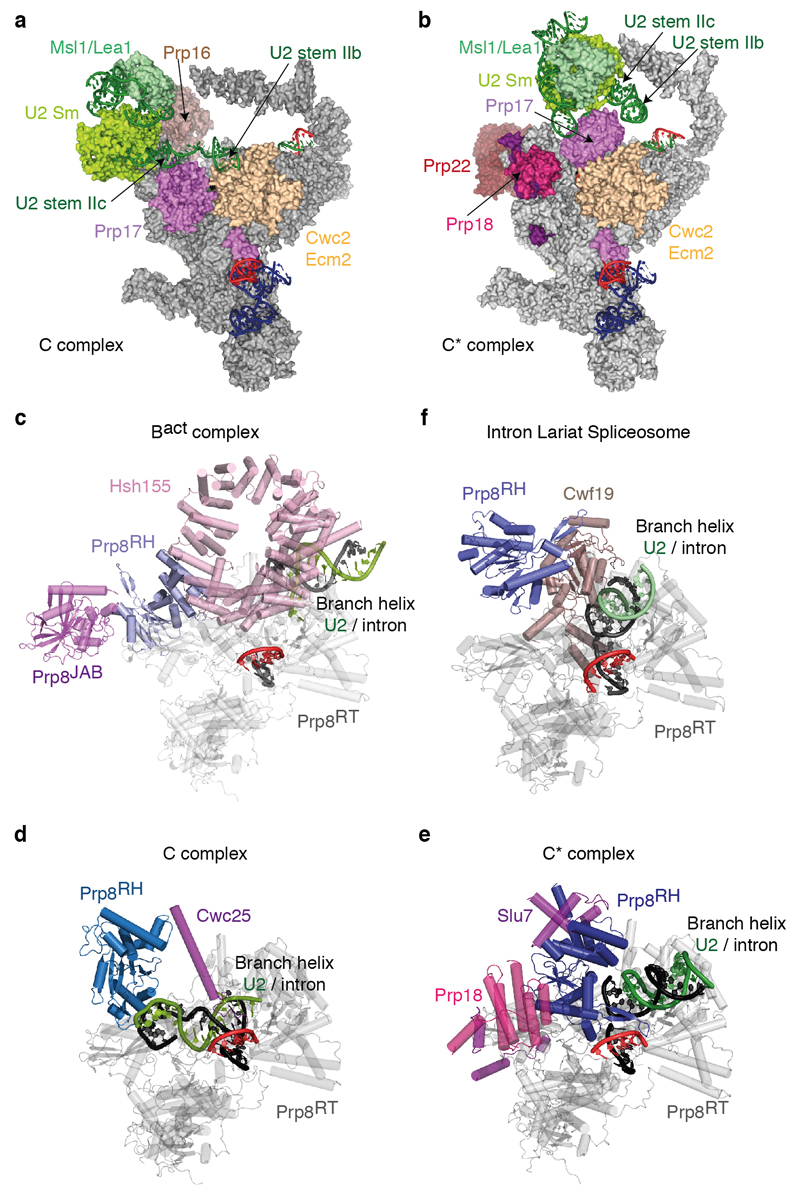
Extended Data Figure 5. U2 snRNP rearrangement between C and C* complexes and repositioning of the Prp8 RNaseH-like domain during splicing.
a-b, Movement of the U2 snRNP domain between C complex (a) and the C* complex (b). Note that U2 stem IIa switches from an interaction with Prp17 in C complex to a position adjacent to the U2 Sm ring in C*; the binding of U2 stem IIb by Ecm2/Cwc2 is disrupted and Prp17 changes its binding surface on Ecm2/Cwc2. In a, Brr2 was omitted for clarity. c, Prp8 RNaseH domain conformation in the Bact complex (PDB 5GM6; ref. 52). Note how Hsh155 sequesters the branch helix away from the RNaseH domain, which projects its β-hairpin into solvent and is stabilized by the Prp8 Jab1/MPN domain and Brr2 (not shown). d-e, RNaseH conformation in C (d) and C* (e) complexes. Note that the RNaseH domain undergoes a dramatic inward rotation towards the body of the complex and is stabilized in alternative conformations by factors specific for branching (C) or exon ligation (C*). f, RNaseH conformation in the S. pombe ILS. Note that the de-branching specific factor Cwf19 is now wedged between the RNaseH domain and the branch helix. All structures were aligned on the Prp8 endonuclease domain (Prp8EN), shown in grey; complex-specific factors are coloured in magenta shades; Prp8RH, Prp8 RNaseH-like domain.