Abstract
Molecular targeting using ligands specific to disease markers has shown great promise for early detection and directed therapy. Bead-based combinatorial libraries have served as powerful tools for the discovery of novel targeting agents. Screening platforms employing magnetic capture have been used to achieve rapid and efficient identification of high-affinity ligands from one-bead-one-compound (OBOC) libraries. Traditional manual methodologies to isolate magnetized “hit” beads are tedious and lack accuracy, and existing instruments to expedite bead sorting tend to be costly and complex. Here, we describe the design and construction of a simple and inexpensive microfluidic magnetic sorting device using standard photolithography and soft lithography approaches to facilitate high-throughput isolation of magnetized positive hit beads from combinatorial libraries. We have demonstrated that the device is able to sort magnetized beads with superior accuracy compared to conventional manual sorting approaches. This chip offers a very convenient yet inexpensive alternative for screening OBOC libraries.
Keywords: one-bead-one-compound combinatorial library, microfluidics, magnetic-activated sorting
Graphical abstract
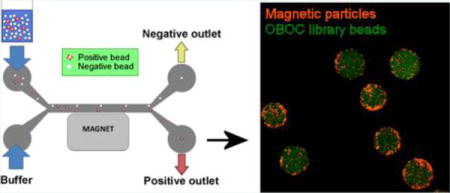
Molecular targeting using ligands specific to disease markers has driven significant advancement in molecular imaging and drug delivery for various pathologies. One-bead-one-compound (OBOC) combinatorial libraries, together with rigorous screening strategies are powerful tools for ligand discovery and have been successfully used for the identification of homing agents for numerous biological targets.1–3 OBOC libraries are chemically synthesized to produce tens of thousands to millions of unique compounds on a solid-phase support, and they confer distinct advantages over biological peptide library methods (i.e., phage display libraries) because their synthetic nature allows for the incorporation of D-amino acids, non-natural amino acids, and even non-amino acid building blocks to produce ligands that are more resistant to proteolytic degradation.1
Despite the importance and value of OBOC libraries, conventional screening methods taking after the notion of searching for “a needle in a haystack” are tedious and challenging. High-throughput screening approaches that exploit magnetic labeling have been developed to facilitate efficient identification and sorting of high-affinity ligands.4,5 Small magnetic particles conjugated with a target protein are able to associate with the ligand-coated library beads in an affinity-dependent manner, resulting in the magnetization of high-affinity beads (positive hits), which can then be isolated through magnetic sorting. The most common sorting method involves placing a strong magnet at the side of a tube containing the library to separate the magnetized hit population from the nonmagnetic (negative) beads. One can then either manually remove the negative beads or collect the positive hit population using a micropipette.4,6 Although this bulk sorting technique is quick and inexpensive, sorting accuracy is extremely low, resulting in a high false positive rate. Automated sorting based on measuring optical parameters using the Complex Object Parametric Analyzer and Sorter (COPAS) instrument (Union Biometrica) has been adopted to improve the precision of the conventional manual screening method.7,8 Nonetheless, the utility of this instrument is limited to sorting library beads based on fluorescence. Furthermore, the COPAS large-particle sorter is exceedingly costly and remains unaffordable to many laboratories.
Microfluidics has proven to be extremely useful in facilitating rapid and precise particle sorting, highlighting its value for application in high-throughput drug screening. Several miniaturized continuous flow systems have been described for magnetophoresis or magnetic isolation of cells and small particles.9–12 For example, Tan et al. have presented a “trap- and-release” microfluidic device, where beads are immobilized inside a channel and selectively retrieved based on fluorescence labeling through microbubble displacement.13 Although this design is elegant, the authors have expressed that the hits were manually selected for heating/displacement, which can be tedious and greatly limits the throughput of this system to the speed of the operator. Pamme and co-workers described a free-flow chip containing a separation chamber with multiple laminar flow paths,10 and Donolato et al. demonstrated an on-chip magnetic particle conveyor coupled with permalloy microstripes to separate magnetic particles based on magnetophoretic mobilities.9 Similarly, others have used high-gradient magnetic concentrators12,14 or microfabricated ferromagnetic strips11 to obtain continuous sorting of magnetic particles or cells with high purity and throughput. Although all of these microfluidic devices have served as valuable tools to accelerate magnetic sorting, their designs tend to be rather complex and require multiple construction steps.
In this article, we describe a simple-to-construct, inexpensive, and reliable microfluidic device that offers a convenient method to screen large OBOC libraries with high-throughput. The device is made entirely out of polydimethylsiloxane (PDMS), and its fabrication is appealing for its simplicity, entailing very few steps. We show that this device can perform high-throughput and rapid bead sorting with great accuracy. In addition, the excellent recovery rate permits for comprehensive retrieval of beads for multiple additional rounds of screening.
Design of a Simple and Cost-Effective Microfluidic Magnetic-Activated Bead Sorter
To achieve automated high-throughput screening of OBOC libraries without a complex and expensive setup, we have fabricated a simple and reproducible on-chip magnetic-activated bead separator. The microfluidic design and dimensions are illustrated in Figure 1a–c. The total width and height of the channel are 0.6 and 0.4 mm, respectively, with ample space to accommodate a single file of 90 μm beads. We rationalized that shifting of a flowing bead from its original trajectory within the channel could compromise sorting accuracy. To prevent negative beads from exiting through the positive outlet, we designed the width of the source path (0.4 mm) to be larger than the collection path (0.2 mm) (Figure 1a) to accommodate for bead oscillation orthogonal to the flow by means of diffusion. Additionally, we expect that the narrower positive outlet would select for the most positive/highly magnetized beads drawn toward the magnet. The length of the channel was designed to be 20 mm (2 cm) so that the device can easily fit the stage of a microscope to allow for real-time analysis of the sorting process. Indeed, these dimensions can be scaled up to increase sorting throughput.
Figure 1.
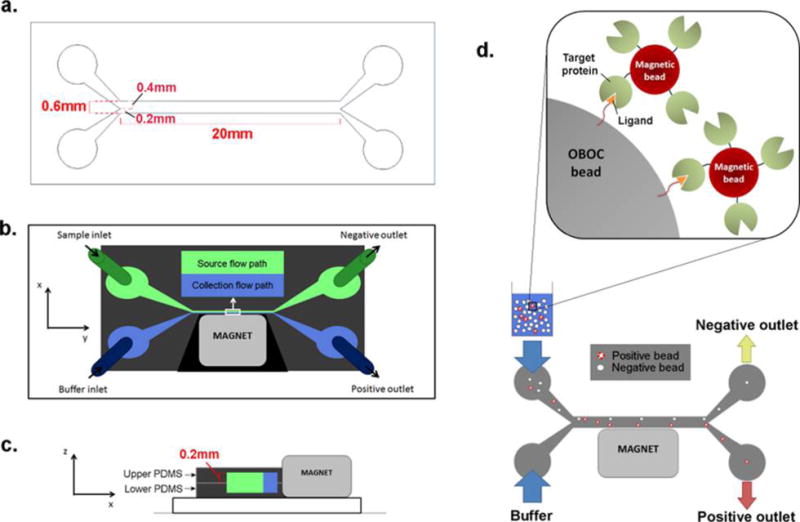
Design of the microfluidic magnetic sorter. (a) Pattern and dimensions of the microfluidic channel (generated in AutoCAD, version 2014) for photomask printing. (b) Microfluidic channel constructed from PDMS (dark gray) is composed of the source flow path (green) and the collection flow path (blue). A magnet is placed adjacent to the collection flow path. (c) The microfluidic channel is made by aligning and bonding two identical open-faced PDMS channels to form one closed channel. (d) The target protein is conjugated onto magnetic particles and mixed with the OBOC library beads. Beads containing ligand that binds to the target protein will become magnetized. The bead mixture is then inserted through the sample inlet, whereas buffer is administered through the buffer inlet simultaneously at the same flow rate to produce two laminar flow paths. Control (nonmagnetized) beads should remain in the source flow path and exit through the negative outlet, whereas the magnet diverts positive (magnetized) beads into the collection flow path and into the positive outlet.
Library beads are introduced into the device through the “sample inlet”, and flow buffer is inserted through the “buffer inlet” simultaneously at a constant flow rate to generate two continuous laminar streams: (1) source flow path and (2) collection flow path (Figure 1b). Once inside the channel, beads should remain in the source path and exit through the negative outlet when no external force is present. When an external magnet is placed along the channel wall adjacent to the collection flow path, magnetized beads should be diverted from the source path into the collection stream toward the positive outlet (Figure 1d).
Fabrication of the Microfluidic Device
To construct the microfluidic device, we first prepared a master mold of the channels by soft-lithography (see Experimental Procedures). The tendency of library beads to stick and aggregate on a glass surface but not on PDMS (data not shown) presents the necessity to construct a channel that is surrounded entirely by PDMS. For this, two identical pieces of PDMS channel were oxidized in Piranha solution, resulting in increased surface hydrophilicity.18 Both channels were then aligned and bonded to form a closed PDMS channel (Supplementary Figure 1a). A neodymium–iron-boron (NdFeB) magnet was placed adjacent to the collection path, and the inlet and outlet tubing were assembled to complete the setup of the device (Supplementary Figure 1b). The field strength and size of the magnet can be varied to achieve the desired screening stringency (for example, a weak magnet can be utilized to isolate the most highly magnetized beads, and a stronger magnet may be required for sorting beads with modest magnetic labeling). Because this device is primarily made out of PDMS and requires very few steps of production, the fabrication cost and time would be substantially lower than those of other existing microfluidic devices for OBOC library screening.13,14,19,20
Magnetic-Activated Sorting
As a proof-of-concept, we generated a mock “positive” magnetized bead population using biotin-coated red-fluorescent magnetic particles (2–4 μm) to capture streptavidin-coated polystyrene beads (90 μm) (Figure 2a) as well as a “negative” green-fluorescent control population by mixing the streptavidin-coated polystyrene beads with biotinylated fluorophore (Biotin-488) (Figure 2b). For generation of the positive beads, precautions were taken to ensure that the magnetic particles were evenly distributed for unbiased association and that the polystyrene beads were resting in a single layer without touching each other to avoid cross-linking of magnetic particles with several polystyrene beads. Unconjugated streptavidin sites on the polystyrene beads were subsequently blocked with an excess of biotin to prevent further cross-reaction with biotinylated magnetic particles bound onto other polystyrene beads. For the control beads, all streptavidin sites on the polystyrene beads should be saturated with the fluorophore, thereby preventing any potential association with biotinylated magnetic beads when mixed together during the screen.
Figure 2.
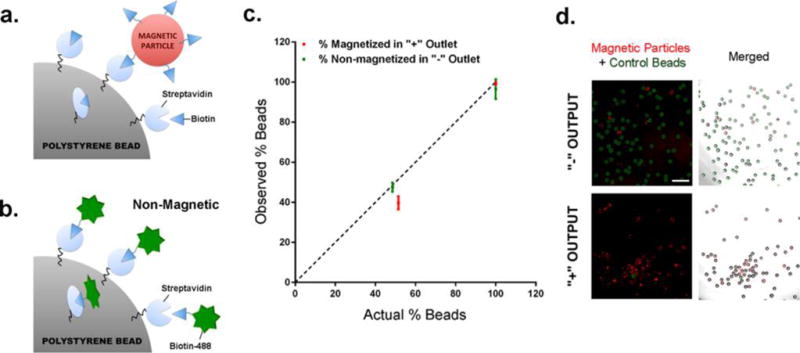
Determining sorting accuracy of the microfluidic device. (a) Generation of magnetized positive hits: streptavidin-coated polystyrene beads were incubated with biotin-coated red-fluorescent magnetic particles, and unconjugated streptavidin sites were saturated with free biotin to block nonspecific cross-linking of magnetic particles with surrounding polystyrene beads. (b) Generation of control green-fluorescent beads: streptavidin-coated polystyrene beads were incubated with biotinylated PromoFluor-488 dye. (c) Graph depicting the percentage of magnetized positive hits (red) in the positive output and control nonmagnetized beads (green) in the negative output. The dashed line represents perfect sorting, where the percentage of positive/negative beads in each output matches exactly with the actual positive/control bead input. The device was tested for sorting control beads alone, magnetized beads alone, and a mixed population of control:magnetized beads (48.5:51.5 ratio). (d) Fluorescence images showing the bead population in the negative and positive output following sorting of a mixed population of control and magnetized beads. (red: magnetic particles; green: control beads). Scale bar, 0.5 mm.
We determined the accuracy of the device by calculating its sorting sensitivity and specificity. Sensitivity signifies the device’s ability to successfully isolate positive magnetized hits, and specificity is characterized by its capability to separate/remove the nonmagnetic control population. We analyzed the device by sorting the following bead combinations: magnetized beads alone (Figure 2a), control beads alone (Figure 2b), and a mixed population of magnetized and control beads. All beads from each output were collected and imaged under a fluorescent microscope. The number of non-green-fluorescent (positive) vs. green-fluorescent (negative) beads from each population was counted, and the accuracy (specificity and sensitivity) of each run is summarized in Table 1 and Figure 2c.
Table 1.
Flow Rate and Accuracy of Sorting Device
| bead population | flow rate (mL/h) | # control beads in negative output/total control beads | # magnetized beads in positive output/total magnetized beads | specificity (%) | sensitivity (%) |
|---|---|---|---|---|---|
| control beads | 2 | 530/530 | 100 | ||
| control beads | 3 | 461/462 | 99.8 | ||
| control beads | 4 | 766/781 | 98.1 | ||
| magnetized beads | 3 | 1423/1428 | 99.6 | ||
| 1:1 mix | 3 | 632/642 | 543/705 | 98.6 | 77 |
We observed that this device can sort control or magnetized beads alone with >98% accuracy (Table 1). When sorting green-fluorescent control beads (Figure 2b) mixed with non-green-fluorescent polystyrene beads (Figure 2a), we saw approximately 48 ± 2% of control beads in the negative output (actual input = 48.5%), whereas 40 ± 3% of positive beads were observed in the positive output (actual input = 51.5%) (Figure 2c and d). These values represent 99 ± 1% specificity (control beads in negative output), and 77 ± 3% sensitivity (positive beads in positive output) when sorting a mixed bead population. Real-time imaging revealed that highly magnetized beads “rolled” gradually along the wall of the collection stream (adjacent to the magnet) and into the positive outlet, whereas control beads traveled rapidly without resistance into the negative outlet (Supplementary Figure 2).
The false positive rate (control beads in the positive output) was determined to be approximately 2% using the calculation
A higher false negative rate (positive beads in the negative output) was observed at 20% as determined by
Sorting errors were mostly noted when bead “clumping” was observed. Therefore, it is imperative to ensure a continuous flow of beads in a single file to preserve high sorting accuracy. We found the optimal bead concentration to be no more than 5000 beads/mL. The false negative rate could also be attributed to the dissociation of magnetic particles from the polystyrene beads during the washing and sorting processes. Nevertheless, the high recovery rate of the device (>99%) allows for a comprehensive retrieval of beads for multiple rounds of screening and sorting for further enrichment of the positive hit population.
All in all, this instrument demonstrates great potential for facilitating large combinatorial library screens with great accuracy and recovery compared to the widely used manual sorting technique, where a magnet is typically placed at the side of a tube containing the beads to separate magnetized positive hits from negative beads.21,22 Although convenient, this bulk sorting approach is very crude and tends to yield very low sorting accuracy. To test this, we used this method to sort a bead population containing a 1:1 mixture of magnetized beads (Figure 2a) and control beads (Figure 2b). In fact, we sought to improve sorting sensitivity by allowing the beads to rest in PBS at the bottom of a 6-well plate, then gently inserting a magnet directly into the solution and holding the magnet in the closest possible proximity above the beads. Magnetized beads should be pulled onto the magnet, and the associated beads can then be retrieved from the magnet by washing with PBS using a micropipette. Within the sorted population, we observed only 49% of true positive hits with a false positive rate of 51% (Table 2). Meanwhile, the remaining population in the 6-well plate consisted of 41% positive beads and 59% control beads. We calculated that only less than 2% of the positive hits were successfully isolated manually (Table 2), indicating that sorting accuracy is dramatically compromised using this method compared to our automated device. The low accuracy rate attributed to this technique emphasizes the value of automated microfluidic sorting. Additionally, the manual sorting strategy, which implements an open system concept (one that interfaces and interacts with its environment) resulted in a substantially lower recovery rate of 80% (2650/3300 beads recovered) compared to our device (>99%).
Table 2.
Accuracy of the Microfluidic Device in Sorting Magnetized Beads from Control Beads in a Mixed Population of Beads
| bead population | sorting methods | sensitivity (%) | false positive (%) | false negative (%) |
|---|---|---|---|---|
| 1:1 mix | automated (3 mL/h) | 77 | 1.8 | 20.4 |
| 1:1 mix | manual sorting | 1.6 | 51 | 41 |
Numerical Simulation to Characterize Device Performance
Computational simulations further confirmed that beads in the fluidic channel can be efficiently and accurately separated within a magnetic field (Figure 3a). The flow rate value indicates the flow speed at each inlet, and the trajectories of the magnetized positive population are highlighted in blue. We varied the following two experimental parameters: (1) the flow rate within the channel and (2) the number of magnetic particles attached to each library bead. We used six different magnetization levels in combination with four flow rates and characterized the separation efficiency of the positive bead population. The percentage of beads collected at outlet 2 was determined, and the results are summarized in Figure 3a and b. As expected, sorting efficiency is directly related to the number of magnetic particles present on each bead. We also observed that lower flow rates resulted in better sorting accuracy, particularly for beads with low levels of magnetic particles. A typical positive hit bead that we would select for isolation from an OBOC library screen is usually associated with >100 magnetic particles. Our simulation studies revealed that the device is most efficient at the lowest flow rate tested (3 mL/h) with the capability to sort 100% of positive beads containing as few as 30 magnetic particles (Figure 3b). This result is comparable with our experimental data (Figure 2c and Table 1). As we increased the flow rates in our simulation analysis, we observed that separation sensitivity decreased accordingly. Nonetheless, we demonstrated that our system has the potential to sort with great consistency and accuracy even at a high flow rate of 10 mL/h (Figure 3b and c). Our simulation results indicate that, at a working concentration of 5000 beads/mL, it is possible to achieve a throughput of 105 beads within 2 h, although further studies will be required to fully evaluate the capacity of the system at higher flow rates.
Figure 3.
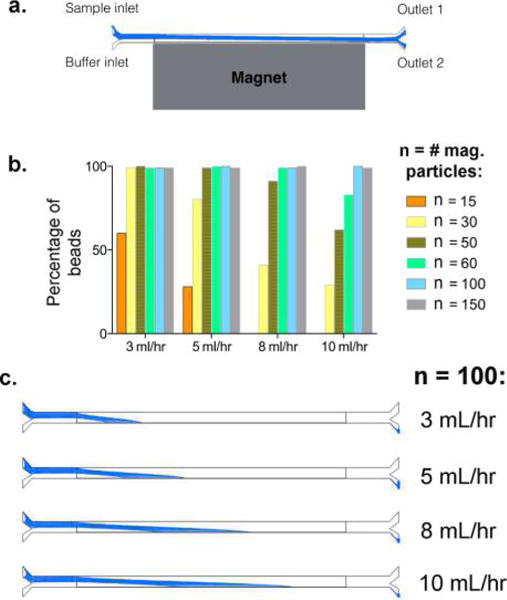
Magnetophoresis simulation of the microfluidic device. (a) Schematic of the system in our simulation analysis. The trajectories of the magnetized positive population are highlighted in blue. (b) Graph depicting the percentage of positive beads (associated with different numbers of magnetic particles) isolated at outlet 2 under varying flow rates. The flow rate value indicates the flow speed at each inlet. (c) The trajectories of a positive bead bound to 100 magnetic particles (representing a typical positive hit in a library screen) within the fluidic channel under varying flow rates, showing the potential of this device to consistently and accurately sort positive hits even under a high flow rate of 10 mL/h.
OBOC Library Screen
An OBOC library was synthesized using the “split-mix” synthesis method1 to produce random 8-amino acid peptides on 90 μm Tentagel beads. When streptavidin-coated magnetic particles (red) were mixed with the library beads, we observed that less than 1% of the library beads appeared to be “positive” (binding >100 magnetic particles), although the level of magnetization across all beads varied greatly. We demonstrated that our microfluidic system was able to isolate highly magnetized positive hits (Figure 4a) from the great bulk of negative library beads containing little to no bound magnetic particles. Analysis of beads in the negative output revealed the presence of a minor population of highly magnetized beads (16%) (Figure 4a). This is expected given our earlier observation of a false negative rate of ∼20% (Figure 2c). Nevertheless, the extraordinary recovery rate of this device allows multiple rounds of sorting for further enrichment of the positive hits population.
Figure 4.
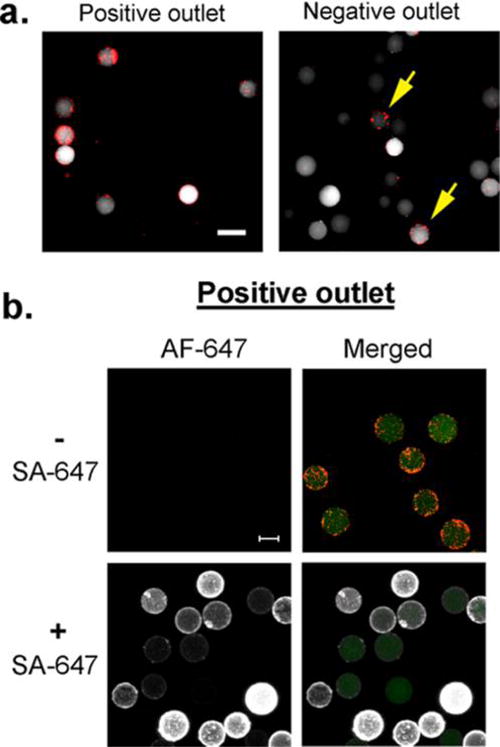
OBOC library screen. (a) OBOC library beads (90 μm, white) were mixed with small streptavidin-coated magnetic particles (2 μm, red) and subjected to sorting in the microfluidic device. Beads from the positive output were highly magnetized (associating with more than 100 magnetic beads), whereas the majority of the beads in the negative output had little to no association with magnetic particles. A minor population (16%) of highly magnetized beads was observed in the negative output (yellow arrows). (b) Beads from the positive output were subjected to Streptavidin-Alexa Fluor 647 (SA-647) at 5 μg/mL concentration (white: Alexa Fluor 647 (AF-647); red: magnetic particles; green: intrinsic autofluorescence of OBOC beads). The majority of the OBOC beads displayed high affinity for SA-647. Scale bar, 0.1 mm.
Next, we incubated the positive bead population (from Figure 4a) with Alexa Fluor 647-conjugated streptavidin (SA-647) to validate the affinity of these hits. We found that approximately 95% (142/149) of the hit beads were labeled with SA-647, although the level of fluorescence labeling varied between each OBOC bead (Figure 4b). The remaining 5% of the beads displayed a similar fluorescence level to that of unlabeled beads, suggesting that these were likely false positives that could have arisen from nonspecific association, such as avidity-driven bead–bead interactions or hydrophobic aggregation. Nonetheless, these results provide further affirmation that the majority of the positive output population indeed displayed “true” binding to their target protein that they were screened against.
As with any screening methodology, specificity and affinity of a ligand to its target must be further validated once it has been identified (e.g., using kinetic analyses and/or cellular uptake assays). In fact, prior to meticulous sequencing and characterization, a secondary screen of the positive hits isolated above using cells expressing the target on their surface can be included as a means to further select for genuine homing agents4 while reducing the burden of cost and tedious workload associated with sequencing and ligand synthesis/purification. We expect that our device can also facilitate hits sorting from a cell-based screen if the cells are labeled with magnetic particles.23 Overall, although this article focuses on the value of the device for sorting combinatorial library beads, its capacity is certainly not limited to this application. We foresee that this device could confer potential benefits in magnetic sorting of other large objects, including microparticles,24,25 cellular spheroids,26 or even small embryos/larvae.27
The versatility and flexibility of our instrument allows diverse screening conditions to be fashioned. PDMS is a stable polymer that is compatible with most screening conditions (i.e., flow buffer, temperature, etc.). Furthermore, magnets of various field strengths can be used interchangeably to achieve desired screening stringency (magnets with low field strength would result in high screening stringency). In fact, block magnets can be readily replaced with an electromagnet28 if desired so that precise regulation of magnetic field strength can be accomplished. In addition, this miniature instrument can also be readily integrated with standard laboratory equipment (i.e., syringe pumps and inverted microscopes). Simulation studies revealed the potential of our system to screen with great accuracy and with a throughput that is comparable with other microfluidic systems. However, in contrast to existing technology,13,14,19,20 our chip does not require complex fabrication steps, thus substantially simplifying its construction, leading to reduced cost and time of production. This makes our device an attractive and valuable screening tool for researchers interested in drug discovery.
Without the need for elaborate and costly equipment, we have established a high-throughput and reliable microfluidic large-particle magnetic-activated sorter. This automated “lab-on-a-chip” platform is inexpensive, reproducible and simple-to-construct, making it practical for most laboratories to devise. Our results here highlight the precision and efficiency of this device for sorting large magnetized particles and demonstrate its potential in facilitating large OBOC library screens.
EXPERIMENTAL PROCEDURES
Microfluidic Magnetic Sorter Design
A microfluidic channel with dimensions of 20 × 0.6 mm (L × W) (source flow path: W = 0.4 mm; collection flow path: W = 0.2 mm) was designed using AutoCAD (Automated Computer-Aided Design) and printed on a photomask in dark-field. The microfluidic system was generated using soft lithography. Briefly, a master mold was prepared on a silicon wafer (University Wafer) using the SU-8 2150 photoresist (Microchem) to produce a height of 0.2 mm. PDMS was poured onto the master and allowed to cure overnight at 65 °C. PDMS was peeled off the master to produce a single layer of open-faced channels. Each channel was cut out individually, yielding an open-faced PDMS channel. Two open-faced channels are required to generate a closed channel. To create the inlets and outlets, holes were created on the upper PDMS channel using a 0.75 mm hole-puncher. Both the upper and lower PDMS surfaces were then exposed to Piranha solution (composed of 2:3 H2O2:H2SO4) for 50 s. After rinsing with H2O and drying with N2 gas, both open-faced channels were aligned precisely under a magnifying glass to form one closed channel completely surrounded by PDMS with a final height of 0.4 mm. The PDMS layers were allowed to bond overnight in a 65 °C incubator, and the device was then adhered onto a glass slide (either by incubating at 65 °C overnight or using high-adhesive glue) for structural support.
Device Operation
Four 21-gauge dispensing needles were disconnected from their hubs using a pair of pliers and fitted into four separate silicone tubing pieces (0.6 mm inner diameter, 1.5 mm outer diameter, 30 cm (inlet) or 10 cm (outlet) length). The tubing was then connected to the microfluidic channel via the needle through the inlet and outlet holes in the upper PDMS layer. The tubing and flow channel were filled with flow buffer (PBS with 18% (v/v) glycerol) while ensuring no air bubbles were present within the system. Collection tubes were placed at the outlets and a strong neodymium magnet (5251 G; NdFeB, grade N52, K&J Magnetics Inc.) was placed along the collection path of the channel. Polystyrene beads (90–100 μm, Spherotech) were inserted into a BD Luer-Lok syringe, which was then connected to the sample inlet. A syringe of the same size was filled with flow buffer and attached to the buffer inlet. Both “beads” and “buffer only” syringes were mounted on a multisyringe pump to regulate the flow rate. Beads and buffer were injected simultaneously at a constant flow rate. It is imperative to ensure that no air bubbles are formed during the assembly of the device and throughout the sorting process as air bubbles will cause disruption of the laminar flow and therefore diminish sorting accuracy.
Generation of Magnetized (Positive) vs Fluorescent Control Beads
For generating a mock positive population, streptavidin-coated polystyrene beads (90–100 μm, Spherotech) were mixed with biotin-coated red-fluorescent magnetic particles (2–4 μm, 0.1% w/v, Spherotech). Approximately 20,000 polystyrene beads (in 500 μL of PBS) were added to a 6-well plate. One milligram of magnetic particles was added into the well, and the beads were incubated for at least 1 h at room temperature, ensuring that the magnetic particles were evenly distributed and that the polystyrene beads were not touching each other to avoid cross-linking of magnetic particles with several polystyrene beads. Unconjugated streptavidin sites on the polystyrene beads were blocked with an excess of biotin (10 μg/mL). Binding of magnetic particles to the polystyrene beads was confirmed under a fluorescent microscope, and the mixture was then washed gently with PBS to remove unbound magnetic particles. As a negative control, streptavidin-coated polystyrene beads were incubated with 10 μg/mL of biotinylated PromoFluor-488 (PromoKine) to generate a population of nonmagnetized green-fluorescent beads.
Accuracy of the Microfluidic Magnetic Bead Sorter
The accuracy (specificity and sensitivity) was calculated as detailed below. Specificity is represented by the percentage of control beads in the “negative output”, whereas sensitivity is characterized by the population of magnetized beads in the “positive output”. We mixed the magnetized and control polystyrene bead populations described in the previous section (in known ratios) and subjected them to sorting using our device.
The device was operated as described in the previous section. Beads were resuspended in PBS (at a concentration of 5000 beads/mL) containing 18% glycerol (v/v) to increase buffer density and prevent bead aggregation. Insertion of beads (through the sample inlet) and flow buffer (through the buffer inlet) into the channel was maintained at a constant flow rate by the syringe pump. All beads collected from both the positive and negative outlets were imaged under a Nikon Eclipse Ti inverted microscope. The numbers of green-fluorescent (control) and nonfluorescent (positive hit) beads in each outlet were counted manually from the images. Specificity was calculated by
While sensitivity was calculated by
Recovery of beads was determined by
Magnetophoresis Simulation to Characterize System Performance
To further evaluate the performance of our device, we carried out a series of numerical simulations.15,16 We used the COMSOL Multiphysics software to calculate the trajectories of beads labeled with varying numbers of magnetic particles. The dimensions of the fluidic channel were those of an actual device (described above). The fluidic field U in the channel was first obtained by solving the Navier–Stokes equation for low Reynolds number
where ρf and μ are fluidic density and viscosity, respectively, and p is the pressure. The magnetophoretic force FMP on a particle by the external magnet was calculated using the equation
where n is the number of magnetic particles on a polystyrene bead, mp is the magnetic moment of individual magnetic particles, and B is the magnetic field. We used mp and B values provided in the manufacturers’ data sheets. We then solved the velocity field V of a magnetic object from the coupled equation
where ρp and rp are the density and the radius of a polystyrene bead, respectively.
OBOC Library Screen
A one-bead-one-compound (OBOC) combinatorial library was synthesized on 90 μm ANP-TentaGel resin using a “split and mix” strategy so that each bead carries multiple copies of a unique ligand, as described previously.1,17 Initially, a photolabile linker, 3-amino-3-(2-nitrophenyl) propionic acid (ANP), was manually added to Tentagel resin beads (1.0 g of Tentagel S –NH2, 90 μm particle size, 0.33 mmol/g loading) using standard Fmoc peptide synthesis procedures. The resin was kept in the dark during the synthesis process while conducted in an automated peptide synthesizer (Biotage Syro Wave, Charlotte, NC).
In the automated synthesizer, the Fmoc group was removed using a 20% piperidine in DMF solution (800 μL/well, ×2). A different D-amino acid was used in each well (3 equiv) together with the reagents HCTU (3 equiv) and DIPEA (6 equiv) in DMF. A total of 18 wells were used, one for each of the common amino acids (as the D isomer), excluding Cys and Met to avoid oxidation products. After each coupling step, the resin was rinsed with DMF and DCM multiple times, recombined in a peptide vessel, and shaken thoroughly before being split again into the synthesizer wells for another round of Fmoc deprotection and coupling. The process of deprotection and coupling was then repeated until the library reached the desired length of eight amino acids.
Library beads (500 mg) were then washed using DMF (×2), MeOH (×2), 5% DIPEA in DMF, DMF (×3), DCM (×3), and finally 50% DMF in water to completely remove all unbound reagents. Ethanol (70%) was later added to the library beads to remove traces of organic solvents, and the beads were then resuspended in phosphate buffer saline (PBS). Library beads were blocked with 3% BSA (w/v) (Sigma-Aldrich) in PBS containing 0.025% Tween-20 (v/v) for 1 h at RT and then mixed with 50 μg of red-fluorescent streptavidin-coated magnetic beads (2 μm, Spherotech Inc.) for 5 h. Beads were washed gently with PBS containing 0.025% Tween-20 by gravity sedimentation to remove unbound magnetic particles. Library beads were sorted as described in previous sections. All beads collected from both the positive and negative outlets were imaged under a Nikon Eclipse Ti inverted microscope. Hits can be easily identified from their interaction with red fluorescent magnetic particles. Beads associating with ≥100 magnetic particles, as counted manually, were considered positive.
Beads from the positive output were collected and incubated with Alexa Fluor 647-conjugated Streptavidin (ThermoFisher Scientific) overnight (final concentration = 5 μg/mL) in PBS containing 3% BSA (w/v) and 0.025% Tween-20 (v/v). Beads were then washed gravity sedimentation (×3) with PBS containing 0.025% Tween-20 and imaged under a Zeiss Axio Observer.Z1 confocal microscope.
Supplementary Material
Acknowledgments
This study was supported by institutional funds from the Department of Neurosurgery, Brigham and Women’s Hospital (E.A.C.), fellowship funds from the Canadian Institute of Health Research (CIHR) (C.-F.C.), Natural Sciences and Engineering Research Council of Canada (NSERC) (L.G.L.) and B*Cured funds (S.E.L.). We acknowledge Calixto Saenz (Harvard Medical School) for providing us with access to the microfluidics facility. We thank Bradley Pentelute for his support and Emily Avery (Princeton University) for her technical assistance.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscombsci.5b00180.
Images of the microfluidic device (PDF)
Time-lapse video microscopy showing bead sorting within the channel (AVI)
Notes
The authors declare no competing financial interest.
References
- 1.Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM, Knapp RJ. A new type of synthetic peptide library for identifying ligand-binding activity. Nature. 1991;354(6348):82–4. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- 2.Chen X, Gambhir SS. Significance of one-bead-one-compound combinational chemistry. Nat Chem Biol. 2006;2(7):351–2. doi: 10.1038/nchembio0706-351. [DOI] [PubMed] [Google Scholar]
- 3.Reddy MM, Wilson R, Wilson J, Connell S, Gocke A, Hynan L, German D, Kodadek T. Identification of candidate IgG biomarkers for Alzheimer’s disease via combinatorial library screening. Cell. 2011;144(1):132–42. doi: 10.1016/j.cell.2010.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho CF, Amadei GA, Breadner D, Luyt LG, Lewis JD. Discovery of novel integrin ligands from combinatorial libraries using a multiplex ″beads on a bead″ approach. Nano Lett. 2012;12(11):5957–65. doi: 10.1021/nl3034043. [DOI] [PubMed] [Google Scholar]
- 5.Pei D. On-bead library screening made easier. Chem Biol. 2010;17(1):3–4. doi: 10.1016/j.chembiol.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Astle JM, Simpson LS, Huang Y, Reddy MM, Wilson R, Connell S, Wilson J, Kodadek T. Seamless bead to microarray screening: rapid identification of the highest affinity protein ligands from large combinatorial libraries. Chem Biol. 2010;17(1):38–45. doi: 10.1016/j.chembiol.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho CF, Behnam Azad B, Luyt LG, Lewis JD. High-throughput screening of one-bead-one-compound peptide libraries using intact cells. ACS Comb Sci. 2013;15(8):393–400. doi: 10.1021/co4000584. [DOI] [PubMed] [Google Scholar]
- 8.Hu BH, Jones MR, Messersmith PB. Method for screening and MALDI-TOF MS sequencing of encoded combinatorial libraries. Anal Chem. 2007;79(19):7275–85. doi: 10.1021/ac070418g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donolato M, Dalslet BT, Hansen MF. Microstripes for transport and separation of magnetic particles. Biomicrofluidics. 2012;6(2):24110–241106. doi: 10.1063/1.4704520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pamme N, Eijkel JCT, Manz A. On-chip free-flow magnetophoresis: Separation and detection of mixtures of magnetic particles in continuous flow. J Magn Magn Mater. 2006;307(2):237–244. [Google Scholar]
- 11.Adams JD, Kim U, Soh HT. Multitarget magnetic activated cell sorter. Proc Natl Acad Sci U S A. 2008;105(47):18165–70. doi: 10.1073/pnas.0809795105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia N, Hunt TP, Mayers BT, Alsberg E, Whitesides GM, Westervelt RM, Ingber DE. Combined microfluidic-micromagnetic separation of living cells in continuous flow. Biomed Microdevices. 2006;8(4):299–308. doi: 10.1007/s10544-006-0033-0. [DOI] [PubMed] [Google Scholar]
- 13.Tan WH, Takeuchi S. A trap-and-release integrated microfluidic system for dynamic microarray applications. Proc Natl Acad Sci U S A. 2007;104(4):1146–51. doi: 10.1073/pnas.0606625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang W, Wei Z, Wang Z, Ma H, Bu X, Hu Z. A continuous flow microfluidic-MS system for efficient OBOC screening. RSC Adv. 2014;4(106):61767–61770. [Google Scholar]
- 15.Chung J, Issadore D, Ullal A, Lee K, Weissleder R, Lee H. Rare cell isolation and profiling on a hybrid magnetic/size-sorting chip. Biomicrofluidics. 2013;7(5):54107. doi: 10.1063/1.4821923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee K, Shao H, Weissleder R, Lee H. Acoustic purification of extracellular microvesicles. ACS Nano. 2015;9(3):2321–7. doi: 10.1021/nn506538f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amadei GA, Cho CF, Lewis JD, Luyt LG. A fast, reproducible and low-cost method for sequence deconvolution of ’on-bead’ peptides via ’on-target’ maldi-TOF/TOF mass spectrometry. J Mass Spectrom. 2010;45(3):241–51. doi: 10.1002/jms.1708. [DOI] [PubMed] [Google Scholar]
- 18.Koh KS, Chin J, Chia J, Chiang CL. Quantitative studies on PDMS-PDMS interface bonding with piranha solution and its swelling effect. Micromachines. 2012;3:427–441. [Google Scholar]
- 19.Cha J, Lim J, Zheng Y, Tan S, Ang YL, Oon J, Ang MW, Ling J, Bode M, Lee SS. Process automation toward ultrahigh-throughput screening of combinatorial one-bead-one-compound (OBOC) peptide libraries. J Lab Autom. 2012;17(3):186–200. doi: 10.1177/2211068211433503. [DOI] [PubMed] [Google Scholar]
- 20.Fu J, Lee T, Qi X. The identification of high-affinity G protein-coupled receptor ligands from large combinatorial libraries using multicolor quantum dot-labeled cell-based screening. Future Med Chem. 2014;6(7):809–23. doi: 10.4155/fmc.14.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu T, Qian Z, Xiao Q, Pei D. High-throughput screening of one-bead-one-compound libraries: identification of cyclic peptidyl inhibitors against calcineurin/NFAT interaction. ACS Comb Sci. 2011;13(5):537–46. doi: 10.1021/co200101w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao Y, Amar S, Pahwa S, Fields G, Kodadek T. Rapid lead discovery through iterative screening of one bead one compound libraries. ACS Comb Sci. 2015;17(1):49–59. doi: 10.1021/co500154e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi X, Astle J, Kodadek T. Rapid identification of orexin receptor binding ligands using cell-based screening accelerated with magnetic beads. Mol BioSyst. 2010;6(1):102–7. doi: 10.1039/b915611a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu X, Xia HS, Sun ZD, Lin Y, Wang K, Yu J, Tang H, Pang DW, Zhang ZL. On-chip dual detection of cancer biomarkers directly in serum based on self-assembled magnetic bead patterns and quantum dots. Biosens Bioelectron. 2013;41:129–36. doi: 10.1016/j.bios.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 25.Zhou Y, Wang Y, Lin Q. A Microfluidic Device for Continuous-Flow Magnetically Controlled Capture and Isolation of Microparticles. J Microelectromech Syst. 2010;19(4):743–751. doi: 10.1109/JMEMS.2010.2050194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernandez LA, Hatch EW, Armann B, Odorico JS, Hullett DA, Sollinger HW, Hanson MS. Validation of large particle flow cytometry for the analysis and sorting of intact pancreatic islets. Transplantation. 2005;80(6):729–37. doi: 10.1097/01.tp.0000179105.95770.cd. [DOI] [PubMed] [Google Scholar]
- 27.Marois E, Scali C, Soichot J, Kappler C, Levashina EA, Catteruccia F. High-throughput sorting of mosquito larvae for laboratory studies and for future vector control interventions. Malar J. 2012;11:302. doi: 10.1186/1475-2875-11-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi JW, Liakopoulos TM, Ahn CH. An on-chip magnetic bead separator using spiral electromagnets with semi-encapsulated permalloy. Biosens Bioelectron. 2001;16(6):409–16. doi: 10.1016/s0956-5663(01)00154-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


