Abstract
Retroviral integrase (IN) functions within the intasome nucleoprotein complex to catalyze insertion of viral DNA into cellular chromatin. Using cryo-electron microscopy, we now visualize the functional maedi-visna lentivirus intasome at 4.9 Å resolution. The intasome comprises a homo-hexadecamer of IN with a tetramer-of-tetramers architecture featuring eight structurally distinct types of IN protomers supporting two catalytically competent subunits. The conserved intasomal core, previously observed in simpler retroviral systems, is formed between two IN tetramers, with a pair of C-terminal domains from flanking tetramers completing the synaptic interface. Our results explain how HIV-1 IN, which self-associates into higher order multimers, can form a functional intasome, reconcile the bulk of early HIV-1 IN biochemical and structural data, and provide a lentiviral platform for design of HIV-1 IN inhibitors.
Integrase (IN) acts on the ends of the linear double stranded viral DNA (vDNA) molecule produced by reverse transcription of the retroviral RNA genome. Initially, IN catalyzes 3′-processing to expose 3′ hydroxyl groups attached to invariant CA dinucleotides at the vDNA ends. Following entry into the nuclear compartment IN inserts the processed vDNA 3′ termini across the major groove of chromosomal target DNA using the 3′ hydroxyls as nucleophiles in the strand transfer reaction. These events take place within the intasome, a stable synaptic complex comprising a multimer of IN assembled on vDNA ends (1). Characterization of prototype foamy virus (PFV, belonging to the spumavirus genus), Rous sarcoma virus (RSV, an α-retrovirus), and mouse mammary tumor virus (MMTV, a β-retrovirus) intasomes illuminated the conserved intasome core (CIC) structure minimally comprising a pair of IN dimers, as in the case of the PFV intasome (2, 3), or decorated by flanking IN dimers in RSV (4) and MMTV (5). The architecture of the lentiviral intasome, the genus that includes HIV-1 and HIV-2 along with highly pathogenic animal viruses, has remained elusive.
Unfavorable biochemical properties of HIV-1 IN necessitate the use of hyperactive and/or solubilizing mutations (6–8), which, by their nature, dramatically change the properties of the protein. Taking a more holistic approach, we sought to identify a lentiviral IN that is amenable for structural studies as a wild type protein. We discovered that the IN from maedivisna virus (MVV), an ovine lentivirus, displays robust strand transfer activity when supplied with oligonucleotides mimicking the vDNA ends in the presence of the common lentiviral integration host factor LEDGF (9, 10) (fig. S1). MVV IN assembled into a functional nucleoprotein complex that could be isolated by size exclusion chromatography (fig. S2A). In the presence of the essential Mg2+ cofactor, the purified nucleoprotein complex catalyzed strand transfer activity and could be inhibited by the HIV-1 IN strand transfer inhibitor (INSTI) dolutegravir (11) (fig. S2B). Sequence analysis of reaction products ascertained that they were formed by full-site integration – coordinated insertion of pairs of vDNA ends across the major groove in target DNA – leading to short duplications of target DNA sequences (fig. S2C). To confirm that the most commonly observed duplication size – 6 bp – is representative of MVV integration, we sequenced 2,526 unique integration sites in primary sheep cells infected with pathogenic MVV and compared them to in vitro integration sites obtained with purified intasomes and deproteinized sheep or bacterial plasmid DNA. Aligning the three sets of integration site sequences revealed symmetric and highly similar sequence preferences that are fully consistent with integration of vDNA ends across 6 bp in target DNA (fig. S3). As expected for a lentivirus (9), MVV displayed a strong preference for transcription units, with 70.2% of integration sites found within predicted sheep genes, compared to 43.7% in the in vitro generated sample (p<10-150).
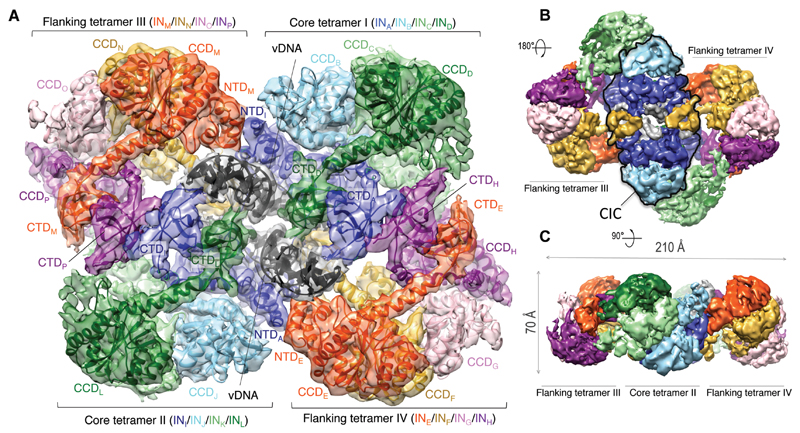
Inspection of the intasome by negative stain electron microscopy (EM) revealed a planar, two-fold symmetric assembly measuring over 20 nm in the widest dimension (fig. S4), which is much larger than any of the previously characterized retroviral intasomes. To determine its structure, we acquired images of single particles in vitreous ice using a transmission electron microscope equipped with a direct detector. The final structure was refined using a dataset of 94,283 single particles to an overall resolution of 4.94 Å, with local resolution varying from 9 Å in the periphery of the structure to ~4 Å throughout the core region (Fig. 1, figs S5-7). A crystal structure spanning the N-terminal and the catalytic core domains (NTD and CCD) of MVV IN is available (12). In addition, we determined two crystal structures spanning the MVV IN C-terminal domain (CTD, table S1). Sixteen MVV IN subunits and two double stranded DNA oligonucleotides representing the synapsed vDNA ends could be unambiguously placed in the electron density map (Fig. 1A, fig. S8, movie S1), consistent with the observed molecular mass of ~0.5 MDa for the complex (fig. S2D).
Fig. 1. Cryo-EM reconstruction of the MVV intasome.
A. Fitted intasome model color-coded to highlight IN subunits including 12 NTDs, 16 CCDs, and 14 CTDs. Molecules of vDNA in dark grey are surrounded by core tetramers I and II (colored in green, light green, sky blue, and blue), and flanking tetramers III and IV (red, yellow, pink, and purple). B-C. Views of the map in two alternative orientations. The CIC structure is highlighted with a black outline in panel B.
The intasome represents a tetramer of tetramers, each comprising a pair of imperfectly symmetric IN dimers with CCD-CTD linkers in extended α-helical configurations that are strikingly similar to the HIV-1 IN dimer observed in crystals (7) (figs S9, S10A). Although intasome formation required the presence of LEDGF, only traces of the host factor remained after purification by two-stage chromatography (fig. S2A). Consequently, no density could be attributed to LEDGF in the structure. It is possible that the remaining LEDGF molecules are distributed over 16 possible binding positions on the intasome.
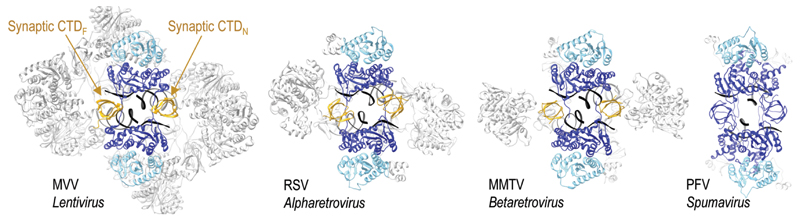
The CIC, analogous to those found in other retroviral systems (2, 4, 5), is located at the center of the assembly (Figs. 1B, 2, fig. S7C), and each of the four MVV IN tetramers is involved in its formation: core tetramers I and II contribute a CCD dimer each, while flanking tetramers III and IV provide a pair of synaptic CTDs that join the halves of the CIC structure. Approximately 20 bp of each vDNA end are well-defined in the electron density. The vDNA ends pass through the CIC structure approaching each other at an angle of 60°, with their terminal base pairs separated by IN CCD α4 helix (Fig. S10C). Each recessed 3′ vDNA end is placed in the active site of a catalytic IN subunit (chains A and I), while the complementary non-transferred strand is threaded between the CCD and the synaptic CTD (Fig. S10C). The catalytic subunits intertwine by exchanging a pair of NTDs, with CCD-NTD linkers crossing the synaptic interface and contacting vDNA minor grooves (Fig. 2, fig. S10B, movie S1).
Fig. 2. CIC structure in MVV and previously characterized synaptic complexes.
The CIC in each structure is shown in color with the remainder in grey; yellow CTDs indicate domains donated by flanking IN subunits.
A layer of CTDs bridges the flanking and core tetramers of the intasome (fig. S8). Four pairs of CTDs belonging to the inner- and outermost IN chains of each lobe stack to form dimers nearly identical to those observed in MVV IN crystals and formed by the isolated HIV-1 CTD in solution (13) (figs S11). The β1-β2 loops of the CTD dimers from tetramers I and II insert into minor grooves of vDNA (fig. S10D) close to the end engaged by the active site of the opposing main tetramer (Fig. 1A). The interactions made across the synaptic interface likely ensure that a stable intasome forms only when the enzyme engages both vDNA ends. The NTDs from the inner core IN and one of the flanking IN lobes interact with the vDNA backbone (fig. S10D), forming an interface previously observed in crystals of the HIV-1 IN NTD-CCD construct (14) (fig. S12).
Due to the role of DNA in synaptic interface formation, retroviral INs and closely related DNA transposases tend to assemble into functional multimers only in the presence of their DNA substrates (2, 4, 5, 15, 16). The intasome would thus be expected to contain a multiple of the minimal multimeric species found in solution, and this conjecture holds true for characterized intasomes containing tetramers of monomers (PFV) or dimers (RSV and MMTV) (2, 4, 5). In contrast, HIV-1 IN forms higher-order multimers in the absence of vDNA (12, 17–19), and crosslinked HIV-1 IN tetramers are functional in vitro (20). Similarly, MVV IN also forms tetramers and higher-order multimers in solution (fig. S2E). Thus, our structure explains how lentiviral INs, which are highly prone to self-associate, combine into the CIC structure. In lieu of the remarkable differences between intasome structures it would be of interest to compare quantitative proteomes of retroviral genera, although the number of IN molecules carried by the virus is unlikely to be limiting (21, 22).
The structural basis for α- and β- retroviral intasomes to comprise more than the minimal IN dimer-of-dimers architecture is relatively short IN CCD-CTD linkers (4, 5), which prohibit the CTD from the core subunits to insert into the synaptic interface. In HIV-1 and MVV IN, the CCD-CTD linkers assume α-helical conformations (7) (figs S9B, S11B), which likewise make it impossible for core tetramer subunits to provide the synaptic CTDs. Strikingly, although the linker region is the least conserved among lentiviral INs, it is invariably predicted to form an extended α helix (fig. S9C), arguing for conservation of the higher-order state of IN within lentiviral intasomes. The high stoichiometry of IN within the lentiviral intasome may help explain the notoriously pleiotropic phenotypes of HIV-1 IN mutant viruses (23). Because the 2-fold symmetric assembly contains eight structurally distinct IN subunits, each IN residue could play as many as eight distinct functions. The CTD plays the most functionally diverse roles within the intasome, contributing to intra- and inter-tetramer interactions, as well as DNA binding.
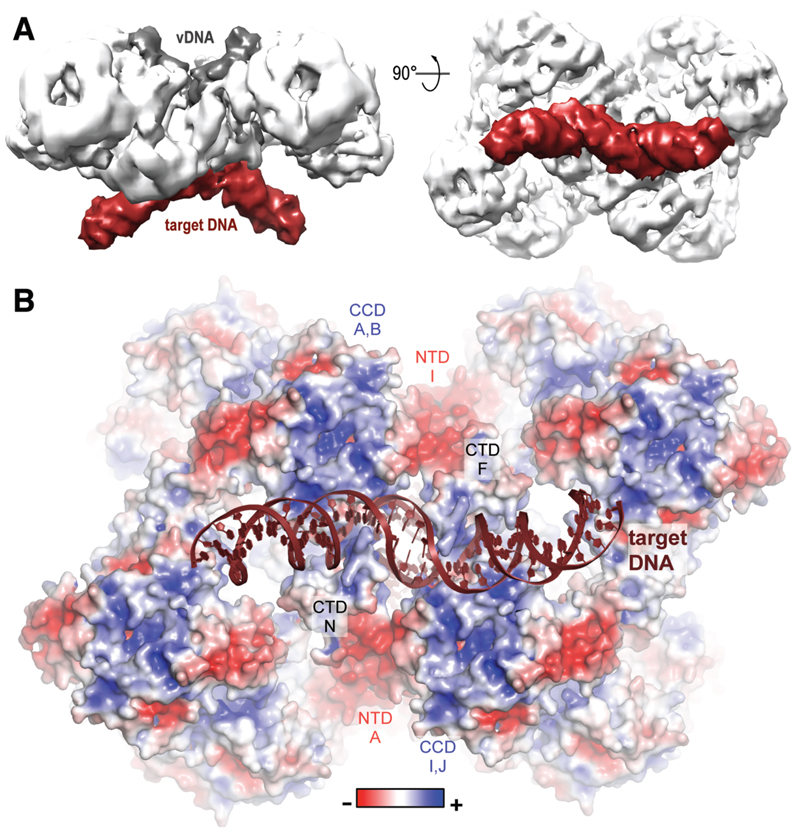
To visualize how the lentiviral intasome engages target DNA, we determined a cryo-EM structure of the MVV strand transfer complex to 8.6 Å resolution (Fig. 3A, fig. S13). In agreement with the analogous PFV and RSV structures (3, 4), target DNA binds between the halves of the CIC structure. The synaptic CTDs insert their β1-β2 loops into expanded major grooves, which contributes to target DNA bending (Fig. 3B). Inspection of the surface potential distribution on the target DNA side of the complex highlighted several patches of positive charge, each corresponding to the cleft at the IN CCD dimerization interface (6) (Fig. 3B), which was recently implicated in non-catalytic interactions with nucleosomal DNA in the PFV system (24). Lentiviral integration is exquisitely selective towards highly active and gene-rich genomic loci, a property that is explained, at least in part, by the direct interaction between IN and chromatin-associated LEDGF (9). The MVV intasome structure seems to be compatible with binding as many as 16 molecules of the host factor (fig. S14). The ability to form such super-multivalent interactions may facilitate the viral integration machinery to locate chromatin highly enriched in LEDGF and possibly other marks associated with transcriptional activity. The MVV intasome system described here should be applicable to studies of HIV-1 INSTIs (fig. S2B). Moreover, the complexity of the lentiviral intasome, presenting multiple IN-IN interfaces, may be exploitable in anti-HIV/AIDS drug development.
Fig. 3. Target DNA binding and surface electrostatic potential distribution.
A. Cryo-EM reconstruction of the MVV strand transfer complex at 8.6 Å resolution viewed in two orientations. Protein, vDNA, and target DNA are shown in white, dark grey and bordeaux, respectively. B. Pseudo-atomic model of the STC with the protein portion of the structure in space-fill mode and colored by charge.
Supplementary Material
One Sentence Summary.
Lentiviral integration is catalyzed by a higher-order multimeric assembly.
Acknowledgements
We thank G. Schoehn for help with preliminary cryo-EM, the staff of the Diamond beamlines I04 and I04-1 for assistance with X-ray data collection, P. Afonine for generous advice on real-space refinement, L. Collinson, R. Carzaniga, T. Pape, P. Walker and A. Purkiss for EM, X-ray crystallography and software support, L. Heck for expert assistance with the computer cluster, and G. Maertens for comments on the manuscript. Data presented in this manuscript are tabulated in the main paper and in the supplementary materials. The cryo-EM maps, pseudo-atomic models and the X-ray structures were deposited with the PDB and the EM databank with accessions EMD-4138, EMD-4139, PDB-5M0Q, PDB-5M0R, PDB-5LLJ, and PDB-5T3A; the integration site sequencing data were deposited with NCBI GEO repository under accession GSE87786. This work was supported by NIH grants GM082251 and AI070042 (PC and ANE), The Francis Crick Institute (PC and AC), The Wellcome Trust (AK), and Icelandic Research Fund (VA, SRJ). We acknowledge the use of the Durham University DiRAC Data Centric system, which is supported by grants ST/K00042X/1, ST/K00087X/1 and ST/K003267/1. The Division of Structural Biology Particle Imaging Center EM Facility at University of Oxford was founded by The Wellcome Trust JIF award 060208/Z/00/Z and is supported by equipment grant 093305/Z/10/Z.
References and Notes
- 1.Lesbats P, Engelman AN, Cherepanov P. Retroviral DNA Integration. Chem Rev. 2016;116:12730–12757. doi: 10.1021/acs.chemrev.6b00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature. 2010;464:232–236. doi: 10.1038/nature08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maertens GN, Hare S, Cherepanov P. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature. 2010;468:326–329. doi: 10.1038/nature09517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yin Z, Shi K, Banerjee S, Pandey KK, Bera S, Grandgenett DP, Aihara H. Crystal structure of the Rous sarcoma virus intasome. Nature. 2016;530:362–366. doi: 10.1038/nature16950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballandras-Colas A, Brown M, Cook NJ, Dewdney TG, Demeler B, Cherepanov P, Lyumkis D, Engelman AN. Cryo-EM reveals a novel octameric integrase structure for betaretroviral intasome function. Nature. 2016;530:358–361. doi: 10.1038/nature16955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dyda F, Hickman AB, Jenkins TM, Engelman A, Craigie R, Davies DR. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 7.Chen JC, Krucinski J, Miercke LJ, Finer-Moore JS, Tang AH, Leavitt AD, Stroud RM. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc Natl Acad Sci U S A. 2000;97:8233–8238. doi: 10.1073/pnas.150220297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li M, Jurado KA, Lin S, Engelman A, Craigie R. Engineered hyperactive integrase for concerted HIV-1 DNA integration. PLoS One. 2014;9:e105078. doi: 10.1371/journal.pone.0105078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craigie R, Bushman FD. Host Factors in Retroviral Integration and the Selection of Integration Target Sites. Microbiol Spectr. 2014;2 doi: 10.1128/microbiolspec.MDNA1123-0026-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cherepanov P. LEDGF/p75 interacts with divergent lentiviral integrases and modulates their enzymatic activity in vitro. Nucleic Acids Res. 2007;35:113–124. doi: 10.1093/nar/gkl885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johns BA, Kawasuji T, Weatherhead JG, Taishi T, Temelkoff DP, Yoshida H, Akiyama T, Taoda Y, Murai H, Kiyama R, Fuji M, et al. Carbamoyl pyridone HIV-1 integrase inhibitors 3. A diastereomeric approach to chiral nonracemic tricyclic ring systems and the discovery of dolutegravir (S/GSK1349572) and (S/GSK1265744) J Med Chem. 2013;56:5901–5916. doi: 10.1021/jm400645w. [DOI] [PubMed] [Google Scholar]
- 12.Hare S, Di Nunzio F, Labeja A, Wang J, Engelman A, Cherepanov P. Structural basis for functional tetramerization of lentiviral integrase. PLoS Pathog. 2009;5:e1000515. doi: 10.1371/journal.ppat.1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lodi PJ, Ernst JA, Kuszewski J, Hickman AB, Engelman A, Craigie R, Clore GM, Gronenborn M. Solution structure of the DNA binding domain of HIV-1 integrase. Biochemistry. 1995;34:9826–9833. doi: 10.1021/bi00031a002. [DOI] [PubMed] [Google Scholar]
- 14.Wang JY, Ling H, Yang W, Craigie R. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J. 2001;20:7333–7343. doi: 10.1093/emboj/20.24.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies DR, Goryshin IY, Reznikoff WS, Rayment I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science. 2000;289:77–85. doi: 10.1126/science.289.5476.77. [DOI] [PubMed] [Google Scholar]
- 16.Montano SP, Pigli YZ, Rice PA. The mu transpososome structure sheds light on DDE recombinase evolution. Nature. 2012;491:413–417. doi: 10.1038/nature11602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 18.McKee CJ, Kessl JJ, Shkriabai N, Dar MJ, Engelman A, Kvaratskhelia M. Dynamic modulation of HIV-1 integrase structure and function by cellular lens epithelium-derived growth factor (LEDGF) protein. J Biol Chem. 2008;283:31802–31812. doi: 10.1074/jbc.M805843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee SP, Xiao J, Knutson JR, Lewis MS, Han MK. Zn2+ promotes the self-association of human immunodeficiency virus type-1 integrase in vitro. Biochemistry. 1997;36:173–180. doi: 10.1021/bi961849o. [DOI] [PubMed] [Google Scholar]
- 20.Faure A, Calmels C, Desjobert C, Castroviejo M, Caumont-Sarcos A, Tarrago-Litvak L, Litvak S, Parissi V. HIV-1 integrase crosslinked oligomers are active in vitro. Nucleic Acids Res. 2005;33:977–986. doi: 10.1093/nar/gki241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacks T, Power MD, Masiarz FR, Luciw PA, Barr PJ, Varmus HE. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 22.Briggs JA, Simon MN, Gross I, Krausslich HG, Fuller SD, Vogt VM, Johnson MC. The stoichiometry of Gag protein in HIV-1. Nat Struct Mol Biol. 2004;11:672–675. doi: 10.1038/nsmb785. [DOI] [PubMed] [Google Scholar]
- 23.Engelman A. In vivo analysis of retroviral integrase structure and function. Adv Virus Res. 1999;52:411–426. doi: 10.1016/s0065-3527(08)60309-7. [DOI] [PubMed] [Google Scholar]
- 24.Maskell DP, Renault L, Serrao E, Lesbats P, Matadeen R, Hare S, Lindemann D, Engelman AN, Costa A, Cherepanov P. Structural basis for retroviral integration into nucleosomes. Nature. 2015;523:366–369. doi: 10.1038/nature14495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andresson OS, Elser JE, Tobin GJ, Greenwood JD, Gonda MA, Georgsson G, Andresdottir V, Benediktsdottir E, Carlsdottir HM, Mantyla EO. Nucleotide sequence and biological properties of a pathogenic proviral molecular clone of neurovirulent visna virus. Virology. 1993;193:89–105. doi: 10.1006/viro.1993.1106. [DOI] [PubMed] [Google Scholar]
- 26.Valkov E, Gupta SS, Hare S, Helander A, Roversi P, McClure M, Cherepanov P. Functional and structural characterization of the integrase from the prototype foamy virus. Nucleic Acids Res. 2009;37:243–255. doi: 10.1093/nar/gkn938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winter G, Lobley CM, Prince SM. Decision making in xia2. Acta Crystallogr D Biol Crystallogr. 2013;69:1260–1273. doi: 10.1107/S0907444913015308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ellison V, Brown PO. A stable complex between integrase and viral DNA ends mediates human immunodeficiency virus integration in vitro. Proc Natl Acad Sci U S A. 1994;91:7316–7320. doi: 10.1073/pnas.91.15.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petursson G, Nathanson N, Georgsson G, Panitch H, Palsson PA. Pathogenesis of visna. I. Sequential virologic, serologic, and pathologic studies. Lab Invest. 1976;35:402–412. [PubMed] [Google Scholar]
- 36.Serrao E, Cherepanov P, Engelman AN. Amplification, Next-generation Sequencing, and Genomic DNA Mapping of Retroviral Integration Sites. J Vis Exp. 2016 doi: 10.3791/53840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, S. Genome Project Data Processing The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quinlan AR. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr Protoc Bioinformatics. 2014;47:11–34. doi: 10.1002/0471250953.bi1112s47. 11 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bell JM, Chen M, Baldwin PR, Ludtke SJ. High resolution single particle refinement in EMAN2.1. Methods. 2016;100:25–34. doi: 10.1016/j.ymeth.2016.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheres SH. Semi-automated selection of cryo-EM particles in RELION-1.3. J Struct Biol. 2015;189:114–122. doi: 10.1016/j.jsb.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 44.Zheng SQ, Palovcak E, Armache J-P, Cheng Y, Agard DA. Anisotropic Correcton of Beam-induced Motion for Improved Single-particle Electron Cryo-microscopy. BioArxiv. 2016 doi: 10.1101/061960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Z, Lasker K, Schneidman-Duhovny D, Webb B, Huang CC, Pettersen EF, Goddard TD, Meng EC, Sali A, Ferrin TE. UCSF Chimera, MODELLER, and IMP: an integrated modeling system. J Struct Biol. 2012;179:269–278. doi: 10.1016/j.jsb.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.