Abstract
Carboxyl-ester lipase is a digestive pancreatic enzyme encoded by the highly polymorphic CEL gene1. Mutations in CEL cause maturity-onset diabetes of the young (MODY) with pancreatic exocrine dysfunction2. Here we identified a hybrid allele (CEL-HYB), originating from a crossover between CEL and its neighboring pseudogene CELP. In a discovery cohort of familial chronic pancreatitis cases, the carrier frequency of CEL-HYB was 14.1% (10/71) compared with 1.0% (5/478) in controls (odds ratio [OR] = 15.5, 95% confidence interval [CI] = 5.1-46.9, P = 1.3 × 10−6). Three replication studies in non-alcoholic chronic pancreatitis cohorts identified CEL-HYB in a total of 3.7% (42/1,122) cases and 0.7% (30/4,152) controls (OR = 5.2, 95% CI = 3.2-8.5, P = 1.2 × 10−11; formal meta-analysis). The allele was also enriched in alcoholic chronic pancreatitis. Expression of CEL-HYB in cellular models revealed reduced lipolytic activity, impaired secretion, prominent intracellular accumulation and induced autophagy. The hybrid variant of CEL is the first chronic pancreatitis gene identified outside the protease/antiprotease system of pancreatic acinar cells.
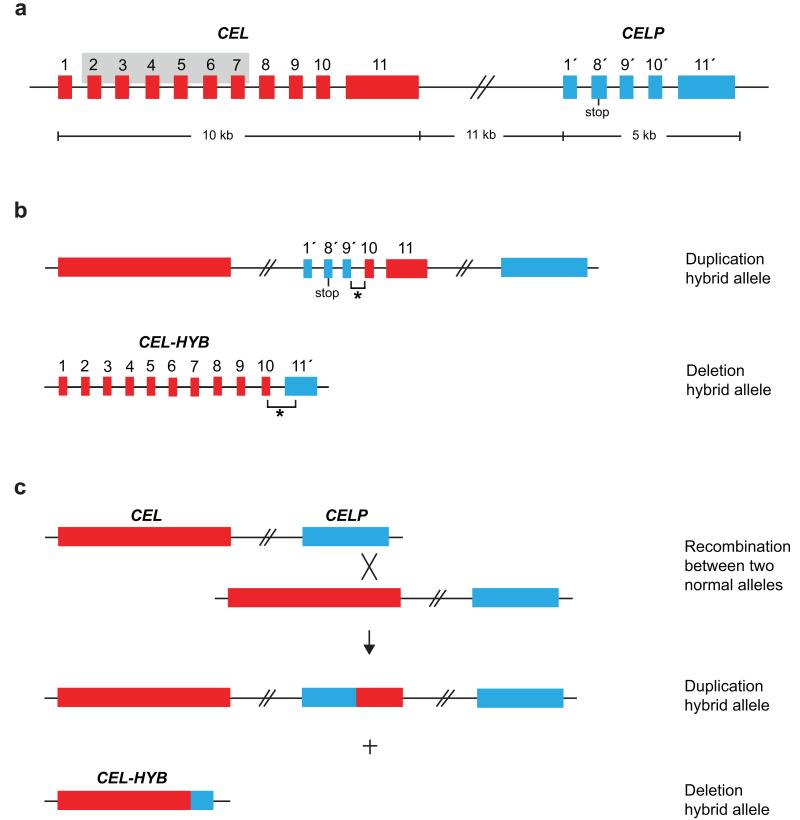
Carboxyl-ester lipase (encoded by CEL, MIM114840) is mainly expressed in the acinar cells of the pancreas and the lactating mammary gland1,3. The enzyme is activated by bile salts in the duodenum and participates in the hydrolysis and absorption of cholesterol and lipid-soluble vitamins1. The human CEL locus on chromosome 9q34.3 also includes CELP, a tandemly arranged CEL pseudogene. The main difference between the two genes is that CELP lacks exon 2-7 of CEL and harbors a stop codon in its second exon (i.e. exon 8′, Fig. 1a), otherwise the two genomic sequences are highly similar4,5.
Figure 1.
Copy number variants of the human CEL gene. The large red and blue boxes without numbers correspond to the complete CEL and CELP genes, respectively. Smaller boxes represent the individual exons and are numbered 1-11 (CEL) and 1′, 8′-11′ (CELP). The stop codon in exon 8′ of CELP is indicated. (a) Structure of the CEL-CELP gene locus. The grey-shaded area in CEL marks the exon 2-exon 7 region missing in CELP. The size of the locus is shown below the figure; exon and intron sizes are not drawn to scale. (b) Schematic structure of the duplication hybrid allele and the deletion hybrid allele (CEL-HYB) identified in this study. The breakpoint regions (not drawn to scale) are indicated by asterisks. (c) Proposed mechanism of the generation of CEL-HYB by non-allelic homologous recombination between CEL and CELP. The X symbolizes the cross-over event in the exon 10-exon 11 region.
The last of the eleven CEL exons contains a variable number of tandem repeat (VNTR) region, consisting of nearly identical 33-bp segments3,4. The segment number fluctuates between 7 and 23, with 16 repeats being the most common in all populations studied so far6-10. We have previously reported that single-base deletions in the CEL VNTR cause maturity-onset diabetes of the young, type 8 (MODY8, MIM609812), an autosomal dominantly inherited pancreatic disease characterized by diabetes and exocrine dysfunction accompanied by morphological changes of the gland2,11. Therefore, we surmised that the highly polymorphic CEL gene might be a candidate for influencing the risk of chronic pancreatitis (CP), a complex, longstanding and progressive inflammatory disease resulting in gradual replacement of the pancreatic parenchyma by fibrous tissue. The clinical picture includes abdominal pain as well as exocrine and endocrine pancreatic insufficiency manifesting as maldigestion and diabetes, respectively.
The constellation of a gene and a nearby homologous sequence is well known to enhance the probability of genomic rearrangements, and can sometimes lead to new fusion genes that have altered functional properties and are associated with disease12. Copy number variants (CNVs) of CEL have been noted, both by others and us9,10,13. Fine-mapping the CEL locus of a subject known to carry an extra copy of CEL exon 11 revealed the presence of a duplication hybrid allele (Fig. 1b, top panel). This allele apparently resulted from non-allelic homologous recombination (Fig. 1c), the crossover occurring within an identical region of 522 bp in introns 9 of CEL and CELP (positions 12780-13301 in CEL and 28549-29070 in CELP; GenBank accession AF072711.1). The part consisting of CELP-CEL fusion sequence was, however, unlikely to encode a functional protein by virtue of its structural similarity to CELP (Fig. 1b, top panel).
In contrast, the reciprocal recombination product of the duplication hybrid allele, namely a deletion hybrid allele, would potentially encode a novel CEL-CELP fusion protein based upon its predicted gene structure (Fig. 1b, lower panel; Fig. 1c). We developed a long-range PCR assay for its detection (Supplementary Fig. 1), and among 190 Norwegian blood donors we found one positive subject. Subsequent DNA sequencing revealed that the identified deletion hybrid allele (denoted CEL-HYB) was not exactly reciprocal to the aforementioned duplication allele; the crossover occurred instead in a 548-bp identical region covering intron 10 and the adjacent exon boundaries (Supplementary Fig. 2a). This might indicate that different recombination events have taken place in the intron 9-exon 11 region of the CEL-CELP locus.
We decided to investigate the role of CEL-HYB in the genetic etiology of CP. Screening a discovery cohort of 71 German subjects with familial CP (Table 1) revealed a carrier frequency of 14.1% (10/71) compared with 1.0% (5/478) in controls (odds ratio [OR] = 15.5; 95% confidence interval [CI] = 5.1-46.9; P = 1.3 × 10−6). To replicate this finding, we screened three independent cohorts (two German, one French) of non-alcoholic CP cases, as well as controls from each contributing laboratory. In each cohort, CEL-HYB was significantly overrepresented among CP cases (Table 1). For the three replication cohorts combined, CEL-HYB was present in 3.7% (42/1,122) of patients and 0.7% (30/4,152) of controls. Formal meta-analysis of the replication cohorts was highly significant (OR = 5.2, 95% CI = 3.2-8.5, P = 1.2 × 10−11). Among the 1,122 cases there were 57 individuals carrying rare PRSS1 variants known to associate with hereditary CP. All PRSS1 carriers tested negative for CEL-HYB. If these carriers were excluded from the study, the combined CEL-HYB frequency for cases increased to 3.9%. Upon meta-analysis of the three replication cohorts the OR was now 5.5 (95% CI = 3.4-8.9, P = 2.6 × 10−12).
Table 1.
Heterozygous carrier frequencies of the CEL-HYB variant in German and French subjects with chronic pancreatitis
| Patient materials* | Cases |
Controls |
|||||
|---|---|---|---|---|---|---|---|
| +/N | % | +/N | % | OR | CI | P value** | |
| Discovery cohort | 10/71 | 14.1 | 5/478 | 1.0 | 15.5 | 5.1 – 46.9 | 1.3 × 10-6 |
|
| |||||||
| Non-alcoholic CP cohort 1 | 11/260 | 4.2 | 5/569 | 0.9 | 5.0 | 1.7 – 14.5 | 0.002 |
| Non-alcoholic CP cohort 2 | 22/510 | 4.3 | 16/2362 | 0.7 | 6.6 | 3.4 – 12.7 | 1.3 × 10−8 |
| Non-alcoholic CP cohort 3 | 9/352 | 2.6 | 9/1221 | 0.7 | 3.5 | 1.4 – 9.0 | 0.009 |
| Meta-analysis *** | - | - | - | - | 5.2 | 3.2 - 8.5 | 1.2 × 10−11 |
|
| |||||||
| Alcoholic CP cohort | 15/853 | 1.8 | 26/3409 | 0.8 | 2.3 | 1.2 – 4.4 | 0.016 |
Cohorts are described in the Methods section
Two-tailed Fisher’s exact test
Fixed effect model (in replication cohorts only). Z= 6.78; heterogeneity chi-squared = 1.18 (d.f. = 3), P = 0.553; I-squared
We also examined the frequency of CEL-HYB in individuals with alcohol-related CP. Again, we found a significant enrichment of the CEL-HYB variant with a carrier frequency of 1.8% in cases versus 0.8% in controls (OR = 2.3, 95% CI = 1.2-4.4, P = 0.016) (Table 1). All CEL-HYB-positive individuals detected in the five cohorts studied were heterozygous carriers of the allele, except one homozygous case among the French patients (non-alcoholic CP cohort 3).
In the discovery cohort, family members from two of the CEL-HYB positive subjects were available for screening. Their pedigrees showed an autosomal dominant inheritance pattern of CEL-HYB although the allele did not fully co-segregate with pancreatitis (Supplementary Fig. 3). Furthermore, when analyzing the CEL-HYB variant in 228 parents of patients from the German CP cohort 2, we detected six heterozygous parents. All of them had transmitted the risk allele to their affected child. Two parents had a diagnosis of pancreatitis. These observations confirm the high, but not absolute, risk associated with CEL-HYB.
To exclude the possibility that alterations in the exon 1-10 region of CEL-HYB could explain the disease association, we performed DNA sequencing of twelve CEL-HYB-positive cases. No predicted amino acid changes were observed in exons 1-9. In one CP patient, however, a premature stop codon was observed in exon 10 of the CEL-HYB reading frame. The corresponding mRNA may not be translated because of nonsense-mediated RNA decay and, if translated, the protein will not include the VNTR. All initially positive cases and controls were then screened for this minor CEL-HYB variant (found altogether in one case and four controls; not included in the carrier frequencies of Table 1).
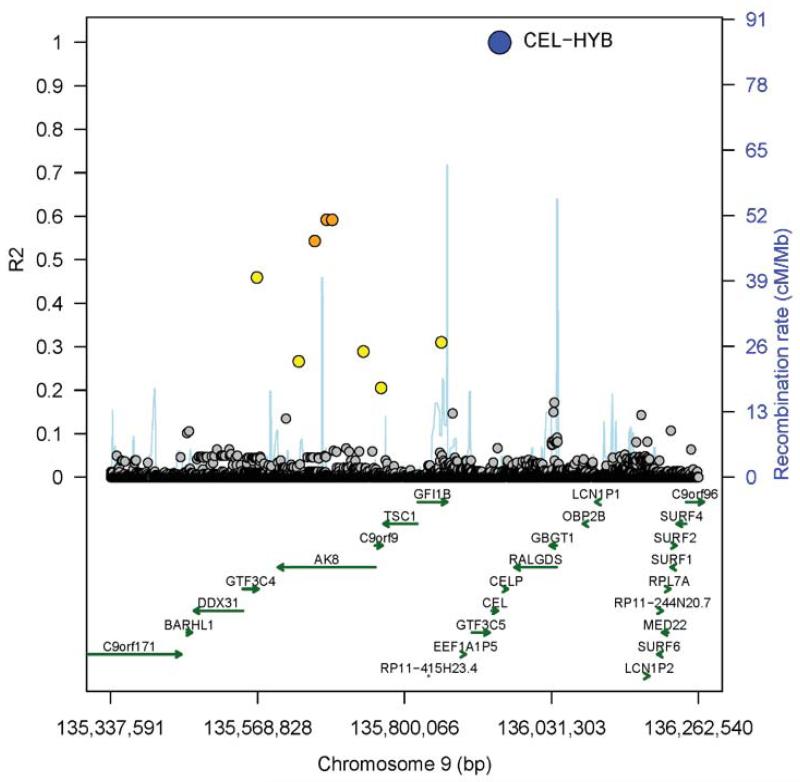
To test whether the CEL-HYB risk variant might be identifiable indirectly through linkage disequilibrium (LD) with surrounding SNPs, we performed a detailed examination of genome-wide association study (GWAS) data in a subset of 787 individuals with known CEL-HYB status (25 carriers, 762 non-carriers). Proxy SNP analysis within the CEL-CELP LD region did not identify variants in strong LD with CEL-HYB. However, some distant correlating variants were found peaking at about 255 kb upstream of the CEL-CELP locus (Fig. 2). No other SNPs correlating with the CEL-HYB variant at R2 ≥ 0.2 were identified on chromosome 9 outside the region shown in Fig. 2. The best R2 was around 0.6 for a few imputed, highly correlated SNPs with low minor allele frequencies (Supplementary Table 1). Moreover, to evaluate the origin of CEL-HYB we sequenced the exon 10-11 breakpoint region of 74 carriers of the allele. We observed five CEL-HYB-associated haplotypes (Supplementary Fig. 2b), indicating that the variant has arisen by multiple independent recombination events. Thus, it is unlikely that a standard GWAS will be able to thoroughly tag and assess the CEL-HYB risk variant.
Figure 2.
Regional plot showing proxy SNPs of the CEL-HYB variant. R2 represents correlation between the CEL-HYB variant and SNPs based on measured and imputed gene doses. Recombination rate is according to 1,000 Genomes haplotypes Phase I (v3), and reported genes are from Ensembl release 70, January 2013 (see URLs). Genes were limited to biotypes ‘protein-coding’ and ‘pseudogene’. Colors represent levels of R2 with CEL-HYB.
We also asked whether SNPs or rare variants within or adjacent to CEL exons 1-10 could be associated with CP outside the context of the CEL-HYB allele. Sequencing of 364 non-alcoholic CP cases and 238 controls revealed no variants that were overrepresented in the CP cohort (Supplementary Table 2).
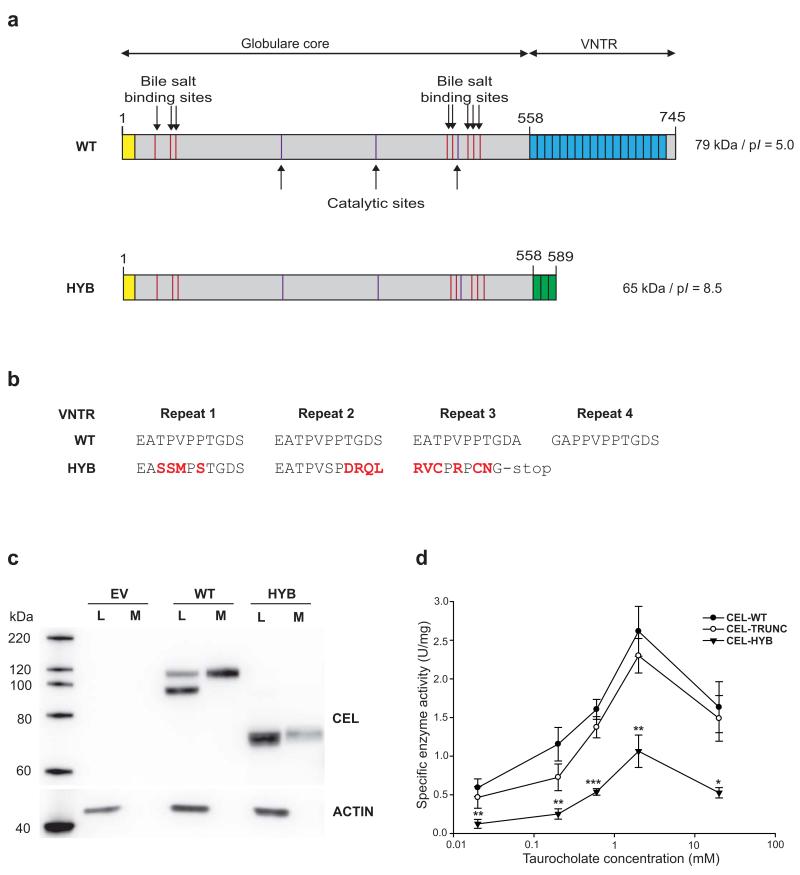
The CEL-HYB allele was predicted to encode a chimeric CEL protein of 589 amino acids, containing a functional enzyme region that derived from CEL but with a shorter and different VNTR originating from the CEL pseudogene (Fig. 3a, b; Supplementary Fig. 4a). To examine the functional consequences of CEL-HYB, we stably expressed the variant in human embryonic kidney (HEK) 293 cells, which do not have a detectable level of endogenous CEL mRNA or protein expression14,15. We found that secretion of the CEL-HYB protein was reduced relative to the wild-type protein (CEL-WT) (Fig. 3c). A similar result was observed in transiently transfected mouse acinar (266-6) cells (Supplementary Fig. 4b).
Figure 3.
Altered structure, expression and enzyme activity of the CEL-HYB protein compared with wild-type CEL (CEL-WT). (a) Predicted structure of the two protein variants. The yellow box indicates the N-terminal signal peptide. The blue and green boxes correspond to the C-terminal VNTR region of the normal CEL protein and CEL-HYB, respectively. (b) Alignment of the amino acid sequence encoded by the first VNTR repeats of the CEL-WT and CEL-HYB alleles. The altered reading frame of CEL-HYB predicts a change of amino acid sequence starting in the first repeat and a premature stop-codon in repeat 3. Substituted amino acids are shown in red. (c) Expression of CEL-WT and CEL-HYB in stably transfected HEK293 cells. Cell lysates (L) and media (M) were analyzed by Western blotting. Transfection of empty vector (EV) was included as negative control. The actin bands indicate the amount of loaded protein in the lysate lanes only. The data are representative of more than three experiments giving similar result. (d) CEL enzyme activity in conditioned media from stably transfected HEK293 cells. Lipase activity was measured at 37°C in the presence of increasing concentrations of sodium taurocholate as activator. CEL-TRUNC, a variant of the CEL protein completely lacking the C-terminal VNTR, was included for evaluating the effect of the VNTR on enzyme activity. Data are shown as means of seven independent experiments, with statistical comparisons performed between CEL-HYB and CEL-WT activity. Error bars, SEM; *, P = 0.01-0.05; **, P = 0.001-0.01; ***, P < 0.001.
Next, we investigated whether CEL-HYB might encode an overactive lipase. Carboxyl-ester lipase activity was measured using conditioned media from the CEL-expressing HEK293 cells, normalizing specific activity to the amount of CEL in the growth medium. When stimulated by bile salts, the CEL-HYB enzyme reached only about 40% of CEL-WT activity (Fig. 3d). An artificial, truncated variant of CEL (CEL-TRUNC) that was missing the whole VNTR, showed about 90% of WT enzyme activity, a finding also reported by others16,17. We therefore propose that the impaired activity of CEL-HYB may relate to effects of the new protein C-terminus rather than to the lack of a normal VNTR. Further, our results strongly suggest that increased lipase activity is not involved in the disease mechanism.
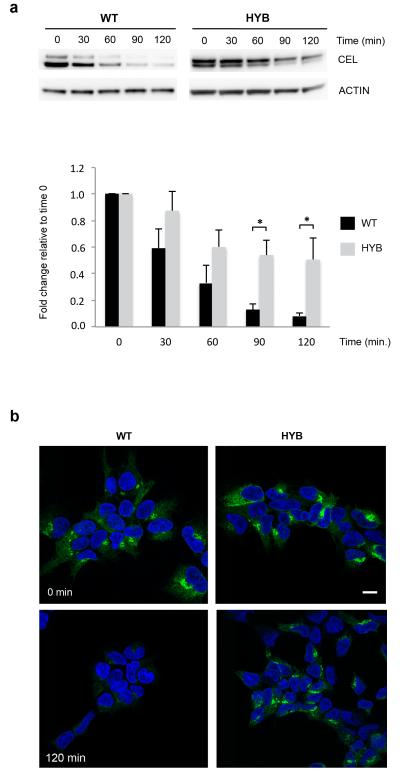
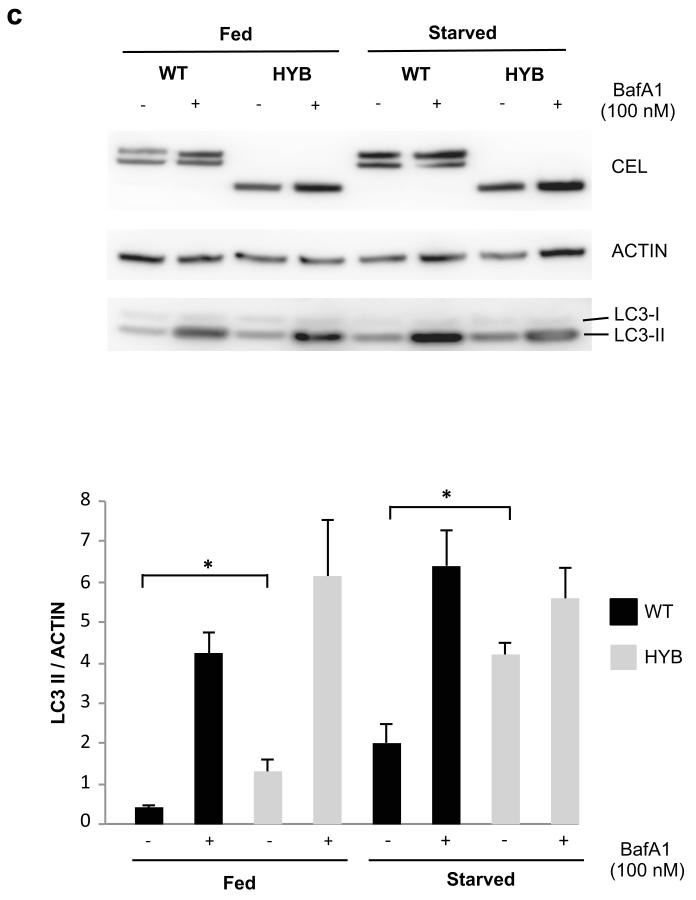
Finally, we explored the intracellular fate of the protein. When protein synthesis was inhibited by cycloheximide, CEL-HYB accumulated in the cells to a much higher degree than CEL-WT (Fig. 4a,b). This intracellular retention of CEL-HYB as well as its impaired secretion (Fig. 3c) could indicate that the pathological effect involves injury inside the cell. We hypothesized that intracellular CEL-HYB might induce compensatory autophagy for removal of defective and excessive secretory proteins. Both when grown in rich and starvation medium there was a significant increase of the autophagy marker LC3-II in CEL-HYB-expressing cells (Fig. 4c). Notably, there are reports on increased levels of LC3-II and impaired autophagic flux in pancreatitis, and it has been proposed that autophagic dysfunction might be a key initiating event in this condition18.
Figure 4.
Intracellular properties of CEL-HYB. (a) Reduced cellular clearance of CEL-HYB. Stably transfected HEK293 cells were treated with the protein synthesis inhibitor cycloheximide (1 μg/ml) and examined at the indicated time points. Lysates from cells expressing CEL-WT or CEL-HYB were analyzed for CEL and actin by Western blotting (upper panel). The bands were quantified and CEL level normalized to actin (lower panel). CEL expression at time 0 was arbitrarily set to 1.0. After 120 min exposure to cycloheximide, 50% of CEL-HYB protein was detected in the cell lysate compared with 9% for CEL-WT. Data are shown as means of three independent experiments. Error bars, SEM; *, P = 0.01-0.05. (b) Effect of cycloheximide confirmed by immunostaining and confocal imaging. Cells expressing CEL-HYB showed a much stronger fluorescence signal than CEL-WT-expressing cells after 120 min of cycloheximide treatment. Scale bar, 10 μm. (c) Autophagy induced by CEL-HYB expression. Stably transfected HEK293 cells were incubated in rich (fed) or starvation medium with or without bafilomycin A1 (BafA1) for 3 hours. Cell lysates were analyzed by Western blotting for CEL, actin and the autophagy marker protein LC3 (upper panel). The bands were quantified and LC3–II normalized to actin (lower panel). CEL-HYB-expressing cells grown in full or starved medium showed increased LC3-II levels as compared with CEL-WT-expressing cells. Data are shown as means of three independent experiments. Error bars, SEM; *, P = 0.01-0.05.
Almost all CP genes identified so far encode proteins of the protease/antiprotease system of the pancreatic acinar cells, such as cationic trypsinogen (PRSS1, MIM276000)19,20, serine protease inhibitor Kazal type 1 (SPINK1, MIM167790)21, chymotrypsinogen C (CTRC, MIM601405)22,23, carboxypeptidase A1 (CPA1, MIM114850)24, and anionic trypsinogen (PRSS2, MIM601564)25. Other genetic risk factors are CFTR (MIM602421)26,27 and SNPs in the CLND2-MORC428,29, ABO and FUT2 loci30. Here we have identified CEL as a new gene for CP. This is the first lipase gene to be directly associated with pancreatitis. Different from most genes listed above, our functional studies suggest a pathogenic mechanism that does not involve premature activation of pancreatic digestive enzymes to initiate CP.
The odds ratio (5.2) of CEL-HYB in the non-alcoholic CP cohorts is comparable with that of the CTRC gene22. However, disease-associated CTRC variants tend to occur together with heterozygous PRSS1 or SPINK variants31,32. This contrasts our findings for CEL-HYB: among the 67 positive cases of Table 1 there were none with a PRSS1 mutation and only one SPINK1 p.N34S/CEL-HYB trans-heterozygous patient.
We also detected a significant enrichment for CEL-HYB in a cohort of alcohol-related CP (OR = 2.3). Alcohol abuse is the leading cause of CP, accounting for about 70% of the cases33. Only a minority of long-term alcohol abusers develops CP, suggesting the presence of genetic risk factors, but the role of such factors is less understood than in non-alcoholic CP. So far, it has been reported that SPINK1 and CTRC variants as well as SNPs in the PRSS1-PRSS2, CLDN2-MORC4, ABO and FUT2 loci are associated with alcoholic CP22,28-30,34, all with modest odds ratios. Our findings suggest that CEL-HYB might be included among the relatively few genes known to interact with alcohol consumption to amplify CP risk. The data must, however, be interpreted with caution as the role of alcohol may sometimes be overestimated in CP and therefore alcoholic CP materials would be expected to include also some cases of idiopathic disease.
Finally, we note that classical molecular genetic techniques (long-range PCR, cloning, Sanger DNA sequencing) were necessary to map and analyze the genomic rearrangements of the CEL-CELP locus. Our detailed SNP analysis of the CEL-CELP region (Fig. 2) indicates that current high-throughput methodologies such as GWAS of common variants and exome sequencing would be unlikely to uncover CEL-HYB and its role in pancreatitis. When genetic risk loci involve complex and highly repetitive regions, these techniques have some inherent limitations, as recently illustrated by the failure of detecting mutations in the VNTR of MUC1 by massively parallel sequencing35. The identification of CEL-HYB as a novel pancreatitis gene therefore nicely highlights that in-depth analysis of the more complex regions of our genome still can play an important role in elucidating the genetic etiology of human disorders.
URLs
FinchTV, http://www.geospiza.com/ftvdlinfo.html; 1000 Genomes, http://mathgen.stats.ox.ac.uk/impute/data_download_1000G_phase1_integrated.html; Ensembl release 70, http://Jan2013.archive.ensembl.org/biomart/martview/3e14905c6880c3043c1.
METHODS
Methods and any associated references are available in the online version of the paper.
METHODS
Subjects
The medical ethical review committees of all participating study centers approved this study. For the discovery phase, we screened 71 unrelated German patients with a diagnosis of familial, non-alcoholic CP, recruited by the University of Greifswald. These patients had unknown genetic etiology as they had previously tested negative for well-established pancreatitis risk genes. For the replication studies, we investigated 1,122 unrelated non-alcoholic CP subjects from University of Leipzig (cohort 1; n=260), Technische Universität München (cohort 2; n=510), and University of Bretagne, Brest (cohort 3; n=352). All cases in the replication cohorts had been screened for rare PRSS1 variants associated with hereditary CP as well as the SPINK1 p.N34S risk variant. The three German patient cohorts were compared to exclude any duplicate samples. In addition, we investigated 853 unrelated German subjects with alcoholic CP from Greifswald (n=214), Leipzig (n=296) and Munich (n=343). Controls were healthy blood donors (Greifswald, n=478; Leipzig, n=569; Munich, n=2,362; Brest, n=1,221). Norwegian blood donors (n=190) were used for the initial identification of CEL-HYB.
The diagnosis of CP was based on the presence of a typical history of recurrent pancreatitis (≥ 2 acute pancreatitis attacks) and/or other typical findings upon imaging like pancreatic calcifications. Familial CP (discovery cohort) was diagnosed when two or more family members in more than one generation suffered from pancreatitis. The patients of the replication cohorts were classified as having non-alcoholic CP when common risk factors such as alcohol abuse or other predisposing disorders were absent. Patients who reported a history of regular alcohol consumption of at least 60 g per day (women) or 80 g per day (men) were included in the alcoholic CP cohort.
Reference sequences
The following reference sequences have been used for primer design, sequence verification and SNP analysis of the CEL-CELP locus: GenBank accession number AF072711.1 (Homo sapiens carboxyl ester lipase gene, complete cds; and carboxyl ester lipase pseudogene, complete sequence); Ensembl database accession numbers OTTHUMG00000020855 (CEL) and OTTHUMG00000020857 (CELP).
Characterization of the CEL duplication hybrid allele
The presence of a CEL duplication was originally inferred by the DNA fragment analysis method of Torsvik et al., 20109. Its allelic structure was worked out by PCR walking, using LA Taq polymerase (TaKaRa Bio) as described by the manufacturer. For mapping of the CEL-CELP recombination site, PCR products were verified by agarose gel electrophoresis, cloned into the pCR2.1-TOPO vector (Invitrogen), and analyzed by sequencing.
Screening for the CEL deletion hybrid allele
A long-range, duplex PCR assay was developed for the postulated deletion hybrid allele (CEL-HYB) as described in Supplementary Fig. 1. We performed the reaction in a total volume of 10 μl, containing 1x GC buffer, 0.4 μM of each of the primers L11F, IAR and CELP-VNTR-R (Supplementary Table 3), 1 M betaine solution, 0.4 mM of each dNTP, 0.5 U LA Taq polymerase, and 10-50 ng genomic DNA. Amplification started with a denaturation step at 94 °C for 1 min followed by 14 cycles of 94 °C for 20 sec and 60 °C for 6 min; then 16 cycles of 94 °C for 20 sec and 62 °C for 6 min, and a final elongation step of 72 °C for 10 min, followed by cooling to 4 °C. After visualization on a 1% agarose gel, all CEL-HYB-positive samples were finally verified by Sanger DNA sequencing.
For screening of CEL-HYB in larger materials, a high-capacity assay on the LightCycler 480 platform (Roche Diagnostics) was developed. We performed PCR using with forward and reverse primers as given in Supplementary Table 3 at a ratio of 5:1 (0.5 μmol/l and 0.1 μmol/l, respectively), 0.75 U OneTaq polymerase (New England BioLabs) and 0.4 mM of each dNTP, in a total volume of 25 μl. The cycle conditions were as follows: initial denaturation for 12 min at 95°C; 40 cycles of 20 s denaturation at 95°C, 40 s of annealing at 60 °C and 90 s of primer extension at 72 °C; and a final extension for 2 min at 72 °C. The amplified PCR products were then tested by melting curve analysis on the LightCycler 480. We used a SimpleProbe (TIB MOLBIOL) that allowed for discrimination between the CEL and CELP sequences by creating different melting points (Supplementary Table 3). A representative image of screening results from the LightCycler assay is shown in Supplementary Fig. 5.
Because the LightCycler assay was based on discrimination of SNPs in the breakpoint region some false positives were detected. All samples positive in the LightCycler assay were therefore subjected to the long-range PCR method above and sequenced for final verification. This approach was used when screening cases and controls recruited by the Leipzig and Munich groups. The other cohorts underwent primary screening directly by long-range PCR. The genotyping concordance for the long-range PCR assay was 100% for 533 duplicates tested.
Sanger DNA sequencing
Sequencing was performed by standard protocols on an ABI 3730 capillary sequencer (Applied Biosystems) using M13 forward or reverse primers, or CEL- and CELP-specific primers (Supplementary Table 3).
When sequencing PCR products directly they were treated with ExoSAP (USB Corporation) as described by the manufacturer, and 2 μl of the treated PCR products were used as template. Sequencing results were analyzed by FinchTV (see URLs) or SeqScape (Applied Biosystems) software.
SNP mapping of the CEL-CELP region
Analysis of linkage disequilibrium between the CEL-HYB variant and SNPs on chromosome 9 was done in pancreatitis patients with available genetic data from Illumina Omni Human Express 700k SNP-chips. Genotypes of 2,824 patients were called with Illumina GenomeStudio. Quality control was done using PLINK. Thereby, the data were filtered for missingness per marker ≤ 0.05, missingness per individual ≤ 0.05, Hardy-Weinberg equilibrium p-value ≥ 10−6, cryptic relatedness filter PI_HAT ≤ 0.185, autosomal heterozygosity within 3 standard deviations, and population stratification to be not more extreme than ± 3 standard deviations. Following quality control, 2,712 individuals (2,065 male, 630 females, 17 of unknown sex) and 32,138 SNPs from chromosome 9 were used for imputation. Imputation was done using SHAPEIT v2 and IMPUTE 2.3.0, applying the 1000 Genomes reference phase 1, release 3 (see URLs). Blocks used in imputing were chosen to include the CEL-CELP locus ± 2 Mb in a single block. For post-imputation quality control, we filtered for SNPs with info-score smaller than 0.3.
For all further analyses, the cohort was limited to individuals with known CEL-HYB status, i.e. 787 individuals. Among these, 25 individuals (3.2 %) were carriers of the CEL-HYB allele. To identify proxies for CEL-HYB, we followed several strategies. First, we calculated Pearson’s correlation of the CEL-HYB variant with allelic doses from imputed and directly measured genotypes (“gene dose” analysis). Additionally, we calculated haplotype-based R2 of directly typed SNPs and best-guess imputed genotypes. Here, we set all imputed genotypes with posterior probability smaller than 90% to missing. Confidence intervals of haplotype-based R2 were calculated as recommended36, applying jack-knife methodology to estimate standard errors. Aggressive two- and three-marker tagging was done using Tagger as implemented in Haploview 4.2 on measured and best-guess imputed genotypes.
CEL-HYB plasmid construction
Human CEL wild-type (CEL-WT) cDNA was inserted into the pcDNA3.1/V5-His vector (Invitrogen). For the enzyme activity experiment, cDNA of a truncated variant of human CEL (CEL-TRUNC), lacking the VNTR after nucleotide 1686, was inserted into the pcDNA3.1/V5-His vector. Both CEL variants were cloned in-frame with the V5-His tag of the pcDNA3.1/V5-His vector as described14,15.
We used the CEL-WT-V5/His plasmid as template for creating the CEL-HYB construct. BamHI and XhoI restriction sites flanked the VNTR sequence of the WT plasmid. A synthetic VNTR sequence of the CEL-HYB allele, flanked by the same restriction sites, was ordered (DNA 2.0). The VNTR of the template was then replaced by the CEL-HYB VNTR, creating a CEL-HYB-V5/His plasmid. The C substitution (rs77696629) observed in exon 10 of all CEL-HYB-positive subjects (Supplementary Fig. 2a) was introduced by using the QuickChange II XL Site- directed Mutagenesis kit (Stratagene). The final structure of the CEL-HYB construct was confirmed by DNA sequencing. Before being employed in cellular experiments, the plasmid was tested in vitro by expression in the TNT T7 Quick-coupled Transcription/Translation System (Promega) (Supplementary Fig. 4a).
Cell cultures and transfection
HEK293 cells (Clontech) were cultured in α-minimal essential medium (Sigma) supplemented with 10% fetal bovine serum (Invitrogen), 4 mM L-glutamine (Sigma), and 1x Antibiotic-Antimycotic (Invitrogen). Hank’s balanced salt solution (PAA - GE Healtcare) was used in starvation experiments. The 266-6 mouse pancreatic acinar cell line (ATCC) was grown in Dulbecco’s Modified Eagle’s Medium (ATCC) containing 10% fetal bovine serum (Invitrogen) and 1x Antibiotic-Antimycotic (Invitrogen). Both cell lines were maintained at 37°C in a 5% CO2-humidified atmosphere.
Transient transfection of CEL-WT-V5/His, CEL-HYB-V5/His, and the pcDNA3.1/V5-His empty vector in 266-6 cells was performed in 6-well plates using METAFECTENE PRO (Biontex) with a lipid (μl) to DNA (μg) ratio of 1:1. HEK293 cells stably transfected with CEL-WT-V5/His, CEL-TRUNC-V5/His, or the pcDNA3.1/V5-His vector were available in the lab14,15. For stable transfection of the CEL-HYB-V5/His plasmid, HEK293 cells were transfected with the FuGENE 6 Transfection Reagent (Roche Diagnostics) with a 3:1 ratio of transfection reagent (μl) to DNA (μg). Cells were then grown under the selection of 500 μg/ml geneticin 418 (Invitrogen) for 18 days. Protein blotting and immunostaining confirmed expression of the CEL-HYB protein. All stable cell lines were maintained under constant geneticin selection.
Preparation of cell medium and lysate
After transient transfection (48 h), 1 ml conditioned medium was isolated and centrifuged for 5 min at 13,000 rpm and 4°C. The supernatant was collected and analyzed. Total cell lysate was prepared by washing the cells in PBS before adding ice-cold Ripa buffer (Millipore) with 1x Complete Mini, EDTA-free protease inhibitor cocktail (Roche Diagnostics). The cells were added 150 μl Ripa buffer per well and incubated for 30 min at 4°C. The lysate was centrifuged for 15 min at 13,000 rpm and 4°C, and the supernatant was further analyzed as total cell lysate. Stably transfected cells were plated in 6-well plates (3 × 105 cells per well) and grown for 48 h before isolation of cell medium and lysate, as described above. Quantification of total protein in cell lysates was measured using the BCA Protein Assay (Pierce).
Western blotting
Lysates of 3 or 5 μg total protein were subjected to SDS-PAGE and Western blotting. For analysis of CEL protein secretion, the volume of medium loaded on the SDS gel was the same as for the corresponding lysate. The proteins were separated by 10% SDS-PAGE at 180 V for 1.5-2 hours and transferred to PVDF membranes by standard methods. The membranes were incubated with primary antibodies at 4°C overnight, washed, and incubated with secondary antibodies for 1 hour at room temperature. The blots were developed with the Enhanced Chemiluminiscence Plus Western Blotting Detection Kit (GE Healthcare) and further analyzed on a LAS-1000 imager (Fujifilm).
CEL activity assay
Aliquots of conditioned media from stably transfected HEK293 cells expressing CEL-WT, CEL-TRUNC or CEL-HYB were used. HEK293 cells stably transfected with empty vector were included as negative control. The cells were grown to 80% confluency in normal growth medium. Subsequently, the medium was substituted with serum-free medium and the cells were incubated for additional 18 hours. Conditioned medium was collected and centrifuged at 300 × g and 4°C to remove detached cells, and kept on ice for immediate use.
CEL enzyme activity was measured using 4-nitrophenyl valerate (4-npv; Sigma) as substrate, monitoring the hydrolysis of 4-npv spectrophotometrically in the presence of sodium taurocholate (Sigma) as activator. The assay was performed at 37°C in a buffer containing 100 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM 4-npv (solubilized in ethanol), and with sodium taurocholate concentrations varying between 0.02 and 20 mM. Reactions were run in triplicate in a 96-well microplate containing 115 μl buffer and 10 μl conditioned medium using an Infinite M200 Pro microplate reader (Tecan) with iControl Tecan Software, continuously monitoring the release of para nitrophenyl at 405 nm. The molar extinction coefficient of 4-npv was 11500 M−1cm−1. Enzyme activity was normalized against the amount of CEL in the conditioned media. CEL protein concentration was determined by SDS-PAGE and immunoblotting using titration of recombinant mouse CEL (5658-CE, R&D Systems) as a standard.
Treatments of stably transfected cells
HEK293 cells were seeded in triplicate in 6-well plates (3 × 105 cells per well), coated with poly-L-lysine (Sigma), and cultured for 48 hours before treatment. For investigating the rate of cellular CEL clearance during inhibition of protein synthesis, the cells were treated with 1 μg/ml cycloheximide (Sigma). Cell lysates were harvested for Western blot analysis after 0, 30, 60, and 90 min of treatment. For autophagy analysis, cells were incubated in full or starvation medium with or without Bafilomycin A1 (100 nM) for 3 hours, followed by harvesting of cell lysates for Western blotting.
Immunofluorescence and confocal microscopy
Cells were seeded in 12-well plates (1.5 × 105 cells per well), and grown on poly-L-lysine coated coverslips for 48 hours. Upon cycloheximide treatment, as described above, the cells were fixed for 30 min with 3% paraformaldehyde before they were immunostained as described previously37. After antibody staining, the coverslips were inverted onto objective glasses in a drop of ProLong Gold antifade mounting reagent (Molecular Probes) with DAPI (Invitrogen). The samples were examined using Leica TCS SP5 confocal microscope (Leica Microsystems) with a 63×/NA 1.4 Plan-Apochromat oil-immersion objective, ~1.2 Airy unit pinhole aperture, and the appropriate filter combinations. Images were acquired with Diode 405 and Argon lasers and processed using Adobe Photoshop CS5 image software (Adobe Systems). This experiment was repeated three times.
Antibodies
Protein expression in HEK293 and 266-6 cells was measured by Western blotting using the mouse anti-V5 primary antibody (R960-25, Invitrogen) or a rabbit polyclonal antibody (Vanko) for CEL detection. The Vanko antibody was raised against recombinant CEL lacking the VNTR-region and was a generous gift from Dr. Olle Hernell (Umeå University, Sweden). For detection of the autophagosome marker LC3, a rabbit polyclonal LC3B antibody (NB600-1384, Novus Biologicals) was employed. An antibody detecting beta-actin (sc-1615, Santa Cruz Biotechnology) was included to control for loading. Immunofluorescent staining of CEL was performed using the mouse monoclonal antibody As20.1, also kindly provided by Dr. Olle Hernell. AlexaFluor488-conjucated goat anti-mouse (A-11017, Invitrogen) was used as secondary antibody.
Statistical analyses
The significance of differences between allele frequencies in affected individuals and controls were tested by two-tailed Fisher’s exact test, and OR and CI were calculated using Two-by-Two Table analysis in Stata/IC 13.0 (StataCorp). A fixed-effect model was used for the formal meta-analysis, which was performed by the method of Mantel and Haenszel, as implemented with the Metan version 9 module in Stata/IC 13.0, treating each study site as separate groups. For the functional protein analyses, significance of change was analyzed using a two-tailed, unpaired t-test. The difference was regarded significant when the P-value was ≤ 0.05. Results are given as mean ±SEM.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank all study participants and the members of the Gesellschaft für Pädiatrische Gastroenterologie und Ernährung (GPGE) for providing clinical data and blood samples. The confocal imaging was performed at the Molecular Imaging Center, Dept. of Biomedicine, University of Bergen. We are also grateful to prof. Geir Bjørkøy for commenting the manuscript. This work was supported by grants and fellowships to P.R.N. and A.M. from the Translational Fund of Bergen Medical Research Foundation, KG Jebsen Foundation, University of Bergen, Research Council of Norway and Western Norway Regional Health Authority (Helse Vest), and to P.R.N from the European Research Council. Work performed in the German, French and Belgian laboratories was supported by grants from the Federal Ministry of Education and Research (BMBF GANI-MED 03152061A; BMBF 0314107), European Union Framework Programme 7 (EPC-TM; REGPOT-2010-1; BetaBat), EFRE-State Ministry of Economics MV (V-630-S-150-2012/132/133), Deutsche Forschungsgemeinschaft (RO 3929/1-1; RO 3939/2-1 1; Wi 2036/2-3; SFB 1052 C01; SPP 1629 TO 718/2-1), Colora Stiftung gGmbH, Leipzig Interdisciplinary Research Cluster of Genetic Factors, Clinical Phenotypes and Environment (LIFE Center, Universität Leipzig), Institut National de la Santé et de la Recherche Médicale (INSERM), the French Association des Pancréatites Chroniques Héréditaires (APCH), Actions de Recherche Concertée de la Communauté Française (ARC), and Fonds National de la Recherche Scientifique (FNRS), Belgium.
Footnotes
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
References
References for online methods: 14, 15, 36, 37.
- 1.Lombardo D. Bile salt-dependent lipase: its pathophysiological implications. Biochim Biophys Acta. 2001;1533:1–28. doi: 10.1016/s1388-1981(01)00130-5. [DOI] [PubMed] [Google Scholar]
- 2.Ræder H, et al. Mutations in the CEL VNTR cause a syndrome of diabetes and pancreatic exocrine dysfunction. Nat Genet. 2006;38:54–62. doi: 10.1038/ng1708. [DOI] [PubMed] [Google Scholar]
- 3.Nilsson J, et al. cDNA cloning of human-milk bile-salt-stimulated lipase and evidence for its identity to pancreatic carboxylic ester hydrolase. Eur J Biochem. 1990;192:543–50. doi: 10.1111/j.1432-1033.1990.tb19259.x. [DOI] [PubMed] [Google Scholar]
- 4.Lidberg U, et al. Genomic organization, sequence analysis, and chromosomal localization of the human carboxyl ester lipase (CEL) gene and a CEL-like (CELL) gene. Genomics. 1992;13:630–40. doi: 10.1016/0888-7543(92)90134-e. [DOI] [PubMed] [Google Scholar]
- 5.Madeyski K, Lidberg U, Bjursell G, Nilsson J. Structure and organization of the human carboxyl ester lipase locus. Mamm Genome. 1998;9:334–8. doi: 10.1007/s003359900762. [DOI] [PubMed] [Google Scholar]
- 6.Lindquist S, Blackberg L, Hernell O. Human bile salt-stimulated lipase has a high frequency of size variation due to a hypervariable region in exon 11. Eur J Biochem. 2002;269:759–67. doi: 10.1046/j.0014-2956.2001.02666.x. [DOI] [PubMed] [Google Scholar]
- 7.Higuchi S, Nakamura Y, Saito S. Characterization of a VNTR polymorphism in the coding region of the CEL gene. J Hum Genet. 2002;47:213–5. doi: 10.1007/s100380200027. [DOI] [PubMed] [Google Scholar]
- 8.Bengtsson-Ellmark SH, et al. Association between a polymorphism in the carboxyl ester lipase gene and serum cholesterol profile. Eur J Hum Genet. 2004;12:627–32. doi: 10.1038/sj.ejhg.5201204. [DOI] [PubMed] [Google Scholar]
- 9.Torsvik J, et al. Mutations in the VNTR of the carboxyl-ester lipase gene (CEL) are a rare cause of monogenic diabetes. Hum Genet. 2010;127:55–64. doi: 10.1007/s00439-009-0740-8. [DOI] [PubMed] [Google Scholar]
- 10.Ragvin A, et al. The number of tandem repeats in the carboxyl-ester lipase (CEL) gene as a risk factor in alcoholic and idiopathic chronic pancreatitis. Pancreatology. 2013;13:29–32. doi: 10.1016/j.pan.2012.12.059. [DOI] [PubMed] [Google Scholar]
- 11.Ræder H, et al. Carboxyl-ester lipase maturity-onset diabetes of the young is associated with development of pancreatic cysts and upregulated MAPK signaling in secretin-stimulated duodenal fluid. Diabetes. 2014;63:259–69. doi: 10.2337/db13-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–22. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- 13.McCarroll SA, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–74. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 14.Johansson BB, et al. Diabetes and pancreatic exocrine dysfunction due to mutations in the carboxyl ester lipase gene-maturity onset diabetes of the young (CEL-MODY): a protein misfolding disease. J Biol Chem. 2011;286:34593–605. doi: 10.1074/jbc.M111.222679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torsvik J, et al. Endocytosis of Secreted Carboxyl-ester Lipase in a Syndrome of Diabetes and Pancreatic Exocrine Dysfunction. J Biol Chem. 2014;289:29097–111. doi: 10.1074/jbc.M114.574244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansson L, et al. Recombinant human milk bile salt-stimulated lipase. Catalytic activity is retained in the absence of glycosylation and the unique proline-rich repeats. J Biol Chem. 1993;268:26692–8. [PubMed] [Google Scholar]
- 17.Downs D, Xu YY, Tang J, Wang CS. Proline-rich domain and glycosylation are not essential for the enzymic activity of bile salt-activated lipase. Kinetic studies of T-BAL, a truncated form of the enzyme, expressed in Escherichia coli. Biochemistry. 1994;33:7979–85. doi: 10.1021/bi00192a001. [DOI] [PubMed] [Google Scholar]
- 18.Gukovskaya AS, Gukovsky I. Autophagy and pancreatitis. Am J Physiol Gastrointest Liver Physiol. 2012;303:G993–G1003. doi: 10.1152/ajpgi.00122.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitcomb DC, et al. Hereditary pancreatitis is caused by a mutation in the cationic trypsinogen gene. Nat Genet. 1996;14:141–5. doi: 10.1038/ng1096-141. [DOI] [PubMed] [Google Scholar]
- 20.Le Marechal C, et al. Hereditary pancreatitis caused by triplication of the trypsinogen locus. Nat Genet. 2006;38:1372–4. doi: 10.1038/ng1904. [DOI] [PubMed] [Google Scholar]
- 21.Witt H, et al. Mutations in the gene encoding the serine protease inhibitor, Kazal type 1 are associated with chronic pancreatitis. Nat Genet. 2000;25:213–6. doi: 10.1038/76088. [DOI] [PubMed] [Google Scholar]
- 22.Rosendahl J, et al. Chymotrypsin C (CTRC) variants that diminish activity or secretion are associated with chronic pancreatitis. Nat Genet. 2008;40:78–82. doi: 10.1038/ng.2007.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masson E, Chen JM, Scotet V, Le Marechal C, Ferec C. Association of rare chymotrypsinogen C (CTRC) gene variations in patients with idiopathic chronic pancreatitis. Hum Genet. 2008;123:83–91. doi: 10.1007/s00439-007-0459-3. [DOI] [PubMed] [Google Scholar]
- 24.Witt H, et al. Variants in CPA1 are strongly associated with early onset chronic pancreatitis. Nat Genet. 2013;45:1216–20. doi: 10.1038/ng.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Witt H, et al. A degradation-sensitive anionic trypsinogen (PRSS2) variant protects against chronic pancreatitis. Nat Genet. 2006;38:668–73. doi: 10.1038/ng1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharer N, et al. Mutations of the cystic fibrosis gene in patients with chronic pancreatitis. N Engl J Med. 1998;339:645–52. doi: 10.1056/NEJM199809033391001. [DOI] [PubMed] [Google Scholar]
- 27.Cohn JA, et al. Relation between mutations of the cystic fibrosis gene and idiopathic pancreatitis. N Engl J Med. 1998;339:653–8. doi: 10.1056/NEJM199809033391002. [DOI] [PubMed] [Google Scholar]
- 28.Whitcomb DC, et al. Common genetic variants in the CLDN2 and PRSS1-PRSS2 loci alter risk for alcohol-related and sporadic pancreatitis. Nat Genet. 2012;44:1349–54. doi: 10.1038/ng.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Derikx MH, et al. Polymorphisms at PRSS1–PRSS2 and CLDN2–MORC4 loci associate with alcoholic and non-alcoholic chronic pancreatitis in a European replication study. Gut. 2014 doi: 10.1136/gutjnl-2014-307453. doi: 10.1136/gutjnl-2014-307453. [DOI] [PubMed] [Google Scholar]
- 30.Weiss FU, et al. Fucosyltransferase 2 (FUT2) non-secretor status and blood group B are associated with elevated serum lipase activity in asymptomatic subjects, and an increased risk for chronic pancreatitis: a genetic association study. Gut. 2014 doi: 10.1136/gutjnl-2014-306930. doi: 10.1136/gutjnl-2014-306930. [DOI] [PubMed] [Google Scholar]
- 31.Rosendahl J, et al. CFTR, SPINK1, CTRC and PRSS1 variants in chronic pancreatitis: is the role of mutated CFTR overestimated? Gut. 2013;62:582–92. doi: 10.1136/gutjnl-2011-300645. [DOI] [PubMed] [Google Scholar]
- 32.Masson E, Chen JM, Audrezet MP, Cooper DN, Ferec C. A conservative assessment of the major genetic causes of idiopathic chronic pancreatitis: data from a comprehensive analysis of PRSS1, SPINK1, CTRC and CFTR genes in 253 young French patients. PLoS One. 2013;8:e73522. doi: 10.1371/journal.pone.0073522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Witt H, Apte MV, Keim V, Wilson JS. Chronic pancreatitis: challenges and advances in pathogenesis, genetics, diagnosis, and therapy. Gastroenterology. 2007;132:1557–73. doi: 10.1053/j.gastro.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 34.Witt H, et al. Mutation in the SPINK1 trypsin inhibitor gene, alcohol use, and chronic pancreatitis. JAMA. 2001;285:2716–7. doi: 10.1001/jama.285.21.2716-a. [DOI] [PubMed] [Google Scholar]
- 35.Kirby A, et al. Mutations causing medullary cystic kidney disease type 1 lie in a large VNTR in MUC1 missed by massively parallel sequencing. Nat Genet. 2013;45:299–303. doi: 10.1038/ng.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scholz M, Hasenclever D. Comparison of estimators for measures of linkage disequilibrium. Int J Biostat. 2010;6 doi: 10.2202/1557-4679.1162. Article 1. [DOI] [PubMed] [Google Scholar]
- 37.Sannerud R, Marie M, Hansen BB, Saraste J. Use of polarized PC12 cells to monitor protein localization in the early biosynthetic pathway. Methods Mol Biol. 2008;457:253–65. doi: 10.1007/978-1-59745-261-8_19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.