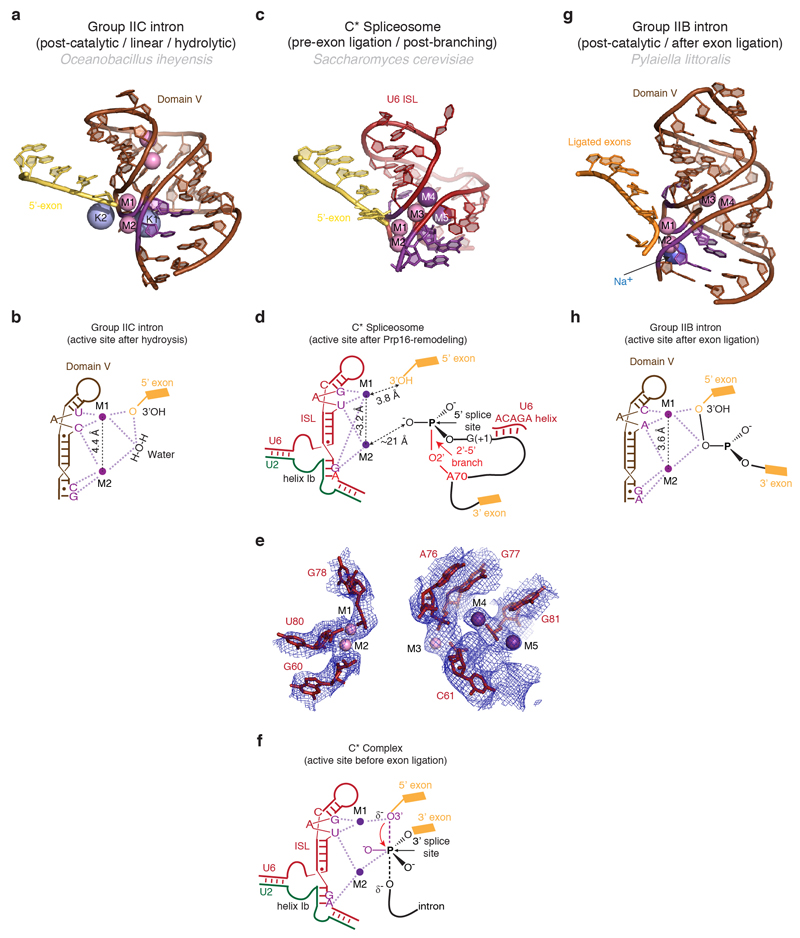
Extended Data Figure 6. Metals in the RNA core of the C* complex.
a,b, Structure (a) and schematic representation (b) of the active site of a group IIC intron trapped in the post-catalytic state in the presence of Mg2+ and K+ (PDB 4FAR, ref. 52). The 5’ exon 3’-hydroxyl interacts with M1, while a water molecule bridges the two catalytic metals. Two additional non-catalytic Mg2+ and two K+ close to the active site are also shown. c-d, Structure of the RNA at the active site of spliceosomal C* complex, with putative metal binding (c), schematic of catalytic metal binding (M1 and M2) (d), and comparison of the putative metal binding model with the EM density (e). Note conservation of the metal binding residues compared to the group II intron and proximity of the cleaved G(-1) 3’-hydroxyl to M1. Besides the two catalytic Mg2+, additional divalent and monovalent metals were observed in the group IIB structure53. Density observed at analogous position in C* complex may be attributable to a Mg2+ (M3) and two K+ (M4 and M5). f, Proposed interactions between U6 snRNA and the two catalytic Mg2+ during the transition state for exon ligation, as inferred from biochemistry (ref. 2). g, h, Structure (g) and schematic (h) of the RNA core of a group IIB intron in a post-catalytic configuration, following both branching and exon ligation (PDB 4R0D, ref. 53). Residues that position the catalytic metals are shown in magenta.