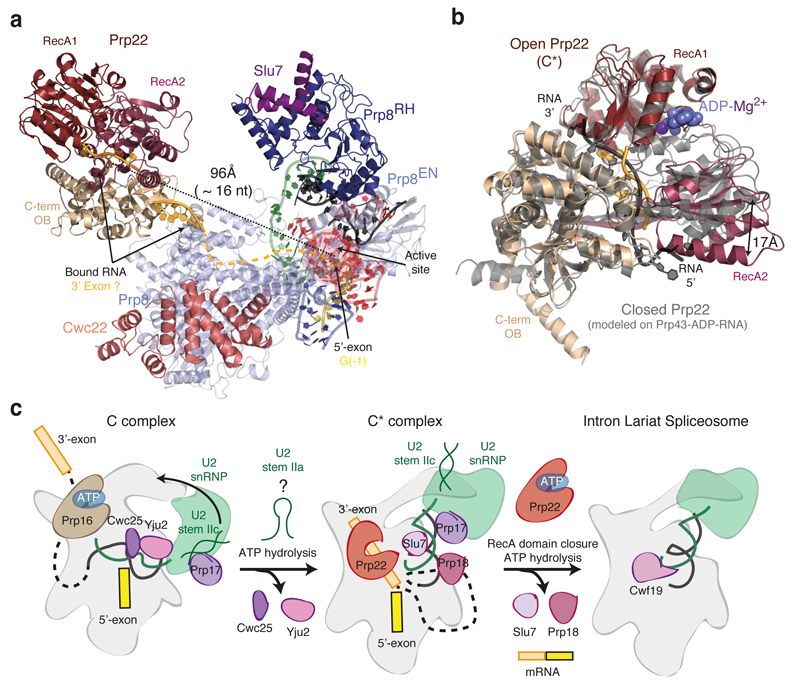
Figure 4. Prp22 and ATP-mediated transitions at the catalytic stage of splicing.
a, Position of the helicase Prp22 in the C* complex. Focussed classification allowed us to obtain an improved map for Prp22 into which individual secondary structural elements could be modelled (Extended Data Fig. 4). Helicase domains are coloured individually and RNA (shown in pale orange) was modelled in density observed at the entrance and in the centre of the helicase RNA channel. An RNA path (shown as dashed line) can be modelled from the active site of the spliceosome to Prp22. The distance from the end of the 5’-exon to the centre of the Prp22 RNA channel could be spanned by 16 nucleotides. b, Open conformation of Prp22 observed in C*. The model of the closed conformation, based on the structure of Prp43 (PDB 3KX2; ref. 29), is in grey. c, Model for Prp16-mediated remodelling. U2 stem IIa could form at an intermediate stage in remodelling to allow a binding switch of Prp17 from stem IIc to the branch helix (cf. ref. 30). Following domain closure and ATP binding, Prp22 would translocate the 3’-exon and release the mRNA.