proteins by a strain-promoted azide-alkyne cycloaddition. Chembiochem. 2010;11:2092–2095. doi: 10.1002/cbic.201000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beatty KE, Szychowski J, Fisk JD, Tirrell DA. A BODIPY-cyclooctyne for protein imaging in live cells. Chembiochem. 2011;12:2137–2139. doi: 10.1002/cbic.201100277. [• This set of papers describes the first steps towards fluorescent tags for live, cytoplasmic labeling using small, membrane-permeable Coumarin and BODIPY fluorophores.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Link AJ, Vink MK, Agard NJ, Prescher JA, Bertozzi CR, Tirrell DA. Discovery of aminoacyl-tRNA synthetase activity through cell-surface display of noncanonical amino acids. Proc Natl Acad Sci U S A. 2006;103:10180–10185. doi: 10.1073/pnas.0601167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanrikulu IC, Schmitt E, Mechulam Y, Goddard WA, 3rd, Tirrell DA. Discovery of Escherichia coli methionyl-tRNA synthetase mutants for efficient labeling of proteins with azidonorleucine in vivo. Proc Natl Acad Sci U S A. 2009;106:15285–15290. doi: 10.1073/pnas.0905735106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ngo JT, Champion JA, Mahdavi A, Tanrikulu IC, Beatty KE, Connor RE, Yoo TH, Dieterich DC, Schuman EM, Tirrell DA. Cell-selective metabolic labeling of proteins. Nat Chem Biol. 2009;10:715–717. doi: 10.1038/nchembio.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ngo JT, Schuman EM, Tirrell DA. Mutant methionyl-tRNA synthetase from bacteria enables site-selective N-terminal labeling of proteins expressed in mammalian cells. Proc Natl Acad Sci U S A. 2013;13:4992–4997. doi: 10.1073/pnas.1216375110. [• Here researchers express E. coli mutant MetRS in mammalian cells to genetically restrict labeling of newly synthesized proteins. They find that the E. coli mutant MetRS is capable of only replacing the terminal methionine in mammalian cells, thereby making the label site-selective.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ngo JT, Babin BM, Champion JA, Schuman EM, Tirrell DA. State-selective metabolic labeling of cellular proteins. ACS Chem Biol. 2012;8:1326–1330. doi: 10.1021/cb300238w. [•• In this study, mutant MetRS expression is driven by state-selective promoters to enable labeling of newly synthesized proteins only under certain physiological conditions, such as oxidative stress.] [DOI] [PMC free article] [PubMed] [Google Scholar]