Abstract
Microbial colony growth can serve as a useful readout in assays for studying complex genetic interactions or the effects of chemical compounds. Although computational tools for acquiring quantitative measurements of microbial colonies have been developed, their utility can be compromised by inflexible input image requirements, non-trivial installation procedures, or complicated operation. Here, we present the Spotsizer software tool for automated colony size measurements in images of robotically arrayed microbial colonies. Spotsizer features a convenient graphical user interface (GUI), has both single-image and batch-processing capabilities, and works with multiple input image formats and different colony grid types. We demonstrate how Spotsizer can be used for high-throughput quantitative analysis of fission yeast growth. The user-friendly Spotsizer tool provides rapid, accurate, and robust quantitative analyses of microbial growth in a high-throughput format. Spotsizer is freely available at https://data.csiro.au/dap/landingpage?pid=csiro:15330 under a proprietary CSIRO license.
Keywords: colony array, yeast, bacteria, synthetic genetic array, microorganism, large-scale screen
Microbial growth can be used as a measure of fitness as a function of environmental and genetic constraints. This information is useful in basic and clinical research for phenotyping bacterial or yeast strains, characterizing chemical compounds, or determining the genetics that underlie biological phenomena including drug sensitivity or resistance (1–5). The generation of growth curves and extraction of quantitative parameters are traditionally achieved using regular liquid cultures or micro-cultures that are grown in multi-well plates. However, acquisition of such data on solid media using colony-based measurements may prove easier for both traditional and high-throughput approaches as no shakers are needed and less space is used (6). Bacterial and yeast species may mainly grow on solid surfaces in the wild, showing characteristics and behaviors of a community when faced with adverse environmental situations (7). Fitness measurements, therefore, in the context of a colony rather than in a homogeneous shaking flask culture might better reflect real environmental conditions and processes.
Besides phenotyping and growth profiles, colony measurements in yeast have been used for the characterization of genome-wide genetic interactions that provide information on synthetic lethality and functional relationships between all non-essential genes within the cell (8). Such studies have been rendered possible in recent years by systematic mutant strain collections, the design and utilization of spotting and arraying robots, and the development of computational and statistical tools for the quantification of colony growth data (9–12). Notably, the last aspect, quantification, is currently one of the main bottlenecks for such approaches.
Multiple software tools to determine colony sizes from culture plate images have been developed in recent years. Examples are CellProfiler (a multipurpose image analysis tool) (13), YeastXtract (14), HT Colony Grid Analyzer (9), Colonyzer (15), ScreenMill (an ImageJ macro) (16), Balony (17), SGAtools (web-based) (18) and gitter (R package) (19). These tools utilize different paradigms, such as single time-point versus time-lapse analysis (i.e., growth curve construction) of colony size, definition of colony size by colony area versus colony volume (i.e., pixel intensity integration), or utilization of manually replica-plated arrays of large colonies versus robotically pinned arrays of small colonies; some tools also include modules for specialized downstream analyses of colony sizes, such as detection of genetic interactions (summarized in Table 1). In some cases, the methods are outdated and not used much by microbiological research communities (e.g., YeastXtract, HT Colony Grid Analyzer). Importantly, for some of these tools, the installation and setup can be technically challenging, and/or their operation requires the use of a command line environment (gitter) that can be intimidating and less intuitive for wet-lab researchers. Another important aspect is the performance of these methods in finding colony spots in an accurate manner. Some tools have quite strict requirements with respect to the image file format and/or resolution (e.g., Balony has strict image naming requirements impractical for processing existing data sets and SGAtools does not seem to support TIFF images). Performance can also be hampered by suboptimal image quality, such as minor image distortions frequently introduced by digital cameras when used under suboptimal conditions. To illustrate the point with our own use case, several of these tools (HT Colony Grid Analyzer, ScreenMill, Balony, and SGAtools) failed to process the test image data set we used in this study (see “Materials and methods” section). Namely, in our hands HT Colony Grid Analyzer 1.1.7 (9) ran without any error messages but produced no output data; ScreenMill (CM engine 1.63) (16) was unable to properly automatically crop the input images and thus could not identify colony positions; Balony (17) was not compatible with the image file naming scheme we employed; SGAtools (18) did not accept TIFF image files (as of 14 February 2016).
Table 1. Main features of Spotsizer and selected colony image analysis tools.
| Tool name | Capabilities | GUI | Supported grids | Supported colony arrays | Accepted file formats | Colony size definition | Framework | Operating system | License | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| Spotsizer | colony size | yes | 96, 384, 1536 | pinned | TIFF, JPEG, PNG | area | Workspace | Windows, Mac, Linux | CSIRO Binary Software License | this study |
| Balony | colony size, genetic interactions, data visualization | yes | 96, 384, 1536 + user-defined | pinned | JPEG | area | Java | cross-platform | GNU GPL | (17) |
| CellProfiler | modular multi-purpose image analysis | yes | user-defined | pinned, stamped | TIFF, JPEG, PNG, other | user-defined | MATLAB | Windows, Mac, Linux | GNU GPL | (13) |
| Colonyzer | colony size, time-lapse | no | 48, 96, 384, 768 (diagonal), 1536 | pinned, stamped | JPEG, ? | area, total pixel intensity | R, Python | cross-platform | GNU GPL | (15) |
| gitter | colony size, data visualization | no | 96, 384, 768 (diagonal), 1536 | pinned | TIFF, JPEG | area | R | cross-platform | GNU LGPL | (19) |
| HT Colony Grid Analyzer | colony size | yes | 96, 384, 768 (diagonal) | pinned | JPEG | area | Java | cross-platform | GNU GPLv2 | (9) |
| ScreenMill | colony size, genetic interactions, data visualization | yes | 384, 1536 | pinned, stamped | TIFF, JPEG, PNG, GIF, other | area, mean pixel intensity | imageJ, web-based | cross-platform | GNU GPL | (16) |
| SGAtools | colony size, genetic interactions, data visualization, GO enrichment | yes | 96, 384, 768 (diagonal), 1536 | pinned | JPEG | area | web-based (uses gitter) | cross-platform | ? | (18) |
| YeastXtract | colony size, time-lapse | yes | user-defined | pinned, stamped | TIFF | total pixel intensity | Java | cross-platform | CC BY-NC-SA 2.5 | (14) |
Summary of main features and capabilities of Spotsizer and a selection of other software tools for colony image analysis.
Here, we describe the development of Spotsizer, a proprietary software tool for collecting high-throughput measurements of microbial colony size in robotically pinned arrays of different densities. Spotsizer has a graphical user interface (GUI), is fast and simple to use for the biologist/clinician/student, and can analyze plate images either one-by-one or in a batch mode. We compare the quality of colony size measurements obtained by the Spotsizer automated method with a curated approach that we have developed in parallel, as well as with gitter, a recently introduced non-graphical package for the R environment (19). Our validation data indicate that Spotsizer is an accurate and highly robust tool for microbial fitness analyses that should prove useful in diverse research settings.
Materials and methods
Fission yeast handling and growth
All strains were grown and maintained in standard rich YES medium (20). For growth on solid media, strains were first inoculated in liquid YES medium in 96-well plates. Using the bench-top RoToR HDA robot (Singer Instruments, Roadwater, UK), 4 initial 96-well plates were condensed in a single 384-spot YES agar plate that was fur ther re-arrayed at a 1536-spot density, each strain being represented by 4 spots that served as technical replicates. Strains were then re-spotted on plates with or without diverse stressors at 96-, 384-, or 1536-spot density. To ensure that only a minimum amount of biomass of all strains is spotted across the whole plate area, we experimented with a range of pickup and spotting pressures on the RoToR. We decided to use 5% pressure in all cases as this ensured that: (i) all colonies, including those located at plate corners, are replicated; and (ii) the amount of transferred biomass remains as small as possible. Strains were grown at 32°C for 48 h and then plate photographs were taken.
The particular cultivation conditions and suitable times of colony image acquisition will likely differ in each use case, depending on the organism, robotics, biological question, etc., and will therefore need to be optimized. For fission and budding yeasts, readers can consult established cultivation procedures for high-throughput genetic interaction mapping (10,12).
Image acquisition
High-resolution images of 96-, 384-, and 1536-spot plates were acquired using a Multi-DocIt transillumination system (UVP, Cambridge, UK). The image data set (121 images, TIFF format, 2592 × 1944 pixels) is available for download from the Spotsizer homepage.
Curated spot finding and quantification
Plate images were segmented into colony area/background using white/black thresholding in the ImageMagick package (www.imagemagick.org). Images were then saved as 16-bit TIFF files (a format required by the GenePix Pro image analysis software). Spot finding and colony size quantification was performed using GenePix Pro 5.0 (Molecular Devices, Wokingham, UK). This software is intended for microarray analysis but can, in principle, analyze any circular objects arrayed in a regular grid. A GenePix Array List (GAL) file describing the plate layout (required for analysis in GenePix Pro) was created according to the manufacturer’s instructions (http://mdc.custhelp.com/app/answers/detail/a_id/18883#gal). The correct identification of all colonies in all plate images was ensured by visual inspection and manual adjustment as needed. Colony area sizes in pixels were derived from the Foreground Total Intensity parameter of each spot by dividing it by 65535 (i.e., the intensity contribution of 1 white foreground pixel at 16-bit color depth). The resulting data set is available for download from the Spotsizer homepage. Median strain colony sizes were then calculated for each plate using an in-house Python script (www.python.org). Data visualizations were performed in R (www.rproject.org).
Workspace implementation of Spotsizer
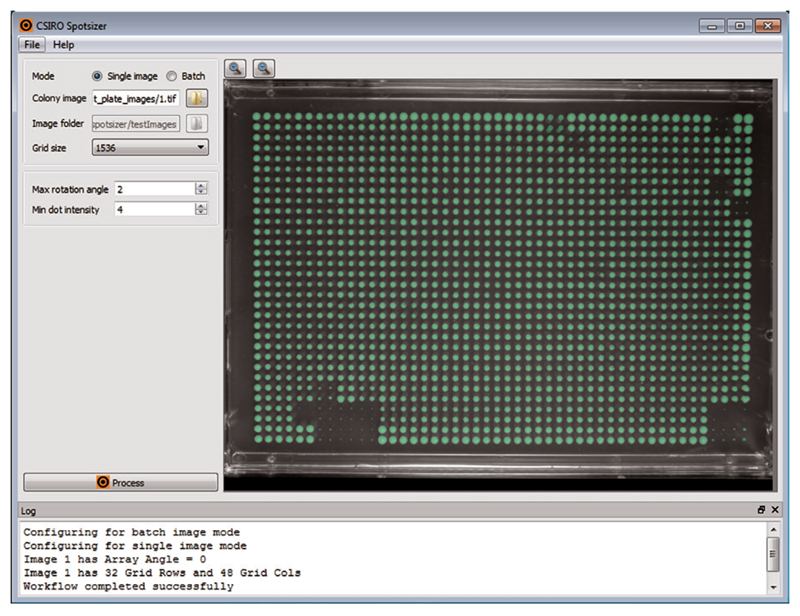
Spotsizer is built using CSIRO’s Workspace package (www.csiro.au/workspace)—a framework in which one can construct workflows, user interfaces, and complete applications quickly and easily—and the image analysis plugin and a custom workflow to analyze colony images was developed by CSIRO’s Quantitative Imaging Group. Spotsizer is provided as binary packages for the Windows 64-bit, Mac OSX, and Linux 64-bit operating systems. Spotsizer has a simple user interface (Figure 1): The left panel is used to set up input data and workflow parameters, the input and output images are displayed in the central panel, and the bottom Log panel displays log and error messages.
Figure 1. The user interface.
Screenshot of Spotsizer analyzing a 1536-colony image.
By default, the software operates in a batch image mode: it processes all images in the folder selected as the input folder in the Image folder field. An operator can switch Spotsizer to process only a particular image by selecting the Single mode option. An input image in this case should be selected using the Colony image field. The operation workflow is executed by clicking the Process button. It is possible to abort the execution by clicking the Stop processing button in the information window that appears while the workflow is running. Output images and text files are saved in a Results subfolder inside the input folder. For each input image, three output files are produced: a binary image of spots (colonies) found by the workflow, an overlay image where the mask of colonies is superimposed with the input image, and a text file with the location and size measurements for each colony.
The workflow algorithm has three steps: background removal, finding the grid, and measuring the spot area at each grid location. Background removal is necessary because uneven lighting and surface reflections can produce variations in background intensity across the image. We use connected component morphological filtering to suppress the background and retain only the spots. The filtering requires an estimate of the maximum grid spacing, which is derived from the operator input parameter, Grid size. Grid finding on the background-suppressed image must be able to handle slight tilts of the grid and also slightly non-parallel grid lines caused by camera lens distortion. We detect the rotation angle of the grid array with an operator input parameter, Max rotation angle, defining the bounds of the angle search to limit the search time. The principle is to use directional mean filtering, that is a mean filter with a linear neighborhood whose size is equal to the width of the image. Directional mean filtering works by taking the mean of an image over a line structuring element. In a filtered image, a pixel value is replaced by the mean value of all pixels along the line centered on a given pixel. The direction of the line is defined by a rotation angle, where a horizontal line has a zero rotation angle. Integral angles between -1*Max rotation angle and Max rotation angle, with a step size of 1, are tested, which amounts to a total of (2 * Max rotation angle + 1) potential rotation angles being tested. When the angle of the linear neighborhood aligns with the angle of rotation of the grid array, the intensity variance, measured along the image edge, will be increased relative to filtering at other angles. The angle of maximum variance is the rotation angle. Directional mean filtering with a narrower linear neighborhood is then performed at the rotation angle, and orthogonally, to locate the rows and columns of the grid (without requiring that these lines be perfectly straight). The intersection of the rows and columns define the grid locations. Spot finding should not be based on simple thresholding because this would underestimate the size of small spots with lower brightness levels. We use seeded region growing (21) (with seeds for the spots at the center of each grid location and seeds for the background at the boundaries between grid regions) to find the boundary between each spot and the background. Because the seeded region growing algorithm preferentially directs region growth outwards from the seed points according to the difference between a pixel’s value and the running mean of values in an adjoining region, the boundary for a spot is the mid-point intensity for each individual spot. Some imaging systems have surface reflections that produce ghost spots alongside the real spot; therefore, a test for the validity of a detected spot must be applied before reporting the size for that spot. This test requires that the mean intensity of the spot be greater than the mean plus two times the standard deviation of the background intensity. For some images with severe ghosting, this test is insufficient, so we provide an operator input parameter, Min dot intensity, to remove falsely detected “ghost” spots.
Spotsizer takes colony array images in TIFF, JPEG, or PNG formats with no specific requirements for image resolution apart from that required to resolve the smallest colonies (a colony should be at least three pixels in diameter). Higher resolution than the minimum will require more memory and, hence, longer processing times. Using the Grid size parameter, the operator can process any colony array with the standard geometry of number of rows = 2 * sqrt(Grid size / 6) and number of columns = 3 * sqrt(Grid size / 6). We have tested the software on 96, 384, and 1536 colony arrays produced by robotic pinning. Currently, Spotsizer is not capable of processing images with 6144 colonies, but in principle, such a modification is possible in the future. Note that Spotsizer is not appropriate for analysis of cultures plated from liquid using a manual replicaplating tool (stamper), but is optimized for rectangular (i.e., not diagonal) arrays of pinned single colonies. Processing time is 6 –12 s per image (of resolution 2929 × 2042 pixels) on a laptop with an Intel i7–3540 CPU (3 GHz with 8 GB RAM) running the Windows 7 64-bit operating system. While Spotsizer does not include a module for growth curve construction from time-lapse colony images, it can in principle be used to obtain colony size measurements from individual time-lapse images that then serve as input for suitable external tools.
Spotsizer can be downloaded freely for academic use from the CSIRO Data Access Portal (https://data.csiro.au/dap/landingpage?pid=csiro:15330). The project page consists of the Description tab and the Data tab. The Spotsizer installer and test data sets are available from the Data tab. The Description tab provides basic information about the project and software IP, and includes the Spotsizer license and user guide files. By downloading and installing Spotsizer, a user automatically agrees with the terms and conditions provided in the license. The Spotsizer user guide is also available from the Help menu in the Spotsizer application.
Results and discussion
Growth assays with fission yeast
To obtain images of microbial colonies suitable for the development and validation of methods for high-throughput quantitative analysis of colony growth, we used a collection of fission yeast strains available in our laboratory. These included natural isolates showing various growth phenotypes (5), the Bioneer deletion collection of ~3000 non-essential genes (22), and individual diagnostic deletion mutants with altered sensitivity to various stresses, such as the sty1 MAP kinase regulating the response of fission yeast cells to a wide range of adverse conditions (23). Cells were grown on solid YES media in the presence of various stressors to obtain a range of growth phenotypes, and plate images were taken (data not shown). In total, we have assembled a set of 121 plate images (see Figure 2A for a representative example).
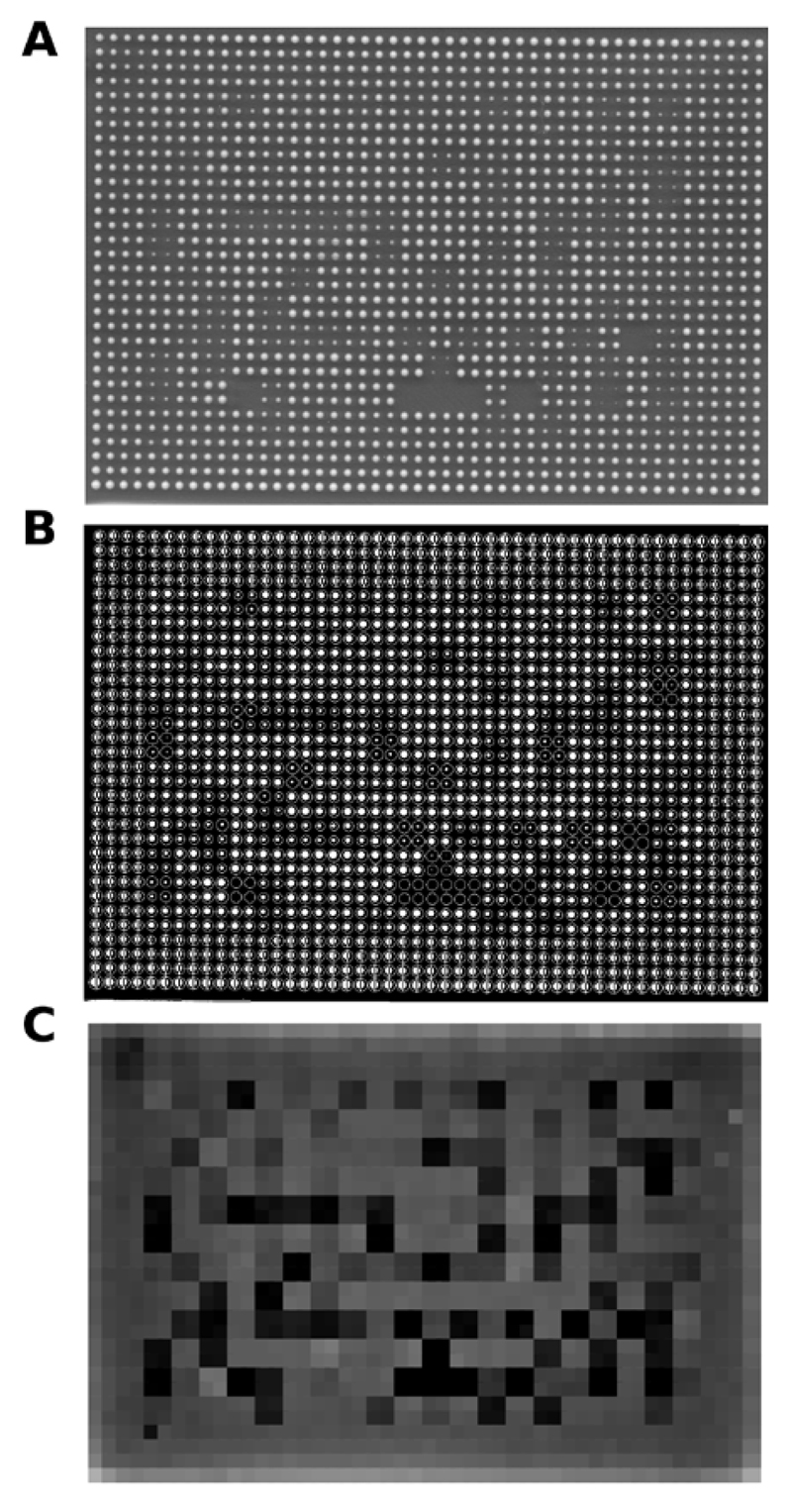
Figure 2. Curated spot-finding procedure.
(A) Sample plate image containing 1536 fission yeast colonies (each strain spotted in quadruplicate). (B) Segmented black-and-white image of the plate being analyzed in GenePix Pro. A grid has been semi-automatically overlaid to mark the positions of all colonies. (C) A plate-like image created using the values extracted by GenePix Pro.
Curated spot-finding and quantification
Plate images (121 plates in total) were first processed by a curated procedure employing the GenePix Pro 5.0 micro-array analysis software as described in the “Materials and methods” section. Unlike some existing tools that require a perfect horizontal/vertical alignment of colonies in the image, this approach is robust to tilting and minor distortions of plate images, which often occur during non-robotic image acquisition using regular digital cameras, scanners, or gel documentation systems. A sample plate analysis is shown in Figure 2. A plot of colony size values obtained this way corresponds well to the original plate image (Figure 2C). These high-quality data were then used as the gold standard for assessing the performance of the Spotsizer automated colony size determination algorithm.
Automated spot-finding and quantification with Spotsizer
Next, plate images were analyzed by our fully automated spot-finding and quantification algorithm implemented in Spotsizer. The resulting colony size data were in excellent agreement with the data obtained by the manually curated procedure described above over the whole range of growth conditions analyzed (typical Pearson’s r = 0.955) (Figure 3A). Analysis by Spotsizer typically only takes a few seconds per image, compared with >10 min per image for the manual pipeline described above, allowing for a large number of plate images to be processed quickly and easily.
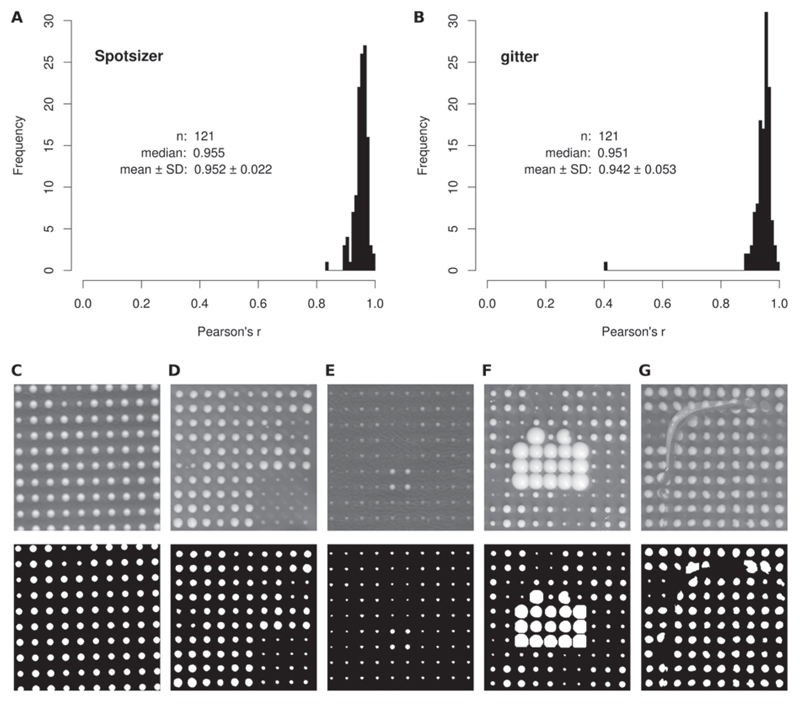
Figure 3. Accuracy and robustness of Spotsizer.
A total of 121 plates, each containing 1536 colonies, were analyzed both in GenePix Pro 5.0 (curated) and using 2 automated tools (Spotsizer and gitter) (19). The colony sizes obtained by the manually curated approach were compared with the results from Spotsizer (A) and gitter (B), with distributions of Pearson’s correlation coefficients shown. (C–G) Analysis of challenging colony images by Spotsizer. The top panel shows part of the original image; the bottom panel shows the colony area mask determined by Spotsizer. (C) A tilted image. (D) Colony reflections (ghosting). (E) Poor colony growth. (F) Overgrown colonies. (G) A cracked agar plate.
To compare the performance of Spotsizer with existing tools, we carried out an analogous analysis using the R package gitter (batch mode, default settings) (19), which is one of the latest tools to be introduced in the field. This command-line software performed comparably well (typical Pearson’s r = 0.951) (Figure 3B) on our test set of 121 images, with the exception of 1 image where gitter failed to position the colony grid properly (Pearson’s r = 0.408). Visual inspection of the plate image in question suggested that the image was tilted to a degree that gitter could not compensate for when run with default settings (image rotation functionality is optional in gitter). Importantly, Spotsizer identified the colony grid in this image correctly. When image rotation was enabled in gitter, the tilted image was analyzed successfully; however, gitter failed in this case to position the grid properly with two other images from the test data set.
To further demonstrate the robustness of Spotsizer, we also successfully analyzed potentially problematic images of plates featuring some common image acquisition ar tifacts or non-standard colony growth (Figure 3, C–G). Thus, our results indicate that Spotsizer is both highly accurate and robust to issues with image quality.
Differential growth analysis using Spotsizer output data
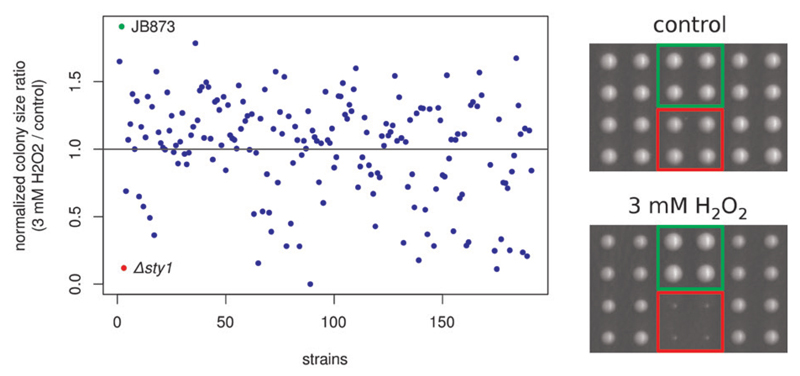
To further demonstrate the utility of Spotsizer, we used it to find yeast strains showing sensitivity or resistance to oxidative stress. Colonies were grown on YES agar plates with or without 3 mM hydrogen peroxide (H2O2). Individual colony sizes were determined automatical ly using Spotsizer, and median colony size (from four replicate colonies) was calculated for each strain. Next, colony size was normalized to the growth of the 972h- fission yeast reference strain under the respective conditions. Finally, colony size ratios between the H2O2 and unstressed conditions were calculated for each strain (Figure 4). Reassuringly, this analysis identified the known Δsty1 master stress-response regulator mutant among the most H2O2-sensitive strains. The analysis also picked up resistant strains, such as the wild isolate JB873 (NCYC3442; National Collection of Yeast Cultures; www.ncyc.co.uk). Spotsizer has also been used successfully for large-scale phenotyping of wild fission yeast isolates, where the accuracy of the data enabled heritability analysis, and for genome-wide association studies with as few as 57 strains (5).
Figure 4. Identification of differential growth using Spotsizer.
Median colony sizes were normalized to the 972h- reference yeast strain, and growth was then compared between control conditions (rich YES medium) and the presence of 3 mM hydrogen peroxide (H2O2). Example sensitive (Δsty1; red) and resistant (JB873; green) strains are indicated. The actual colonies of the example strains are shown in the right panel.
Here, we developed and validated a novel, fully automated algorithm for the quantification of microbial growth on plates. Spotsizer is a free, wet-lab, biologist-friendly tool that features an intuitive GUI, high analysis accuracy and speed, and robustness to the quality of input image data. Spotsizer works with multiple colony grid types and image file formats and can be run in a batch mode, allowing large numbers of images to be processed rapidly and easily. We have demonstrated that Spotsizer is highly effective for finding differential growth phenotypes in fission yeast.
Method Summary
Spotsizer is a biologist-, clinician-, and student-friendly software tool for automated colony size measurements using images of arrayed microbial colonies. Spotsizer features a convenient graphical user interface, has both single-image and batch-processing capabilities, and works with multiple input image formats and different colony grid types.
Acknowledgments
This work was supported in part by the Czech Science Foundation grant P305/12/P040 and Charles University in Prague project UNCE 204013 (M.P.), The Commonwealth Scientific and Industrial Research organisation of Australia (L.B. and Y.A.), the BBSRC Research Grant BB/I012451/1 and Wellcome Trust Senior Investigator Award 095598/Z/11/Z (J.B.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author contributions
C.R., M.P., and D.J. conceived and designed the experiments. C. R. performed the experiments, and M.P. carried out data analyses. L.B. and Y.A. developed Spotsizer with testing and feedback from M.P., C.R. ,and D.J. The paper was written by M.P., C.R., D.J., L.B., Y.A., and J.B.
Competing interests
The authors declare no competing interests.
References
- 1.Nichols RJ, Sen S, Choo YJ, Beltrao P, Zietek M, Chaba R, Lee S, Kazmierczak KM, et al. Phenotypic landscape of a bacterial cell. Cell. 2011;144:143–156. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown WR, Liti G, Rosa C, James S, Roberts I, Robert V, Jolly N, Tang W, et al. A geographically diverse collection of Schizosaccharomyces pombe isolates shows limited phenotypic variation but extensive karyotypic diversity. G3 (Bethesda) 2011;1:615–626. doi: 10.1534/g3.111.001123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parsons AB, Brost RL, Ding H, Li Z, Zhang C, Sheikh B, Brown GW, Kane PM, et al. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat Biotechnol. 2004;22:62–69. doi: 10.1038/nbt919. [DOI] [PubMed] [Google Scholar]
- 5.Jeffares DC, Rallis C, Rieux A, Speed D, Převorovský M, Mourier T, Marsellach FX, Iqbal Z, et al. The genomic and phenotypic diversity of Schizosaccharomyces pombe. Nat Genet. 2015;47:235–241. doi: 10.1038/ng.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartman JL, Tippery NP. Systematic quantification of gene interactions by phenotypic array analysis. Genome Biol. 2004;5:R49. doi: 10.1186/gb-2004-5-7-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palková Z. Multicellular microorganisms: laboratory versus nature. EMBO Rep. 2004;5:470–476. doi: 10.1038/sj.embor.7400145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JLY, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins SR, Schuldiner M, Krogan NJ, Weissman JS. A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome Biol. 2006;7:R63. doi: 10.1186/gb-2006-7-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roguev A, Wiren M, Weissman JS, Krogan NJ. High-throughput genetic interaction mapping in the fission yeast Schizosaccharomyces pombe. Nat Methods. 2007;4:861–866. doi: 10.1038/nmeth1098. [DOI] [PubMed] [Google Scholar]
- 11.Dixon SJ, Costanzo M, Baryshnikova A, Andrews B, Boone C. Systematic mapping of genetic interaction networks. Annu Rev Genet. 2009;43:601–625. doi: 10.1146/annurev.genet.39.073003.114751. [DOI] [PubMed] [Google Scholar]
- 12.Baryshnikova A, Costanzo M, Dixon S, Vizeacoumar FJ, Myers CL, Andrews B, Boone C. Synthetic genetic array (SGA) analysis in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Methods Enzymol. 2010;470:145–179. doi: 10.1016/S0076-6879(10)70007-0. [DOI] [PubMed] [Google Scholar]
- 13.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah NA, Laws RJ, Wardman B, Zhao LP, Hartman JL. Accurate, precise modeling of cell proliferation kinetics from time-lapse imaging and automated image analysis of agar yeast culture arrays. BMC Syst Biol. 2007;1:3. doi: 10.1186/1752-0509-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawless C, Wilkinson DJ, Young A, Addinall SG, Lydall DA. Colonyzer: automated quantification of micro-organism growth characteristics on solid agar. BMC Bioinformatics. 2010;11:287. doi: 10.1186/1471-2105-11-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dittmar JC, Reid RJ, Rothstein R. ScreenMill: a freely available software suite for growth measurement, analysis and visualization of high-throughput screen data. BMC Bioinformatics. 2010;11:353. doi: 10.1186/1471-2105-11-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young BP, Loewen CJR. Balony: a software package for analysis of data generated by synthetic genetic array experiments. BMC Bioinformatics. 2013;14:354. doi: 10.1186/1471-2105-14-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagih O, Usaj M, Baryshnikova A, Vandersluis B, Kuzmin E, Costanzo M, Myers CL, Andrews BJ, et al. SGAtools: one-stop analysis and visualization of array-based genetic interaction screens. Nucleic Acids Res. 2013;41:W591–W596. doi: 10.1093/nar/gkt400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagih O, Parts L. gitter: A robust and accurate method for quantification of colony sizes from plate images. G3 (Bethesda) 2014;4:547–552. doi: 10.1534/g3.113.009431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 21.Adams R, Bischof L. Seeded region growing. IEEE Trans Pattern Anal Mach Intell. 1994;16:641–647. [Google Scholar]
- 22.Kim D-U, Hayles J, Kim D, Wood V, Park H-O, Won M, Yoo H-S, Duhig T, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe . Nat Biotechnol. 2010;28:617–623. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen D, Toone WM, Mata J, Lyne R, Burns G, Kivinen K, Brazma A, Jones N, Bähler J. Global transcriptional responses of fission yeast to environmental stress. Mol Biol Cell. 2003;14:214–229. doi: 10.1091/mbc.E02-08-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]