Abstract
Self-assembly and aggregation of amyloid peptides, such as Aβ(1–40) and Aβ(1–42), lead to the development of Alzheimer disease and similar neurodegenerative disorders associated with protein aggregation. The structures of large aggregates, specifically fibrils, are well characterized. However, our understanding about the structure of oligomeric forms of amyloids is incomplete and needs to be expanded, particularly given the finding that oligomeric rather than fibrillar amyloid morphologies are neurotoxic. This lack of knowledge is primarily due to the existence of transient oligomeric forms that require the use of non-traditional approaches capable of probing transiently existing amyloid forms. We have recently developed the Single-Molecule Force Spectroscopy (SMFS) approach enabling us to probe dimeric forms of amyloids. These studies suggest that the assembly of amyloid proteins into dimers leads to extremely stabilized amyloids in non-native, misfolded states [1]. Herein, we applied the SMFS approach to probe amyloid trimers. We used the Aβ(14–23) segment of Aβ42 protein that is responsible for full-size protein aggregation. The dimerization of this peptide was recently characterized [2]. The dimeric form of Aβ (14–23) was assembled by the use of a tandem Aβ(14–23)-YNGK-Aβ(14–23), in which the YNGK motif between the two Aβ(14–23) monomers makes a β turn to form a hairpin loop with an antiparallel arrangement of Aβ(14–23) monomers[3]. The Aβ(14–23) monomer was tethered to the AFM tip, and trimers were formed by approaching the tip to the mica surface on which the Aβ(14–23)-YNGK-Aβ(14–23) dimer was immobilized via a polyethylene glycol tether. We identified trimers by rupture forces that were considerably larger than those for dimers. Models for the trimer assembly process are discussed.
Keywords: Dynamic force spectroscopy, Dimer-Monomer Interaction, F-D curve, Amyloid Peptide
Introduction
The deposition of amyloid fibrils is a hallmark of many types of human neurodegenerative diseases, including Alzheimer’s disease (AD)[1, 4–6]. Aβ(1–40) and Aβ(1–42) are two of the most common proteins generated from amyloid precursor protein by enzymatic (β-secretase and γ-secretase) cleavage, and are primarily responsible for amyloid fibril formation [7–9]. In AD, the self-assembly propensity of these amyloid peptides causes insoluble amyloid fibrils to be deposited into the extracellular space, which is a hallmark of the disease. However, recent data showed that much smaller assemblies of oligomers are neurotoxic rather than larger insoluble aggregates such as fibrils (review and references therein) [10–12]. Therefore, a detailed study of oligomers is crucial to improve our understanding of the molecular mechanism of amyloid aggregation and for the rational design of new therapeutic strategies to prevent Aβ aggregation and possibly treat AD. Traditional structural techniques, including NMR [13,14] and X-ray fibril diffraction [15, 16], were instrumental in deciphering the amyloid protein structure within fibrils, but the structure of distinct oligomers is unknown. Transient states exist along the Aβ aggregation pathway; therefore, the structures of oligomers depend on their sizes. In the majority of published studies, only mixtures of relatively large aggregates with different morphologies have been analyzed by different methods such as NMR [17–19], EPR [20], mass spectroscopy [21,22] and X-ray crystallography [23,24]. However, these data do not provide information on how the aggregation process is initiated and how the growth of oligomers progresses. Computational approaches were very useful to model the process of how monomers assemble into fibril segments, and these approaches provided the structure and dynamics of aggregates at the atomic level [25–27]. However, these powerful simulations require the knowledge of the initial structure.
Significant progress has occurred in the characterization of dimers, which are the very first amyloid oligomers. A critical factor in this advancement was the application of single-molecule probing [28–30], including our Atomic Force Microscopy (AFM) spectroscopy [31–37]. Previously, we used this technique to study protein misfolding and intermolecular interactions to demonstrate that the strength of interprotein interactions correlates with the propensity of proteins to aggregate [36,38]. Extension of this approach to the single-molecule level enabled us to employ the dynamic force spectroscopy (DFS) methodology [39] to characterize properties of transient dimeric states of misfolded α-synuclein (α-Syn) [33,37] and Aβ peptides [35,38,40]. These studies led to the discovery that transiently assembled dimers are very stable, suggesting that during appropriate conditions, dimers can be used as transient seeds for additional aggregation processes. Recently, Aβ dimers capable of causing neuritic degeneration were discovered as the predominate oligomeric species isolated from the brains of humans with AD [10,11]. This finding points to an important role of these small oligomeric species in neurodegenerative disease development. It was also recently discovered that α-Syn aggregation, a hallmark of Parkinson’s Disease (PD), exists in the cytosol of neurons as metastable tetramers and related oligomers with varying amounts of free monomers. These observations suggest that a careful, detailed analysis is required for higher oligomer structures [41].
In our previous study, we applied SMFS and Molecular Dynamic (MD) simulation to investigate the dimerization process of Aβ(14–23) [2]. This 13–23 segment (HHQKLVFFAED) of Aβ contains the Aβ42 region that is crucial for Aβ fibril formation and mediates the strongest Aβ-Aβ interaction within fibrils [42, 43]. The MD simulations revealed that the structural rearrangement of monomers within the dimer assembly leads to a dimer structure with extremely long lifetimes compared to the short lifetimes of other assemblies. In a subsequent paper, [44] we validated the simulated structures of dimers using a novel computational approach enabling one to identify the structure by comparing the simulated force spectroscopy results with experimental results. We showed that between two different structures of dimers discovered during MD simulations, only one structure containing an out-of-register assembly has a similar strength to that observed during experimental conditions.
Herein, we continued the study of the amyloid assembly mechanism using Aβ(14–23) peptide as an experimental system to determine whether the assembly of trimers alters the stability of the system. To address this question, we assembled Aβ(14–23) into a preformed dimer by the use of Aβ(14–23)-YNGK-Aβ(14–23) tandem peptide. According to Tjernberg et al., the YNGK motif forms a β turn, thereby arranging Aβ(14–23) into an antiparallel type hairpin [3].Trimer formation was analyzed by performing AFM probing experiments in which Aβ(14–23) monomers were immobilized onto the AFM tip, and the Aβ(14–23)-YNGK-Aβ(14–23) dimer was tethered to the functionalized mica surface, as described in our prior studies [32,45]. Cysteinyl was added to amyloid peptide sequences to facilitate the covalent attachment of peptides via thiolmaleimide chemistry. Probing revealed that the force necessary to unravel trimers is considerably higher than the force for dimers, suggesting that monomers are arranged differently in trimers.
Materials and Methods
Materials
Cysteinyl peptides CHQKLVFFAED [Aβ(14–23)] and CHQKLVF-FAED-YNGK-HQKLVFFAED [Aβ(14–23)-YNGK-Aβ(14–23)] were synthesized and HPLC purified by Peptide 2.0 company (VA, USA). Similar to our previous publications, [35–40] the 10 mM stock solution of β-mercaptoethanol (Sigma-Aldrich, USA) was prepared in sodium phosphate buffer (10 mM, pH=7.0) and stored at −20°C. 1.0 mM Tris-(2-Carboxyethyl)phosphine, Hydrochloride (TCEP; Hampton Research Inc., CA, USA) was prepared in the same buffer. The 2.92 mM stock solution of maleimide-polyethylene glycol-silatrane (MAS, ref [38,45]) was prepared in water and stored at −20 °C. A stock solution of MAL-PEG-NHS (PEG M. Wt 3400) (Laysan Bio. Inc, USA) was prepared in DMSO (Sigma-Aldrich, USA) at a concentration of 1.67 mM and stored at −20 °C. Deionized water (18.2 MΩ, 0.22 µm pore size filter, APS Water Services Corp., Van Nuys, CA) was used for buffer preparation and washing.
Preparation of peptide stock solution
A measured amount of peptide was first dissolved and sonicated for 5 min in 100 µL of 1,1,1,3,3,3 Hexafluoroisopropanol (HFIP) to destroy pre-aggregated oligomers. The solvent was then evaporated in a vacuum for 2h to completely remove HFIP from the sample. The stock solution of peptide was prepared in DMSO (concentration 2 µM) and stored at −20 °C until needed.
Tip modification
The procedure was the same as described in ref [35–40]. Briefly, silicon nitride (Si3N4) AFM tips (MLCT, Bruker AFM Probes) were immersed in 98% ethanol for 30 min, rinsed thoroughly with deionized water and dried under gentle flow with dry argon, followed by UV treatment for 40 min (CL-1000 Ultraviolet Cross-linker, UVP, Upland, CA). AFM tips were then immersed in an aqueous solution of 167 µM maleimide-polyethylene glycol-silatrane (MAS) for 3h followed by multiple rinses with deionized water. The peptide was diluted with sodium phosphate buffer (10 mM, pH=7.0) to 10 nM and treated with a few microliters of 1.0 mM TCEP for 10 min to reduce any disulfide bonds that formed. The MAS functionalized AFM tips were then dipped into the peptide solution for 1h and washed with deionized water. Any unreacted maleimide moieties on the tips were quenched with 10 mM β-mercaptoethanol in PBS for 10 min. The tips were then washed several times with water and stored in sodium phosphate buffer (10 mM, pH=7.0) at 4°C until needed.
Mica surface modification
We followed the protocols described in ref [32–37] including Parkinson’s disease (PD]. Mica sheets (Asheville-Schoon-maker Mica Co., Newport News, VA) were cut into ~1.5 cm × 1.5 cm squares and glued onto glass slides using epoxy glue EPO-TEK 353ND (Epoxy Technology, Inc., Billerica, MA). The upper surface of the mica was cleaved with scotch tape, and the surface was immediately treated with 167 µM of 1-(3-Aminopropyl) Silatrane (APS) in water for 30 min. The surface was washed with fresh water several times to remove any unbound APS. The surface was then dried with argon, followed by treatment with 167 µM MAL-PEG-NHS (PEG M. Wt = 3400) in DMSO for 2h. The surface was washed with fresh DMSO and water to remove unreacted compound. 20 nM Aβ(14–23)-YNGK-Aβ(14–23) in phosphate buffer (10 mM, pH = 7.0) pretreated with a few microliters of 1.0 mM TCEP was then added to the surface and incubated for 1h in a humid environment. The surface was then washed with fresh buffer several times and unre-acted maleimide groups were blocked by treating the surface with 10 mM β-mercaptoethanol in PBS for 10 min. Finally, the surface was washed with deionized water, covered with phosphate buffer and stored at 4°C until needed.
Force measurement
The force-distance (F–D) measurements were performed in sodium phosphate buffer (10 mM, pH = 7.0) at room temperature in MFP 3D AFM (MFP-3D, Asylum Research, Santa Barbara, CA), as described previously [35,38,40]. AFM probes with nominal spring constants of 0.03 N/m were used throughout the experiments. The thermal noise analysis method (Igor Pro 6.31) was used for calculating the spring constant of the AFM probes. The ramp size was set to 200 nm throughout the experiments. A low trigger force (100 pN) was applied to the AFM probes to facilitate contact between the two types of pep-tides under observation. For the DFS study, F-D curves were acquired at different retraction velocities in a range from 100 nm/s to 2500 nm/s. F-D curves were acquired by probing different points on the surface set by a grid (5 × 5 µm), with points separated from each other by 100 nm. The dwell time was set at 0.5 second. Approximately 1500 curves were acquired during each retraction speed for statistical analysis.
Data analysis
The F-D curves were analyzed with the data processing software Igor Pro 6.31, provided by Asylum Research. The force curves were fitted with the Worm-like chain (WLC) Model [46], as shown below
| (1) |
where F(x) is the force at the distance of x, kB is the Boltzmann constant, T is the absolute temperature, and Lp and Lc are the persistence length and the contour length, respectively. The WLC model was chosen because it properly describes the elasticity of a polypeptide chain [47, 48]. The persistence length was allowed to be varied for the best fitting curve and evaluated as a variable parameter along with the contour length. A set of rupture forces (F), contour lengths (Lc), and persistence lengths (Lp) were obtained from fitting. The contour length (Lc) values were used to construct histograms for contour length that were fitted with the Gaussian function.
Apparent loading rates at different retraction speeds were calculated using the following equation [36]
| (2) |
where Fp =kBT/Lp, kc is the spring constant (N/m), v is the tip velocity, F is the rupture force, and r is the apparent loading rate (pN/s). The calculated apparent loading rates were taken in the form of mean ± SEM. These apparent loading rates were used to fit force histograms with the probability density function [49].
| (3) |
where p(F) is the most probable force, koff is off-rate constant, F is the measured force, and xb is the distance of energy barrier.
The most probable rupture forces were obtained from each histogram with its corresponding apparent loading rates. For each speed, we obtained a value for the most probable rupture force and apparent loading rate. In DFS experiments, we have plotted the most probable rupture force (F) against the logarithmic apparent loading rate (r) and fitted the data points with the Bell-Evan equation [50–51].
| (4) |
From the extrapolation of the best fit, the zero pulling rate, the off-rate constant koff and the distance of the energy barrier xb for monomerdimer interactions are obtained.
The energy landscape profile was constructed according to our previous papers [37,40] using the following equation
| (5) |
where ΔG is the height of the energy barrier, kB is the Boltzmann constant, T is temperature, koff is the off-rate constant, and h is Plancks constant.
Results and Discussion
Experimental design and approach
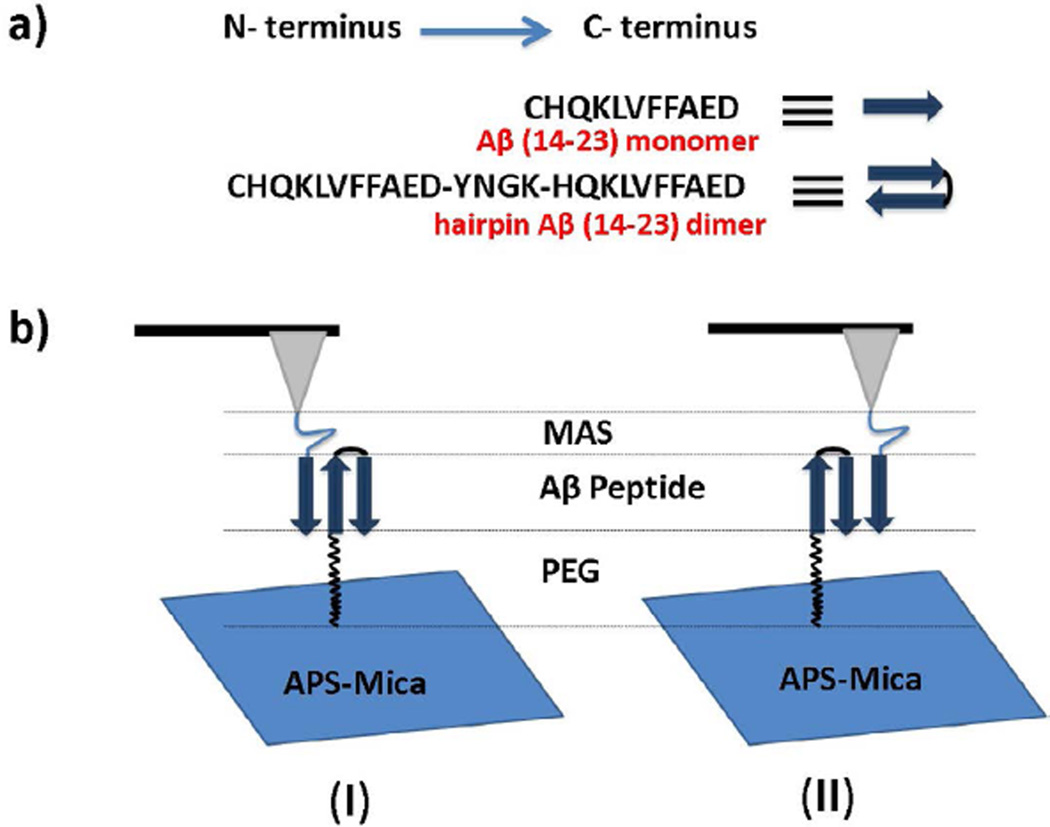
The schematics of the experimental set up and SMFS experiments are shown in Figure 1. The Aβ(14–23) trimer is formed when the monomer immobilized on the AFM tip is brought into contact with the hairpin construct tethered to the surface. The dimer was assembled by the intramolecular folding of the tandem peptide consisting of two Aβ(14–23) monomers connected by the YNGK motif. According to NMR studies, [3] this motif forms a U-turn structure, arranging the Aβ(14–23)-YNGK-Aβ(14–23) monomers in a tandem antiparallel fashion. The rationale for this design comes from our recent computational analysis showing that Aβ(14–23) monomers are arranged in an antiparallel orientation in the dimer [44].
Figure 1.
(a) Schematic representation of Aβ(14–23) monomer and hairpin Aβ(14–23) dimer; (b) Schematic representation of the experimental set up for the force measurements, where (I) and (II) depict two types of possible monomer-dimer interactions.
The cysteinyl peptides were covalently anchored to either the AFM tip or the mica surface via flexible tethers (MAS or PEG) terminated with maleimide groups. According to NMR and hydrogen-deuterium exchange experiments, conjugation of cysteine to the N-termini of Aβ should not effect its intrinsic amyloid character [52,53]. The amyloid peptides were pretreated with TCEP before coupling to the maleimide functionalized surface or tip to minimize S-S bond formation and reduce S-S bonds to free SH groups. The propensity of self-aggregation was minimized by using a very low peptide concentration (10 nM for the AFM tip and 20 nM for the mica surface) during the covalent attachment of peptides to the maleimide groups. A peptide concentration of several orders of magnitude is required for aggregation [54]. The specific interactions between Aβ(14–23) monomers and hairpin Aβ (14–23) dimers were measured by multiple approach-retraction cycles over various spots of the mica surface.
Probing of trimers
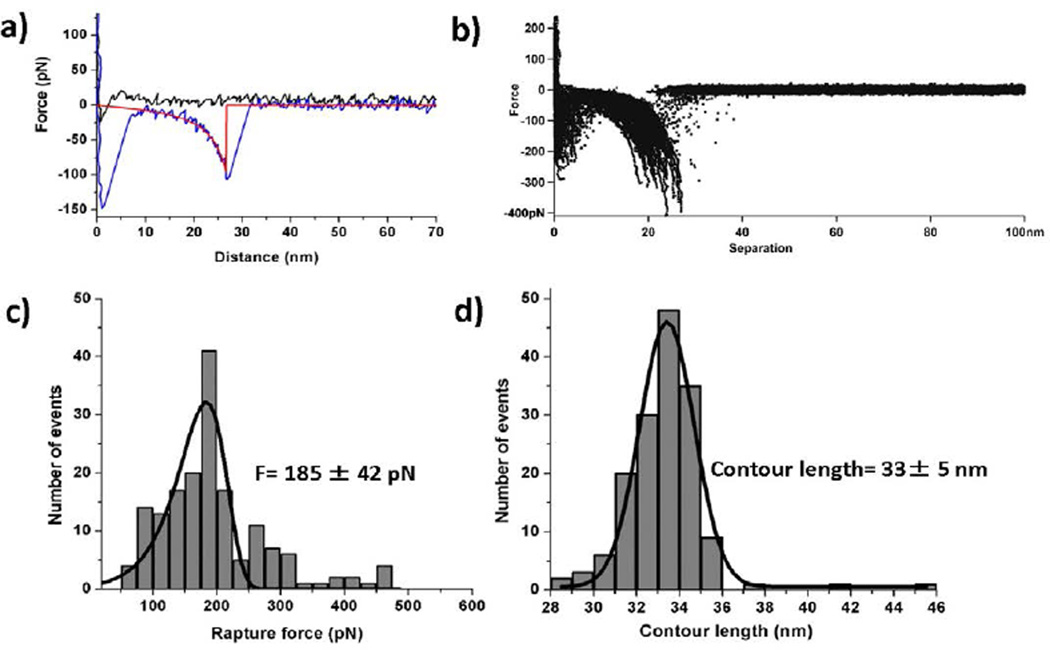
SMFS was performed with the Molecular Force Probe 3D AFM system (MFP-3D, Asylum Research, Santa Barbara, CA) in sodium phosphate buffer at room temperature (25°C). Figure 2a shows typical approach (black) and retraction force curves. The rupture event appears on the retraction force curve as a drop of the force after which the system dissociates and no force changes are detected. The rupture force value was calculated after fitting each force curve with the WLC approximation using equation 1. An example is shown in Figure 2a, in which the fit is depicted by a red curve with the rupture event evident as a sharp transition. A large peak appears at the beginning of the retraction force curve that corresponds to non-specific interactions that typically appear in AFM experiments [37, 36]. The specific interaction peak is identified by its position on the rupture profile and appears after stretching the PEG and unstructured segments of the peptide [38].
Figure 2.
(a) A typical force-distance curves; approach curve (black), retraction curve (blue), and WLC fit (red); (b) Overlay plot of retraction force-distance curves at a speed of 500 nm/s, showing the superposition of specific interactions, with the number of curves taken = 165. (c) Force histogram (grey bars) at 500 nm/s retraction speed with the PDF fit curve (black); the most probable rupture force is 185 ± 42 pN [peak point of PDF fitting ± standard deviation]. (d) Contour length histogram (grey bars) at 500 nm/s, speed of retraction with Gaussian fit curve (black); the most probable contour length is 33 ± 5 nm [peak point of Gaussian fitting ± width of fitting]. The number of F-D curves used for making histograms for the force and contour length is 165. The bin sizes are: force histogram = 20 pN; contour length histogram = 1 nm.
Similar force curves were obtained by multiple probing cycles at various positions on the substrate. The superposition of the force curves is shown in Figure 2b and illustrates the overall visual reproducibility of the probing experiments. Typically, ~1500 curves were acquired at a speed of 500 µm/s at different positions on the mica surface. Among them, 9–10% of the curves showed specific monomerdimer interactions. This is a high yield, given the requirement to measure single-molecule interactions between Aβ(14–23) monomers and hairpin Aβ(14–23) dimers [55].
The force curves were used to measure two major parameters, the rupture force and the contour length. The histograms for both parameters are shown in Figure 2c and d, respectively. The force histogram was fitted with the probability density function (eq. 3; black curve), demonstrating a good fit. The most probable force for the rupture of the Aβ(14–23) monomer and hairpin Aβ(14–23) complex is 185 ± 42 pN. This value is twice as large as the value obtained for the rupture of Aβ(14–23) dimers in monomer-monomer probing experiments [2].
Figure 2D shows the contour length distribution. The histogram is symmetric, and approximation of the distribution with the Gaussian equation produces a contour length corresponding to the maximum distribution of 33 ± 5 nm. Given the fact that flexible tethers are stretched prior to the rupture of the complex, it is important to compare this value with the known lengths of the flexible tethers (Figure 1b). The corresponding lengths are: ~3 nm for MAS, ~4 nm for the peptide, and ~25 ± 5 nm for PEG, yielding 32 ± 5 nm for the total length of the tethers. This value is very close to that obtained during the experiments. In fact, these estimates were made for the peptide position at the apex of the tip. During tip functionalization, various positions are possible, leading to contour length values that are shorter than expected [45,56]. This is also illustrated by the data in Table 1, which shows the contour length values obtained from four independent experiments.
Table 1.
Experimental contour lengths for Aβ (14–23) monomer and hairpin Aβ (14–23) dimer complex formation during four different experiments.
| Experiment number | 1st | 2nd | 3rd | 4th |
|---|---|---|---|---|
| Contour length (nm) | 27 ± 6 | 33 ± 5 | 23 ± 5 | 28 ±6 |
Dynamic Force Spectroscopy
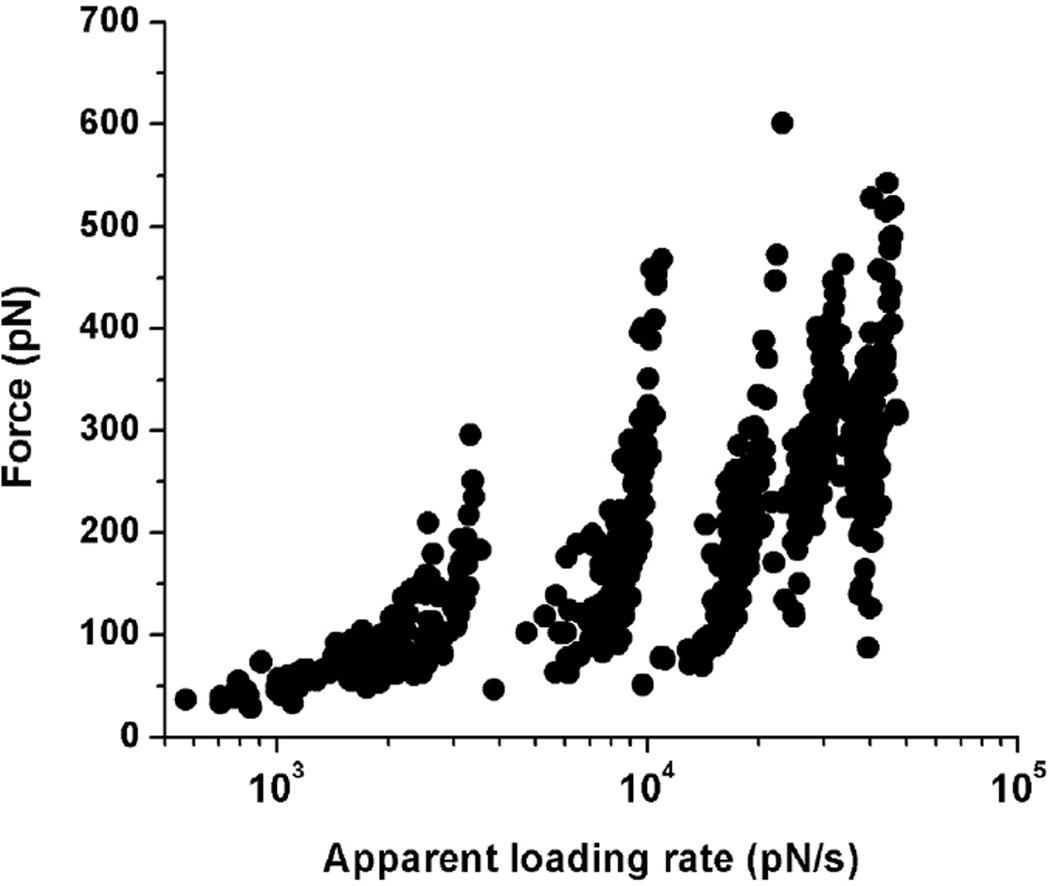
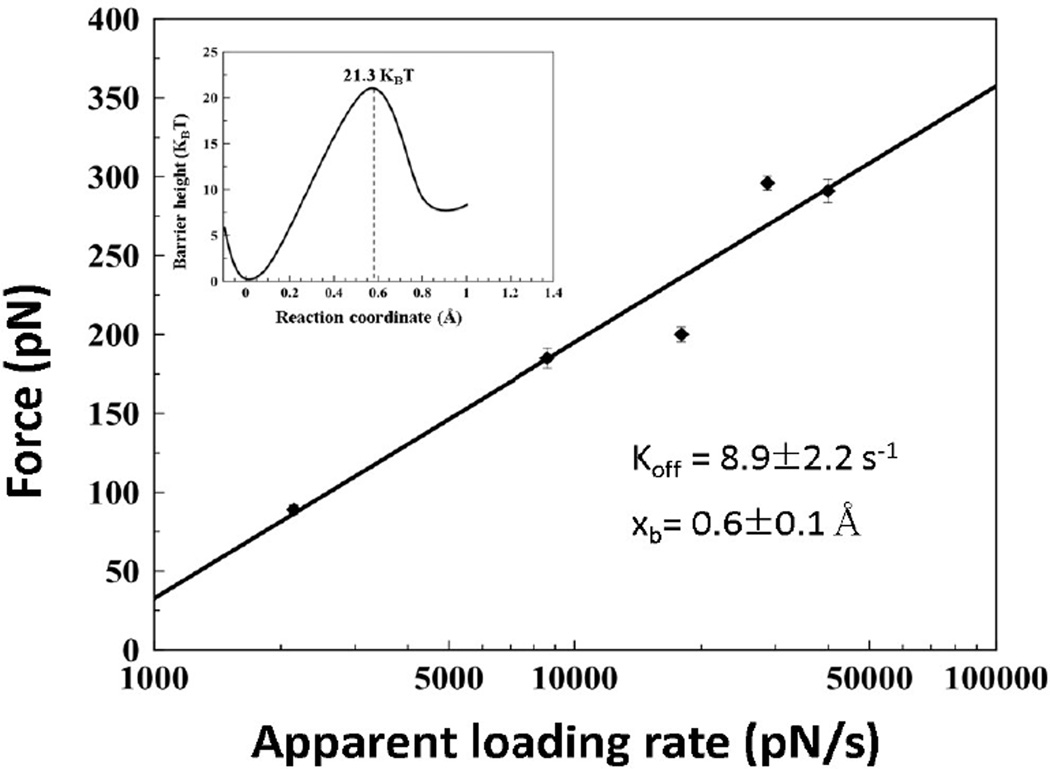
Next, we applied the DFS (Dynamic Force Spectroscopy) approach to estimate the stability of the trimers (eq. 4). We performed force spectroscopy experiments with pulling rates ranging from 100–2500 nm/s that correspond to the apparent loading rates in the range of 500–50000 pN/s. The full data set showing the dependence of rupture forces on the logarithm of apparent loading rates [57] is shown in Figure 3. The entire set was divided into five different groups of loading rates. The force and loading rate distributions were obtained and maxima values were used to generate the DFS plot as shown in Figure 4. The bar sizes in the plot correspond to the SEM values for the force distributions. The linear fit, according to equation 4, produced kinetic parameters for the complexes, including their stability, off-rate constant (koff = 8.9 ± 2.2 s−1), and the energy barrier position (xb = 0.6 ± 0.1 Å) [58]. The off-rate constant corresponds to the complex lifetime of 0.11 ± 0.02 seconds that is similar to the Aβ(14–23) dimer lifetime of 1.06 ± 0.95s obtained in ref [2]. The height of the energy barrier was calculated from the off-rate constant using Eq. 5, ΔG= 21.3 kBT, and the energy landscape profile corresponding to this ΔG value is shown as an inset in Figure 4.
Figure 3.
The dataset of the rupture force vs. loading rate (raw data) used for dynamic force spectroscopy analysis.
Figure 4.
Dynamic force spectroscopy data for Aβ(14–23) monomer and hairpin Aβ(14–23) dimer interactions. The black straight line represents the fit with the Bell-Evans model. The inset shows the energy profile diagram.
Assembly and stability of the trimer
Monomer-monomer Aβ(14–23) interactions within a dimer were recently analyzed in ref [44]. The experimental value was 53 pN, which is substantially less than the value 185 pN obtained in this analysis of the trimer. According to the computational analysis performed in ref [44], monomers can adopt two different conformations, out-of-register and in-register. The dissociations of these different conformations produce rupture force values of 46 pN and 178 pN, respectively. The experimental value for the trimer rupture is close to the value obtained for the in-register arrangement of the monomer relative to another monomer within the dimer.
According to Figure 1, two possible orientations of the monomer relative to the dimer are possible, either parallel or anti-parallel orientations. A recent analysis [59] showed that the parallel orientation of monomers produces rupture forces in the range of 30 pN that are substantially lower that the values obtained for antiparallel orientations even in the out-of-register arrangement. Therefore, we conclude that the antiparallel orientation is associated with the trimer, thereby rejecting model II.
In our model for the trimer depicted in Figure 1, we assume that the dimer is assembled as a hairpin, with both monomers aligned in the in-register antiparallel orientation. Although this assumption is supported by the NMR analysis of the YNGK containing peptides, additional evidence comes from the force spectroscopy studies and computational analysis [44]. Indeed, the 185 pN rupture force value obtained in this paper is comparable to the 178 pN value obtained in the simulations for the in-register assembly of the monomers. If the monomer in the dimer did not assemble as a long β-hairpin, the rupture force between the monomer and the dimer would be less than that value. The 7 pN higher rupture force value obtained in this paper and the force for the in-register assembly of the Aβ(14–23) peptides indicates that the interactions within the trimer are higher than that for the dimer, suggesting that the dimer structure is more dynamic than the trimer. However, this hypothesis requires additional justification and represents a goal of our future research.
Conclusion
In summary, we report on a novel approach using SMFS that uses oligopeptides with pre-assembled structures to provide insight into amyloid trimerization. The analysis revealed that the Aβ(14–23) trimer has elevated stability, providing a quantitative measure for subsequent steps of oligomer assembly. This approach can be extended to explore mechanisms of molecular interactions for higher oligomerization steps, such as tetramers, pentamers, hexamers, and so on. Performing computational analyses with recently developed approaches can provide details of these mechanisms.
Acknowledgments
This work was supported by grants to Y.L.L. from the National Institutes of Health (NIH: 5R01 GM096039-02) and the National Science Foundation (EPS-1004094). The authors thank the Lyubchenko lab members for the results discussion and useful inputs.
Abbreviations
- AFM
Atomic Force Microscopy
- F-D curve
Force-Distance Curve
- PEG
Polyethylene Glycol
- WLC
Worm-like Chain
- DFS
Dynamic Force Spectroscopy
- APS
1-(3-aminopropyl) silatrane
- NHS-PEG-MAL
N-Hydroxysuccinimide- Polyethyleneglycol-Maleimide
- MAS
Maleimide- Polyethylene Glycol-Silatrane
- NMR
Nuclear Magnetic Resonance
- RP-HPLC
Reverse Phase High Performance Liquid Chromatography
- SEM
Standard Error of Mean
References
- 1.Lyubchenko YL, Kim BH, Krasnoslobodtsev AV, Yu J. Nano-imaging for protein misfolding diseases. Wiley Interdiscip Rev Nanomedicine Nanobiotechnology. 2010;2(5):526–543. doi: 10.1002/wnan.102. [DOI] [PubMed] [Google Scholar]
- 2.Lovas S, Zhang Y, Yu J, Lyubchenko YL. Molecular mechanism of misfolding and aggregation of Aβ(13–23) J Phys Chem B. 2013;117(20):6175–6186. doi: 10.1021/jp402938p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tjernberg LO, Tjernberg A, Bark N, Shi Y, Ruzsicska BP, et al. Assembling amyloid fibrils from designed structures containing a significant amyloid beta-peptide fragment. Biochem J. 2002;366(Pt 1):343–351. doi: 10.1042/BJ20020229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelly JW. The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways. Curr Opin Struct Biol. 1998;8(1):101–106. doi: 10.1016/s0959-440x(98)80016-x. [DOI] [PubMed] [Google Scholar]
- 5.Dobson CM. Getting out of shape. Nature. 2002;418(6899):729–730. doi: 10.1038/418729a. [DOI] [PubMed] [Google Scholar]
- 6.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 7.Benilova I, Karran E, De Strooper B. The toxic Aβ oligomer and Alzheimer’s disease: an emperor in need of clothes. Nat Neurosci. 2012;15(3):349–357. doi: 10.1038/nn.3028. [DOI] [PubMed] [Google Scholar]
- 8.Shioi J, Georgakopoulos A, Mehta P, Kouchi Z, Litterst CM, et al. FAD mutants unable to increase neurotoxic Aβ 42 suggest that mutation effects on eurodegeneration may be independent of effects on Aβ. J Neurochem. 2007;101(3):674–681. doi: 10.1111/j.1471-4159.2006.04391.x. [DOI] [PubMed] [Google Scholar]
- 9.Paravastu AK, Leapman RD, Yau W-M, Tycko R. Molecular structural basis for polymorphism in Alzheimer’s beta-amyloid fibrils. Proc Natl Acad Sci U S A. 2008;105(47):18349–18354. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilcox KC, Lacor PN, Pitt J, Klein WL. Aβ Oligomer-Induced Synapse Degeneration in Alzheimer’s Disease. Cel Mol Neurobiol. 2011:1–10. doi: 10.1007/s10571-011-9691-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakono M, Zako T. Amyloid oligomers: Formation and toxicity of Aβ oligomers. FEBS J. 2010;277(6):1348–1358. doi: 10.1111/j.1742-4658.2010.07568.x. [DOI] [PubMed] [Google Scholar]
- 12.Savage MJ, Kalinina J, Wolfe A, Tugusheva K, Korn R, et al. A sensitive aβ oligomer assay discriminates Alzheimer’s and aged control cerebrospinal fluid. J Neurosci. 2014;34(8):2884–2897. doi: 10.1523/JNEUROSCI.1675-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hou L, Shao H, Zhang Y, Li H, Menon NK, et al. Solution NMR Studies of the Aβ(1–40) and Aβ(1–42) Peptides Establish that the Met35 Oxidation State Affects the Mechanism of Amyloid Formation. J Am Chem. Soc. 2004;126(7):1992–2005. doi: 10.1021/ja036813f. [DOI] [PubMed] [Google Scholar]
- 14.Tycko R. Solid-state NMR studies of amyloid fibril structure. Annu. Rev Phys Chem. 2011;62:279–299. doi: 10.1146/annurev-physchem-032210-103539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gras SL, Squires AM. Dried and hydrated X-ray scattering analysis of amyloid fibrils. Methods Mol Biol. 2011;752:147–163. doi: 10.1007/978-1-60327-223-0_10. [DOI] [PubMed] [Google Scholar]
- 16.Madine J, Jack E, Stockley PG, Radford SE, Serpell LC, et al. Structural insights into the polymorphism of amyloid-like fibrils formed by region 20–29 of amylin revealed by solid-state NMR and X-ray fiber diffraction. J Am Chem Soc. 2008;130(45):14990–15001. doi: 10.1021/ja802483d. [DOI] [PubMed] [Google Scholar]
- 17.Ahmed M, Davis J, Aucoin D, et al. Structural conversion of neurotoxic amyloid-beta(1–42) oligomers to fibrils. Nat Struct Mol Biol. 2010;17(5):561–567. doi: 10.1038/nsmb.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colvin MT, Silvers R, Frohm B, Su Y, Linse S, et al. High Resolution Structural Characterization of Aβ42 Amyloid Fibrils by MAS NMR. J Am Chem Soc. 2015 doi: 10.1021/jacs.5b03997. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fawzi NL, Ying J, Torchia DA, Clore GM. Kinetics of amyloid β monomer-to-oligomer exchange by NMR relaxation. J Am Chem Soc. 2010;132(29):9948–9951. doi: 10.1021/ja1048253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu L, Liu C, Guo Z. Structural insights into Aβ 42 oligomers using site-directed spin labeling. J Biol Chem. 2013;288(26):18673–18683. doi: 10.1074/jbc.M113.457739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young LM, Cao P, Raleigh DP, Ashcroft AE, Radford SE. Ion Mobility Spectrometry-Mass Spectrometry Defines the Oligomeric Intermediates in Amylin Amyloid Formation and the Mode of Action of Inhibitors. J Am Chem Soc. 2014;136(2):660–670. doi: 10.1021/ja406831n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Rempel DL, Zhang J, Sharma AK, Mirica LM, et al. Pulsed hydrogen-deuterium exchange mass spectrometry probes conformational changes in amyloid beta (Aβ) peptide aggregation. Proc Natl Acad Sci U S A. 2013;110(36):14604–14609. doi: 10.1073/pnas.1309175110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spencer RK, Li H, Nowick JS. X-ray crystallographic structures of trimers and higher-order oligomeric assemblies of a peptide derived from Aβ(17–36) J Am Chem Soc. 2014;136(15):5595–5598. doi: 10.1021/ja5017409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pham JD, Demeler B, Nowick JS. Polymorphism of oligomers of a peptide from β-amyloid. J Am Chem Soc. 2014;136(14):5432–5442. doi: 10.1021/ja500996d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urbanc B, Cruz L, Ding FD, Sammond S, Khare, et al. Molecular dynamics simulation of amyloid beta dimer formation. Biophys J. 2004;87(4):2310–2321. doi: 10.1529/biophysj.104.040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buchete N-V, Tycko R, Hummer G. Molecular dynamics simulations of Alzheimer’s beta-amyloid protofilaments. J Mol Biol. 2005;353(4):804–821. doi: 10.1016/j.jmb.2005.08.066. [DOI] [PubMed] [Google Scholar]
- 27.Chebaro Y, Mousseau N, Derreumaux P. Structures and thermodynamics of alzheimer’s amyloid-β Aβ(16–35) monomer and dimer by replica exchange molecular dynamics simulations: Implication for full-length Aβ fibrillation. J Phys Chem B. 2009;113(21):7668–7675. doi: 10.1021/jp900425e. [DOI] [PubMed] [Google Scholar]
- 28.Yu H, Liu X, Neupane K, Gupta AN, Brigley AM, et al. Direct observation of multiple misfolding pathways in a single prion protein molecule. Proc Natl Acad Sci. 2012;109(14):5283–5288. doi: 10.1073/pnas.1107736109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffmann A, Neupane K, Woodside MT. Single-molecule assays for investigating protein misfolding and aggregation. Phys Chem Chem Phys. 2013;15(21):7934–7948. doi: 10.1039/c3cp44564j. [DOI] [PubMed] [Google Scholar]
- 30.Yu H, Dee DR, Woodside MT. Single-molecule approaches to prion protein misfolding. Prion. 2013;7(2):140–146. doi: 10.4161/pri.23303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kransnoslobodtsev AV, Shlyakhtenko LS, Ukraintsev E, Zai-kova TO, Keana JFW, et al. Nanomedicine and protein misfolding diseases. Nanomedicine. 2005;1(4):300–305. doi: 10.1016/j.nano.2005.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu J, Malkova S, Lyubchenko YL. alpha-Synuclein misfold-ing: single molecule AFM force spectroscopy study. J Mol Biol. 2008;384(4):992–1001. doi: 10.1016/j.jmb.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 33.Yu J, Lyubchenko YL. Early stages for Parkinson’s development: α-Synuclein misfolding and aggregation. J Neuroimmune Pharmacol. 2009;4(1):10–16. doi: 10.1007/s11481-008-9115-5. [DOI] [PubMed] [Google Scholar]
- 34.Lyubchenko YL, Sherman S, Shlyakhtenko LS, Uversky VN. Nanoimaging for protein misfolding and related diseases. J Cell Biochem. 2006;99(1):53–70. doi: 10.1002/jcb.20989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim BH, Palermo NY, Lovas S, Zaikova T, Keana JFW, et al. Single-molecule atomic force microscopy force spectroscopy study of Aβ-40 interactions. Biochemistry. 2011;50(23):5154–5162. doi: 10.1021/bi200147a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu J, Warnke J, Lyubchenko YL. Nanoprobing of α-synuclein misfolding and aggregation with atomic force microscopy. Nanomedicine. 2011;7(2):146–152. doi: 10.1016/j.nano.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 37.Krasnoslobodtsev AV, Volkov IL, Asiago JM, Hindupur J, Rochet JC, et al. α-Synuclein misfolding assessed with single molecule AFM force spectroscopy: Effect of pathogenic mutations. Biochemistry. 2013;52(42):7377–7386. doi: 10.1021/bi401037z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lv Z, Condron MM, Teplow DB, Lyubchenko YL. Nanoprobing of the effect of Cu2+ cations on misfolding, interaction and aggregation of amyloid β peptide. J Neuroimmune Pharmacol. 2013;8(1):262–273. doi: 10.1007/s11481-012-9416-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evans E. Probing the relation between force--lifetime--and chemistry in single molecular bonds. Annu Rev Biophys Biomol Struct. 2001;30:105–128. doi: 10.1146/annurev.biophys.30.1.105. [DOI] [PubMed] [Google Scholar]
- 40.Lv Z, Roychaudhuri R, Condron MM, Teplow DB, Lyubchenko YL. Mechanism of amyloid β-protein dimerization determined using single-molecule AFM force spectroscopy. Sci Rep. 2013;3:2880. doi: 10.1038/srep02880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Selkoe D, Dettmer U, Luth E, Kim N, Newman A, et al. Defining the native state of α-synuclein. Neurodegener Dis. 2014;13(2–3):114–117. doi: 10.1159/000355516. [DOI] [PubMed] [Google Scholar]
- 42.Santini S, Wei G, Mousseau N, Derreumaux P. Pathway complexity of Alzheimer’s β-amyloid Aβ16–22 peptide assembly Structure. 2004;12(7):1245–1255. doi: 10.1016/j.str.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 43.Balbach JJ, Ishii Y, Antzutkin ON, Leapman RD, Rizzo NW, et al. Amyloid Fibril Formation by Aβ 16–22 , a Seven-Residue Fragment of the Alzheimer’s β-Amyloid Peptide, and Structural Characterization by Solid State NMR. Biochemistry. 2000;39(45):13748–13759. doi: 10.1021/bi0011330. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Lyubchenko YL. The Structure of Misfolded Amyloidogenic Dimers: Computational Analysis of Force Spectroscopy Data. Biophys J. 2014;107(12):2903–2910. doi: 10.1016/j.bpj.2014.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim BH, Lyubchenko YL. Nanoprobing of misfolding and interactions of amyloid β 42 protein. Nanomedicine. 2014;10(4):871–878. doi: 10.1016/j.nano.2013.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ludwig M, Rief M, Schmidt L, Li H, Oesterhelt F, et al. AFM, a tool for single-molecule experiments. Appl Phys A Mater Sci Process. 1999;68(2):173–176. [Google Scholar]
- 47.Tskhovrebova L, Trinick J, Sleep JA, Simmons RM. Elasticity and unfolding of single molecules of the giant muscle protein titin. Nature. 1997;387(6630):308–312. doi: 10.1038/387308a0. [DOI] [PubMed] [Google Scholar]
- 48.Stirnemann G, Giganti D, Fernandez JM, Berne BJ. Elasticity, structure, and relaxation of extended proteins under force. Proc Natl Acad Sci U S A. 2013;110(10):3847–3852. doi: 10.1073/pnas.1300596110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Friedsam C, Wehle AK, Kuhner F, Gaub HE. Dynamic single-molecule force spectroscopy: bond rupture analysis with variable spacer length. J Phys Condens Matter. 2003;15(18):1709–1723. [Google Scholar]
- 50.Evans E, Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys J. 1997;72(4):1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200(4342):618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 52.Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, et al. A structural model for Alzheimer’s beta -amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci U S A. 2002;99(26):16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lührs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, et al. 3D structure of Alzheimer’s amyloid-β(1–42) fibrils. Pro Natl Acad Sci U S A. 2005;102(48):17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tjernberg LO, Callaway DJ, Tjernberg A, et al. A molecular model of Alzheimer amyloid beta-peptide fibril formation. J Biol Chem. 1999;274(18):12619–12625. doi: 10.1074/jbc.274.18.12619. [DOI] [PubMed] [Google Scholar]
- 55.Lee H, Scherer NF, Messersmith PB. Single-molecule mechanics of mussel adhesion. Proc Natl Acad Sci U S A. 2006;103(35):12999–13003. doi: 10.1073/pnas.0605552103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Farrance OE, Paci E, Radford SE, Brockwell DJ. Extraction of Accurate Biomolecular Parameters from Single-Molecule Force Spectroscopy Experiments. ACS Nano. 2015;9(2):1315–1324. doi: 10.1021/nn505135d. [DOI] [PubMed] [Google Scholar]
- 57.Wildling L, Rankl C, Haselgrübler T, et al. Probing binding pocket of serotonin transporter by single molecular force spectroscopy on living cells. J Biol Chem. 2012;287(1):105–113. doi: 10.1074/jbc.M111.304873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Merkel R, Nassoy P, Leung A, Ritchie K, Evans E. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature. 1999;397(6714):50–53. doi: 10.1038/16219. [DOI] [PubMed] [Google Scholar]
- 59.Krasnoslobodtsev AV, Zhang Y, Viazovkina E, Gall A, Bertagni C, et al. A Flexible Nanoarray Approach for the Assembly and Probing of Molecular Complexes. Biophys J. 2015;108(9):2333–2339. doi: 10.1016/j.bpj.2015.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]