Abstract
Synthetic recoding of genomes, to remove targeted sense codons, may facilitate the encoded cellular synthesis of unnatural polymers by orthogonal translation systems. However, our limited understanding of allowed synonymous codon substitutions and the absence of methods that enable the stepwise replacement of the E. coli genome with long synthetic DNA, and provide feedback on allowed and disallowed design features in synthetic genomes, have restricted progress on this goal. Here we endow E. coli with a system for efficient, programmable replacement of genomic DNA with long (~100 kb) synthetic DNA, through the in vivo excision of double stranded DNA from an episomal replicon by CRISPR/Cas9, coupled to lambda red mediated recombination and simultaneous positive and negative selection. We iterate the approach, providing a basis for stepwise whole-genome replacement. We attempt systematic recoding in an essential operon using eight synonymous recoding schemes. Each scheme systematically replaces target codons with defined synonyms and is compatible with codon reassignment. Our results define allowed and disallowed synonymous recoding schemes, and enable the identification and repair of recoding at idiosyncratic positions in the genome.
The design and synthesis of genomes provides a powerful approach for understanding and engineering biology1–6. Genome synthesis has the potential to elucidate synonymous codon function7, accelerate metabolic engineering8, and facilitate genetically encoded unnatural polymer synthesis9,10.
Methods that i) replace the genome in sections6, ii) provide feedback on precisely where a given design fails and on how to repair it, and that iii) can be rapidly iterated for whole genome replacement, would accelerate our ability to understand and manipulate the information encoded in genomes.
In E. coli, the workhorse of synthetic biology, progress on replacing large sections of the genome has been slower than in naturally recombinogenic organisms6,11. Sequence specific recombinases, may be introduced into E. coli to direct recombination at defined target sequences, that must be introduced into the genome in advance, and these approaches cannot been iterated12. Lambda red mediated homologous recombination13, using linear ds DNA that is electroporated in to cells, can be programmed to target any region of the genome via short (50 bp) homology regions (HRs) at either end of a linear double stranded (ds) DNA (referred to herein as HR1 and HR2). However, this approach is commonly limited to inserting or replacing only 2-3 kb of genomic DNA, and has not been used to introduce long sequences into the genome.
We are interested in reprogramming the genetic code for the in vivo biosynthesis of unnatural polymers9. Reassigning particular codons in the genome to synonymous codons would enable removal of their cognate tRNAs, compression of the number of synonymous codons used to encode certain natural amino acids, and the reassignment of certain sense codons, and an expanded set of quadruplet codons14,15, to evolved orthogonal translation systems for unnatural polymer synthesis. However, recoding the E. coli genome requires the development of i) methods for efficiently replacing genomic DNA with synthetic DNA and ii) an understanding of the best synonymous codon substitutions, from many possible choices, for recoding.
Nature chooses one triplet codon from up to six potential synonyms to encode each amino acid at each position in the genome; this choice can define transcriptional16 or translational17 regulatory elements, translation speed18,19, mRNA folding7, gene expression, co-translational folding20,21, protein production levels7, and is likely to have further undiscovered roles. Synonymous codons may have distinct roles at different sites in the genome, and there may be epistatic interactions amongst codons within and between genes22–24. Our limited understanding of the factors driving codon choice suggests that the best synonymous codon substitutions to implement for synthetic recoding should be determined empirically.
Here we endow E. coli with a system that enables efficient, programmable, one step introduction of long synthetic DNA into the genome, as insertions or replacements, and iterate the approach for stepwise replacement of longer genomic regions. Using our approach we investigate different synonymous recoding schemes for replacing the same target codons with distinct sets of synonyms, in an operon rich in both target codons and essential genes, providing insight into allowed and disallowed schemes for genome recoding and synonymous codon compression.
Inserting DNA into the genome by REXER
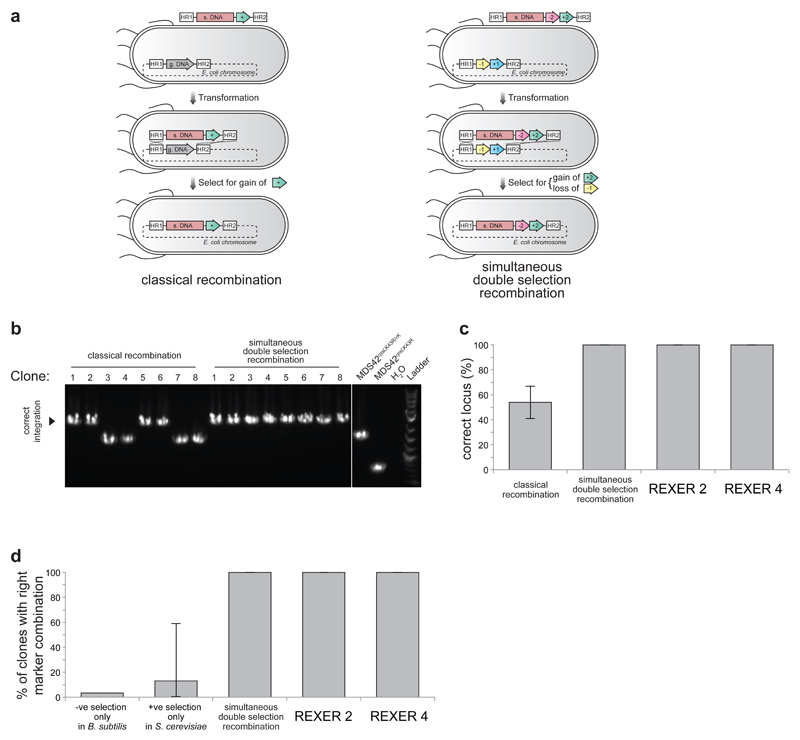
The overall efficiency of lambda red mediated recombination protocols is the product of the transformation efficiency for linear double stranded (ds) DNA and the efficiency with which the linear dsDNA mediates intracellular recombination. The overall efficiency decreases drastically with dsDNA length and we hypothesized that this results primarily from challenges in efficiently delivering intact dsDNA into cells. To address this challenge, we envisioned introducing the DNA of interest into E. coli in an episomal replicon, and excising the dsDNA of interest to facilitate lambda red mediated recombination. To select for the correct integrants we envisioned employing simultaneous positive and negative selections, to select for the integration of a positive selection marker from the replicon into the genome and the loss of a negative selection marker from the genomic locus targeted for replacement; such a double selection strategy substantially enhances integration at the target locus by lambda red mediated recombination (Extended Data Figure 1).
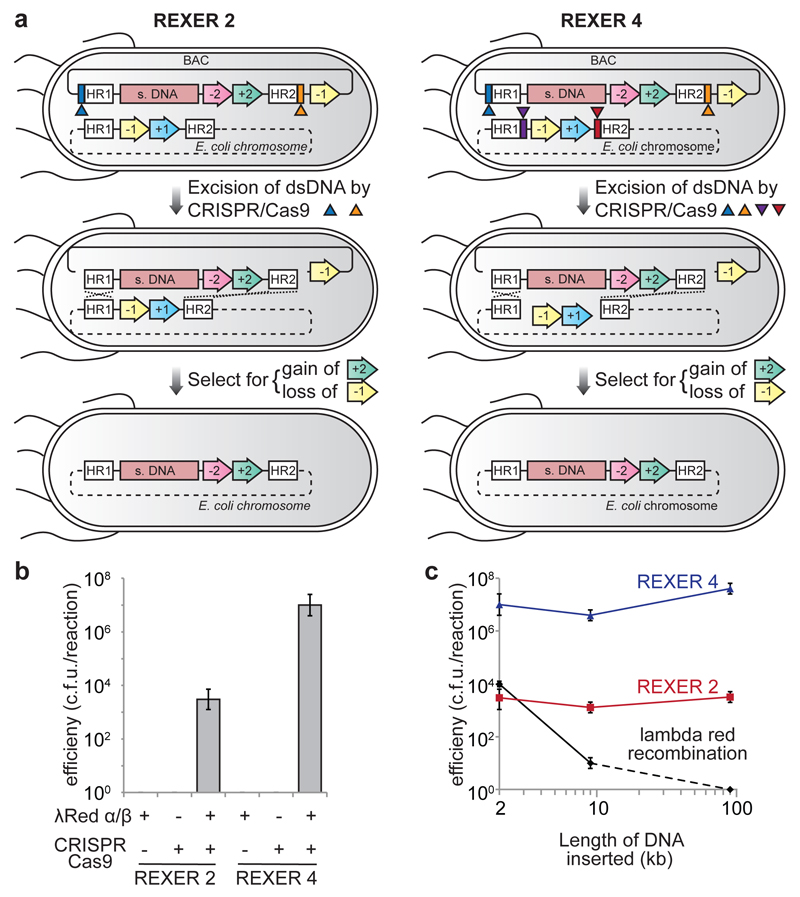
We created E. coli MDS42 rpsLK43R/rK (a genome minimized strain of E. coli25 in which the genomic copy of rpsL contains a K43R mutation conferring resistance to streptomycin, and the -1/+1 selection cassette encoding an rpsL-KanR fusion inserted between cra and mraZ) containing a bacterial artificial chromosome (BAC) in which the -2/+2 cassette (encoding sacB-CmR) is flanked by HR1 and HR2 sequences and Cas9 target sites (containing protospacer-PAM sequences) and expressing lambda red (alpha/beta/gamma)13, Cas926, and tracrRNA26 (Figure 1a). Addition of a plasmid, encoding spacer RNAs targeting the protospacers26 within the BAC target sites to these cells, and selection for the gain of resistance to both chloramphenicol (gain of +2) and streptomycin (loss of -1 from the genome, and loss of the backbone of the BAC) led to replacement of the sequence between HR1 and HR2 in the genome, with the sequence between HR1 and HR2 from the BAC (Figure 1a, Extended Data Figure 2).
Figure 1.
Efficient, programmable insertion of very long synthetic DNA (s. DNA) into the genome of E. coli. a. REXER 2 and REXER 4. CRISPR protospacer sequences are blue and orange rectangles respectively. Triangles indicate spacer RNAs that program cleavage within colour matched protospacers.. REXER 4 augments REXER 2 by adding two extra protospacers (purple and red rectangles), and triggering cleavage with four spacer RNAs. +1 is KanR, -1 is rpsL, +2 is CmR, -2 is sacB. b. REXER 2 and REXER 4 are dependent on the CRISPR/Cas9 system and recombination. Controls omit either spacer RNA or lambda red beta. Data show mean (n=4-6, ± s.d.). c. The efficiency of REXER 2 and REXER 4 is constant for insertions between 2 kb and 90 kb. C.f.u, colony forming units (c.f.u.). The data show the mean (n=6, 3 biological replicates performed in duplicate, ± s.d.) for 2 kb insertion, and the data for 9 kb and 90 kb insertions (n=4, 2 biological replicates performed in duplicate, ± s.d.). It was not possible to obtain a 90 kb linear dsDNA product in vitro for classical lambda red recombination, and our data reflect this, rather than the efficiency of recombination per se. It is well established that lambda red recombination efficiency falls off rapidly with linear dsDNA length.
Genomic replacement was strictly dependent on CRISPR/Cas9, and the lambda red recombination machinery (Figure 1b), and targeted to the desired genomic locus (Extended Data Figure 1c,d); consistent with the CRISPR/Cas9 mediated excision of the dsDNA between HR1 and HR2 in the BAC, and lambda red mediated integration of this sequence between HR1 and HR2 in the genome. We named our approach REXER 2 (replicon excision for enhanced genome engineering through programmed recombination; 2 indicates the number of CRISPR/Cas9 cuts).
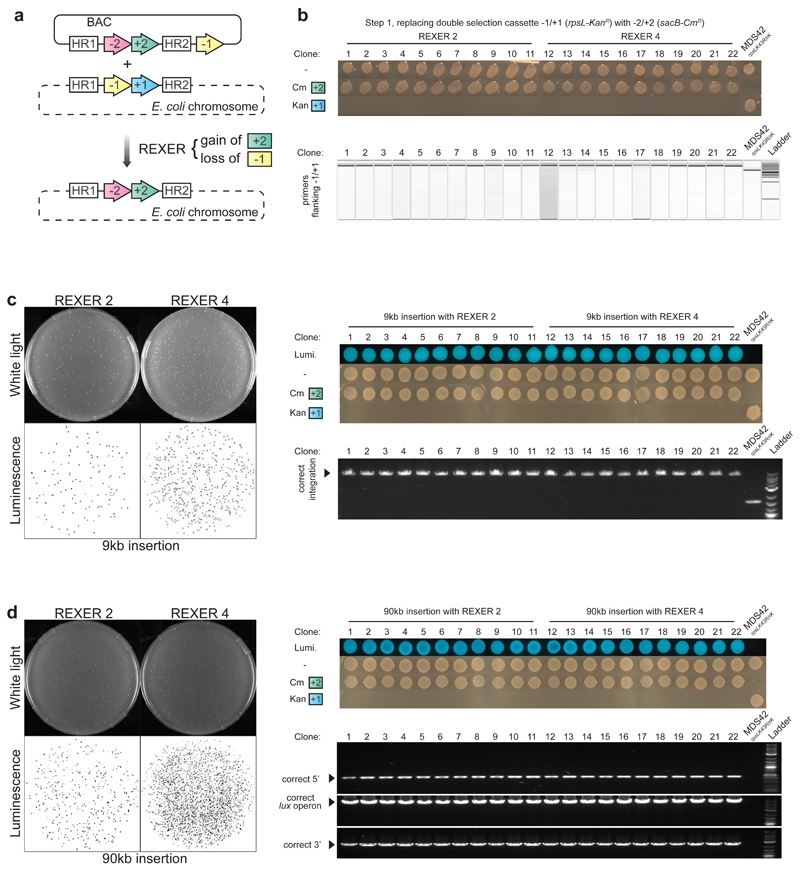
To investigate the dependence of REXER 2 on the length of DNA inserted into the genome, we created BACs with 9 kb or 90 kb of DNA inserted between HR1 and -2/+2 (Figure 1c, Extended Data Figure 2). The insertions contain a luxABCDE operon27, which is sufficient to generate bioluminescence in E. coli. We transformed each BAC into E. coli MDS42 rpsLK43R/rK and implemented the REXER 2 protocol. All cells selected on chloramphenicol and streptomycin integrated the lux operon at the correct locus and were bioluminescent (Extended Data Figure 2). Moreover, while the efficiency of classical recombination, drops dramatically from 104 colony forming units (c.f.u.) for a 2 kb insertion to approximately 10 c.f.u.s for a 9 kb insertion, the efficiency of REXER 2 is constant, at 104 c.f.u. for all insertions (Figure 1c).
Next we asked whether the efficiency of REXER could be improved by making two additional CRISPR Cas9 mediated double strand breaks in the genome between HR1 and HR2 (Figure 1a). The resulting REXER 4 protocol led to replacement of the sequence between HR1 and HR2 in the genome, with the sequence between HR1 and HR2 from the BAC (Extended Data Figure 2), and destruction of all four Cas9 target sites. REXER 4 was strictly dependent on both the CRISPR Cas9 system and the lambda red recombination machinery (Figure 1b), and led to integration at the correct locus (Extended Data Figure 2). Like REXER 2, the efficiency of REXER 4 was length independent for insertions tested (up to 90kb), and REXER 4 further increases the efficiency of synthetic DNA insertion with respect to REXER 2 by 103-fold, while maintaining insertion at the correct locus (Figure 1c and Extended Data Figure 1c,d).
Replacing genome sections by REXER
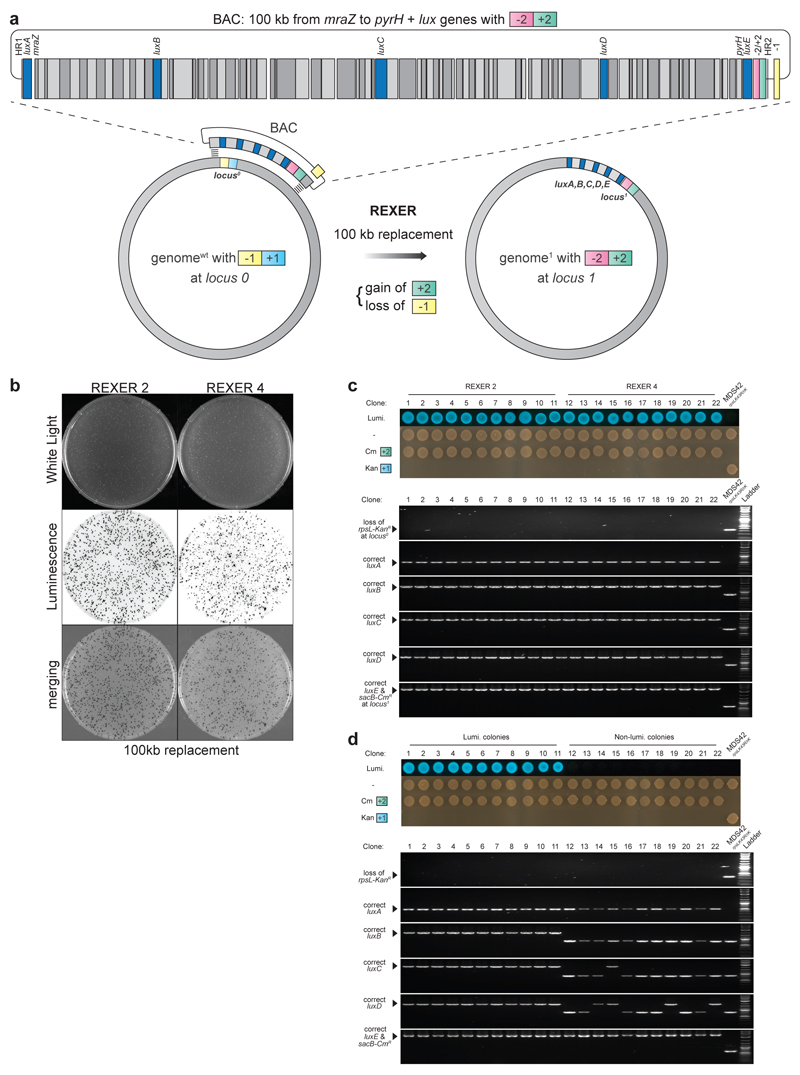
Next, we demonstrated that REXER can be used to efficiently replace 100 kb of the E. coli genome in a single step. We targeted the region from mraZ to pyrH for replacement and inserted the -1/+1 selection cassette at the 5’ end of this region. We assembled a BAC, from DNA fragments in S. cerevisiae28, in which the 100 kb region between mraZ and pyrH is watermarked along its length by genes from the lux operon (Extended Data Figure 3). REXER 2 yielded 2 × 104 c.f.u. per reaction, of which 80% were bioluminescent, while REXER 4 yielded 5 × 106 c.f.u. per reaction, of which 50% were bioluminescent (Extended Data Figure 3). Further characterization confirmed the integration of the lux watermarks at the expected loci for all bioluminescent colonies (Extended Data Figure 3). These results demonstrate that REXER enables the replacement of genomic regions with synthetic DNA.
Iterative REXER for genome replacement
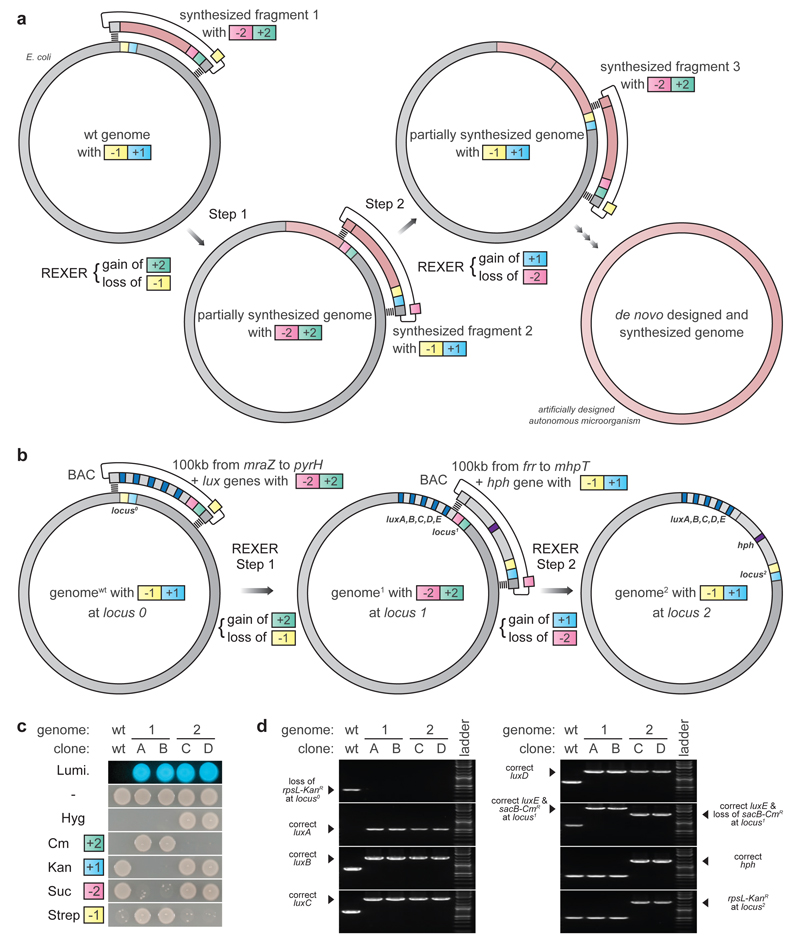
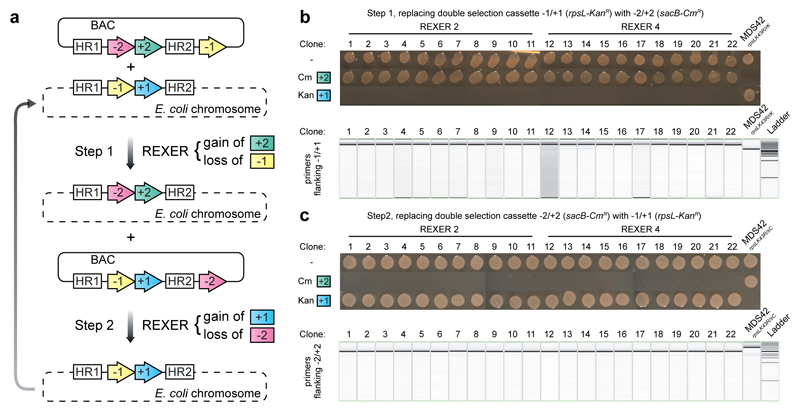
Iteratively replacing large sections of the genome with synthetic DNA via REXER will enable genome stepwise interchange synthesis (GENESIS) for whole genome replacement (Figure 2a). Towards this goal we demonstrated iterative REXER (Extended Data Figure 4). The genome created in a first round of REXER, that introduces -2/+2, provides a direct template for a second round of REXER, using a BAC that contains distinct positive and negative selection markers (-1/+1) (Extended Data Figure 4). This product is a template for third round of REXER; thus the approach can be iterated (Extended Data Figure 4).
Figure 2.
Iterating REXER for genome stepwise interchange synthesis (GENESIS). a. Iterative genomic replacement by REXER will enable genome replacement in less than 40 linear steps. b. Iterative REXER replaces 220 kb of the E. coli genome with 230 kb of synthetic DNA in two steps. LuxA, B, C, D, E (cyan rectangles) are necessary and sufficient for luminescence. hph (violet rectangle) is the hygromycin B phosphotransferase gene, conferring resistance to hygromycin B. c. Cells phenotype correctly through rounds of REXER. The parental cell line (genomewt), independent clones from the 1st round of REXER (clone A and B), and independent clones from the 2nd round of REXER (clone C and D), Lumi (luminescence), Cm (chloramphenicol), Kan (Kanamycin), Suc (Sucrose), Strep (Streptomycin). d. Cells genotype correctly through rounds of REXER. For gel source data, see Supplementary Figure 1.
To explicitly demonstrate the stepwise replacement of long regions of genomic DNA with synthetic DNA for GENESIS, we used cells in which we replaced the 100 kb genomic region between mraZ and pyrH by REXER (Extended Data Figure 3 & Figure 2b) for a second step of REXER. This second step introduced 124 kb of DNA spanning from frr to mhpT and the hygromycin B phosphotransferase gene (hph). We confirmed the replacement of 220 kb of the genome with 230 kb of synthetic DNA in two steps (Figure 2c,d). This compares favourably with the largest replacement in the naturally recombinogenic S. cerevisiae (270 kb, 11 steps)6.
Testing synonymous recoding schemes
We used REXER to define synonymous substitutions that are disallowed and poorly tolerated and synonymous substitutions that are allowed and can be implemented at many positions in the genome. To define a system for experimental investigation we identified i) the codons to target for removal, ii) the codons with which the target codons might be replaced to define recoding rules and iii) a region of the genome in which to test recoding rules.
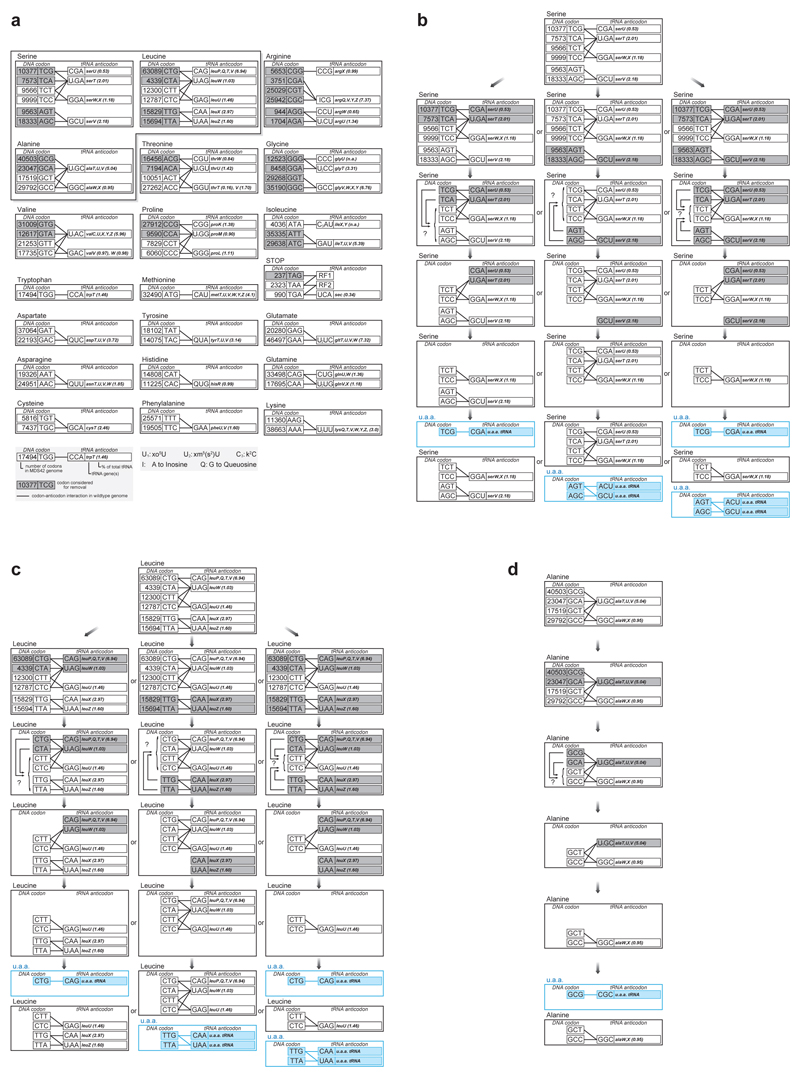
We chose target codons that i) when removed from the genome would enable the removal of all the tRNAs that decode them, and where ii) removal of these tRNAs would not remove all decoding of the remaining synonymous codons in the genome; these are the minimum criteria for removing a sense codon from the genome to enable its unambiguous reassignment (Extended Data Figure 5). We focused on removing serine, leucine and alanine codons that fulfill these criteria, as these are the three codon sets for which the aminoacyl-tRNA synthetases do not recognize the anticodon of their cognate tRNAs29. This will facilitate reassignment of the target codon (or upto four quadruplet derivatives), following deletion of host tRNAs, to orthogonal synthetase/tRNA pairs in orthogonal translation14.
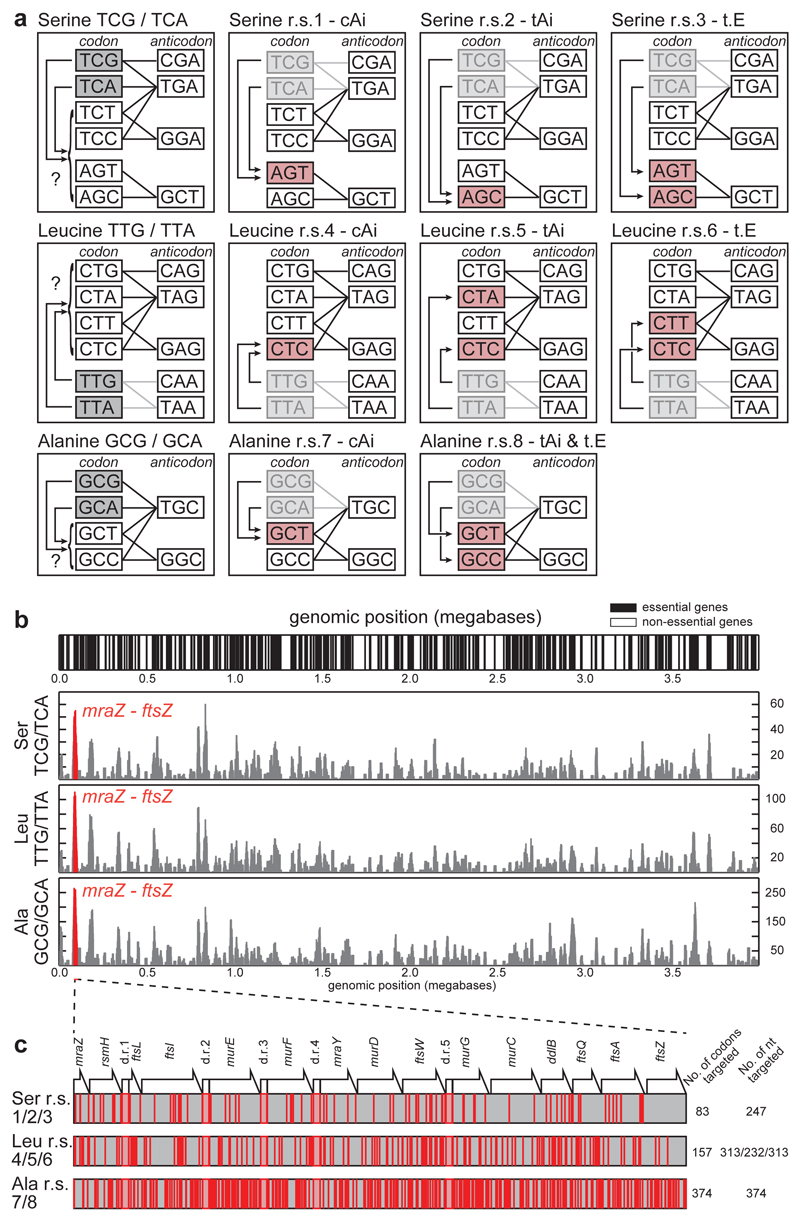
We defined candidate synonymous replacements for the target codons by identifying the closest match for the target codons, as judged by either codon adaptation index (cAi)30, tRNA adaptation index (tAi)31,32, or a third metric we define (translation efficiency, t.E) (Extended Data Table 1, Methods). These considerations led us to investigate eight recoding schemes (Figure 3a).
Figure 3.
Systematic and defined synonymous codon reassignment in an E. coli operon rich in essential genes. a. Identifying codons target for removal (grey) and the synonyms to which they are reassigned (pink) in each recoding scheme. Lines indicate codon-anticodon interactions. Replacements were chosen by cAi, tAi, or t.E. Application of each recoding scheme genome-wide would allow the targeted codons to be completely removed from the E. coli genome and, following deletion of the cognate tRNA genes, codon reassignment to orthogonal translation systems for unnatural polymer synthesis. b. Identifying a target operon rich in target codons and essential genes to test recoding schemes. The top panel indicates the positions of essential genes. In the bottom three panels the y axis scores the number of the indicated target codons in essential genes at the genomic position indicated on the x axis. The mraZ to ftsZ region (coloured in red) was identified in the highest scoring 20 kb region across the E. coli MDS42 genome for all targeted codons. c. Position and density of targeted codons in the mraZ to ftsZ region. The positions of targeted codons (the indicated sense codons plus TAG to TAA) are coloured in red and pink regions with red outlines indicate duplicated regions (d.r.s) which refactor2 overlapping open reading frames to enable independent recoding of the downstream open reading frames.
We identified the E. coli cell division operon as an ideal target to test these synonymous recoding schemes because it i) is rich in essential genes (12 out of 15 genes in the region are essential)33, ii) contains genes expressed at a range of levels34, iii) includes membrane proteins34 (a class of proteins for which co-translational folding is known to be affected by synonymous codon choice), iv) includes several proteins that interact and for which the ratios of proteins expressed are distinct and crucial35,36 (which will favour intergenic epistatic interactions amongst codons), and v) is rich in the target codons (Figure 3b, Extended Data Table 2). We anticipated that these features would ensure that the region captures important properties of the genome, and that the success and failure of synonymous recoding in the region, would be reflected in viability and growth.
Allowed and disallowed recoding schemes
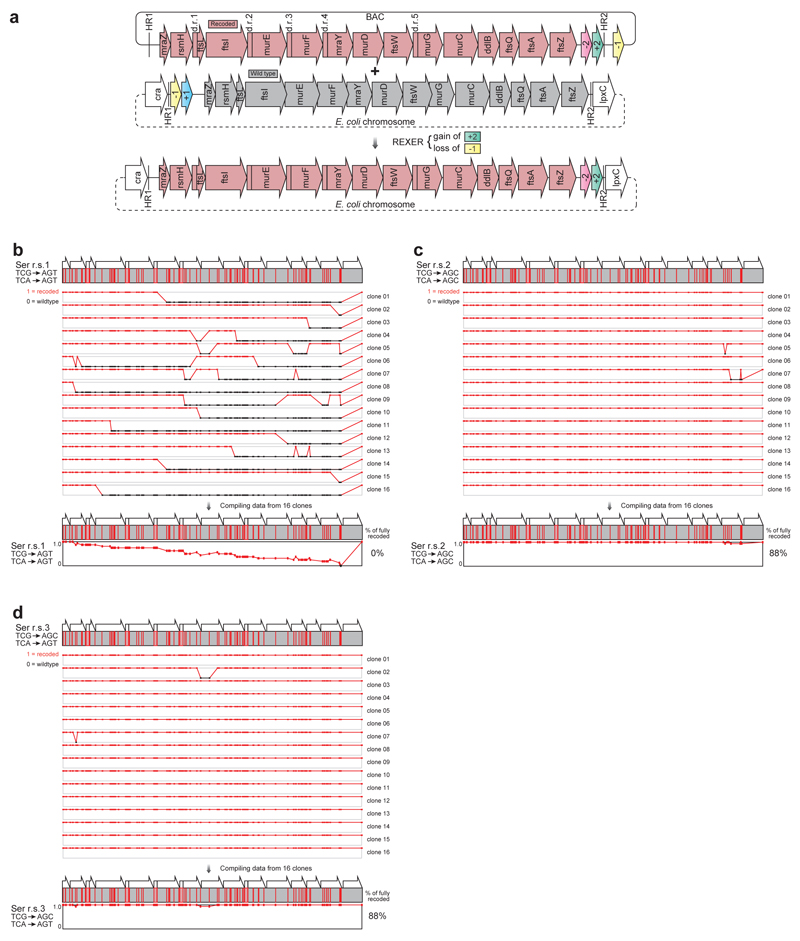
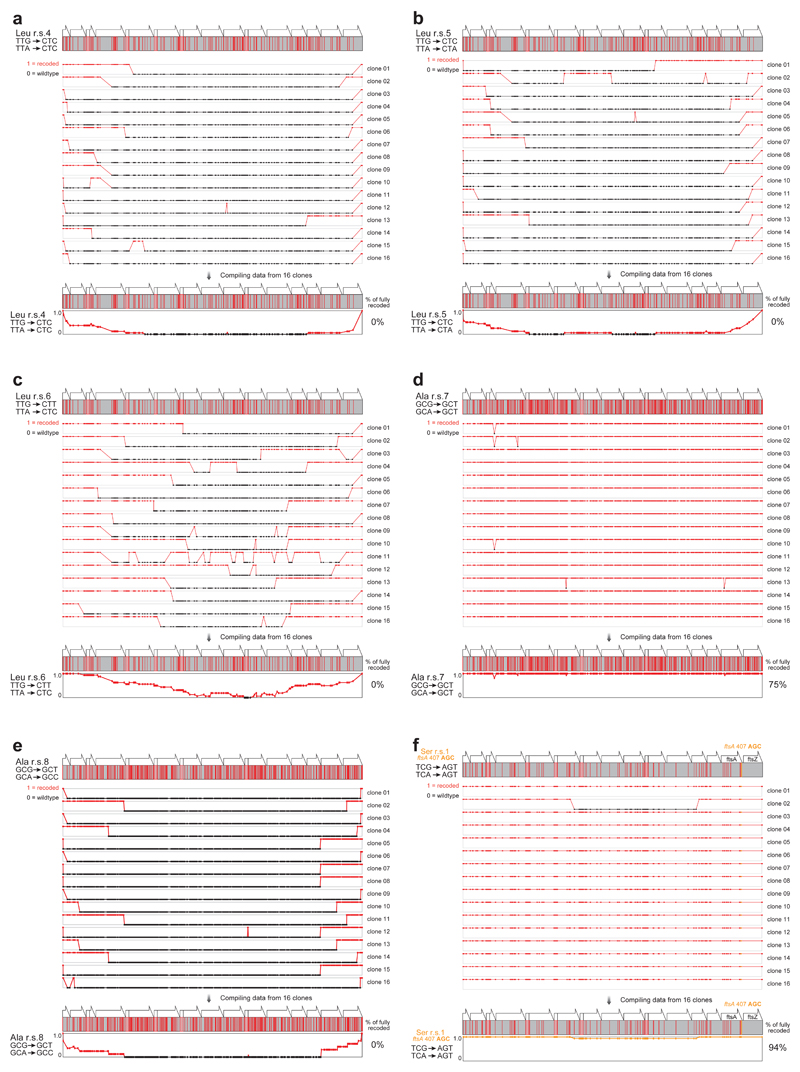
We designed DNA sequences in which each of the recoding schemes is implemented within all of the fifteen genes simultaneously. Overall, the schemes investigate the consequences of 1,468 codon changes and 2,347 nucleotide changes (Figure 3c). The DNA for each scheme was synthesized de novo, assembled into a BAC in S. cerevisiae and genomic recoding via REXER investigated (Extended Data Figure 6, 7).
Following REXER we sequenced 16 independent clones from each recoding scheme. We observed chimeras between the wild-type genomic DNA and the recoded DNA in several cases, consistent with recombination-mediated crossover between the recoded sequence and the starting genome, these chimeras defined a recoding landscape. We aligned the individual recoding landscapes to create a “compiled recoding landscape” (Extended Data Figure 6, 7) that reveals peaks and plateaus for synonymous substitutions that are allowed and valleys or troughs for synonymous substitutions that are consistently disallowed. We observe clear differences in the extent to which replacement of the same target codons by different synonymous codons are tolerated (Figure 4a,b,c and Extended Data Figure 6, 7).
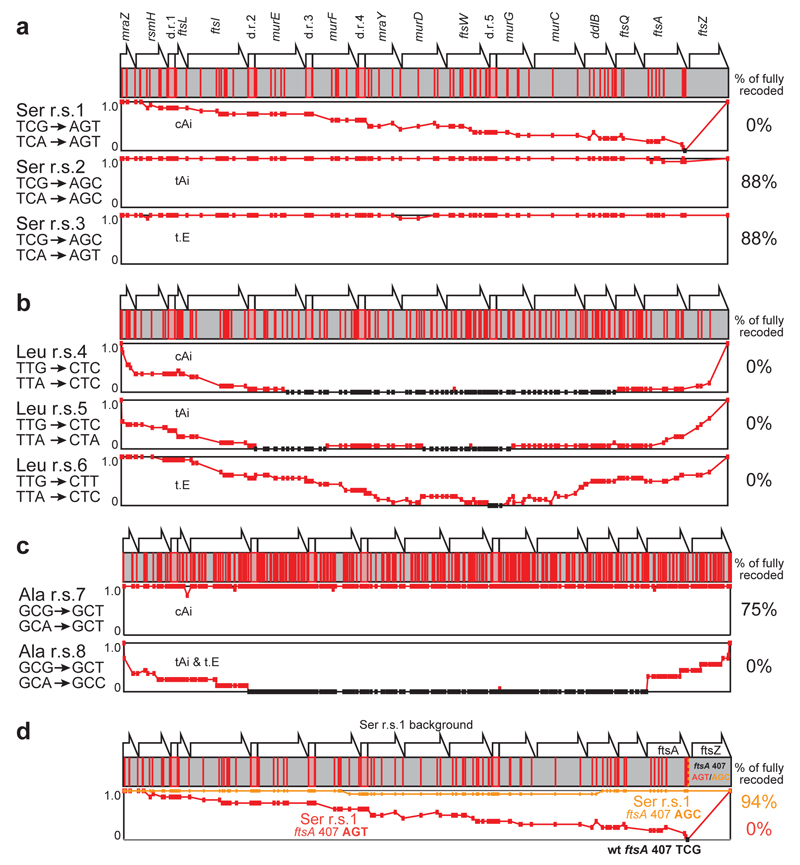
Figure 4.
Compiled recoding landscapes of targeted codons reveal allowed and disallowed synonymous recoding schemes, and enable the identification and repair of idiosyncratic positions in the genome. The fraction of recoding across sixteen independent sequences is indicated on the y axis of the graphs. Codons positions that are not recoded with the indicated scheme are in black. a., b., c. Complied recoding landscapes of targeted serine, leucine and alanine codons respectively. d. Identifying and fixing a deleterious sequence in defined and systematic synonymous recoding. The compiled recoding landscape of serine r.s.1, is plotted in red, revealing the single position at which the wild type sequence is maintained, codon 407 in ftsA. The compiled recoding landscape of serine r.s.1 with ftsA 407 AGT changed to AGC (as in serine r.s.2 and r.s.3) is plotted in orange. This mutation repairs the deleterious effect of ftsA 407 AGT without reintroducing the codons targeted for removal.
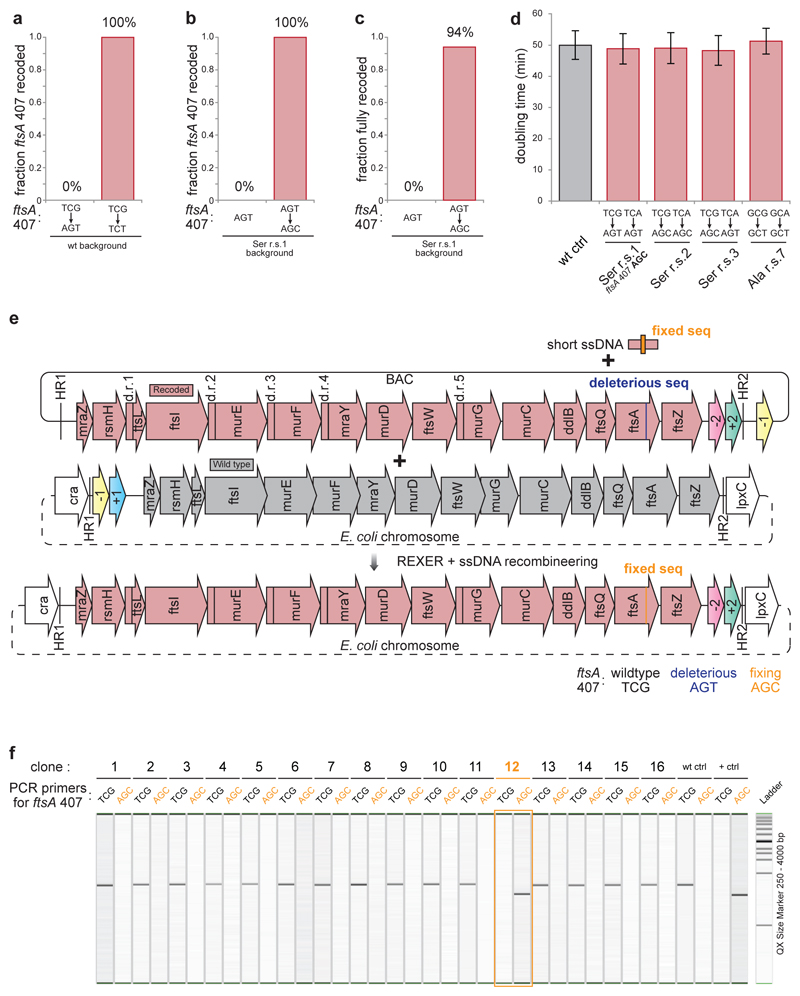
We first investigated the serine recoding schemes 1-3 (Figure 4a). For scheme 1, 0% of clones are completely recoded. In stark contrast, for scheme 2 and scheme 3 88% of clones were fully recoded. In contrast, we find that none of the leucine recoding schemes tested (schemes 4-6) led to complete recoding, and for scheme 4 and 5 recoding fails catastrophically, indicating that the synonymous substitutions have phenotypic consequences at many sites in the operon (Figure 4b). Finally, we find that the two alanine recoding schemes tested (schemes 7-8) have dramatically different outcomes (Figure 4c). Recoding scheme 7 leads to 75% of clones being completely recoded at all 374 positions while no clones are fully recoded by scheme 8. The doubling times for all fully-recoded clones were comparable to each other and to a control E. coli strain (Extended Data Figure 8). Overall, this work successfully removes up to 374 sense codons across 20 kb from an operon rich in essential genes in a single strain. Thus the scale of sense codon removal is much greater than previously reported work that investigated one gene at a time37.
Our data reveal the drastic differences between precisely defined recoding schemes that are obscured when the choice of synonymous substitution is randomized37. For serine recoding, scheme 2 and 3 recoding is allowed, while scheme 1 recoding is not; even though the codons used for replacement in scheme 1 and scheme 2 and 3 differ by only a single base (AGT vs AGC), and are decoded by the same tRNA (with anticodon GCT) via wobble and Watson-Crick decoding, respectively (Figure 3a). Similarly for alanine codons, scheme 7 recoding is allowed while scheme 8 fails catastrophically. These recoding schemes differ only in the conversion of a single base (GCT vs GCC) in the allowed and disallowed substitution for GCA. Again, both of the new codons are decoded by the same set of tRNAs (Figure 3a). cAI, tAi and tE all produce at least one successful recoding, but no single metric predicts which synonymous recoding will be successful. These observations underscore the importance of empirically determining the best systematic and well-defined synonymous recoding scheme for each codon.
E. coli consistently rejects a single codon mutation (TCG to AGT ) at codon 407 of ftsA in recoding scheme 1 (Figure 4a). Attempts to introduce the ftsA 407 TCG to AGT mutation (without additional recoding at other positions in the genome) failed (Extended Data Figure 8). In contrast, we were able to quantitatively recode ftsA 407 TCG to the synonymous TCT codon (Extended Data Figure 8). These results demonstrate that the ftsA 407 TCG to AGT mutation is deleterious.
Mutation of the codon at position 407 in ftsA from AGT to AGC (the codon found at this position in recoding schemes 2 and 3) is sufficient to repair recoding scheme 1 (Figure 4d, Extended Data Figure 7, 8). This mutation dramatically alters REXER mediated recoding, increasing the fraction of fully recoded sequences from 0 % to 94 % and the fraction of recoding at codon 407 of ftsA from 0 % to 100 % (Extended Data Figure 8). We also successfully introduced this mutation into recoding scheme 1 by combining single stranded DNA recombineering with REXER (Extended Data Figure 8). The growth of E. coli was not affected by the successful recoding schemes (Extended Data Figure 8). These results demonstrate that the major defect in recoding scheme 1 results from AGT being disallowed at position 407 of ftsA, though it is tolerated at many other positions. Since TCG, TCT and AGC are allowed at position 407 of ftsA but AGT (which shares nucleotides with allowed codons at each position of the triplet) is disallowed, we conclude that the problem at this codon lies in the entire triplet. These experiments exemplify how REXER may be used to i) identify idiosyncratic positions in the genome that are refractory to recoding by otherwise well-tolerated recoding schemes, and ii) repair the recoding scheme by the introduction of alternative codons at these idiosyncratic positions.
Discussion
In conclusion, we have generated an efficient approach to enable both the programmed insertion of large synthetic DNA sequences into the E. coli genome and the replacement of sections of the E. coli genome with synthetic DNA. The approach can be iterated, and will enable replacement of the entire E. coli genome with synthetic DNA in no more than 40 linear steps (Figure 2a). Each step takes only a few days to implement and convergent syntheses may further accelerate complete genome synthesis. The strategy will enable radical, high-density changes to the genome not accessible through site directed mutagenesis approaches10,38, and enable diverse applications including recoding and metabolic engineering. We anticipate the approach may be extended to facilitate genome engineering in other organisms.
We have simultaneously recoded several genes in an essential operon using eight well-defined synonymous recoding rules that are compatible with codon reassignment for unnatural polymer synthesis. Our results reveal dramatic differences in the extent to which different synonymous replacements for the same target codons are allowed. Our approach also enables both the identification and repair of idiosyncratic positions within the 'recoding landscape' where a precise codon substitution that is allowed at many other positions in the operon is disallowed. Our investigation empirically defines precisely defined schemes for sense codon removal and synonymous replacement for genome-wide application.
Methods
Sequences
Sequences of plasmids, BACs, and modified genomic loci are provided in Supplementary data 1–5.
Construction of selection cassettes, cell strains, and plasmids
Two double selection cassettes were constructed. The -1/+1 is a fusion between the negative selection marker rpsL (-1) encoding the essential ribosomal protein S12 and conferring sensitivity to streptomycin in rpsLK43R genomic background, and the positive selection marker KanR (+1) encoding the kanamycin resistance gene neomycin phosphotransferase II. The rpsL-KanR cassette was expressed as two separate proteins from a single mRNA driven by constitutive transcription from wildtype rpsL promoter. The -2/+2 is a fusion between the negative selection marker sacB (-2) conferring sensitivity to sucrose, and the positive selection marker CmR (+2) encoding the chloramphenicol resistance gene chloramphenicol acetyl transferase. The sacB-CmR cassette was expressed as two separate proteins from a single mRNA driven by constitutive transcription from EM7 promoter. Both selection cassettes were synthesized de novo.
The minimum genome E. coli strain MDS42 was used as the starting strain25. A K43R mutation was introduced into the rpsL gene to create MDS42rpsLK43R to confer resistance to streptomycin in the absence of additional wildtype copy of rpsL, and sensitivity to streptomycin in the presence of any additional copy of wildtype rpsL. The -1/+1 cassette rpsL-KanR was inserted between 89,061 and 89,587 in the MDS42rpsLK43R genome to create MDS42rpsLK43R/rK.
pKW20_CDFtet_pAraRedCas9_tracrRNA was constructed by assembling multiple PCR fragments using Gibson Assembly. The plasmid backbone and replication origin is from pCDFDuet-1 plasmid (Addgene), in which the spectinomycin resistance marker is replaced with a tetracycline resistance marker from pBR322 plasmid (New England BioLab). The araC gene, the arabinose promoter (pAra), and the lambda red (alpha/beta/gamma) genes were PCR amplified from pRed/ET plasmid (GeneBridges). The open reading frame of Cas9 was PCR amplified from pCas9 plasmid26 and placed downstream of the lambda red alpha. The tracrRNA with its endogenous promoter was PCR amplified from pCas9 plasmid26, and placed in the same orientation downstream of the araC gene. The pKW20_CDFtet_pAraRed(Δβ)Cas9_tracrRNA was derived from pKW20_CDFtet_pAraRedCas9_tracrRNA by inserting GTAC between the 314th and 315th nucleotide of lambda red beta open reading frame, which leads to translational frame shifting and thus inactivation of lambda red beta.
pKW21_MB1amp_Spacer0 was constructed by assembling two PCR fragments using Gibson Assembly Master Mix (from New England BioLab). The pMB1 replication origin and ampicillin resistance marker were PCR amplified from pBR322 plasmid (from New England BioLab). The CRISPR array with no functional spacer RNA (hence the nomenclature 0) between BamHI and EcoRI was PCR amplified from pCRISPR26. pKW21_MB1amp_Spacer0 was verified by sequencing. CRISPR arrays with two or four different spacer RNA sequences for directing REXER 2 or REXER 4 respectively with interspaced direct repeats were commercially synthesized. The arrays were cloned into pKW21_MB1amp_Spacer0, replacing the empty CRISPR array to create different pKW21_MB1amp_Spacers×2 or pKW21_MB1amp_Spacers×4 plasmids. The final pKW21_MB1amp_Spacers plasmids were sequence verified. A related version of pKW21_MB1erm_Spacers plasmids was prepared replacing the ampicillin resistance marker in pKW21_MB1amp_Spacers with an erythromycin resistance marker.
The BAC holding the synthetic DNA was constructed by assembling multiple fragments. The BAC backbone is based on pBeloBAC11 (New England BioLabs) from nucleotide 1542 to 7041 with the addition of the double selection cassette -2/+2 and the negative selection marker -1, and assembled using Gibson Assembly Master Mix (New England BioLabs). An alternative arrangement utilises -1/+1 coupled with -2. The synthetic DNA was always flanked by AvrII sites, which also function as PAM and part of protospacer for CRISPR/Cas9.
Assembling short synthetic DNA onto BAC using Gibson Assembly
The pBAC_HR(89,061)-sC-HR(89,587)_r was constructed by assembling three PCR fragments using Gibson Assembly: the first fragment being the 2.2 kb long -2/+2 sacB-CmR cassette flanked with HR1 (89,012 - 89,061, all numbering is from the MG1655 reference sequence) and HR2 (89,587 – 89,636) and further flanked with two AvrII sites, the second fragment being the -1 rpsL gene with rrnC terminator, and the third fragment being the pBeloBAC11 backbone from nucleotide 1542 to 7041. The assembled pBAC_HR(89,061)-sC-HR(89,587)_r was selected on LB agar plates with 18μg/ml chloramphenicol and sequence verified. The pBAC_HR(89,061)-rK-HR(89,587)_s was similarly constructed using -1/+1 rpsL-KanR cassette flanked with HR1(89,012 - 89,061) and HR2(89,587 – 89,636) and further flanked with two AvrII sites, -2 sacB gene with rrnC terminator, and the pBeloBAC11 backbone. The pBAC_HR(89,061)-T5Lux-sC-HR(89,587)_r was constructed by inserting a PCR product of an artificial lux operon between the HR1 and the -2/+2 sacB-CmR cassette in the pBAC_HR(89,061)-sC-HR(89,587)_r.
Assembling long synthetic DNA onto BAC using recombination in S. cerevisiae
Long synthetic DNA fragments (≥ 20 kb) were assembled in S. cerevisiae. The pBeloBAC11 backbone was converted into a BAC/YAC shuttle vector by introducing a S. cerevisiae replication centromere CEN and URA3 selection marker (from S. cerevisiae vector pRS316, Addgene). The BAC/YAC shuttle vector holding long synthetic DNA was assembled from 5-16 DNA fragments in S. cerevisiae28.
Classical recombination and simultaneous double selection recombination protocol
The sacB-CmR cassette was PCR amplified using primers containing HR1 and HR2. In classical recombination, 3 μg of this purified PCR product was transformed into 100 μl of electro-competent MDS42rpsLK43R/rK cells, which are pre-transformed with the pRed/ET plasmid and induced to express the λ Red components. The cells were recovered in 4 ml SOB media for 1 hour at 37°C and then diluted to 100 ml LB and incubated for 4 hours at 37°C with shaking. The culture was then spun down and re-suspended in 4 ml of LB and spread in serial dilutions on selection plates of LB agar with 18μg/ml chloramphenicol.
In simultaneous double selection recombination (DOSER), 3 μg of the same PCR product was transformed into 100 μl of electro-competent MDS42rpsLK43R/rK cells following the same transformation and recovery protocol as above. The culture was then spun down and re-suspended in 4 ml LB and spread in serial dilutions on selection plates of LB agar with 18 μg/ml chloramphenicol and 50 μg/ml streptomycin.
Multiple colonies from classical recombination and DOSER were picked for phenotyping. Colony-PCRs of multiple clones from classical recombination and DOSER were performed using primer pair flanking the genomic locus 89,061 to 89,587 with MDS42rpsLK43R/rK, MDS42rpsLK43R, and Milli-Q filtered water with no resuspended colony as controls. All PCR products were run in parallel to NEB 2-Log DNA Ladder (from New England BioLab, and sequence verified by Sanger sequencing.
REXER protocol
MDS42rpsLK43R/rK cells were double transformed with pKW20_CDFtet_pAraRedCas9_tracrRNA and pBAC_HR(89,061)-sC-HR(89,587)_r and plated on LB agar plates supplemented with 2% glucose, 10 μg/ml tetracycline and 18 μg/ml chloramphenicol. Individual colonies were inoculated into LB media with 10 μg/ml tetracycline and 18 μg/ml chloramphenicol, and grown overnight at 37°C with shaking. The overnight culture was diluted in LB media with 10 μg/ml tetracycline and 18 μg/ml chloramphenicol to OD600 = 0.05 and grown at 37°C with shaking for around 3 hours until OD600 ≈ 0.3. Arabinose powder was added to the culture to reach a final concentration of 0.5% and the culture was incubated for one additional hour at 37°C with shaking. The cells were harvested at OD600 ≈ 0.6, and made electro-competent in 1/500th of the culture volume.
3 μg pKW21_MB1amp_Spacers×2 or pKW21_MB1amp_Spacers×4 plasmid was electroporated into 100 μl of competent cells. The cells were recovered in 4 ml SOB media for 1 hour at 37°C and then diluted to 100 ml LB supplemented with 50 μg/ml ampicillin and 10 μg/ml tetracycline and incubated for 4 hours at 37°C with shaking. The culture was spun down and re-suspended in 4 ml LB and spread in serial dilutions on selection plates of LB agar with 18 μg/ml chloramphenicol and 50 μg/ml streptomycin. The plates were incubated at 37°C overnight, and the efficiency was calculated by counting colonies. Multiple colonies were picked, resuspended in Milli-Q filtered water, and arrayed on LB agar plates or LB agar plates supplemented with 18 μg/ml chloramphenicol, or supplemented with 50 μg/ml kanamycin. Colony-PCR was also performed from resuspended colonies using the primer pair flanking the genomic locus 89,061 to 89,587.
The resulting colonies with the -2/+2 sacB-CmR cassette replacing the -1/+1 rpsL-KanR cassette at the genomic locus 89,062 to 89,586 were incubated in LB without ampicillin, to lose the pKW21_MB1amp_Spacers×2 or pKW21_MB1amp_Spacers×4 plasmid. The resulting cells were double transformed with pKW20_pCDFtet_pAraRedCas9_tracrRNA and pBAC_HR(89,061)-rK-HR(89,587)_s. An individual colony was picked, inoculated into LB, and prepared into electro-competent cell as previously described. 3μg pKW21_MB1erm_Spacers×2 or pKW21_MB1erm_Spacers×4 plasmid was electroporated into the pre-induced cell. The cells were recovered in 4 ml SOB media for 1 hour at 37°C and then diluted to 100 ml LB supplemented with 25 μg/ml erythromycin and 5 μg/ml tetracycline and incubated for 4 hours at 37°C with shaking. The culture was spun down and re-suspended in 4 ml LB and spread in serial dilutions on selection plates of LB agar with 3% sucrose and 25 μg/ml kanamycin. After incubating the selection plate at 37°C overnight, multiple colonies were picked, resuspended in Milli-Q filtered water, and arrayed on LB agar plates, or LB agar plates supplemented with 18 μg/ml chloramphenicol or 50 μg/ml kanamycin. Colony-PCR was performed from resuspended colonies using the primer pair flanking the genomic locus 89,061 to 89,587.
The pBAC_HR(89,061)-T5Lux-sC-HR(89,587)_r, pBAC_HR(89,061)-90kb/Lux-sC-HR(89,587)_r, pBAC_HR(89,061)-100kb/Lux-sC-HR(192,744)_r, and pBAC_HR(89,061)-20kb-sC-HR(106,508)_r with matching pKW21_MB1amp_Spacers×2 or pKW21_MB1amp_Spacers×4 plasmids were used in the other REXER experiments following the same protocol. Colony PCR of the lux operon and the coupled -2/+2 sacB-CmR cassette inserted at the genomic locus 89,061 to 89,587 using the primer pair flanking the genomic locus generated a 9 kb band for successful insertion and 1.5 kb for the MDS42rK control. Primer pairs flanking the 5’ or 3’ end of the inserted/replaced DNA were used, which generates a PCR band for correct insertion/replacement, and no band or band of the wrong size with MDS42rK control. Colony PCR using primers for the internal watermarks were also performed. The 20 kb recoded region (from 89,062 to 106,507) was PCR amplified from purified genomic DNA (QIAGEN DNeasy Blood & Tissue Kit) using primer pair flanking the whole region. The 20 kb PCR product was purified using Bio-Rad PCR Kleen Columns and fully sequenced by Sanger sequencing.
Choice of region for systematic and defined synonymous recoding
We applied a sliding window approach, in which we counted the number of target codons within all essential genes within a 10 kb region of the MDS42 genome. Starting from the first 10 kb of the genome sequence, we iteratively shifted the window by 100 nt and performed the codon analysis until the end of the MDS42 genome sequence. Gene essentiality was defined by transposon insertion densities from a genome-scale genetic footprinting study in E. coli33, which led to comparable results as when we used the KEIO collection data39.
Choice of recoding rules
We characterised all serine, leucine, and alanine codons using the codon adaptation index (cAi)30 and the tRNA adaptation index (tAi)31,32. In case of cAi, we used the relative adaptiveness of each codon i (expressed as cAiwi) as a metric. In case of tAi, we used the relative adaptiveness value of each codon i (expressed as tAiwi) in Table S231,32. We defined ideal synonymous substitutions for targeted codons by minimising the difference in cAiwi and tAiwi. Comparing cAiwi and tAiwi for all codons, we noticed that the two metrices do not correlate well (Pearson’s R2 = 0.24) and decided to propose a third metric. In particular, we assumed that translation efficiency increases proportionally with increasing isoacceptor tRNA concentration and decreases proportionally with increasing numbers of competing codons that are translated by the same isoacceptor tRNA. On this basis we defined the translation efficiency (t.E) of codon i as follows:
where codon i is translated by tRNAs j, kij denotes the interaction strength between codon i and tRNA j, m denotes each codon translated by tRNA j, and kmj denotes the interaction strength between codon m and tRNA j. The interaction strengths were defined in five groups: i) "cognate" for codons that are reverse complements to the respective tRNA anticodon as well as AUAIle-k2CAUtRNA, ii) "G-U / U-G wobble" for codons where a third position G or U interacts with a (modified) tRNA U or G, respectively, iii) "C/U-xo5U" for codons where a third position C or U interacts with an xo5-modified uridine in the tRNA anticodon, iv) "C/U-inosine" where a third position C or U in the codon interacts with an inosine in the tRNA anticodon (an interaction shown to be 3-8-fold weaker than G-U wobbling40), and v) "A-inosine" for the reportedly weak interaction between the third position A in a codon with an inosine in the tRNA anticodon41. We obtained the tRNA concentrations [tRNAj] from reported measurements performed on E. coli cultures, expressed as a fraction of tRNA out of total tRNA in percent42. To determine the relative transcriptomic codon frequency q for each codon i we first calculated the codon's absolute transcriptomic frequency ri:
where gix is the frequency of codon i in gene x and tx is the transcript abundance of gene x according to empirical data (DNA array data for wild type E. coli grown at 0.5 h-1)43. Finally ri was transformed into qi by dividing ri by the maximal value found for r across all codons:
Using the three coding metrics, we constructed Extended Data Table 1 by assigning the closest substitutions (in pink) for synonymous recoding of TCASer and TCGSer (in grey).
Individual recoding landscapes and compiled recoding landscapes
Based on the complete sequence of the recoded region for each clone, the identity of codon at each of the attempted recoding position, and the d.r.1 to 5, was identified either as recoded with a binary value of 1 and coloured in red, or wildtype with a value of 0 and coloured in black. The distribution of targeted positions that are recoded and that remain wildtype across the refactored mraZ to ftsZ region gives an “individual recoding landscape”.
The 16 individual recoding landscape of the 16 individual clones of each of the recoding schemes were compiled to generate the “compiled recoding landscape” of each recoding scheme by counting the fraction of clones being recoded at each targeted position across the whole refactored mraZ to ftsZ region. When the recoding fraction at a given position is greater than 0 (coloured in red), it indicates that there is at least one sequenced clone being recoded at this position. When the recoding fraction reaches 0 (coloured in black), it indicates that the wildtype codon always remains and that the recoded codon may not be tolerated at these positions.
REXER + ssDNA recombineering protocol
A single stranded oligo of a total length of 90 nt was designed and synthesized to change the deleterious sequence of AGT in ftsA codon position 407 on the synthetic sequence of r.s.1 to a fixing sequence of AGC. The oligo sequence was designed based on the reverse strand of the synthetic sequence to bind the forward strand with the single nucleotide change positioned in the middle (position 45 from 5’ end). The last two nucleotides in the 5’ end of the oligo were substituted with phosphorothioate backbone to protect the oligo from unspecific exonuclease degradation.
3 μg of matching pKW21_MB1amp_Spacers×2 plasmid and 0.2 nmol of the fixing oligo was co-electroporated into the pre-induced MDS42rpsLK43R/rK cell with pCDFtet_pAraRedCas9_tracrRNA and pBAC_HR(89,061)-20kb/r.s.1-sC-HR(106,508)_r. The normal REXER procedure was carried out without any modification. SNP-PCRs from re-suspended survival colonies were performed44 using the SNP-PCR primer pairs either specific for the ftsA codon position 407 wildtype sequence TCG or the fixed sequence AGC45 with KAPA 2G fast multiplex mix, and analyzed on QIAGEN QIAxcel Advanced using QIAxcel DNA Screening Kit with QX Alignment Marker 15 bp/5 kb and QX Size Marker 250 - 4000 bp. Clones with the correct genotype following REXER + ssDNA recombineering were verified by Sanger sequencing through the entire 20 kb region.
Growth rate measurements and analysis
Glycerol stocks of the assayed bacterial clones were used to inoculate 5 mL LB in absence of antibiotics for overnight incubation at 37°C with shaking. The overnight cultures were used to inoculate triplicates of 1 mL of LB in a deep-well pre-culture plate at a ratio of 1:100, followed by incubation at 37°C for 6 hours with shaking. Each replicate on the pre-culture plate was used to inoculate 200 μL of LB in a 96-well measurement plate with a dilution factor of 100. The measurement plate was incubated at 37 °C for 16 hours with shaking at 400 rpm in an M200 Pro Plate Reader (Tecan). Readings of OD600 were taken for each well every 10 min. Plate reader absorbance data was adjusted to correspond to spectrophotometer readings by collecting measurements from a dilution series of bacterial cultures and fitting the plate reader data y with a polynomial to the spectrophotometer values x: y = 2.053 x2 + 2.2 x + 0.061.
To determine doubling times, the growth curves were log2-transformed, and the first derivative was determined (d(log2(x))/dt). During exponential growth, the log2-derivative is maximal and constant. The ten time-points with the maximal log2-derivatives were identified, and used to calculate the average value with standard deviation for each of the replicate. A total of 12 replicates (three independent clones, each independently repeated four times) were used to calculate the doubling time for each fully recoded scheme. For each doubling time, the average across the n = 12 replicates was determined and the error σ was propagated using the following formula:
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Extended Data
Extended Data Figure 1.
Simultaneous double selection and recombination enhances integration at a target locus. a. Classical recombination and double selection recombination. In classical recombination, a linear double stranded DNA with a synthetic DNA (s. DNA) sequence and a positive selection marker (+, CmR) flanked by homologous region 1 (HR1) and homologous region 2 (HR2) is transformed into the cell. Recombinants are selected by expression of the positive selection marker. By simultaneous double selection recombination, s. DNA containing double selection marker -2/+2 (sacB-CmR) is integrated in place of the double selection marker -1/+1 (rpsL-KanR) on the genome. Double selection for the gain of +2 and loss of -1 selects for simultaneous gain of s. DNA and loss of genomic sequence, and improves recombination at the target genomic locus. b. Colony PCR of clones from classical recombination and simultaneous double selection and recombination. c. All of the clones isolated by simultaneous double selection and recombination have s. DNA integrated at the target locus. The data show the mean of three independent experiments, the error bars represent the standard deviation (n=6). d. Both simultaneous double selection recombination (n = 8), and REXER 2 and REXER 4 (n = 296) result in the right combination of markers. A previously reported method integrating foreign DNA into B. subtilis genome only using negative selection gave 3% of selected clones with right combination of markers3,11. A previously reported method replacing S. cerevisiae chromosome III fragments with s. DNA only using positive selection gave 0.5% (replacement of 55 kb) to 59% (replacement of 9 kb) of selected clones with right combination of markers (a 13% mean of all reported value plotted with error bar representing the range)6. For gel source data, see Supplementary Figure 1.
Extended Data Figure 2.
REXER enables site-specific integration of large DNA fragments into the genome. a. The use of two distinct double selection cassettes -1/+1 (rpsL-KanR) and -2/+2 (sacB-CmR) allows for simultaneous selection for the loss of the negative selection marker on the genome and the gain of the positive selection marker from the BAC, upon integration of synthetic DNA. b. Efficient replacement of genomic rpsL-KanR with BAC bound sacB-CmR using REXER 2 and REXER 4. All colonies contained the correct combination of selection markers after REXER 2 or REXER 4 as analysed by phenotyping, colony PCR, and DNA sequencing (not shown) (n = 22). c. Efficient insertion of 9 kb synthetic DNA. Genomic rpsL-KanR was replaced with a synthetic lux operon coupled to sacB-CmR using REXER 2 and REXER 4. All colonies on the 10-fold dilution double selection plates for REXER 2 and the 104-fold plates for REXER 4 show bioluminescence. 11 colonies each from REXER 2 and REXER 4 showed correct integration by phenotyping, colony PCR, and DNA sequencing (not shown). d. Efficient insertion of 90kb synthetic DNA. The 90 kb DNA consisted of the lux operon in the middle of 80 kb DNA (previously deleted from the MDS42 genome) and followed by sacB-CmR, carried on a BAC. For gel source data, see Supplementary Figure 1.
Extended Data Figure 3.
Replacement of 100 kb of genomic DNA via REXER. a. The synthetic DNA contain the 100 kb wildtype DNA (open reading frames in grey) with five genes of the lux operon (blue) and sacB-CmR. Complete replacement leads to integration of all five lux genes (luxA, B, C, D, E) resulting in bioluminescent cells, while partial replacement confers loss of one or more lux genes hence loss of bioluminescence. b. After REXER 2, 80 % of 2x102 colonies examined were bioluminescent while for REXER 4 yields 50 % of 2x102 colonies examined were bioluminescent. c. Bioluminescent colonies from REXER 2 and REXER 4 that were analysed (n = 11) had all five lux genes correctly integrated indicating complete replacement of the 100 kb genomic region. All clones contained the right combination of selection markers. d. While bioluminescent colonies contain all five lux watermarks, the non-bioluminescent colonies are missing one or more lux genes indicating partial replacement of the genomic region. All clones contained the right combination of selection markers. For gel source data, see Supplementary Figure 1.
Extended Data Figure 4.
Iterative REXER. a. The product of REXER shown in Extended Data Figure 2a was used as a template for the next round of REXER. b. The phenotypes of clones from the first round of REXER. c. The phenotypes of clones from the second round of REXER. For gel source data, see Supplementary Figure 1.
Extended Data Figure 5.
Synonymous codon compression strategies. a. Codon and anticodon interactions in the E. coli genome. 28 sense codons are highlighted in grey, along with the amber stop codon. The genome wide removal of these sense codons, but not other sense codons, would enable all their cognate tRNA to be deleted without removing the ability to decode one or more sense codons remaining in the genome. This is necessary but not sufficient for the reassignment of sense codons to unnatural monomers. Serine, leucine and alanine codon boxes are highlighted because the endogenous aminoacyl-tRNA synthetases for these amino acids do not recognize the anticodons of their cognate tRNAs. This may facilitate the assignment of codons within these boxes to new amino acids through the introduction of tRNAs bearing cognate anticodons that do not direct mis-aminocylation by endogenous synthetases. The number of total codon counts for all 64 triplet codons in the MDS42 genome (GenBank accession no. AP012306), all known codon-anticodon interactions through both Watson-Crick base-paring and wobbling, base modification on tRNA anticodons, tRNA genes, and measured in vivo tRNA relative abundance are reported. This analysis identifies 10 codons from the serine, leucine, and alanine groups (serine codon TCG, TCA, AGT, AGC; leucine codon CTG, CTA, TTG, TTA; and alanine codon GCG, GCA) satisfy both the codon-anticodon interaction and aminoacyl-tRNA synthetases recognition criteria for codon reassignment. b., c. , d. Serine, leucine and alanine codon removal and tRNA deletion strategies compatible with codon reassignment to unnatural amino acids (u.a.a).
Extended Data Figure 6.
Recoding landscapes for compression of serine codons by REXER. a. The sequences for the systematically recoded mraZ to ftsZ region were de novo designed, synthesized and assembled into BAC and used for REXER. b-d. The recoding landscapes for serine recoding schemes r.s.1-3, and the resulting compiled recoding landscape.
Extended Data Figure 7.
Recoding landscapes. a-e. r.s.4-8. f. r.s.1 with ftsA codon 407 changed from AGT to AGC (highlighted in orange).
Extended Data Figure 8.
Identifying and fixing a deleterious sequence in defined and systematic synonymous recoding. a. Recoding codon 407 in ftsA in the wildtype genomic background. The wildtype codon at ftsA codon position 407 is the serine codon TCG. We sequenced 16 post-REXER clones for TCG to AGT and 20 post-REXER clones for TCG to TCT. b. Changing ftsA 407 AGT to AGC in the serine r.s.1 background. We sequenced 16 AGT clones and 16 AGT to AGC clones. c. Changing ftsA 407 AGT to AGC in the serine r.s.1 background dramatically improved the fraction of fully recoded clones across the entire 20 kb region from 0% to 94% (16 clones sequenced). d. The fixed serine r.s.1 with ftsA 407 AGC yielded clones with no measurable growth defect. The doubling times of fully recoded clones from serine r.s.1 with ftsA 407 AGC, from serine r.s.2, serine r.s.3, and alanine r.s.7 are measured, and show no measurable growth defects when compared to the wildtype MDS42 E. coli control with the second double selection cassette integrated at the same genomic locus. n=12 biological replicates ± s.d.. e. Combining single stand DNA recombineering with REXER to fix short a deleterious stretch within the synthetic sequence of r.s. 1. A 90 nt. single stranded oligo was designed to change the deleterious sequence of AGT in ftsA codon position 407 in r.s.1 to a tolerated sequence, AGC. The oligo sequence was designed based on the reverse strand of the synthetic sequence to bind the forward strand with the single nucleotide change positioned in the middle (45 from nt 5’ end). The oligo was co-transformed into E. coli during a REXER experiment which introduces r.s. 1 into the genome.. f. Fixing short deleterious sequence on synthetic DNA with REXER + ssDNA recombineering. 16 clones from REXER double selection described in (e) were randomly picked and subject to single nucleotide polymorphism (SNP) genotyping using primers specific for either the wildtype sequence in ftsA codon position 407 (TCG) or the fixed sequence (AGC). MDS42 rpsLK43R/rK was used as the wildtype control and a fully recoded clone from serine r.s.3 with verified ftsA 407 AGC as the positive control. SNP genotyping at ftsA codon position 407 identified one clone (clone 12, highlighted in orange) out of a total of 16 clones tested with fixed sequence AGC, which was then fully sequenced across the entire 20 kb recoding region and confirmed as fully recoded at all 83 targeted codon positions. For gel source data, see Supplementary Figure 1.
Extended Data Table 1.
Defining recoding rules by codon adaptation index (cAi), tRNA adaptation index (tAi), and translation efficiency (t.E). We defined the best synonymous replacements for the target serine (a), leucine (b), and alanine codons (c) by identifying the closest match for the target codons, as judged by either codon adaptation index (cAi), tRNA adaptation index (tAi), or a third metric that combines codon abundance and measured tRNA concentrations to estimate translation efficiency (t.E) (see Methods). The table assigns the closest substitutions (in pink) for synonymous recoding of targeted codons (in grey) using the three coding metrics. Where two substitutions are comparable the one that conserves G, C content is chosen. The number in bold is the value of the best matching substitution in a given coding metric.
| a | Codon | cAi | tAi | t.E | |||
| Metric | Substitution | Metric | Substitution | Metric | Substitution | ||
| TCGSer | 0.017 | AGTSer | 0.165 | AGCSer | 0.086 | AGCSer | |
| TCASer | 0.077 | AGTSer | 0.125 | AGCSer | 0.049 | AGTSer | |
| TCTSer | 1.000 | 0.110 | 0.034 | ||||
| TCCSer | 0.744 | 0.250 | 0.057 | ||||
| AGTSer | 0.085 | 0.055 | 0.044 | ||||
| AGCSer | 0.410 | 0.125 | 0.088 | ||||
| b | Codon | cAi | tAi | t.E | |||
| Metric | Substitution | Metric | Substitution | Metric | Substitution | ||
| CTGLeu | 1 | 0.540 | 0.098 | ||||
| CTALeu | 0.007 | 0.125 | 0.010 | ||||
| CTTLeu | 0.042 | 0.055 | 0.036 | ||||
| CTCLeu | 0.037 | 0.125 | 0.069 | ||||
| TTGLeu | 0.02 | CTCLeu | 0.165 | CTCLeu | 0.034 | CTTLeu | |
| TTALeu | 0.02 | CTCLeu | 0.125 | CTALeu | 0.068 | CTCLeu | |
| c | Codon | cAi | tAi | t.E | |||
| Metric | Substitution | Metric | Substitution | Metric | Substitution | ||
| GCGAla | 0.424 | GCTAla | 0.120 | GCTAla | 0.020 | GCTAla | |
| GCAAla | 0.586 | GCTAla | 0.375 | GCCAla | 0.040 | GCCAla | |
| GCTAla | 1 | 0.110 | 0.019 | ||||
| GCCAla | 0.122 | 0.250 | 0.027 | ||||
Extended Data Table 2.
Protein functions, localizations, expression levels (in parts per million), and lengths (both ORF length in bp and peptide length in amino acid count) of the genes in the essential cell division operon all simultaneously recoded in this work (a), and of individually recoded ribosomal and release factor 2 genes reported previously (b)37. The numbers of codons targeted for removal according to different recoding schemes are also reported. The expression level data is from www.pax-db.org.
| a | Number of target codons | |||||||||||||
| Gene | Function | Localisation | Protein level ppm | ORF length | Peptide length | r.s.1 | r.s.2 | r.s.3 | r.s.4 | r.s.5 | r.s.6 | r.s.7 | r.s.8 | |
| mraZ | Transcription factor | cytosol, nucleoid | 11.3 | 459 | 153 | 4 | 4 | 4 | 9 | 9 | 9 | 4 | 4 | |
| rsmH | Methyltransferase | cytosol | 122.0 | 942 | 314 | 8 | 8 | 8 | 4 | 4 | 4 | 13 | 13 | |
| ftsL | Cell division | membrane | 1.9 | 366 | 122 | 2 | 2 | 2 | 5 | 5 | 5 | 4 | 4 | |
| ftsI | Cell division | membrane | 9.7 | 1767 | 589 | 9 | 9 | 9 | 15 | 15 | 15 | 38 | 38 | |
| murE | Cell division | cytosol | 121.3 | 1488 | 496 | 5 | 5 | 5 | 10 | 10 | 10 | 47 | 47 | |
| murF | Cell division | cytosol | 67.1 | 1359 | 453 | 7 | 7 | 7 | 12 | 12 | 12 | 34 | 34 | |
| mraY | Cell division | membrane | 13.7 | 1083 | 361 | 5 | 5 | 5 | 9 | 9 | 9 | 16 | 16 | |
| murD | Cell division | cytosol | 67.5 | 1317 | 439 | 3 | 3 | 3 | 12 | 12 | 12 | 36 | 36 | |
| ftsW | Cell division | membrane | 2.7 | 1245 | 415 | 13 | 13 | 13 | 19 | 19 | 19 | 27 | 27 | |
| murG | Cell division | membrane | 21.5 | 1068 | 356 | 5 | 5 | 5 | 11 | 11 | 11 | 34 | 34 | |
| murC | Cell division | cytosol | 83.4 | 1476 | 492 | 2 | 2 | 2 | 12 | 12 | 12 | 29 | 29 | |
| ddlB | Cell wall synthesis | cytosol | 33.1 | 921 | 307 | 7 | 7 | 7 | 15 | 15 | 15 | 24 | 24 | |
| ftsQ | Cell division | membrane | 5.4 | 831 | 277 | 3 | 3 | 3 | 12 | 12 | 12 | 15 | 15 | |
| ftsA | Cell division | membrane | 113.6 | 1263 | 421 | 10 | 10 | 10 | 9 | 9 | 9 | 23 | 23 | |
| ftsZ | Cell division | cytosol | 633.6 | 1152 | 384 | 0 | 0 | 0 | 3 | 3 | 3 | 30 | 30 | |
| Total number of target codons: | 83 | 83 | 83 | 157 | 157 | 157 | 374 | 374 | ||||||
| b | Number of target codons | |||||||||||||
| Gene | Function | Localisation | Protein level ppm | ORF length | Peptide length | r.s.1 | r.s.2 | r.s.3 | r.s.4 | r.s.5 | r.s.6 | r.s.7 | r.s.8 | |
| rpmH | Protein translation | cytosol | 4075.7 | 141 | 47 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 9 | |
| rpmD | Protein translation | cytosol | 3046.3 | 180 | 60 | 0 | 0 | 0 | 2 | 2 | 2 | 20 | 20 | |
| rpmC | Protein translation | cytosol | 5980.0 | 192 | 64 | 0 | 0 | 0 | 1 | 1 | 1 | 12 | 12 | |
| rpsR | Protein translation | cytosol | 5806.5 | 228 | 76 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | |
| rpmB | Protein translation | cytosol | 5502.1 | 237 | 79 | 0 | 0 | 0 | 1 | 1 | 1 | 5 | 5 | |
| rpsP | Protein translation | cytosol | 6611.2 | 249 | 83 | 3 | 3 | 3 | 2 | 2 | 2 | 16 | 16 | |
| rpsQ | Protein translation | cytosol | 2179.3 | 255 | 85 | 1 | 1 | 1 | 0 | 0 | 0 | 3 | 3 | |
| rpmA | Protein translation | cytosol | 4380.7 | 258 | 86 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 3 | |
| rpsS | Protein translation | cytosol | 3094.6 | 279 | 93 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 3 | |
| rplW | Protein translation | cytosol | 2091.7 | 303 | 101 | 0 | 0 | 0 | 1 | 1 | 1 | 7 | 7 | |
| rpsN | Protein translation | cytosol | 4612.5 | 306 | 102 | 2 | 2 | 2 | 0 | 0 | 0 | 9 | 9 | |
| rplU | Protein translation | cytosol | 1856.1 | 312 | 104 | 0 | 0 | 0 | 4 | 4 | 4 | 10 | 10 | |
| rpsJ | Protein translation | cytosol | 3472.7 | 312 | 104 | 0 | 0 | 0 | 1 | 1 | 1 | 9 | 9 | |
| rplX | Protein translation | cytosol | 4456.0 | 315 | 105 | 1 | 1 | 1 | 2 | 2 | 2 | 5 | 5 | |
| rplV | Protein translation | cytosol | 7848.2 | 333 | 111 | 0 | 0 | 0 | 1 | 1 | 1 | 5 | 5 | |
| rplS | Protein translation | cytosol | 3859.3 | 348 | 116 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 4 | |
| rplR | Protein translation | cytosol | 6367.3 | 354 | 118 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 8 | |
| rplT | Protein translation | cytosol | 3291.4 | 357 | 119 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 9 | |
| rpsM | Protein translation | cytosol | 5733.1 | 357 | 119 | 1 | 1 | 1 | 0 | 0 | 0 | 10 | 10 | |
| rplL | Protein translation | cytosol | 14543.5 | 366 | 122 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 8 | |
| rplN | Protein translation | cytosol | 8866.6 | 372 | 124 | 1 | 1 | 1 | 2 | 2 | 2 | 3 | 3 | |
| rpsL | Protein translation | cytosol | 5532.8 | 375 | 125 | 0 | 0 | 0 | 1 | 1 | 1 | 10 | 10 | |
| rplQ | Protein translation | cytosol | 4272.8 | 384 | 128 | 0 | 0 | 0 | 1 | 1 | 1 | 5 | 5 | |
| rpsK | Protein translation | cytosol | 2900.5 | 390 | 130 | 1 | 1 | 1 | 0 | 0 | 0 | 5 | 5 | |
| rpsH | Protein translation | cytosol | 3828.3 | 393 | 131 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | |
| rpsI | Protein translation | cytosol | 3410.8 | 393 | 131 | 0 | 0 | 0 | 2 | 2 | 2 | 5 | 5 | |
| rplP | Protein translation | cytosol | 3778.1 | 411 | 137 | 0 | 0 | 0 | 2 | 2 | 2 | 9 | 9 | |
| rplM | Protein translation | cytosol | 4268.0 | 429 | 143 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 9 | |
| rplO | Protein translation | cytosol | 5111.6 | 435 | 145 | 1 | 1 | 1 | 1 | 1 | 1 | 3 | 3 | |
| rplJ | Protein translation | cytosol | 7731.6 | 498 | 166 | 1 | 1 | 1 | 2 | 2 | 2 | 3 | 3 | |
| rpsE | Protein translation | cytosol | 7657.3 | 504 | 168 | 0 | 0 | 0 | 1 | 1 | 1 | 11 | 11 | |
| rplF | Protein translation | cytosol | 5012.1 | 534 | 178 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 5 | |
| rpsG | Protein translation | cytosol | 8660.2 | 540 | 180 | 0 | 0 | 0 | 2 | 2 | 2 | 11 | 11 | |
| rplE | Protein translation | cytosol | 3489.1 | 540 | 180 | 0 | 0 | 0 | 2 | 2 | 2 | 5 | 5 | |
| rplD | Protein translation | cytosol | 3469.9 | 606 | 202 | 0 | 0 | 0 | 1 | 1 | 1 | 5 | 5 | |
| rpsD | Protein translation | cytosol | 5187.4 | 621 | 207 | 1 | 1 | 1 | 3 | 3 | 3 | 9 | 9 | |
| rplC | Protein translation | cytosol | 4460.3 | 630 | 210 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 6 | |
| rpsC | Protein translation | cytosol | 5755.0 | 702 | 234 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | |
| rpsB | Protein translation | cytosol | 4324.5 | 726 | 242 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | |
| rplB | Protein translation | cytosol | 5658.4 | 822 | 274 | 1 | 1 | 1 | 1 | 1 | 1 | 16 | 16 | |
| prfB | Protein translation | cytosol | 570.9 | 1099 | 366 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 8 | |
| rpsA | Protein translation | cytosol | 2649.1 | 1674 | 558 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 3 | |
| Total number of target codons: | 14 | 14 | 14 | 36 | 36 | 36 | 292 | 292 | ||||||
Supplementary Material
Acknowledgements
Work was supported by the Medical Research Council, UK (MC_U105181009 and MC_UP_A024_1008, J.W.C.), the Danish Council for Independent Research (DFF – 4090-00289, to J.F.), a Boehringer Ingelheim Fonds PhD fellowship, S.F.B. The Gates-Cambridge Scholarship (S.H.K), and by an ERC Advanced Grant (SGCR to J.W.C.). We thank Neil Grant MRC-LMB Visual Aids for photography.
Footnotes
Author contributions
J.W.C. defined the direction of research. K.W., J.F., S.F.B., S.H.K., and T.C. performed experiments. All authors analysed the data. K.W. and J.W.C. wrote the paper with input from all authors. K.W. designed and constructed the REXER system. J.F. implemented DNA assembly methods in S. cerevisiae. K.W. and S.F.B. identified target codons, developed t.E, designed recoding schemes.
Competing financial interests
The authors declare no competing financial interest.
References
- 1.Cello J, Paul AV, Wimmer E. Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template. Science. 2002;297:1016–1018. doi: 10.1126/science.1072266. [DOI] [PubMed] [Google Scholar]
- 2.Chan LY, Kosuri S, Endy D. Refactoring bacteriophage T7. Molecular Systems Biology. 2005;1 doi: 10.1038/msb4100025. 2005.0018–E10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Itaya M, Tsuge K, Koizumi M, Fujita K. Combining two genomes in one cell: stable cloning of the Synechocystis PCC6803 genome in the Bacillus subtilis 168 genome. Proc Natl Acad Sci USA. 2005;102:15971–15976. doi: 10.1073/pnas.0503868102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gibson DG, et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 5.Gibson DG, et al. Creation of a Bacterial Cell Controlled by a Chemically Synthesized Genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 6.Annaluru N, et al. Total synthesis of a functional designer eukaryotic chromosome. Science. 2014;344:55–58. doi: 10.1126/science.1249252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ro D-K, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 9.Chin JW. Reprogramming the genetic code. Science. 2012;336:428–429. doi: 10.1126/science.1221761. [DOI] [PubMed] [Google Scholar]
- 10.Mukai T, et al. Reassignment of a rare sense codon to a non-canonical amino acid in Escherichia coli. Nucleic Acids Research. 2015;43:8111–8122. doi: 10.1093/nar/gkv787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Itaya M, Fujita K, Ikeuchi M, Koizumi M, Tsuge K. Stable positional cloning of long continuous DNA in the Bacillus subtilis genome vector. Journal of Biochemistry. 2003;134:513–519. doi: 10.1093/jb/mvg168. [DOI] [PubMed] [Google Scholar]
- 12.Krishnakumar R, et al. Simultaneous non-contiguous deletions using large synthetic DNA and site-specific recombinases. Nucleic Acids Research. 2014;42:e111–e111. doi: 10.1093/nar/gku509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang K, et al. Optimized orthogonal translation of unnatural amino acids enables spontaneous protein double-labelling and FRET. Nature Chemistry. 2014;6:393–403. doi: 10.1038/nchem.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neumann H, Wang K, Davis L, Garcia-Alai M, Chin JW. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature. 2010;464:441–444. doi: 10.1038/nature08817. [DOI] [PubMed] [Google Scholar]
- 16.Cho B-K, et al. The transcription unit architecture of the Escherichia coli genome. Nature Biotechnology. 2009;27:1043–1049. doi: 10.1038/nbt.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li G-W, Oh E, Weissman JS. The anti-Shine–Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012;484:538–541. doi: 10.1038/nature10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sørensen MA, Pedersen S. Absolute in vivo translation rates of individual codons in Escherichia coli. The two glutamic acid codons GAA and GAG are translated with a threefold difference in rate. Journal of Molecular Biology. 1991;222:265–280. doi: 10.1016/0022-2836(91)90211-n. [DOI] [PubMed] [Google Scholar]
- 19.Curran JF, Yarus M. Rates of aminoacyl-tRNA selection at 29 sense codons in vivo. Journal of Molecular Biology. 1989;209:65–77. doi: 10.1016/0022-2836(89)90170-8. [DOI] [PubMed] [Google Scholar]
- 20.Kimchi-Sarfaty C, et al. A ‘Silent’ Polymorphism in the MDR1 Gene Changes Substrate Specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 21.Zhang G, Hubalewska M, Ignatova Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol. 2009;16:274–280. doi: 10.1038/nsmb.1554. [DOI] [PubMed] [Google Scholar]
- 22.Quax TEF, et al. Differential translation tunes uneven production of operon-encoded proteins. Cell Rep. 2013;4:938–944. doi: 10.1016/j.celrep.2013.07.049. [DOI] [PubMed] [Google Scholar]
- 23.Quax TEF, Claassens NJ, Söll D, van der Oost J. Codon Bias as a Means to Fine-Tune Gene Expression. Mol Cell. 2015;59:149–161. doi: 10.1016/j.molcel.2015.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li G-W, Burkhardt D, Gross C, Weissman JS. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014;157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pósfai G, et al. Emergent properties of reduced-genome Escherichia coli. Science. 2006;312:1044–1046. doi: 10.1126/science.1126439. [DOI] [PubMed] [Google Scholar]
- 26.Jiang W, Cox D, Zhang F, Bikard D, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nature Biotechnology. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bryksin AV, Matsumura I. Rational Design of a Plasmid Origin That Replicates Efficiently in Both Gram-Positive and Gram-Negative Bacteria. PLoS ONE. 2010;5:e13244. doi: 10.1371/journal.pone.0013244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kouprina N, Noskov VN, Larionov V. Selective isolation of large chromosomal regions by transformation-associated recombination cloning for structural and functional analysis of mammalian genomes. Methods Mol Biol. 2006;349:85–101. doi: 10.1385/1-59745-158-4:85. [DOI] [PubMed] [Google Scholar]
- 29.Giegé R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Research. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharp PM, Li WH. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Research. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reis dos M, Savva R, Wernisch L. Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Research. 2004;32:5036–5044. doi: 10.1093/nar/gkh834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tuller T, Waldman YY, Kupiec M, Ruppin E. Translation efficiency is determined by both codon bias and folding energy. Proceedings of the National Academy of Sciences. 2010;107:3645–3650. doi: 10.1073/pnas.0909910107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerdes SY, et al. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J Bacteriol. 2003;185:5673–5684. doi: 10.1128/JB.185.19.5673-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keseler IM, et al. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Research. 2012;41:D605–D612. doi: 10.1093/nar/gks1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dai K, Lutkenhaus J. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J Bacteriol. 1992;174:6145–6151. doi: 10.1128/jb.174.19.6145-6151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dewar SJ, Begg KJ, Donachie WD. Inhibition of cell division initiation by an imbalance in the ratio of FtsA to FtsZ. J Bacteriol. 1992;174:6314–6316. doi: 10.1128/jb.174.19.6314-6316.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lajoie MJ, et al. Probing the Limits of Genetic Recoding in Essential Genes. Science. 2013;342:361–363. doi: 10.1126/science.1241460. [DOI] [PubMed] [Google Scholar]
- 38.Lajoie MJ, et al. Genomically Recoded Organisms Expand Biological Functions. Science. 2013;342:357–360. doi: 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Molecular Systems Biology. 2006;2 doi: 10.1038/msb4100050. 2006.0008–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grosjean HJ, de Henau S, Crothers DM. On the physical basis for ambiguity in genetic coding interactions. Proc Natl Acad Sci USA. 1978;75:610–614. doi: 10.1073/pnas.75.2.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Curran JF. Decoding with the A:I wobble pair is inefficient. Nucleic Acids Research. 1995;23:683–688. doi: 10.1093/nar/23.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong H, Nilsson L, Kurland CG. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. Journal of Molecular Biology. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- 43.Ishii N, et al. Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science. 2007;316:593–597. doi: 10.1126/science.1132067. [DOI] [PubMed] [Google Scholar]
- 44.Gallagher RR, Li Z, Lewis AO, Isaacs FJ. Rapid editing and evolution of bacterial genomes using libraries of synthetic DNA. Nat Protoc. 2014;9:2301–2316. doi: 10.1038/nprot.2014.082. [DOI] [PubMed] [Google Scholar]
- 45.Newton CR, et al. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS) Nucleic Acids Research. 1989;17:2503–2516. doi: 10.1093/nar/17.7.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.