Abstract
Phosphoinositide-3-kinase (PI3K) plays a key role in cellular metabolism and cancer. Using a mass spectrometry-based metabolomics platform, we discovered that plasma concentrations of 26 metabolites, including amino acids, acylcarnitines and phosphatidylcholines, were decreased in mice bearing PTEN-deficient tumors compared with non-tumor bearing controls and in addition were increased following dosing with Class I PI3K inhibitor pictilisib (GDC-0941). These candidate metabolomics biomarkers were evaluated in a Phase I dose-escalation clinical trial of pictilisib. Time- and dose-dependent effects were observed in patients for 22 plasma metabolites. The changes exceeded baseline variability, resolved after drug washout and were recapitulated on continuous dosing. Our study provides a link between modulation of the PI3K pathway and changes in the plasma metabolome and demonstrates that plasma metabolomics is a feasible and promising strategy for biomarker evaluation. Also, our findings provide additional support for an association between insulin resistance, branched-chain amino acids and related metabolites following PI3K inhibition.
Keywords: metabolomics, PI3K, pharmacodynamics, plasma biomarker, amino acids, acylcarnitines, insulin resistance
Introduction
The successful incorporation into the Pharmacologic Audit Trail of pharmacodynamic biomarkers can significantly increase the scientific rigor, decision-making reliability and hypothesis-testing power in drug development, and in particular help mitigate the risk of attrition by confirming target engagement and informing key decisions regarding optimal dose and schedule (1). In preclinical studies in oncology, solid tumor samples can readily be obtained from mice bearing human tumor xenografts to evaluate the effects of drugs. In the clinical situation where few patients have accessible tumor, this is challenging with pre- and post-treatment biopsies typically obtained in the best case scenarios (2). Consequently, biomarkers are often derived from surrogate tissues, for example platelet-rich plasma or blood mononuclear cells, which can be obtained in a minimally invasive way (3, 4). In patients with metastatic disease, circulating tumor cells may be obtained but they are few and not systematically amenable to studies evaluating target engagement (5). Imaging technologies are very appealing and non-invasive but remain expensive and difficult to deploy across multiple centres (6).
We hypothesised that circulating metabolites may present an attractive strategy to assess biomarker modulation in early phases of clinical drug development. As a minimally invasive assessment, plasma metabolomic analysis has the potential to complement direct tumoral measurements and, particularly where cancer biopsies are unavailable, can report on the integrated output of multiple systems including sites of disease. There is no doubt that the signatures will include output from both normal tissues and cancers. However, given the increasing recognition that there is in fact a key bi-directional interplay between the two, this could be considered an advantage. Moreover, we reasoned that the PI3K axis (which is a key mediator of cellular metabolism) represents a desirable test system to evaluate this approach (7). It is thought to function as a critical logic gate with a nutrient-sensing role and controls the rate of metabolic processes including fatty acid synthesis, β-oxidation, oxidative phosphorylation and glycolysis (8, 9). This pathway is significantly associated with disordered metabolism, including insulin resistance, and is modulated by anti-diabetic interventions (10, 11). The presence of germline PTEN mutations has been linked with constitutively increased insulin sensitivity and glucose tolerance in mice and humans (12, 13). There are thus strong a priori rationales for the study of the PI3K pathway to evaluate the potential of plasma metabolomics as a marker of target engagement in clinical studies. Multiple agents are in development that target key mediators of the PI3K pathway; among them, pictilisib (GDC-0941, Genentech Inc., South San Francisco, CL) is a potent and selective pan-Class I inhibitor of PI3K that is currently being assessed in randomized Phase II clinical trials (14–20).
Using liquid chromatography mass spectrometry (LC-MS), we implemented a non-targeted exploratory screen for plasma metabolites exhibiting consistent changes across genetic and pharmacologic perturbations of the PI3K pathway in mouse models. We next evaluated the identified biomarker candidates in a dose-escalation clinical trial of pictilisib, seeking dose- and time-dependent changes in patients with advanced cancers following drug administration. The confirmation of plasma metabolite changes in patients undergoing pharmacological inhibition of PI3K demonstrates the feasibility and promise of this approach for minimally invasive biomarker development. The metabolite changes we observed are consistent with the development of an insulin resistance phenotype following treatment with a pan-Class I PI3K inhibitor.
Materials & Methods
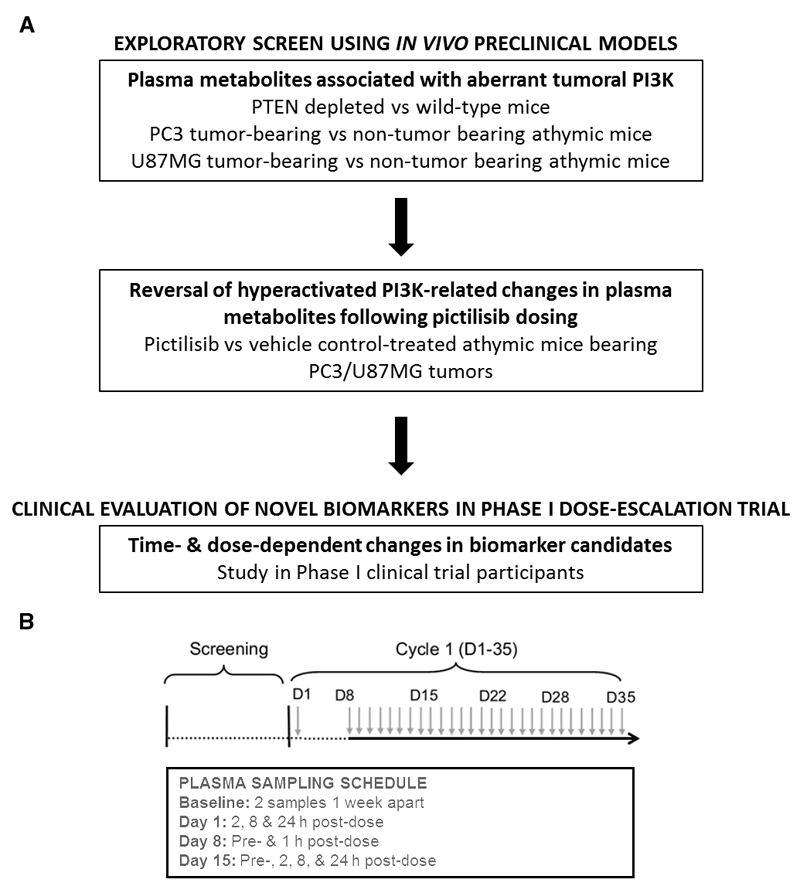
The experimental workflow is presented in Fig. 1A. In the exploratory preclinical screening studies, we compared plasma from: (1) tumor-bearing PTEN (+/-) C57BL/6 euthymic mice with non-tumor-bearing wild-type littermate controls; and (2) athymic mice inoculated subcutaneously with or without PTEN (-/-) U87MG glioblastoma or PC3 prostate human tumor xenografts (21). Some of the mice were treated with pictilisib (100 mg/kg) or vehicle controls at 2, 4, 8 and 24 hours post-dosing. We compared the effects in plasma and tumor of another chemically distinct oral pan-Class I PI3K inhibitor (buparlisib) with those for pictilicib at 8 hours following a single dose. We also determined plasma metabolite profiles of athymic mice bearing U87MG tumor xenografts dosed with vehicle or a representative cytotoxic agent (carmustine; BCNU).
Figure 1.
(A) Experimental workflow. (B) Schema illustrating once-daily dosing schedule of pictilisib (each green arrow representing a dose administered) with the plasma sampling schedule for metabolomic analysis summarized in the sub-table.
At the subsequent clinical evaluation stage, within the framework of a Phase I dose-escalation trial, we first monitored the levels of the candidate plasma metabolite biomarkers on two separate days within a 2-week screening window, prior to treatment, in order to define baseline levels. This was followed by determining the plasma metabolite changes, relative to pre-treatment baseline, at 2, 8 and 24 hours post-treatment with different dose levels of pictilisib following oral administration of an initial single dose on day 1. A longitudinal study was then carried out following continuous once-daily oral dosing of pictilisib from day 8 in patients treated at the maximally tolerated dose (or higher) of pictilisib. Plasma metabolites were assessed pre-dose and at 1 hour post-dose on day 8 and pre-dose and at 2, 8 and 24 hours on day 15.
Preclinical mouse models
All animal experiments were conducted in accordance with local and United Kingdom National Cancer Research Institute guidelines (22). Female mice with heterozygous (+/-) PTEN knockout (23) (n=5) on the C57BL/6 background and 25 week old age-matched wild-type littermates (n=6) were bled synchronously in the same experiment. To generate the tumor xenograft-bearing models, 2 million human cancer cells were injected subcutaneously bilaterally into the flanks of NCr athymic mice of 6 to 8-weeks of age, bred in-house; PC3 human prostate cancer cells and U87MG human glioblastoma cells were administered into male and female mice respectively. Cell lines were obtained from ATCC and used within 6 months of purchase. Authentication of the cell lines was performed in-house. Cell lines were analyzed by short tandem repeat (STR) profiling. Polymorphic STR loci were amplified using a polymerase chain reaction (PCR) primer set. The PCR product (each locus being labelled with a different fluorophore) was analyzed simultaneously with size standards using automated fluorescent detection. The number of repeats at 7-10 different loci was used to define the STR profile and this was cross-referenced with online databases to confirm authenticity.
During the course of the experiment, food pellets (Certified Rodent Diet 5002, Labdiet, Indianapolis, Indiana, USA) and water were available ad libitum. Dosing of each drug and its respective vehicle control was undertaken synchronously in the same experiment when tumors were well-established and measured approximately 8-10 mm in diameter. Carmustine was prepared in 5% ethanol and 0.5% sterile saline, pictilisib or buparlisib in 10% DMSO, 5% Tween 20 and 85% sterile saline. Compounds were dosed in 0.1 mL/10 g body weight of vehicle once or twice daily (carmustine intra-peritoneally at 20 mg/kg; pictilisib by oral gavage at 50 or 100 mg/kg; buparlisib by oral gavage at 60mg/kg); these represented biologically active, well tolerated doses associated with modulation of known downstream pharmacodynamic biomarkers of PI3K inhibitors (17). Control animals received an equivalent volume of appropriate vehicle. Blood samples were collected into bottles using sodium heparin as anticoagulant at 2, 8 and 24 hours post-drug administration. The samples were centrifuged at 13000 rpm for 2 minutes and then transferred onto dry ice; the entire process from collection to storage in dry ice took less than 5 minutes per sample. Three to six mice were used for each time point per treatment. Following completion of the entire experiment, the samples were stored at -80°C until further analyses.
Phase I dose-escalation clinical trial of pictilisib
Plasma samples for metabolomic analysis were obtained from 41 patients with advanced cancers treated at The Royal Marsden NHS Foundation Trust, London, United Kingdom, in a Phase I study of pictilisib with a standard 3+3 dose-escalation design (24). Pictilisib was administered orally on day 1, followed by a one-week washout, and dosing was once-daily on either a 21- or 28-day dose schedule every 28 days. The assessed dose levels ranged from 45 mg to 450 mg (45 mg [n=2], 60 mg [n=4], 80 mg [n=3], 100 mg [n=4], 245 mg [n=1], 330 mg [n=16], 340 mg [n=7], 400 mg [n=1], 450 mg [n=3]). In this study, the maximal tolerated dose was 330 mg administered once daily (20). Of 35 patients with RECIST-measurable disease in whom metabolomic analysis was undertaken, one achieved a partial response, 28 achieved stable disease and 6 sustained progressive disease (20). Study duration in this cohort of patients ranged from 7 to 288 days. Notably, there was marked heterogeneity in the primary tumor type. For all dose levels assessed in this study, metabolomic samples were collected pre-dose and 2, 8 and 24 hours post-dose on day 1. In 17 patients treated with pictilisib at dose levels ≥330 mg, once daily, samples were collected at the following additional time points following a protocol amendment: baseline at screening within 2 weeks prior to day 1, pre-dose and 1 h post-dose on day 8; and pre-dose and 2, 8 and 24 hours post-dose on day 15. All patients were fasted for at least 2 hours prior to each sampled time point but were permitted water ad libitum. Plasma was separated from blood (using sodium heparin as anticoagulant) following centrifugation at 1500 g for 15 minutes at 4°C; it was then stored at -80°C until further analyses. The schedule of sample collection is presented in Fig. 1B. Non-mandated immunohistochemical analysis including phospho-AKT (Ser473) of tumor biopsies was undertaken but these were limited in number and graded on a semi-quantitative scale. The electrochemiluminescense-based assay of phospho-AKT (Luminex xMAP; Luminex Corp.) of surrogate platelet-rich plasma was only conducted late in the trial at the dose expansion phase. These limited our ability to perform exploratory assessments of their relationship with the observed metabolomic changes and additionally highlight challenges in performing pharmacodynamic assessments in Phase I trials. At dose levels ≥100 mg, plasma was obtained pre-dose and at 1 hour after dose on cycle 1 day 1 for analysis of glucose and insulin levels. Estimates of insulin resistance were derived based on the Homeostasis Model Assessment model (25). Non-mandated whole-body 18FDG-PET scans were performed at baseline, and 1 to 4 hours after dose between day 22 and the end of cycle 1. In all, 23 and 13 respective patients who underwent metabolomic analysis also undertook glucose/insulin and 18FDG-PET studies. All aspects of the study were conducted in accordance with the Declaration of Helsinki and the International Conference on Harmonization Good Clinical Practice Guidelines. Written informed consent was obtained from all participants.
Plasma metabolomics analysis
Non-targeted LC-MS metabolomics was initially used in the preclinical screen as described previously (26); significant changes in plasma metabolites were identified. To follow up and confirm these preliminary findings, we carried out targeted, quantitative metabololomic analysis by electrospray ionization tandem MS using the AbsoluteIDQ™ p180 kit (Biocrates Life Sciences AG, Innsbruck, Austria); results from the confirmatory, quantitative platform are presented in this report. Samples were anonymized and analyses were carried out on a Waters Acquity H-class UPLC coupled to Xevo TQ-S triple-quadrupole MS/MS System (Waters Corporation, Manchester, UK). Quantification of the metabolites of the biological sample was achieved by reference to appropriate internal standards. The method conforms with the United States Food and Drug Administration Guidelines ‘Guidance for Industry – Bioanalytical Method Validation (May 2001)’, providing proof of reproducibility within a given error range.
Data analysis
The analytical process to derive metabolite concentrations was performed using the MetIDQ software package. The data matrix obtained was subsequently subjected to multivariate analysis using SIMCA v.13 software (MKS Umetrics AB, Sweden); metabolite features that were differentially expressed between defined groups of mice: (1) PTEN (+/-) mice versus wild type littermates; (2) PC3-bearing versus non-tumor-bearing mice; (3) U87MG-bearing versus non-tumor-bearing mice; and (4) pictilisib-treated (100 mg/kg) versus vehicle control-treated mice bearing PC3 prostate tumors or U87MG glioblastoma). Metabolites responsible for changes were identified using orthogonal partial least square-discriminant analysis (OPLS-DA) (27) with a threshold of variable importance in the projection (VIP) value >0.8. To pass the exploratory preclinical screen, a metabolite was required to change consistently across both xenograft-bearing mouse models treated with pictilisib (passing the OPLS-DA filter) and at least one other preclinical model involving the transgenic PTEN (+/-) or PTEN null tumor xenograft-bearing mice (compared with wild-type and non-tumor-bearing littermates, respectively) with no contradictory change in any other model. Permutation analysis was carried out for model validation. Heat maps were generated using the concentration changes relative to vehicle control animals at each time point. In the clinical study, changes relative to pre-treatment baseline levels on day 1 were calculated for each patient across all time points; the geometric mean, associated 95% confidence intervals and p-values (H0: no different between pre- and post-treatment levels) of relative change were estimated for each time point and dose range (low: 45-60 mg [n=6]; mid: 80-245 mg [n=8]; high: 330-450 mg [n=27]). For unsupervised multivariate analysis, principal component analysis (PCA) was undertaken using SIMCA v.13 software. In addition, the SPSS program (v.21, Chicago, USA) and GraphPad Prism (v.6, California, USA) were used for all other statistical analyses. All p-values presented are two-sided and values <0.05 were considered statistically significant.
Results
Preclinical models
Mice with PTEN-deficient tumors have distinct plasma metabolite profiles
At 8 weeks of age, no significant differences were observed in the plasma metabolite profile between genetically engineered PTEN (+/-) mice with no observable macroscopic tumors compared with wild-type littermates. At 25 weeks, palpable tumors, splenomegaly and thymic enlargements were observed in the PTEN (+/-) mice. Using OPLS-DA, the concentrations of 53 plasma metabolites were found to be significantly decreased in PTEN (+/-) transgenic mice compared with wild-type littermates.
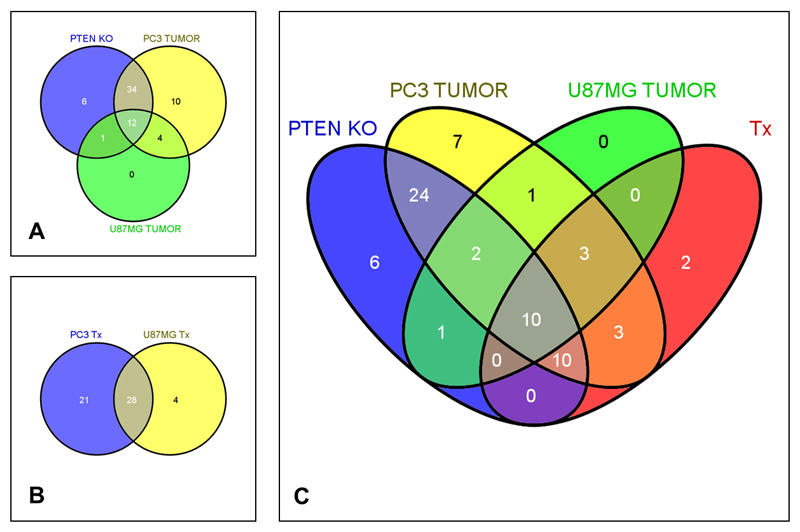
In addition, when mice harboring human tumor xenografts were compared with non-tumor bearing littermates, a decrease was observed in the levels of 60 and 17 plasma metabolites for the PTEN-negative PC3 prostate and U87MG glioblastoma models respectively. The overlap of the changes seen in the human tumor xenografts and the PTEN-depleted transgenic mouse tumors is illustrated in Fig. 2A. Twelve plasma metabolites were consistently altered across all three animal models, with reductions seen in the PI3K pathway-aberrant tumor-bearing mice; metabolites affected included amino acids, acylcarnitines and phospholipids. We note that the greatest overlap was between the PTEN-depleted versus wild-type littermates and the PC3 tumor-bearing versus non-tumor-bearing comparison. Regardless of the number of plasma metabolites identified in each comparison, the majority of changes in each case were common to the three models. Of note, high degrees of separation between the experimental (tumor-bearing, transgenic or treated animals) and the relevant control groups were observed in the OPLS-DA, with all R2>0.8 and Q2>0.6 (representing the explained variance and predictive capability in the internal cross-validation, respectively).
Figure 2.
Venn diagrams showing the overlap in plasma metabolites between preclinical animal models: (A) PTEN +/- (depleted) versus wild-type littermates and athymic mice bearing PTEN null human PC3 prostate carcinoma or U87MG glioblastoma xenografts compared with non-tumor bearing age-matched controls; (B) treatment with pictilisib versus vehicle in PC3- and U87MG-bearing mice; and (C) overlap between changes in all 3 models in (A) and those common to both models in (B). Metabolites that were further evaluated in the clinical setting are enclosed by a thick black line. (Tx: treatment)
In contrast to the number of metabolites noted above showing decreased plasma levels, only a few metabolites across the panel assessed were elevated in the xenograft tumor-bearing mice and none was significantly elevated in the PTEN-depleted model.
Potent and specific pharmacological inhibition of Class I PI3K using pictilisib is associated with changes in the plasma metabolome
We sought to examine the circulating metabolite changes at doses of pictilisib inhibiting the phosphorylation of AKT Ser473 over 24 hours in platelet rich plasma (17). OPLS-DA of the comparisons between plasma metabolites in pictilisib (at 100 mg/kg) and control vehicle-treated mice revealed an increase in the concentrations of 49 and 32 plasma metabolites in mice bearing PC3 and U8MG tumors respectively. Of these, 28 metabolites were common to both models (Fig. 2B). Moreover, these metabolites were similarly increased in mice bearing U87MG human tumor xenografts treated with 100 mg/kg GDC-0941 or 60 mg/kg buparlisib (Supplementary Fig. 1). When these pictilisib treatment-related changes were compared with those illustrated in Fig. 2A, 10 plasma metabolites were observed to exhibit variations consistent across all 5 models: 13 metabolites in 4 of 5 models and 3 metabolites in 3 of 5 models.
Based on these data, a panel of 26 plasma metabolites was selected for further clinical evaluation. The relevant changes observed in this panel of 26 metabolites across the various models along with their patterns of overlap are summarized in Fig. 2 and Supplementary Table 1.
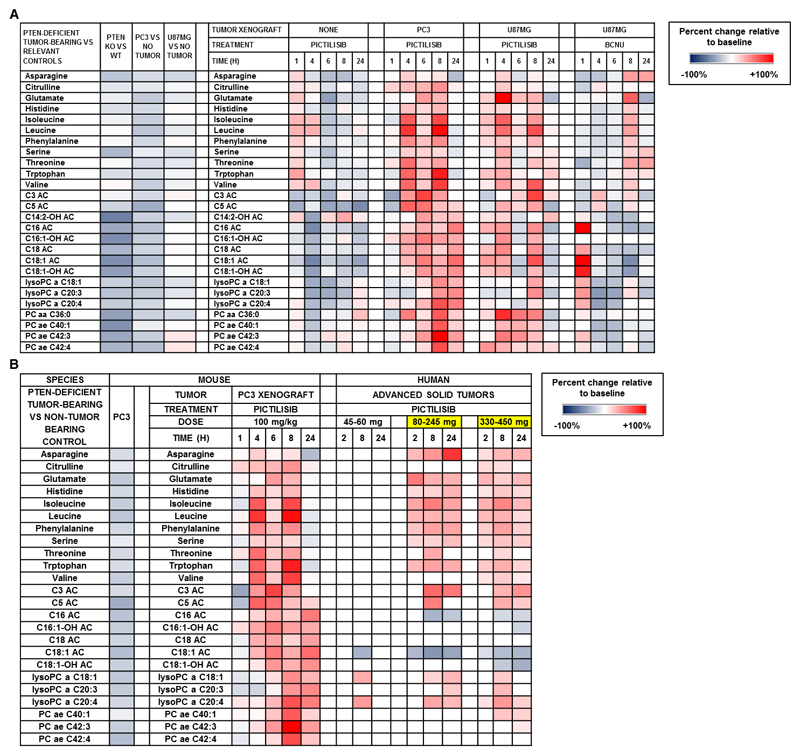
The plasma concentrations of 6 of 9 essential amino acids (histidine, phenylalanine, threonine, valine. isoleucine and tryptophan) were significantly decreased in the genetically engineered PTEN+/- tumor-bearing animals and subsequently increased following treatment with pictilisib in these mice. The levels of non-essential amino acids, including asparagine, citrulline, glutamate and serine, were similarly decreased in the tumor-bearing animals and increased following treatment with pictilisib in tumor-bearing mice. The concentrations of acylcarnitines (with the following acyl-side chains: C3, C5, C16, C14:2-OH, C16:1-OH, C18, C18:1 and C18:1-OH) were lower in the plasma of PTEN (+/-) animals and/or PC3-bearing mice compared with control animals and in addition were increased following pictilisib administration in both PC3- and U87MG tumor-bearing mice. Significant changes in phosphocholine metabolism were observed with lysophosphatidylcholine C18:1, C20:3 and C20:4, acyl-acyl phosphatidylcholine C36:0, and acyl-alkyl phosphatidylcholines C40:1, C42:3 and C42:4 decreased in at least one tumor-bearing model and increased following pictilisib treatment of tumor-bearing mice.
Thus, following the administration of a single dose of pictilisib at doses where AKT phosphorylation is inhibited (17), the observed increase in the plasma concentrations of the candidate biomarkers in both tumor-bearing models represented a reversal of changes observed in the presence of PTEN-deficient tumors (Fig. 3). The post-treatment changes were generally greater in the PC3-bearing mice, occurred mainly after 4 hours, and persisted for up to 24 hours or more post-dose (Fig. 3). The metabolite changes observed in plasma from U87MG-bearing mice following treatment with pictilisib were mirrored following treatment with buparlisib (Supplementary Fig. 1).
Figure 3.
(A) Heat map of differences between transgenic PTEN-deficient tumor-bearing mice compared with their normal PTEN wild-type littermates (change relative to control) and changes across time in candidate plasma metabolite biomarkers following treatment with pictilisib or carmustine (relative to vehicle control) in tumor and non-tumor bearing mice. (a, acyl; aa, acyl-acyl; ae, acyl-alkyl; Cx:y, where x is the number of carbons in the fatty acid side chain; y is the number of double bonds in the fatty acid side chain; AC, acylcarnitine; DC, decarboxyl; M, methyl; OH, hydroxyl; PC, phosphatidylcholine) (B) Heat map (in right panel with header labelled ‘HUMAN’) of statistically significant changes (relative to baseline) in plasma concentrations of metabolite biomarker candidates in patients treated in the dose-escalation trial of pictilisib. Juxtaposed (in left panel with header labelled ‘MOUSE’) are changes seen in selected preclinical models as shown in Figure 3A. Highlighted in yellow are the dose levels of pictilisib at which modulation of mechanism-based pharmacodynamic biomarkers including phosphorylated S6 ribosomal protein and AKT, and 18FDG-PET were observed. (a, acyl; aa, acyl-acyl; ae, acyl-alkyl; Cx:y, where x is the number of carbons in the fatty acid side chain; y is the number of double bonds in the fatty acid side chain; AC, acylcarnitine; DC, decarboxyl; M, methyl; OH, hydroxyl; PC, phosphatidylcholine)
In contrast to pictilisib treatment of tumor-bearing mice, there was a general trend of reduction, rather than an increase, of the metabolites following administration of the PI3K inhibitor to non-tumor-bearing mice. Of note, a transient and slight increase in plasma levels of amino acids was followed by a reduction in their levels from 4 to 6 h post-dose.
The metabolite changes described above are represented in the heat map in Fig. 3A. Taken together, the data define a set of changes in the plasma metabolome that are responsive to genetic and pharmacologic perturbation of the PI3K pathway.
The chemotherapeutic agent carmustine induces plasma metabolite changes distinct from those following PI3K inhibition in U87MG tumor-bearing mice
As an additional control, we additionally investigated changes in the plasma metabolites described above in U87MG tumor-bearing mice following treatment with carmustine as a chemotherapeutic clinical standard of care for glioblastoma. The observed changes were different from those in pictilisib-treated tumor-bearing mice. In the carmustine-treated U87MG-bearing mice, 2 distinct groups of plasma metabolite changes were observed: (1) the amino acids and short chain acylcarnitines showed a reduction in level followed by an increase at 8 hours post-dose; and (2) the long chain acylcarnitines and phosphatidylcholines generally increased acutely and then decreased from 4 hours post-dose (Fig. 3A).
The major differences in plasma metabolome changes for the cytotoxic agent carmustine versus the selective PI3K inhibitor pictilisib provide evidence in support of specificity for the effects seen with the latter agent.
Baseline characteristics of participants in Phase I dose-escalation clinical trial of pictilisib
Plasma samples were obtained from 41 advanced cancer patients with no conventional treatment options who participated in a dose-escalation clinical trial of pictilisib (20, 24). The treatment dosing and plasma sampling schedules are summarized in Fig. 1B. Modulation of mechanism-based pharmacodynamic biomarkers, including phosphorylation of S6 ribosomal protein and AKT as well as 18FDG-PET uptake, was observed at dose levels ≥80 mg once-daily (20, 24).
Clinical evaluation of candidate plasma metabolite biomarkers
For the 26 plasma metabolome biomarkers selected from the non-targeted exploratory preclinical study described above, the mean intra-patient coefficient of variation in baseline clinical plasma samples ranged from 7 to 47%. Apart from C14:2-OH acylcarnitine and acyl-acyl phosphatidylcholine C36:0, all metabolites had a mean coefficient of variation of <20% (Supplementary Table 2). Further clinical evaluation was not carried out for the 2 metabolites with high coefficients of variation, leaving 24 metabolites that we pursued further.
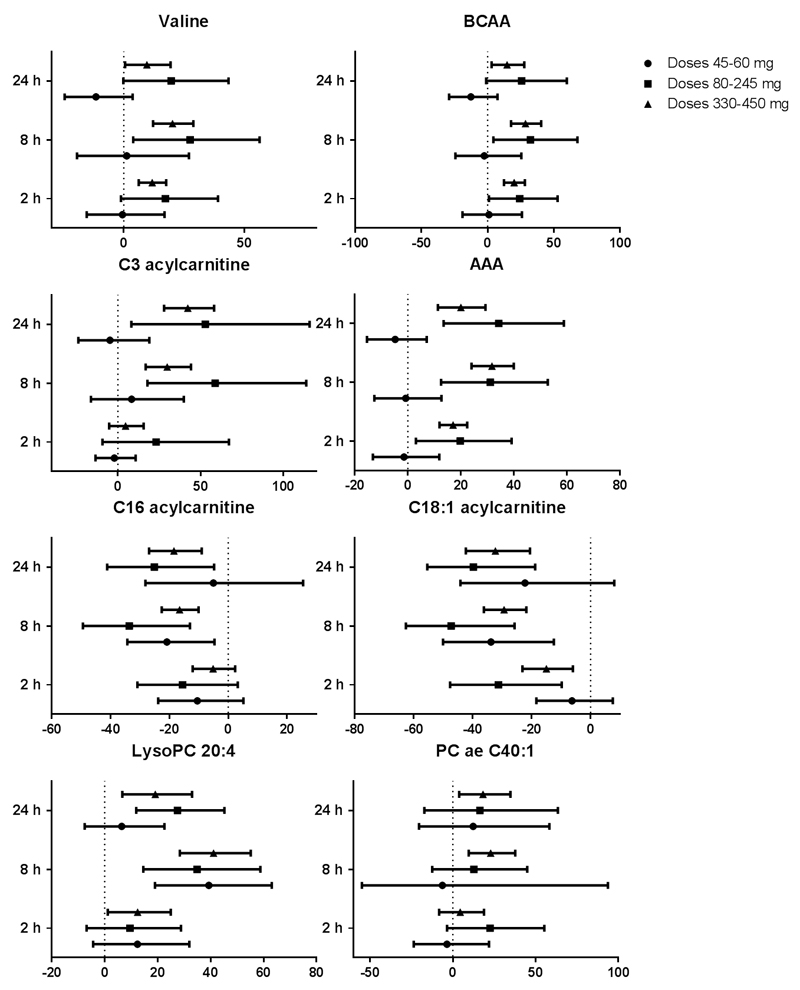
Following single dose pictilisib treatment on day 1, dose- and time-dependent changes were observed in 22 of these 24 candidate biomarkers, with only C18 acylcarnitine and acyl-alkyl phosphatidylcholine C42:4 not changing significantly (Fig. 3B). The maximal changes post-treatment were greater than baseline variability for each of these metabolites (Supplementary Tables 2, 3 and 4). Significant changes were observed 2 hours post-dose in 13 of 24 metabolites and were detected only at doses of pictilisib ≥80 mg. This modulation of circulating biomarker coincides with doses where inhibition of AKT phosphorylation Ser473 was observed in platelet rich plasma (20). The highest number and the most significant changes occurred at 8 hours post-dose with 19 of 24 candidate metabolites showing statistically significant changes; persistence of these changes was seen at 24 hours post-dose for 18 metabolites. Only 3 metabolites demonstrated significant changes following administration of pictilisib in the lowest dose range (<80 mg once-daily); these changes were observed at 8 hours post-dose only. At 8 and 24 hours post-administration, the greatest frequency of statistically significant changes was associated with the highest dose range. The observed changes were generally consistent with that seen in the pictilisib-treated tumor-bearing mice, the only exception being the long chain acylcarnitines for which a decrease was observed in patients following pictilisib treatment. The latter changes were more similar to those seen in the non-tumor bearing as opposed to the tumor xenograft-bearing mice. The various changes are summarized in Figs. 3B and 4 and presented in Supplementary Table 3.
Figure 4.
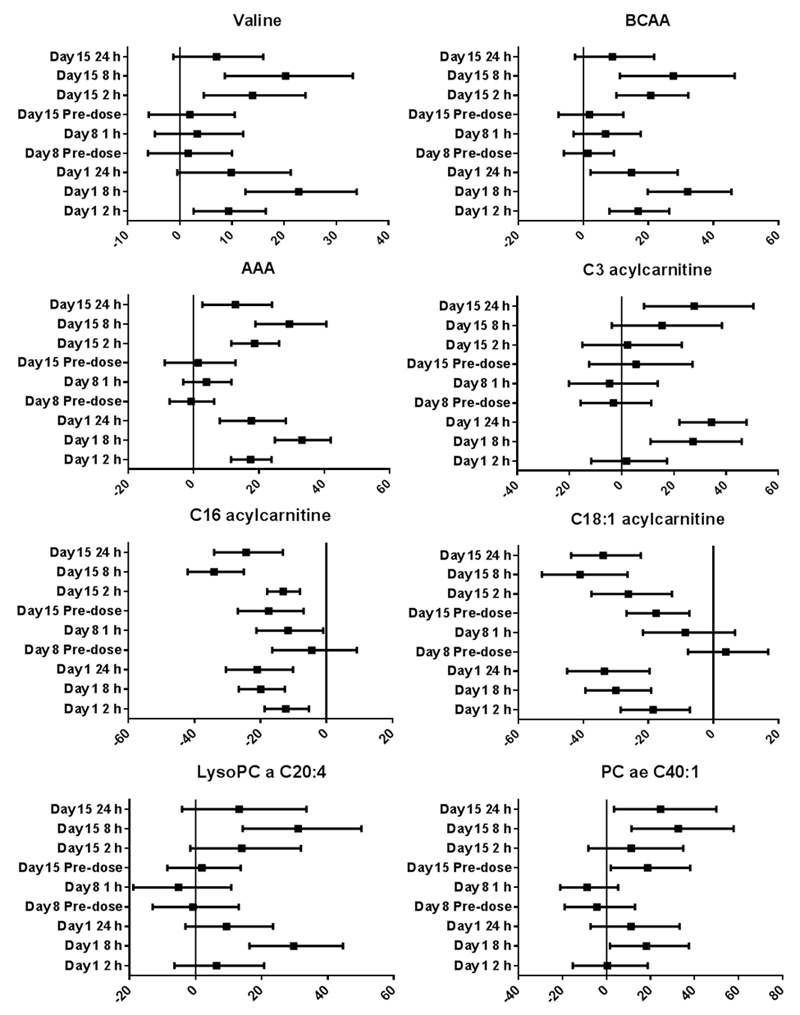
Percentage change relative to baseline of selected plasma metabolites at 2, 8 and 24 hours post-pictilisib dosing in 41 patients treated at different dose levels. Geometric means with 95% confidence intervals are represented in the plots. (BCAA, Branched chain amino-acids; AAA, aromatic amino-acids; PC, phosphatidylcholine)
In 17 patients treated with pictilisib at doses ≥330 mg once-daily, additional plasma samples were obtained on days 8 and 15 of the continuous dosing phase. Note that all patients received a single dose of pictilisib on day 1, followed by a one-week drug holiday; dosing recommenced on day 8 on a continuous once-daily schedule. Importantly, following dosing on day 15, the trends and directionality of pictilisib-induced metabolome changes for the panel of 24 candidate metabolite biomarkers were similar to those observed for day 1 dosing. To illustrate this, plots of selected metabolites and metabolite classes are presented in Fig. 5 and detailed results of these analyses are provided in Supplementary Table 2.
Figure 5.
Percentage change relative to baseline of selected plasma metabolites at planned time points over 2 weeks on pictilisib treatment in 17 patients treated at dose levels ≥330 mg once-daily. Geometric means with 95% confidence intervals are represented in the plots. Other details are as indicated in the legend to Figure 4.
Of the panel of 24 metabolites, none exhibited a significant change from the original baseline concentration prior to recommencement of pictilisib on day 8, indicating a return to baseline after drug washout. Just prior to dosing on day 15, only 4 metabolites were significantly different from baseline levels.
We conclude that for the Phase I cohort of patients studied, we demonstrated successful translation and validation of the targeted panel of plasma metabolites responsive to PI3K inhibition that we initially discovered in the preclinical non-targeted exploratory screen.
Exploratory assessment of relationship between metabolite changes with other pharmacodynamic and clinical parameters
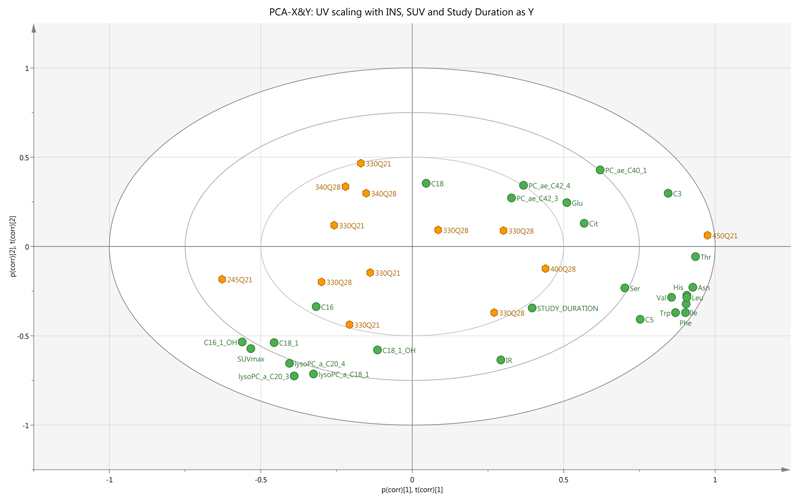
Unsupervised multivariate analysis based on the 22-metabolite signature revealed an association between higher dose levels, longer study durations, greater insulin resistance with increase in amino acid and short chain acylcarnitine levels but decrease in SUVmax and levels of long chain acylcarnitines (Fig. 6).
Figure 6.
PCA biplot comprising superimposed scatter and loading plots showing relationship between plasma metabolite changes (as X components), insulin resistance (IR), study duration and SUVmax (as Y components) as well as dose of pictilisib (labelled for each observation) (n=13 patients). Clustering of changes in levels of branched chain and aromatic amino acids, study duration and insulin resistance is noted; these variables are broadly related to inverse changes in SUVmax and changes in long chain acylcarnitines and lysophosphatidylcholine levels.
Discussion
In this study, we show that a panel of circulating metabolite biomarkers we discovered for assessing target engagement in experimental models have successfully translated to a Phase I clinical trial. Perturbations of the PI3K pathway induced either by genetic or pharmacological means resulted in changes in the plasma metabolome, comprising amino acids, acylcarnitines and phosphatidylcholines. We show that the concentrations of 26 plasma metabolites were decreased in various mouse models of PTEN-deficient tumors and increased following treatment with pictilisib (a potent and specific pan-class I inhibitor of PI3K) in mice bearing PTEN-deficient human prostate and glioblastoma tumor xenografts. Excluding the 2 plasma metabolites with high physiological baseline variability, we found that 22 of 24 of the identified plasma metabolome biomarker candidates exhibited significant dose- and time-dependent changes in response to PI3K inhibition when evaluated in the dose-escalation Phase I clinical trial of pictilisib.
Although an increasing number of novel inhibitors of the PI3K pathway are entering clinical trials, currently available pharmacodynamic biomarkers are restricted by issues including biomarker signal lability, risk of sampling complications, limited sensitivity and throughput, high costs of the associated analytical methods and lack of amenability to repeated sampling (2). In this context, the present study provides a general framework for the rational discovery of pharmacodynamic plasma metabolite biomarkers. In the preclinical exploratory stage, we generated a panel of PI3K-responsive metabolomics plasma biomarker candidates from model systems that are genetically defined and maintained under a well-controlled laboratory environment. These metabolite biomarkers were subsequently assessed and confirmed in the clinical stage where time- and dose-dependencies of the candidate metabolite biomarkers were carefully evaluated. The use of a combination of orthogonal genetic and pharmacologic preclinical pathway perturbation models to screen for mechanism-based metabolite candidates provided increased rigor, decreasing the likelihood of false positive leads, and also reduced the size of the panel of potential metabolite biomarkers for subsequent targeted analysis. In addition, the chosen LC-MS analytical platform is robust, reproducible and high throughput; sample preparation for a 96-well plate takes about 2 hours and MS data acquisition is completed after 12 hours. This is on a background of generally increasing affordability and uptake of LC-MS instrumentation in recent years.
Importantly, the changes observed in the majority of metabolites that we discovered in the preclinical screen were subsequently confirmed in a Phase I clinical trial setting, providing confidence in the preclinical biomarker discovery strategy. Since an analytically validated, quantitative platform was used to generate absolute concentrations of the assessed metabolites, we were therefore able to compare the data obtained in preclinical models and those in patients.
We and others have previously shown that the interpretation of clinical metabolite pharmacodynamic data is challenging and may be confounded by variables such as food and diurnal variation (28–30). To circumvent these potential effects, we carried out multiple clinical measurements including: (1) assessment of baseline pre-treatment variability in metabolites of interest taken on two separate days; (2) examination of the relationship between metabolite changes and dose levels on day 1 of the clinical trial; and (3) comparison of metabolite concentrations in patients treated at the maximal tolerated dose or higher. In addition, plasma levels over 24h on day 15 following a week of continuous once-daily oral dosing were compared with concentrations on day 8 after 1 week washout after drug administration. In our clinical study of 41 patients, the majority (22 of 24) of our identified biomarker candidates demonstrated consistent changes across these assessments and the observed changes post-treatment were significantly greater than pre-treatment baseline variability (Supplementary Tables 2, 3 and 4). These effects occurred at pictilisib doses for which mechanism-based biomarker modulation (plasma glucose and insulin levels, phosphorylated S6 ribosomal protein and AKT as well as 18FDG-PET uptake) has already been observed in the clinical study (20). The concordance of these clinical observations with our preclinical findings increases the likelihood that the plasma metabolite changes that we observed are genuinely associated with systemic PI3K pathway modulation. Furthermore, an exploratory unsupervised multivariate analysis including patients in whom mechanism-based pharmacodynamics was concurrently performed revealed associations between insulin resistance, 18FDG-PET changes and candidate plasma metabolites. Importantly, these observations increased our confidence of the choice of the recommended Phase II dose-schedule.
The present study has provided proof-of-feasibility for the use of a broad, systems-based strategy, translating from mouse models to a Phase I clinical trial setting, to identify effects on the metabolome. We identified numerous amino acids in the plasma, including the branched chain (BCAA) and aromatic (AAA) amino acids, along with the products of their catabolism, the short-chain C3 and C5 acylcarnitines that were down-regulated in the presence of PTEN-depleted tumors. Moreover, we showed that these changes were reversed by PI3K pathway inhibition using the pan-class I PI3K inhibitor pictilisib. Importantly, elevated levels of these plasma metabolites have been consistently associated with insulin resistance, are predictive of the risk of developing diabetes mellitus, and are highly responsive to anti-diabetic interventions (31–35). Furthermore, numerous studies have demonstrated a direct role of BCAA and AAA in contributing to the development of insulin resistance. The infusion of amino acids (particularly BCAA) into humans and animals has been shown to lead to insulin resistance, accompanied by activation of mTOR, S6K1 and phosphorylation of IRS-1 (36). Addition of BCAA or AAA to cultured muscle cells in vitro leads to activation of mTOR and impairment of insulin-stimulated phosphorylated AKT and glucose uptake (37). Amino acids such as leucine and glutamate are also potent insulin secretagogues (38). To put our metabolite findings into pharmacologic context, dose-dependent hyperglycemia and hyperinsulinemia were reported in the Phase I clinical trials of pictilisib and buparlisib (20, 24, 39). The adverse event of hyperglycemia was the dose-limiting toxicity of the latter agent. The acute and chronic pharmacologic inhibition of the PI3K pathway has been shown to impact differentially on glucose homeostasis; specifically, chronic dosing of pan-class I inhibitors was associated with an attenuation of treatment-related hyperglycemia but not insulin sensitivity (40, 41). Furthermore, chronic genetic inhibition of p110α was found to protect mice from age-related reduction in insulin sensitivity, glucose tolerance and fat accumulation (42).
The increase in short chain acylcarnitines that we observed is consistent with the reported PI3K-AKT signaling and is linked to suppression of β-oxidation via inhibition of carnitine palmitoyltransferase 1a (CPT1a) gene expression and a glucose-addicted phenotype (8, 43). Based on this model, one would expect an increase in CPT1a level following pharmacologic PI3K inhibition and hence an increase in long chain acylcarnitine levels. We observed such changes consistently in both of the pictilisib-treated human tumor xenograft mouse models studied here. This has also been reported by others in human cancer cells treated in vitro (43). However, contrary to expectations, a significant decrease in the plasma levels of these particular metabolites was observed in humans, an effect that is similar to the changes we observed in non-tumor-bearing mice following pictilisib treatment. Lower levels of long chain acylcarnitines (in contrast to their C3 and C5 counterparts) have been linked to insulin resistance and may partly explain our findings (34). The regulation of plasma long chain acylcarnitine level is complex and reflects the balance between processes such as fatty acid synthesis and hydrolysis and β-oxidation. The described differences between murine and human mitochondrial β-oxidation of long chain fatty acids may also account for the differences that we observed (44).
Our studies show a significant increase in specific lysophosphatidylcholines and phosphatidylcholine following treatment with pictilisib in plasma and tumor. This is in contrast with observations in glioblastoma cells by NMR where decreases in total glycerophosphocholine and phosphocholine have been reported largely supported by by the decrease in choline kinase (45, 46). However, other studies performed by 31P-NMR in breast cancer models have reported an increase in these metabolites attributed to increased levels of phospholipase A2 group 4A (47). Lysophosphatidylcholine acyltransferase 1 (LPCAT1), has been studied in colorectal, gastric and prostate cancers (48–50) where low plasma levels of these metabolites are associated with the presence of cancer, supporting our preclinical findings. Lysophosphatidylcholines enhance glucose-dependent insulin secretion from pancreatic β-cells and inhibit insulin-induced AKT activation through protein kinase C α in vascular smooth muscle cells (51, 52). Interestingly, the long chain carnitines, lysophosphatidylcholine and phosphatidylcholine that were significantly increased in our studies are even-chain. It has been shown that the associated even-chain saturated fatty acids are positively associated with diabetes mellitus whilst odd-chain derivatives are inversely associated with it (53).
We observed that the plasma levels of the vast majority of our large panel of metabolites in the transgenic PTEN (+/-) mice were considerably lower compared with wild-type littermates. These changes showed a significant overlap with plasma metabolite levels we determined in the presence of PTEN null human tumor xenografts, especially the PC3 prostate carcinoma. Our results are consistent with published clinical data on plasma amino acids being reduced in a large series of patients with advanced cancer although they are contradicted by data from smaller studies (54, 55). We note also that the metabolite changes were greater in mice harbouring PC3 prostate carcinoma compared with U87MG glioblastoma xenografts; pictilisib is a P-glycoprotein substrate and the marked difference in P-glycoprotein expression between the two xenografts (absent in the former and present in the latter) may have contributed to this observation (56,57).
Whilst the assessment of plasma metabolomics in patients cannot distinguish between tumor- and non-tumor-related changes, it is interesting that the plasma metabolite changes that we observed following pictilisib treatment between tumor- and non-tumor-bearing animals were distinct. We propose that this difference may be attributed to the presence of tissue with a hyperactivated PI3K pathway in the former. PTEN null human tumor xenografts were chosen to increase the likelihood of detecting pathway-related metabolic changes, i.e. drug inhibition of a hyperactivated pathway. Further follow-up studies are now required to determine metabolome profiles in a wider range of tumor models with different PI3K pathway activation status and using various PI3K pathway inhibitory drugs and other molecular targeted inhibitors.
In summary, using an analytically validated, highly sensitive, high-throughput, LC-MS-based metabolomics approach, we have identified distinct plasma metabolome changes that were consistently altered across different preclinical tumor models in which the PI3K pathway is activated by PTEN depletion. In the clinical evaluation of the candidate pharmacodynamic plasma metabolome biomarkers, time- and dose-dependent changes were observed for the majority of the metabolites. These findings suggest that under controlled conditions, plasma metabolomics is a valid tool to assess biomarker modulation in early clinical studies.
Supplementary Material
Acknowledgements for Research Support
F.I. Raynaud, P. Workman, S.A. Eccles, A.D.H. Brandon, G. Box, M. Valenti, A. Henley, Y. Asad are supported by a Cancer Research UK programme grant (C309/A11566) at the Cancer Research UK Cancer Therapeutics Unit. P. Workman is a Cancer Research UK Life Fellow (C309/A8992). J.E. Ang was supported by a Wellcome Trust PhD studentship grant (090952/Z/09/Z) as part of the Wellcome Trust PhD programme in mechanism-based drug discovery research project at The Institute of Cancer Research which is directed by P. Workman. R. Pandher was funded by a PhD studentship from The Institute of Cancer Research. The Phase I clinical trial was supported by Genentech Inc., The Drug Development Unit, the Royal Marsden NHS Foundation Trust and The Institute of Cancer Research. Support was also provided by Experimental Cancer Medicine Centre grants to The Institute of Cancer Research and from the National Health Service to the National Institute for Health Research Biomedical Research Centre at The Institute of Cancer Research and the Royal Marsden Hospital. Work in the laboratory of B. Vanhaesebroeck is supported by Cancer Research UK (C2338/A15965).
Footnotes
Disclosures and Potential Conflicts of Interest: J.E. Ang, Y. Asad, A. Henley, M. Valenti, G..Box, A.D.H. Brandon., S.A. Eccles, S.B. Kaye, P. Workman, and F.I. Raynaud are employees of The Institute of Cancer Research which has a financial interest in the development of pictilisib. L. Friedman and M. Derynck are employees of Genentech Inc. which is developing pictilisib. B. Vanhaesebroeck is a consultant to Activiomics (London, UK) and Karus Therapeutics (Oxford, UK). P. Workman is a founder of Piramed Pharma which has been acquired by Roche.
References
- 1.Yap TA, Sandhu SK, Workman P, de Bono JS. Envisioning the future of early anticancer drug development. Nat Rev Cancer. 2010;10:514–23. doi: 10.1038/nrc2870. [DOI] [PubMed] [Google Scholar]
- 2.Ang JE, Kaye S, Banerji U. Tissue-based approaches to study pharmacodynamic endpoints in early phase oncology clinical trials. Curr Drug Targets. 2012;13:1525–34. doi: 10.2174/138945012803530062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sarker D, Workman P. Pharmacodynamic biomarkers for molecular cancer therapeutics. Adv Cancer Res. 2007;96:213–68. doi: 10.1016/S0065-230X(06)96008-4. [DOI] [PubMed] [Google Scholar]
- 4.Workman P. How much gets there and what does it do?: The need for better pharmacokinetic and pharmacodynamic endpoints in contemporary drug discovery and development. Curr Pharm Des. 2003;9:891–902. doi: 10.2174/1381612033455279. [DOI] [PubMed] [Google Scholar]
- 5.Yap TA, Lorente D, Omlin A, Olmos D, de Bono JS. Circulating tumor cells: a multifunctional biomarker. Clin Cancer Res. 2014;20:2553–68. doi: 10.1158/1078-0432.CCR-13-2664. [DOI] [PubMed] [Google Scholar]
- 6.Winfield JM, Payne GS, deSouza NM. Functional MRI and CT biomarkers in oncology. Eur J Nucl Med Mol Imaging. 2015 doi: 10.1007/s00259-014-2979-0. [DOI] [PubMed] [Google Scholar]
- 7.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nature reviews Genetics. 2006;7:606–19. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 8.Deberardinis RJ, Lum JJ, Thompson CB. Phosphatidylinositol 3-kinase-dependent modulation of carnitine palmitoyltransferase 1A expression regulates lipid metabolism during hematopoietic cell growth. J Biol Chem. 2006;281:37372–80. doi: 10.1074/jbc.M608372200. [DOI] [PubMed] [Google Scholar]
- 9.Goberdhan DC, Ogmundsdottir MH, Kazi S, Reynolds B, Visvalingam SM, Wilson C, et al. Amino acid sensing and mTOR regulation: inside or out? Biochem Soc Trans. 2009;37:248–52. doi: 10.1042/BST0370248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foukas LC, Claret M, Pearce W, Okkenhaug K, Meek S, Peskett E, et al. Critical role for the p110alpha phosphoinositide-3-OH kinase in growth and metabolic regulation. Nature. 2006;441:366–70. doi: 10.1038/nature04694. [DOI] [PubMed] [Google Scholar]
- 11.Powis G, Ihle N, Kirkpatrick DL. Practicalities of drugging the phosphatidylinositol-3-kinase/Akt cell survival signaling pathway. Clin Cancer Res. 2006;12:2964–6. doi: 10.1158/1078-0432.CCR-06-0617. [DOI] [PubMed] [Google Scholar]
- 12.Wong JT, Kim PT, Peacock JW, Yau TY, Mui AL, Chung SW, et al. Pten (phosphatase and tensin homologue gene) haploinsufficiency promotes insulin hypersensitivity. Diabetologia. 2007;50:395–403. doi: 10.1007/s00125-006-0531-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pal A, Barber TM, Van de Bunt M, Rudge SA, Zhang Q, Lachlan KL, et al. PTEN mutations as a cause of constitutive insulin sensitivity and obesity. N Engl J Med. 2012;367:1002–11. doi: 10.1056/NEJMoa1113966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8:627–44. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Workman P, Clarke PA, Raynaud FI, van Montfort RL. Drugging the PI3 kinome: from chemical tools to drugs in the clinic. Cancer Res. 2010;70:2146–57. doi: 10.1158/0008-5472.CAN-09-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Folkes AJ, Ahmadi K, Alderton WK, Alix S, Baker SJ, Box G, et al. The identification of 2-(1H-indazol-4-yl)-6-(4-methanesulfonyl-piperazin-1-ylmethyl)-4-morpholin-4-yl-t hieno[3,2-d]pyrimidine (GDC-0941) as a potent, selective, orally bioavailable inhibitor of class I PI3 kinase for the treatment of cancer. J Med Chem. 2008;51:5522–32. doi: 10.1021/jm800295d. [DOI] [PubMed] [Google Scholar]
- 17.Raynaud FI, Eccles SA, Patel S, Alix S, Box G, Chuckowree I, et al. Biological properties of potent inhibitors of class I phosphatidylinositide 3-kinases: from PI-103 through PI-540, PI-620 to the oral agent GDC-0941. Mol Cancer Ther. 2009;8:1725–38. doi: 10.1158/1535-7163.MCT-08-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clarke PA, Workman P. Phosphatidylinositide-3-kinase inhibitors: addressing questions of isoform selectivity and pharmacodynamic/predictive biomarkers in early clinical trials. J Clin Oncol. 2012;30:331–3. doi: 10.1200/JCO.2011.38.7167. [DOI] [PubMed] [Google Scholar]
- 19.Shuttleworth SJ, Silva FA, Cecil AR, Tomassi CD, Hill TJ, Raynaud FI, et al. Progress in the preclinical discovery and clinical development of class I and dual class I/IV phosphoinositide 3-kinase (PI3K) inhibitors. Curr Med Chem. 2011;18:2686–714. doi: 10.2174/092986711796011229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarker D, Ang JE, Baird R, Kristeleit R, Shah K, Moreno V, et al. First-in-human phase I study of pictilisib (GDC-0941), a potent pan-class I phosphatidylinositol-3-kinase (PI3K) inhibitor, in patients with advanced solid tumors. Clin Cancer Res. 2015;21:77–86. doi: 10.1158/1078-0432.CCR-14-0947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–7. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 22.Workman P, Aboagye EO, Balkwill F, Balmain A, Bruder G, Chaplin DJ, et al. Guidelines for the welfare and use of animals in cancer research. Br J Cancer. 2010;102:1555–77. doi: 10.1038/sj.bjc.6605642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Podsypanina K, Ellenson LH, Nemes A, Gu J, Tamura M, Yamada KM, et al. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc Natl Acad Sci U S A. 1999;96:1563–8. doi: 10.1073/pnas.96.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banerjee S, Baird RD, Basu B, Shah K, Tunariu N, Moreno Garcia V, et al. A Phase I Study Evaluating GDC-0941, a Pan-Phosphoinositide-3 Kinase (PI3K) Inhibitor, in Patients with Advanced Solid Tumours, Multiple Myeloma, and PIK3CA Mutant Tumours. European Journal of Cancer. 2011;47(Supplement 1):S159. [Google Scholar]
- 25.Wallace TM, Levy JC, Matthews DR. Use and abuse of HOMA modeling. Diabetes Care. 2004;27:1487–95. doi: 10.2337/diacare.27.6.1487. [DOI] [PubMed] [Google Scholar]
- 26.Pandher R, Ducruix C, Eccles SA, Raynaud FI. Cross-platform Q-TOF validation of global exo-metabolomic analysis: application to human glioblastoma cells treated with the standard PI 3-Kinase inhibitor LY294002. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:1352–8. doi: 10.1016/j.jchromb.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 27.Svensson PA, Olson FJ, Hagg DA, Ryndel M, Wiklund O, Karlstrom L, et al. Urokinase-type plasminogen activator receptor is associated with macrophages and plaque rupture in symptomatic carotid atherosclerosis. Int J Mol Med. 2008;22:459–64. [PubMed] [Google Scholar]
- 28.Ang JE, Revell V, Mann A, Mantele S, Otway DT, Johnston JD, et al. Identification of human plasma metabolites exhibiting time-of-day variation using an untargeted liquid chromatography-mass spectrometry metabolomic approach. Chronobiol Int. 2012;29:868–81. doi: 10.3109/07420528.2012.699122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dallmann R, Viola AU, Tarokh L, Cajochen C, Brown SA. The human circadian metabolome. Proc Natl Acad Sci U S A. 2012;109:2625–9. doi: 10.1073/pnas.1114410109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davies SK, Ang JE, Revell VL, Holmes B, Mann A, Robertson FP, et al. Effect of sleep deprivation on the human metabolome. Proc Natl Acad Sci U S A. 2014;111:10761–6. doi: 10.1073/pnas.1402663111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–53. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newgard CB. Interplay between lipids and branched-chain amino acids in development of insulin resistance. Cell Metab. 2012;15:606–14. doi: 10.1016/j.cmet.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shah SH, Crosslin DR, Haynes CS, Nelson S, Turer CB, Stevens RD, et al. Branched-chain amino acid levels are associated with improvement in insulin resistance with weight loss. Diabetologia. 2012;55:321–30. doi: 10.1007/s00125-011-2356-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laferrere B, Reilly D, Arias S, Swerdlow N, Gorroochurn P, Bawa B, et al. Differential metabolic impact of gastric bypass surgery versus dietary intervention in obese diabetic subjects despite identical weight loss. Sci Transl Med. 2011;3:80re2. doi: 10.1126/scitranslmed.3002043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andujar-Plata P, Pi-Sunyer X, Laferrere B. Metformin effects revisited. Diabetes Res Clin Pract. 2012;95:1–9. doi: 10.1016/j.diabres.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tremblay F, Krebs M, Dombrowski L, Brehm A, Bernroider E, Roth E, et al. Overactivation of S6 kinase 1 as a cause of human insulin resistance during increased amino acid availability. Diabetes. 2005;54:2674–84. doi: 10.2337/diabetes.54.9.2674. [DOI] [PubMed] [Google Scholar]
- 37.Saha AK, Xu XJ, Lawson E, Deoliveira R, Brandon AE, Kraegen EW, et al. Downregulation of AMPK accompanies leucine- and glucose-induced increases in protein synthesis and insulin resistance in rat skeletal muscle. Diabetes. 2010;59:2426–34. doi: 10.2337/db09-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li C, Najafi H, Daikhin Y, Nissim IB, Collins HW, Yudkoff M, et al. Regulation of leucine-stimulated insulin secretion and glutamine metabolism in isolated rat islets. J Biol Chem. 2003;278:2853–8. doi: 10.1074/jbc.M210577200. [DOI] [PubMed] [Google Scholar]
- 39.Bendell JC, Rodon J, Burris HA, de Jonge M, Verweij J, Birle D, et al. Phase I, dose-escalation study of BKM120, an oral pan-Class I PI3K inhibitor, in patients with advanced solid tumors. J Clin Oncol. 2012;30:282–90. doi: 10.1200/JCO.2011.36.1360. [DOI] [PubMed] [Google Scholar]
- 40.Smith GC, Ong WK, Rewcastle GW, Kendall JD, Han W, Shepherd PR. Effects of acutely inhibiting PI3K isoforms and mTOR on regulation of glucose metabolism in vivo. Biochem J. 2012;442:161–9. doi: 10.1042/BJ20111913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith GC, Ong WK, Costa JL, Watson M, Cornish J, Grey A, et al. Extended treatment with selective PI 3-kinase and mTOR inhibitors has effects on metabolism, growth, behaviour and bone strength. FEBS J. 2013 doi: 10.1111/febs.12428. [DOI] [PubMed] [Google Scholar]
- 42.Foukas LC, Bilanges B, Bettedi L, Pearce W, Ali K, Sancho S, et al. Long-term p110alpha PI3K inactivation exerts a beneficial effect on metabolism. EMBO Mol Med. 2013;5:563–71. doi: 10.1002/emmm.201201953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghosh JC, Siegelin MD, Vaira V, Faversani A, Tavecchio M, Chae YC, et al. Adaptive Mitochondrial Reprogramming and Resistance to PI3K Therapy. J Natl Cancer Inst. 2015;107 doi: 10.1093/jnci/dju502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chegary M, Brinke H, Ruiter JP, Wijburg FA, Stoll MS, Minkler PE, et al. Mitochondrial long chain fatty acid beta-oxidation in man and mouse. Biochim Biophys Acta. 2009;1791:806–15. doi: 10.1016/j.bbalip.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Al-Saffar NM, Marshall LV, Jackson LE, Balarajah G, Eykyn TR, Agliano A, et al. Lactate and choline metabolites detected in vitro by nuclear magnetic resonance spectroscopy are potential metabolic biomarkers for PI3K inhibition in pediatric glioblastoma. PLoS One. 2014;9:e103835. doi: 10.1371/journal.pone.0103835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Venkatesh HS, Chaumeil MM, Ward CS, Haas-Kogan DA, James CD, Ronen SM. Reduced phosphocholine and hyperpolarized lactate provide magnetic resonance biomarkers of PI3K/Akt/mTOR inhibition in glioblastoma. Neuro Oncol. 2012;14:315–25. doi: 10.1093/neuonc/nor209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Esmaeili M, Bathen TF, Engebraten O, Maelandsmo GM, Gribbestad IS, Moestue SA. Quantitative (31)P HR-MAS MR spectroscopy for detection of response to PI3K/mTOR inhibition in breast cancer xenografts. Magn Reson Med. 2014;71:1973–81. doi: 10.1002/mrm.24869. [DOI] [PubMed] [Google Scholar]
- 48.Zhao Z, Xiao Y, Elson P, Tan H, Plummer SJ, Berk M, et al. Plasma lysophosphatidylcholine levels: potential biomarkers for colorectal cancer. J Clin Oncol. 2007;25:2696–701. doi: 10.1200/JCO.2006.08.5571. [DOI] [PubMed] [Google Scholar]
- 49.Kikuchi H, Uehara T, Setoguchi T, Yamamoto M, Ohta M, Kamiya K, et al. Overexpression of LPCAT1 and concomitant lipid alterations in gastric cancer. Cancer Research. 2012;72(Supplement 8):3022. [Google Scholar]
- 50.Zhou X, Lawrence TJ, He Z, Pound CR, Mao J, Bigler SA. The expression level of lysophosphatidylcholine acyltransferase 1 (LPCAT1) correlates to the progression of prostate cancer. Exp Mol Pathol. 2012;92:105–10. doi: 10.1016/j.yexmp.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soga T, Ohishi T, Matsui T, Saito T, Matsumoto M, Takasaki J, et al. Lysophosphatidylcholine enhances glucose-dependent insulin secretion via an orphan G-protein-coupled receptor. Biochem Biophys Res Commun. 2005;326:744–51. doi: 10.1016/j.bbrc.2004.11.120. [DOI] [PubMed] [Google Scholar]
- 52.Motley ED, Kabir SM, Gardner CD, Eguchi K, Frank GD, Kuroki T, et al. Lysophosphatidylcholine inhibits insulin-induced Akt activation through protein kinase C-alpha in vascular smooth muscle cells. Hypertension. 2002;39:508–12. doi: 10.1161/hy02t2.102907. [DOI] [PubMed] [Google Scholar]
- 53.Forouhi NG, Koulman A, Sharp SJ, Imamura F, Kroger J, Schulze MB, et al. Differences in the prospective association between individual plasma phospholipid saturated fatty acids and incident type 2 diabetes: the EPIC-InterAct case-cohort study. Lancet Diabetes Endocrinol. 2014;2:810–8. doi: 10.1016/S2213-8587(14)70146-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maeda J, Higashiyama M, Imaizumi A, Nakayama T, Yamamoto H, Daimon T, et al. Possibility of multivariate function composed of plasma amino acid profiles as a novel screening index for non-small cell lung cancer: a case control study. BMC Cancer. 2010;10:690. doi: 10.1186/1471-2407-10-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miyagi Y, Higashiyama M, Gochi A, Akaike M, Ishikawa T, Miura T, et al. Plasma free amino acid profiling of five types of cancer patients and its application for early detection. PLoS One. 2011;6:e24143. doi: 10.1371/journal.pone.0024143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Brussel JP, van Steenbrugge GJ, Romijn JC, Schroder FH, Mickisch GH. Chemosensitivity of prostate cancer cell lines and expression of multidrug resistance-related proteins. European journal of cancer. 1999;35:664–71. doi: 10.1016/s0959-8049(98)00435-3. [DOI] [PubMed] [Google Scholar]
- 57.Vanpouille C, Le Jeune N, Kryza D, Clotagatide A, Janier M, Dubois F, et al. Influence of multidrug resistance on (18)F-FCH cellular uptake in a glioblastoma model. European journal of nuclear medicine and molecular imaging. 2009;36:1256–64. doi: 10.1007/s00259-009-1101-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.