Abstract
The origin and expansion of biological diversity is regulated by both developmental trajectories1,2 and limits on available ecological niches3–7. As lineages diversify an early, often rapid, phase of species and trait proliferation gives way to evolutionary slowdowns as new species pack into ever more densely occupied regions of ecological niche space6,8. Small clades such as Darwin’s finches demonstrate that natural selection is the driving force of adaptive radiations, but how microevolutionary processes scale up to shape the expansion of phenotypic diversity over much longer evolutionary timescales is unclear9. Here we address this problem on a global scale by analysing a novel crowd-sourced dataset of 3D-scanned bill morphology from >2000 species. We find that bill diversity expanded early in extant avian evolutionary history before transitioning to a phase dominated by morphospace packing. However, this early phenotypic diversification is decoupled from temporal variation in evolutionary rate: rates of bill evolution vary among lineages but are comparatively stable through time. We find that rare but major discontinuities in phenotype emerge from rapid increases in rate along single branches, sometimes leading to depauperate clades with unusual bill morphologies. Despite these jumps between groups, the major axes of within-group bill shape evolution are remarkably consistent across birds. We reveal that macroevolutionary processes underlying global-scale adaptive radiations support Darwinian9 and Simpsonian4 ideas of microevolution within adaptive zones and accelerated evolution between distinct adaptive peaks.
The role of adaptive radiations as the source of much of the world’s biological diversity has been widely emphasised10,11. Studies of small clades have provided insights into the role of natural selection as a diversifying force, but cannot illuminate the processes that shape the diversity and discontinuities of radiations over much longer evolutionary timeframes. Indeed, at large taxonomic scales, the diversification of clades11,12 and traits13 shows no evidence of the predicted slowdowns in evolutionary rates, despite there being numerous examples in small clades3,14–16. This apparent paradox is potentially resolved by G. G. Simpson’s model, in which major jumps to new adaptive zones (“quantum evolution”) can occur unpredictably throughout clade history. These jumps give rise to rapid lineage expansion into previously unoccupied niche space as sub-clades continue to radiate within distinct adaptive zones and subzones4. Simpson’s models introduced the concept of ‘mega-evolution’—diversification over large temporal and spatial scales—unifying microevolution with other factors such as ecological opportunity and evolutionary constraints that shape the macroevolutionary trajectories of radiating lineages. However, while phylogenetic studies involving thousands of species have demonstrated heterogeneity in rates of phenotypic evolution13,17, it is unclear whether the processes outlined by Simpson play an important role in large-scale adaptive radiations. This is because previous studies have been unable to specifically assess the macroevolutionary dynamics of ecologically relevant traits. Here we study the evolution of an important ecological trait (bill shape) across an entire Class of organisms (birds) to elucidate the processes shaping the accumulation of phenotypic diversity within a global-scale adaptive radiation.
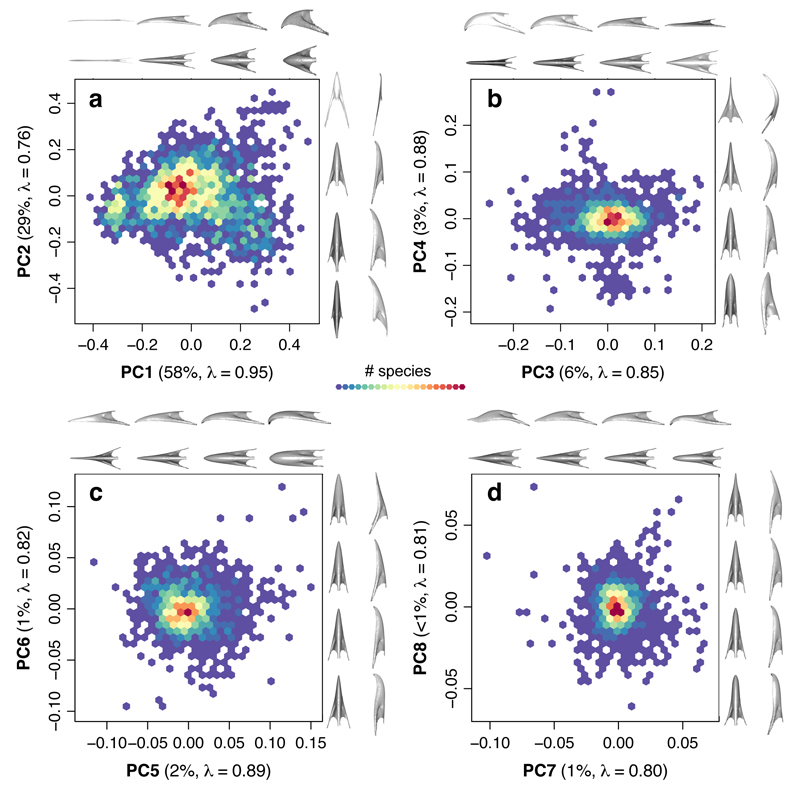
Our approach is based around a novel data set describing avian bill shape. The avian bill is closely associated with species’ dietary and foraging niches16,18,19 and represents a highly-adaptable ecological trait known to play a key role in classic avian adaptive radiations16,18,20. We took 3D scans of museum study skins comprising >2000 species (>97% of extant genera) representing the full range of bill shape diversity. We landmarked bills (Extended Data Fig. 1) using a bespoke crowd-sourcing website, www.markmybird.org, and quantified the bill shape morphospace of extant birds using Procrustes superimposition and Principal Components Analyses (PCA, see Methods). The first eight PC axes explain >99% of the total variation in bill shape (Fig. 1). PC1 (58% of overall shape variation) describes the volumetric aspect ratio from elongated (e.g. sword-billed hummingbird, Ensifera ensifera) to stout bills (e.g. large ground finch, Geospiza magnirostris) and captures the range of shape variation encompassed by standard linear measurements (length, width and depth). Variation in these bill dimensions may relate to fine scale division of the dietary or foraging niche among closely related species, but cannot explain the diversity of shapes observed among extant birds. More complex aspects of shape (42% of total variation) are explained by the remaining PCs (Fig. 1), which retain high phylogenetic signal (Extended Data Table 1). Importantly, although these higher shape axes explain a low proportion of shape variance, they capture large differences in ecologically relevant aspects of bill shape. The narrow (long tail) distributions of higher shape axes, compared to the broad distribution of PC1 (Extended Data Fig. 2, Extended Data Table 1), suggest that the majority of species have relatively simple bill shapes and diversify in densely packed regions of bill morphospace.
Figure 1. Bird bill morphospace density plots.
PC axes 1-8 are shown as pairwise scatterplots, along with warps representing the change in bill shape (n = 2028 species) along each axis in dorsal and lateral views. Each axis is labeled with the proportion of variance explained and estimates of phylogenetic signal (Pagel’s λ). The colour scale refers to the number of species in 20 bins with minimum and maximum richness of a, 1-23 b, 1-72 c, 1-64, and d, 1-98 species, respectively.
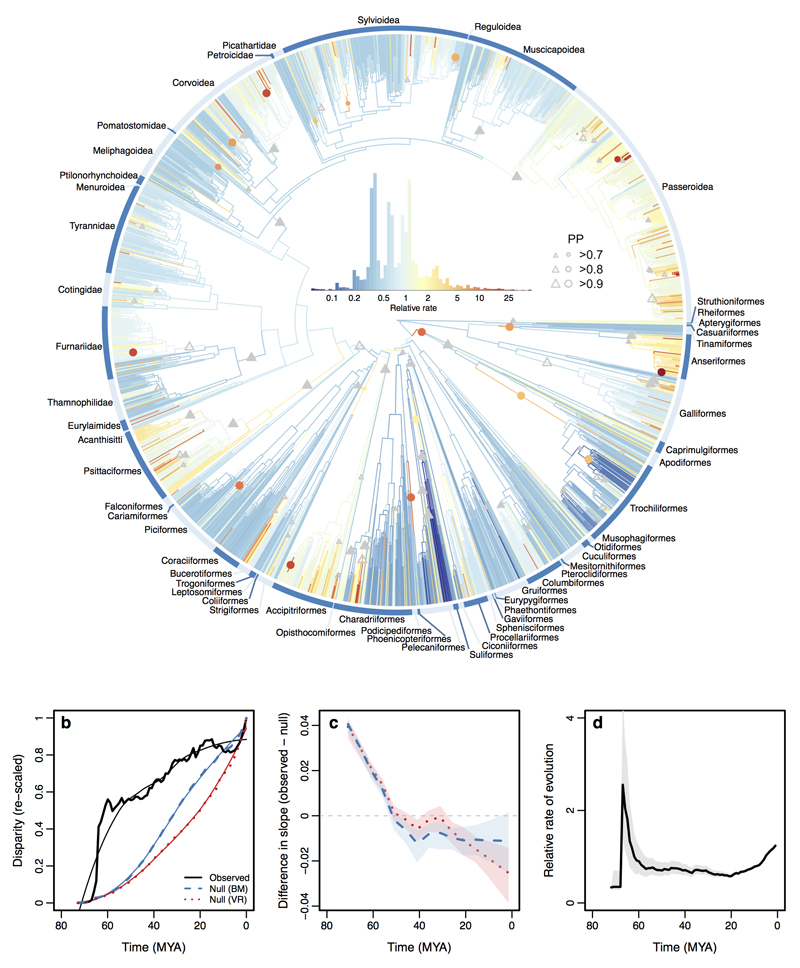
We tested an important prediction of Simpson’s model by evaluating how niche expansion and niche packing have contributed to the accumulation of bill shape disparity throughout avian evolutionary history. We estimated multivariate disparity through time using ancestral state estimates derived from rate heterogeneous models of trait evolution (see Methods)13. In 1 million year time slices, we calculated disparity as the sum of the variances21 from the first eight shape axes. We compared observed disparity through time with two null models—constant-rate (Brownian motion) and rate heterogeneous trait evolution—that are unbiased with respect to niche filling processes (see Methods). Relative to these null expectations, we find that the filling of avian bill morphospace through time shows a striking dominance of niche expansion early in avian history, followed by a more recent transition towards niche packing (Fig. 2a-b, Extended Data Fig. 2). Our data includes only extant taxa due to the poor preservation of bills in the avian fossil record22, although we acknowledge that some extinct taxa had bills that may lie outside the range of extant diversity (e.g. Phorusrhacidae, Gastornithidae, Dromornithidae). This can result in underestimates of disparity particularly if these morphologies arise early in clade history22–24. Our analyses are therefore conservative with respect to transitions from bill morphospace expansion to filling and consistent with recent studies of avian skeletal material22. The transition in the mode of niche filling is consistent with a process of ever-finer divisions of niche space and would be expected to correspond to slowdowns in rates of bill evolution. However, the switch from niche expansion to niche packing does not map onto temporal trends in the rate of bill shape evolution. Plotting evolutionary rates through time reveals an initial low rate followed by a moderate (two to four-fold) increase that is coincident with the divergence of many non-Passerine orders (Fig. 2c, Extended Data Fig. 3, 4). Thereafter average rates dip and then rise gradually with less than 1.5-fold total variation over ~80 million years of evolutionary history, contrasting sharply with >250-fold variation in evolutionary rate among individual lineages (Fig. 3).
Figure 2. Morphospace filling through time.
a, Accumulation of multivariate disparity through time in 1 million time slices (thick black line: observed data; thin black line: after LOESS smoothing; blue lines: constant rate null model; red lines: variable rate null model). b, Comparison of slopes (estimated in 5 million year windows) of the LOESS-smoothed observed data and null models. Differences in slope above and below zero indicate dominance of morphospace expansion versus morphospace packing respectively. Shading indicates 95% confidence intervals. c, Mean relative rates of evolution with 95% confidence intervals (grey) through time.
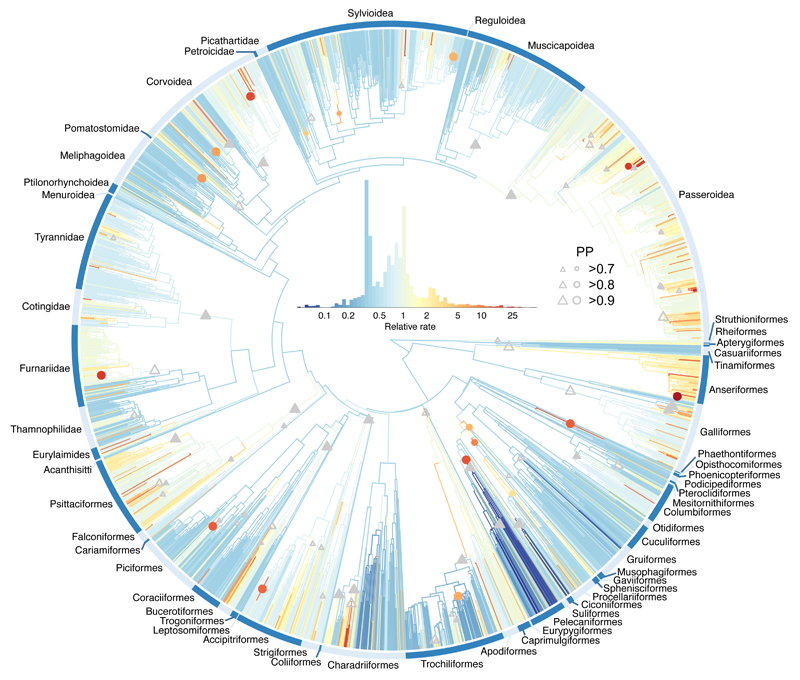
Figure 3. Multivariate rates of bill shape evolution.
The avian phylogeny (n = 2028 species) coloured by estimates of the mean relative multivariate rate of bill shape evolution. Grey triangles show the stem branch of clades with support for whole clade shifts in evolutionary rate. Coloured circles show rate shifts on individual internal branches (colour indicates the rate estimate). The relative size of triangles and circles indicates the posterior probability (PP) of a rate shift. Triangles distinguish shifts on the focal node (filled) and shifts at the focal node or on one of its two daughter nodes (open).
The disjunction between rates of evolution and the accumulation of bill shape disparity suggests that temporal trends in evolutionary rate are not necessarily indicative of the underlying mode of niche filling. This decoupling could arise if some clades diverge rapidly within regions of morphospace that are occupied by other clades, but where the respective clades occur in allopatry. To test this idea, we mapped rates of bill evolution onto the avian phylogeny (Fig. 3, Extended Data Fig. 3-5). We find several instances of clades exhibiting exceptionally high rates of evolution consistent with speciational or phyletic evolution within adaptive subzones (Fig. 3). Some of the fastest rates of bill evolution arise in island radiations of passerine birds, where ecological divergence has been closely linked to ecological opportunity (e.g. Malagasy vangas16, Galapagos finches18, Hawaiian honeycreepers20), suggesting that lineages radiating on isolated island archipelagos can explore morphological space independently of the global avifauna. Notably high rates of bill evolution occur in several large species-rich clades that have high speciation rates, including the Psittaciformes, the Furnariidae, and the Passeroidea. However, these clades occupy regions of morphospace that overlap with other more slowly evolving clades and so, while rapid divergence among close relatives within a subzone leads to locally high rates, they do not contribute uniquely to the global expansion of morphospace. In contrast, some large (Anseriformes) and some smaller clades (Alcidae, Bucerotiformes) that exploit more unusual ecological resources have also evolved rapidly.
Next, we find evidence for several notable instances of exceptionally high rates of evolution along single branches (Extended Data Table 2). Such instances indicate unusually large jumps in bill phenotype and many of the most extreme shifts (e.g. Phoenicopteridae, Musophagidae, Pelecanidae, and Caprimulgiformes; Fig. 3) occur towards the base of the avian radiation, consistent with the idea of early, rapid quantum evolution into new adaptive zones. In some cases (e.g. Pelecanidae and Ciconiidae), the evolution of extreme bill shapes is associated with a subsequent slowdown in the rate of bill shape evolution (Fig. 3), suggesting that ancestral shifts towards a highly specialised bill phenotype may often constrain further opportunities for either bill evolution or speciation25. In contrast, some rapid jumps result in speciose clades occupying more densely packed regions of morphospace. For instance, the Hirundinidae diverge from other Sylvoidea but converge on a swift-like aerial insect hawking form. These latter types of shift do not appear to be restricted to any particular time periods or regions of the avian phylogeny. Similarly, the Trochiliformes diverge rapidly away from the Apodiformes towards a range of bill phenotypes that opened up additional opportunities for continued diversification, consistent with the idea of rapid speciation driven by ecological opportunity following the invasion of an unoccupied adaptive zone4,8.
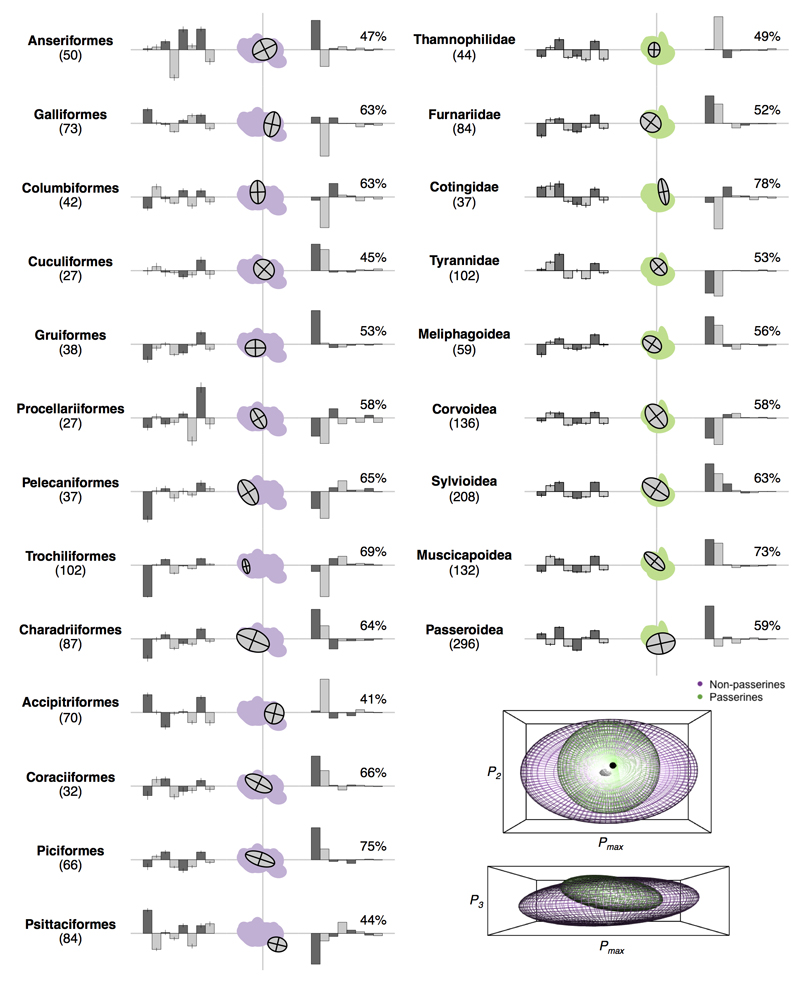
Major phenotypic shifts early in the avian adaptive radiation followed by limited divergence within sub-clades, implies a disconnect between mega-evolutionary radiations on a global scale and adaptive radiations within smaller constituent clades. Although the average phenotypes (morphospace centroids) of some higher taxa diverge from one another (Extended Data Fig. 6, 7), it is unclear whether the primary axes of bill shape variation within sub-clades parallel the major axes of variation across birds as a whole (i.e. higher PCs), or whether evolution within clades occurs along axes of variation that are distinct from the major global axes (i.e. lower PCs). We explored these ideas by quantifying the variances and covariances (termed P matrices, see Methods) of bill shape axes within higher taxa (families, superfamilies and orders)26,27. We find that shape variation within higher taxa is explained by a single significant eigenvector of P, with the exception of the Psittaciformes (two significant eigenvectors). In contrast, the number of significant eigenvectors across all birds combined is three, suggesting that there is low dimensional divergence within clades but high dimensional divergence between clades. We then asked whether the dominant eigenvector within each sub-clade (Pmax) was consistent across higher taxa. We find that bill shape (i.e. PC) axes 1 and 2—those that explain the majority of variation across birds as a whole—also consistently load most heavily onto Pmax within higher taxa (Extended Data Fig. 7). This suggests that bill shape evolution within higher taxa tends to fall back to limited pathways irrespective of the position of the clade in morphospace
The low dimensionality and consistency of bill shape variation within clades, and high dimensionality among clades, demonstrates striking discontinuities between how phenotypic disparity accumulates in the early stages of major radiations, versus how disparity accumulates as younger clades evolve within an already mature and ecologically diverse radiation. This early expansion of morphospace has parallels with observations of peak disparity early in clade history in palaeontological datasets of a wide range of metazoan taxa28. The earliest known fossil assemblages of the ancestors of modern birds, dating from the Early Cretaceous, were functionally and ecologically depauperate29. It is likely that the rise of modern birds from the late Cretaceous onwards occurred in a rapidly changing world30, coinciding with extensive ecological opportunity. Our results imply that this dynamic adaptive landscape may have driven Simpsonian mega-evolution across adaptive zones, later giving way to smaller scale fine-tuning of the bill as avian diversity expanded across the globe.
Methods
Data sampling
We measured 2,028 species, representing 2,028 of 2,091 genera across 194 families. Specimens were obtained primarily from the avian skin collection at the Natural History Museum, Tring, and also from the Manchester Museum. Study skins, rather than skeletal material, were used because they are generally much better represented in museum collections with more species and specimens available than in skeletons, and secondly because the rhamphotheca (the keratinous sheath surrounding the fused premaxilla, maxilla and nasal bones) is often absent from skeletonised specimens. This is the portion of the bill that interacts directly with the environment and is thus the subject of selection. Where available, one mature male per species was selected for scanning. This was necessary to achieve the taxonomic sampling required within a reasonable time frame and because males are generally better represented in the collections than females. Care was taken to select specimens that were undamaged, with all the landmarks visible and unobstructed (see below). When undamaged males were unavailable, females were preferentially chosen over unsexed specimens. Some species (e.g. Strigiformes, Podargidae, and others) have bills that are obscured by protruding feathers or rictal bristles that ‘shade’ the bill from the scanner. For specimens where this was an issue, or for specimens that were not represented in the skins collections, specimens were chosen from the skeletons collection at Tring.
3D scanning and processing
3D scans of the bills were taken using white or blue structured light scanning (FlexScan3D, LMI Technologies, Vancouver, Canada). The use of 3D scans provides a more complete and nuanced estimate of bill diversity than standard linear measures (length, width, depth) that reflect only the relative proportions of the bill and effectively assume that bills are no more than proportional variations on a cone shape. For bills of lengths > 5 cm, a R3X white-light scanner (calibration boards 10 – 25 mm, resolution 0.075 mm) was used, and for bills of lengths < 3 cm a MechScan white-light macro scanner (calibration boards 1.3 – 4 mm, resolution 0.010 mm) was used. For bills intermediate between these lengths, a pre-calibrated HDI blue-light scanner (resolution 0.080 mm) was used. In some cases, larger bills (e.g. those with a high aspect ratio, such as hummingbirds) were scanned on the higher resolution scanner. In order to fully capture 3D geometry, approximately 5 - 25 scans per bill were obtained, and aligned and combined in the FlexScan software before being exported as .ply files. Scans were imported into Geomagic Studio (3D Systems, Rock Hill, SC, USA), automatically decimated to approximately 500,000 faces, and cleaned to remove mesh errors (holes, reversed normals, high aspect ratio spikes). In some specimens, it was necessary to remove feathers or scanning artefacts that had obstructed portions of the geometry by manual cleaning of the mesh. Following cleaning, meshes were exported as .obj files.
Landmark choice
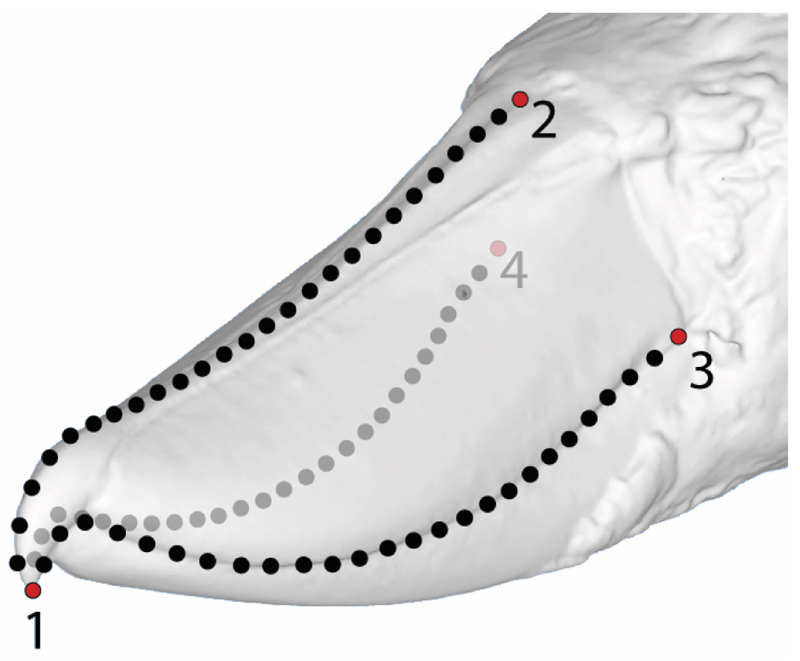
Landmark-based geometric morphometrics (GM) is a method for analysing variation in geometric shape based on the positions of equivalent homologous points (landmarks) placed on every specimen in the study31,32. While homologous in this context is usually taken to mean developmentally homologous, in practice the key to landmark selection is that the points chosen must be easily identifiable, such that they can be accurately placed and repeatable within and between specimens32. This is difficult to do on the rhamphotheca because, other than the tip of the bill, it lacks any obvious landmarks, especially as the nostrils are not exposed in many bird species. We therefore opted to identify four true landmarks: 1) the tip of the beak; and the posterior margin of the keratinous rhamphotheca, along the 2) midline dorsal profile; 3) left; and 4) right tomial edges. Three semilandmark curves joined point 1 to points 2, 3, and 4 to represent the dorsal profile, and the left and right tomial edges respectively (Extended Data Fig. 1).
Crowdsourcing
In order to facilitate landmarking of such a high number of species, a crowdsourcing website, www.markmybird.org, was developed to allow members of the public to participate in the research by placing landmarks on to the bill scans. After registration, volunteers were required to landmark two training bills with easily identifiable (shoebill, Balaeniceps rex) and more challenging (brown-chested alethe, Alethe poliocephala) landmarks. Instructions were shown to all users for every landmark, with links to more detailed instructions provided. Bills were assigned to users by randomly selecting a bill from the 100 scans most recently uploaded. To account for the fact that different users will always place homologous landmarks in slightly different places33, each bill was marked by three to four different users.
Quality control and landmark averaging
Custom R scripts were used to check for common mistakes that may not have been caught by real-time error checks (confusing left and right, large asymmetries in landmark position, incorrect order of semilandmarks, and semilandmarks that deviated from the correct curve due to user failure to rotate the bill and assess their landmark placement in three dimensions). If any landmark configuration failed these tests, the data was manually checked and if necessary removed with the bill made re-available for landmarking. Finally, the three/four repetitions for each bill were averaged to find the mean shape between users, and tested to ensure that all users had placed the landmarks within an acceptable range (Procrustes distance < 0.2) of one another. The average bill shapes were then passed forward for geometric morphometric (GM) analysis. Using ANOVA approaches for assessing measurement error in geometric morphometrics33, we found that repeatability was consistently high among users when comparing among PC axes (see below; Extended Data Table 2).
Geometric morphometrics
All GM analysis was performed in the R package Geomorph34. First, landmark configurations were subjected to a Generalised Procrustes Analysis (GPA) to remove the effects of size and translational and rotational position on the landmark configurations. This is a common first step in GM analyses as it removes all the geometric information from the landmark coordinates that is not related to shape31. During alignment, symmetry was enforced so that slight user-introduced differences in the left/right positions of landmarks were removed. Semilandmarks were slid to minimise bending energy35. The Procrustes aligned coordinates were then assessed using PCA to identify the major axes of shape variation within bird bills, which were plotted as morphospaces. PC scores for the first eight axes are available as supplementary material. As morphospaces are projections of multidimensional Kendall’s shape space into two-dimensional tangent space, they may be prone to distortions the further one moves from the central coordinates of the morphospace. In other words, extreme bill morphologies plotting at the edges of morphospace have the potential to distort the projection such that Procrustes distances at the edges of a morphospace are not equivalent to those at the centre of a morphospace. To assess the extent to which projected tangent space differed from the underlying Kendall’s shape space, the Procrustes aligned coordinates were analysed using tpsSmall 1.3036. We found no evidence of distortion: distance in tangent was very tightly correlated with Procrustes distance (uncentred correlation: 0.999; regression through the origin slope: 0.985; root mean squared error < 0.001). Similarly, Procrustes distances were consistently close to tangent distances (minimum Procustes D: 0.024, minimum Tangent d: 0.024; mean Procustes D: 0.194, mean Tangent d: 0.192; maximum Procustes D: 0.525, maximum Tangent d: 0.501).
Warps of the associated shape changes with each PC were generated by transforming the landmarks of the bill closest to the average shape (rusty-fronted barwing, Actinodura egertoni) to landmarks representing the extremes of a given PC when all other PCs = 0, and interpolating the surface in between.
To assess any possible distortion of PCA by the underlying phylogenetic non-independence among species, we also ran a phylogenetic PCA37,38. As with the standard PCA, the first eight PCs accounted for >99% of total shape variance. We found that the first two pPCs did not correlate with the first two original PCs—pPC1 was more closely correlated with PC2 and pPC2 was more closely correlated with PC1. The remaining PCs and pPCs were closely correlated and retained the same order in terms of the proportion of variance explained. We also re-ran rate variable models on the first eight pPCs (see below). For this analysis we allowed the pPCs to be correlated because a property of pPCA is that the axes are not expected to be orthogonal. The multivariate results are similar regardless of the choice of PCA or pPCA (Extended Data Fig. 3). Recently identified problems inherent with using PCA (or pPCA) that can lead to misidentifying macroevolutionary models are expected to arise when individual PCs are analysed, particularly when the variance explained is distributed fairly evenly across multiple PCs39. Because we use a multivariate approach these problems are minimized.
Phylogenetic framework
We base our analyses on the phylogenetic tree distributions from www.birdtree.org11. For both ‘Hackett’ and ‘Ericson’ backbones, we sampled 10,000 ‘stage 2’ trees (i.e. those containing all 9,993 species) from www.birdtree.org, which were pruned to generate tree distributions for the 2,028 species in our dataset. We also generated similar tree distributions using ‘stage 1’ trees from the same source, which contain only the subset of species placed using genetic data. Of the 2028 species in the full dataset, 1,627 (80%) were represented in stage 1 trees. Based on these distributions, we used TreeAnnotator40 to generate maximum clade credibility (MCC) trees, setting branch lengths equal to ‘Common Ancestor’ node heights. In addition, we constructed a composite of the Jetz et al. trees and the genomic backbone tree of Prum et al.41 (Extended Data Fig. 4) by grafting sub-clades of the Stage 2 Hackett MCC tree onto nodes in the Prum et al. phylogeny at positions where the two trees could be sensibly combined (see Supplementary Material for node matching data and R code to combine the trees). This process resulted in a composite tree combining the genus level resolution afforded by the Jetz et al. tree with the branching topology and age estimates of the Prum et al. backbone, which are notably younger than those in the Jetz et al. trees.
Phylogenetic signal
We calculated the phylogenetic signal of bill shape by estimating Pagel’s λ using the R package MOTMOT42. λ can vary between 0 and 1, with a value of 0 indicating no phylogenetic signal and a value of 1 indicating similar levels of phylogenetic covariance as expected under a BM model.
Models of trait evolution
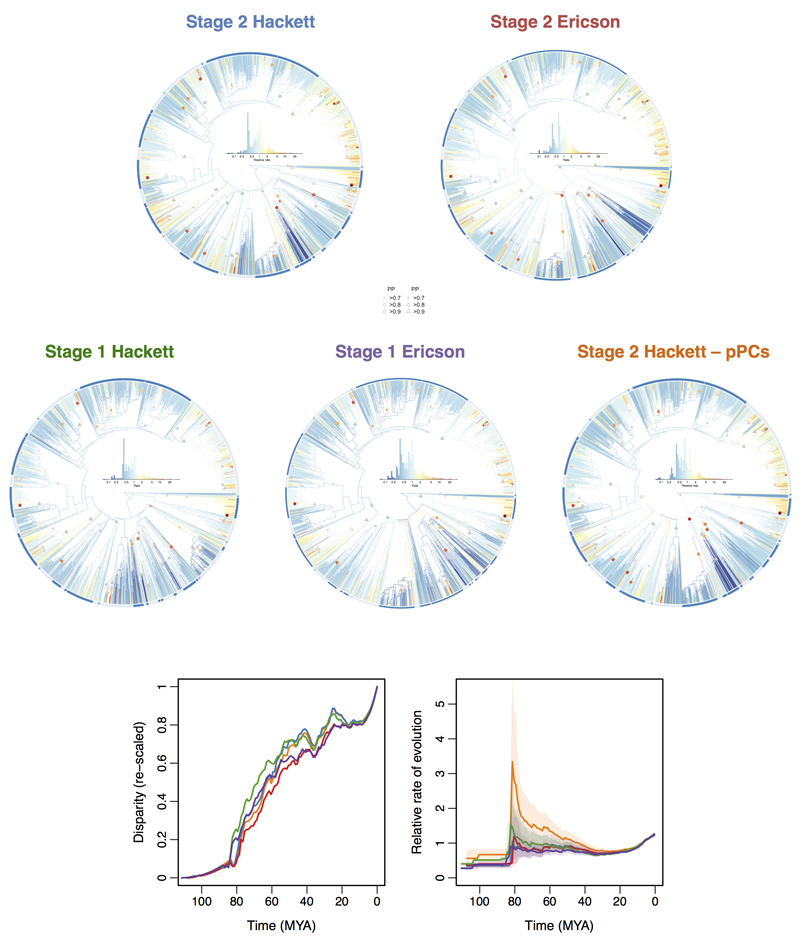
Univariate variable rates models were estimated using the software BayesTraits (available from http://www.evolution.rdg.ac.uk/) using default priors and a single-chain Markov chain Monte Carlo (MCMC) run for at least 1 billion (1,000,000,000) iterations. From each chain we sampled parameters every 100,000 iterations and final parameter estimates for each model were based on 5,000 post-burn in samples. Uncorrelated multivariate models were estimated using the same approach. At each iteration in the MCMC chain, the multivariate models fit a single branch length transformation to the tree across all trait (i.e. PC) axes. An uncorrelated multivariate model is justified because PC axes are inherently orthogonal, however this may limit inference of some forms of rate change. Specifically, the uncorrelated multivariate model is informative with respect to changes in the variances among clades and shifts in the morphospace centroids of clades (i.e. single branch shifts) but cannot detect cases where variances and centroids are similar but covariances among clades differ. We summarised the results of each run by calculating (i) the mean rate and (ii) the probability of a rate shift (branch or clade) over all posterior samples for each node in the tree. It is often challenging to pinpoint the precise location of rate shifts in the tree, particularly when such shifts involve clades of species with short internode intervals at their base. In such cases it becomes difficult to assign the location of a shift to a single node and the inference of a rate shift is then often distributed across two or more nested nodes in the phylogeny. To account for this, we also summarised our results using a second approach in which the posterior probability for a particular rate shift was calculated as the sum of the probability of a shift having occurred on a focal node or on either of the nodes immediately descending from it. We focus on the multivariate analyses because bill shape is a high dimensional trait. In the main text (Fig. 2, 3) we report results from the stage 2 Hackett tree but found comparable results regardless of tree choice (Extended Data Fig. 3, 4).
We checked for biases in rate estimates across the phylogeny by comparing our observed multivariate rate estimates of bill shape evolution to results generated using simulated data. Using the stage 2 Hackett MCC tree, we generated 10 null multivariate data sets (simulated under BM) and estimated rates using runs of 200 million iterations and 1,000 post-burn samples. We found that on average branch-specific rates derived from simulated data sets were uncorrelated with observed rates of bill shape evolution (Spearman’s rho = 0.03; p = 0.34), indicating that our results are unlikely to be affected by underlying biases in rate estimation.
In addition to BayesTraits we compared the fit of three single process models (Brownian motion [BM], early burst [EB] and Ornstein-Uhlenbeck [OU]), fit using the ‘fitContinuous’ function and default settings in the R package Geiger v2.043, as well as alternative formulations of the BAMM model44 that differed in their handling of temporal rate variation (time constant [T constant], time variable [T var] and time flip [T flip]). The BayesTraits, BAMM and single process models are not fitted in common a framework with consistent likelihood calculations. We therefore compared the fit of the alternative models within each shape axis by calculating the likelihood of a BM model fit to the mean rate-transformed Jetz et al. trees derived from each model. In the absence of support for alternative models (Extended Data Table 3), and because BAMM does not currently allow analyses of multivariate data, we focus our interpretation on analyses using BayesTraits.
Disparity and rates through time
Estimating ancestral disparity
We estimated ancestral values for each component axis of bill shape variation using a maximum likelihood approach implemented in the R package phytools38. We estimated ancestral states using the mean rate-transformed trees for each component axis to account for unequal rates of evolution across the tree and among shape axes. To generate estimates of ancestral disparity through time, we took time slices at 1 million year intervals starting at the root of the tree. For each time slice we extracted ancestral state estimates for each component axis for the lineages in the phylogeny existing at that particular time point. We then quantified multivariate disparity in trait values by calculating the sum of the variances across all 8 trait axes21. Unlike other disparity metrics, the sum of the variances is expected to be independent of richness and sensitive to changes in both expansion and packing of trait space, thus providing an indication of the relative strength of these two patterns19.
Null models of morphospace filling
We generated two alternative null models of morphospace filling based on BM models of trait evolution to assess whether the observed patterns of bill shape disparity through time were distinct from unbiased patterns of disparity accumulation. In the first we assumed that trait variation accumulates at a constant rate (‘CR’) that is homogeneous with respect to time and also to a lineage’s position in the phylogeny. In the second we relaxed these assumptions of rate constancy and instead simulated traits using the mean rate-transformed trees for each axis, thereby providing a null model of disparity accumulation incorporating variable rates (‘VR’) of trait evolution. For each model we simulated 500 replicate data sets and used these to calculate two sets of null disparity through time curves using identical approaches to those describe above. Irrespective of whether evolutionary rates are fixed to be constant or allowed to vary, an important feature of both null models is that the underlying balance between morphospace expansion and packing is expected to be effectively equal and constant over time. This is due to the inherently non-directional nature of trait change simulated using the BM model. Consequently, any deviation in the observed rate of disparity accumulation compared to the null rates suggests that one process (either expansion or packing) has dominated over the other.
Summarising evolutionary rates through time
For each 1 million year time slice, we calculated the mean rate of evolution across all branches present at that time point. We repeated this procedure for each tree in the posterior distribution to generate a distribution of average rate estimates in 1 million year intervals.
Estimation of phenotypic variance-covariance (P) matrices
We examined the consistency of bill shape evolution within and among avian clades using Bayesian estimates of phenotypic variance-covariance matrices (P matrices) of bill shape within higher taxa (families, superfamilies and orders)26,27. First, we estimated the number of independent axes (i.e. eigenvectors of P) that are required to adequately explain the total trait variance in P in each higher taxon. We then tested whether the dominant eigenvector of bill shape variation (Pmax) is consistent among clades. Pmax is the first principal component of P and an estimate of the major axis of phenotypic variation. We estimated phenotypic variance-covariance matrices for higher taxa containing ≥20 sampled species. Posterior distributions of variance-covariance matrices were generated using Bayesian MCMC MANOVA models implemented in the R package MCMCglmm27. We used weak uniform priors and ran each model for 80,000 iterations with a burn-in of 40,000 and sampling that produced 1,000 estimates of the posterior distribution. Based on these distributions we used a set of Bayesian matrix quantification approaches26 to extract information on (i) centroid position, (ii) subspace orientation, (iii) individual trait loadings onto and variance explained by Pmax, and (iv) number of significant eigenvectors associated with each P.
Data availability
Scan and landmark data that support the findings of this study have been deposited in the NHM Data Portal with the identifier doi:10.5519/0005413. All other data analysed during this study are included as source and supplementary information files.
Extended Data
Extended Data Figure 1. Positions of landmarks and semilandmarks.
The image shows a 3D scan of a shoebill (Balaeniceps rex) bill marked up with four fixed landmarks (numbered red points) and three semi-landmark curves along the dorsal profile (from points 1 to 2) and tomial edges (left from point 1 to 3 and right from point 1 to 4). Each curve consists of 25 semi-landmarks (black points).
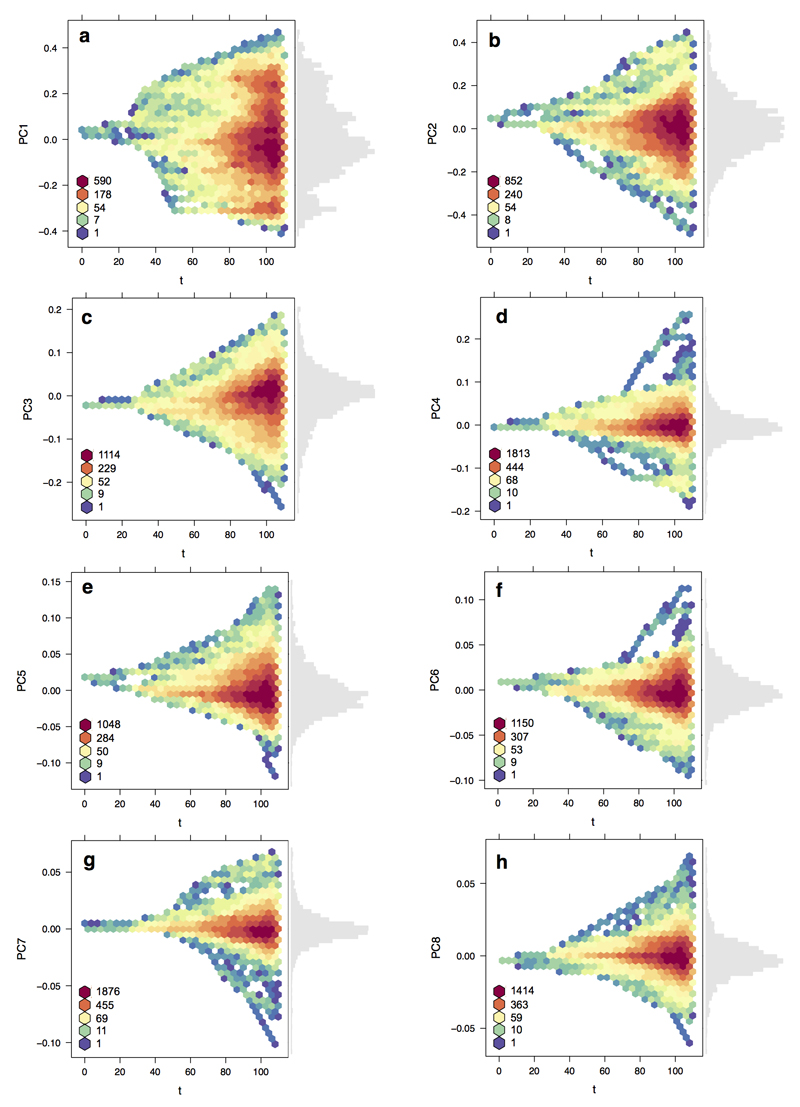
Extended Data Figure 2. Morphospace density through time.
Plots show the filling of avian bill morphospace through time (n = 2028 species) for PCs a,1; b, 2; c, 3; d, 4; e, 5; f, 6; g, 7; and h, 8. Densities were calculated in 1 million year time slices based on univariate rate heterogeneous models of trait evolution using a stage 2 Hackett MCC tree from www.birdtree.org. The scale runs from low density (blue) to high density (red), indicating the extent of niche packing through time in different regions of bill morphospace. For each axis the frequency distribution of PC scores among species is also shown (grey bars).
Extended Data Figure 3. Comparison of multivariate rates of bill shape evolution and disparity through time for alternative datasets.
The plot shows estimates of the mean relative multivariate rate of bill shape evolution for four alternative versions of the avian phylogeny and also when using phylogenetic Principal Components (pPCs) (see Methods). Shown below are plots comparing estimates of disparity and rates through time derived from each dataset. For stage 2 trees n = 2028 species and for stage 1 trees n = 1627 species.
Extended Data Figure 4. Multivariate rates of bill shape evolution for a composite tree based on the Prum et al. backbone.
The avian phylogeny coloured according to estimates of the mean relative multivariate rate of bill shape evolution. Grey triangles show the stem branch of clades with support for whole clade shifts in evolutionary rate. Coloured circles show rate shifts on individual internal branches (colour indicates the rate estimate). The relative size of triangles and circles indicates the posterior probability (PP) of a rate shift. Filled and open triangles distinguish between shifts on the focal node (filled) and shifts that occur either at the focal node or on one of the two immediate daughter nodes (open).
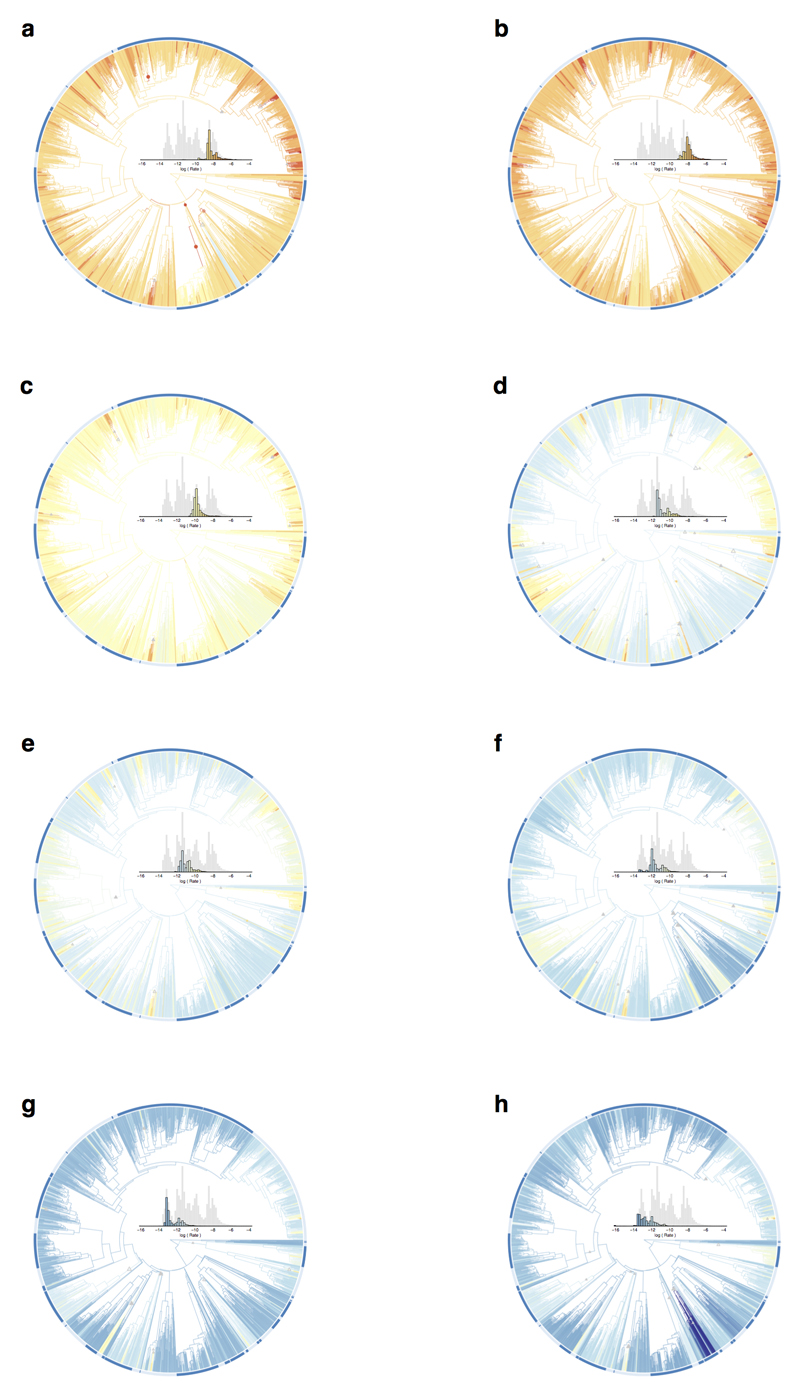
Extended Data Figure 5. Phylogenetic mapping of univariate rates of bill shape evolution.
The plots shows the avian phylogeny of all taxa included in the study (n = 2028 species) with branches coloured on a common scale across panels according to estimates of the univariate rate of bill shape evolution. a, PC1, b, PC2, c, PC3, d, PC4, e, PC5, f, PC6, g, PC7, h, PC8.
Extended Data Figure 6. Morphospaces of avian higher taxa.
Pairwise scatterplots of PCs 1 and 2, 3 and 4, 5 and 6, and 7 and 8 showing focal higher taxa (non-passerines, purple; passerines, green) against total avian morphospace (grey). Values in parentheses show the number of species sampled.
Extended Data Figure 7. Morphological subspaces of the P of avian higher taxa.
The figure shows representations of P for avian higher taxa with ≥20 species sampled. First column: distribution of species values on each of the first eight raw PCs showing variation in morphospace centroid for each higher taxon. Second column: two-dimensional subspace for each taxon with non-passerine (purple) and passerine (green) subspaces. The x- and y-axes follow the global leading (Pmax) and secondary eigenvectors. Third column: percentage of total variance explained and individual PC loadings onto each taxon specific Pmax. Inset: three-dimensional subspace for all non-passerines (purple) and passerines (green). Values in parentheses show the number of species sampled.
Extended Data Table 1. Variance, repeatability and phylogenetic signal of PC axes.
The table shows individual and cumulative variance values, kurtosis values, scores of among user repeatability (R) and repeatability after averaging (Rn), and maximum likelihood estimates and 95% confidence intervals of Pagel’s λ for the first eight PC’s of bill shape. λ was estimated using two different tree topologies based on the Hackett and Ericson backbone trees taken from www.birdtree.org.
| PC axis | Variance (%) | Cumulative (%) | Kurtosis | R | Rn | Stage 2 Hackett λ | Stage 2 Ericson λ |
|---|---|---|---|---|---|---|---|
| 1 | 57.8 | 57.8 | −0.487 | 0.998 | 1.000 | 0.949 (0.931-0.964) | 0.954 (0.936-0.968) |
| 2 | 29.0 | 86.8 | 0.795 | 0.913 | 0.976 | 0.758 (0.704-0.806) | 0.760 (0.706-0.808) |
| 3 | 6.2 | 93.1 | 1.381 | 0.967 | 0.991 | 0.851 (0.813-0.882) | 0.861 (0.824-0.892) |
| 4 | 2.8 | 95.9 | 7.370 | 0.987 | 0.997 | 0.878 (0.845-0.906) | 0.873 (0.838-0.903) |
| 5 | 1.8 | 97.7 | 1.867 | 0.977 | 0.994 | 0.897 (0.863-0.924) | 0.888 (0.851-0.917) |
| 6 | 0.9 | 98.6 | 2.122 | 0.945 | 0.985 | 0.822 (0.774-0.863) | 0.816 (0.766-0.858) |
| 7 | 0.4 | 99.0 | 6.426 | 0.953 | 0.987 | 0.803 (0.756-0.843) | 0.803 (0.756-0.843) |
| 8 | 0.3 | 99.2 | 3.452 | 0.938 | 0.983 | 0.805 (0.752-0.848) | 0.794 (0.739-0.840) |
Extended Data Table 2. Summary of major single-lineage bill evolutionary rate shifts.
Table shows fold-change rate of evolution and posterior probability (PP) for major (PP > 0.7 and fold-increase > 10) ancestral single-lineage shifts in rate of bill shape evolution.
| Order | Family | Genera | N | Fold-increase | PP |
|---|---|---|---|---|---|
| PHOENICOPTERIFORMES | Phoenicopteridae | Phoeniconaias, Phoenicoparrus, Phoenicopterus | 3 | 45.2 | 1.000 |
| APODIFORMES | Trochilidae | Discosura, Lophornis, Sephanoides | 3 | 38.5 | 0.999 |
| PELECANIFORMES | Threskiornithidae | Bostrychia, Cercibis, Eudocimus, Geronticus, Lophotibis, Mesembrinibis, Nipponia, Phimosus, Platalea, Plegadis, Pseudibis, Thaumatibis, Theristicus, Threskiornis | 14 | 29.6 | 0.989 |
| PASSERIFORMES | Dendrocolaptidae | Campylorhamphus, Drymornis, Lepidocolaptes | 3 | 23.5 | 0.994 |
| PASSERIFORMES | Paradisaeidae | Parotia, Pteridophora | 2 | 22.2 | 0.992 |
| PASSERIFORMES | Melanocharitidae | Oedistoma, Toxorhamphus | 2 | 21.4 | 0.914 |
| PASSERIFORMES | Platysteiridae | Batis, Platysteira | 2 | 20.1 | 0.990 |
| PICIFORMES | Ramphastidae | Andigena, Aulacorhynchus, Pteroglossus, Ramphastos, Selenidera | 5 | 18.9 | 0.988 |
| ANSERIFORMES | Anatidae | Lophodytes, Mergellus, Mergus | 3 | 18.4 | 0.974 |
| ACCIPITRIFORMES | Accipitridae | Helicolestes, Rostrhamus | 2 | 18.0 | 0.980 |
| PASSERIFORMES | Hirundinidae | Alopochelidon, Atticora, Cheramoeca, Delichon, Eurochelidon, Haplochelidon, Hirundo, Neochelidon, Notiochelidon, Petrochelidon, Phedina, Progne, Psalidoprocne, Pseudhirundo, Pseudochelidon, Pygochelidon, Riparia, Stelgidopteryx, Tachycineta | 19 | 14.8 | 0.783 |
| PASSERIFORMES | Fringillidae | Loxioides, Telespiza | 2 | 13.0 | 0.842 |
| MUSOPHAGIFORMES | Musophagidae | Corythaeola, Corythaixoides, Crinifer, Musophaga, Ruwenzorornis, Tauraco | 6 | 11.5 | 0.838 |
| PASSERIFORMES | Timaliidae | Jabouilleia, Rimator | 2 | 11.5 | 11.1 |
Extended Data Table 3. Comparison of trait models.
The table shows delta likelihood values for alternative models of trait evolution within each shape axis and for different tree topologies. Values were generated by calculating the likelihoods of a BM model fit to the mean rate-transformed trees derived from each model.
| Tree | PC axis | BayesTraits | BAMM (T var) | BAMM (T flip) | BAMM (T constant) | OU | EB | BM |
|---|---|---|---|---|---|---|---|---|
| Stage 2 Hackett | 1 | 0 | 45.0 | 171.2 | 284.5 | 635.4 | 630.8 | 635.4 |
| 2 | 0 | 85.3 | 171.0 | 280.3 | 591.4 | 496.7 | 591.4 | |
| 3 | 0 | 48.6 | 177.1 | 319.7 | 595.3 | 534.0 | 595.3 | |
| 4 | 0 | 46.0 | 156.2 | 292.2 | 876.3 | 830.0 | 876.3 | |
| 5 | 0 | 65.1 | 169.2 | 294.5 | 598.9 | 557.4 | 598.9 | |
| 6 | 0 | 41.6 | 121.8 | 276.0 | 703.6 | 631.8 | 703.6 | |
| 7 | 0 | 65.1 | 170.2 | 289.3 | 805.3 | 718.8 | 805.3 | |
| 8 | 0 | 56.4 | 134.3 | 281.2 | 826.8 | 725.1 | 826.8 | |
| Stage 2 Ericson | 1 | 0 | 71.3 | 166.5 | 302.2 | 623.6 | 618.8 | 623.6 |
| 2 | 0 | 82.8 | 172.5 | 286.4 | 575.1 | 483.4 | 575.1 | |
| 3 | 0 | 51.2 | 164.3 | 338.7 | 583.6 | 529.3 | 583.6 | |
| 4 | 0 | 65.5 | 157.0 | 283.7 | 875.0 | 824.7 | 875.0 | |
| 5 | 0 | 59.1 | 172.6 | 310.9 | 625.8 | 577.1 | 625.8 | |
| 6 | 0 | 50.2 | 128.5 | 261.3 | 710.8 | 636.7 | 710.8 | |
| 7 | 0 | 58.6 | 159.2 | 297.1 | 805.7 | 720.7 | 805.7 | |
| 8 | 0 | 69.9 | 154.1 | 333.7 | 831.3 | 728.2 | 831.3 | |
| Stage 1 Hackett | 1 | 0 | 56.8 | 134.7 | 227.2 | 479.5 | 473.6 | 479.5 |
| 2 | 0 | 59.8 | 149.8 | 243.1 | 483.8 | 398.0 | 483.8 | |
| 3 | 0 | 26.4 | 135.5 | 271.1 | 493.5 | 439.2 | 493.5 | |
| 4 | 0 | 40.5 | 128.4 | 237.4 | 714.7 | 675.2 | 714.7 | |
| 5 | 0 | 52.0 | 136.7 | 278.4 | 478.6 | 439.8 | 478.6 | |
| 6 | 0 | 22.6 | 95.7 | 219.2 | 579.5 | 517.6 | 579.5 | |
| 7 | 0 | 26.3 | 135.1 | 238.4 | 670.7 | 586.1 | 670.7 | |
| 8 | 0 | 29.1 | 103.4 | 232.2 | 675.3 | 570.4 | 675.2 | |
| Stage 1 Ericson | 1 | 0 | 69.7 | 132.5 | 248.7 | 486.4 | 479.6 | 486.4 |
| 2 | 0 | 59.4 | 143.3 | 239.7 | 488.2 | 400.3 | 488.2 | |
| 3 | 0 | 21.8 | 136.4 | 275.2 | 502.7 | 447.2 | 502.7 | |
| 4 | 0 | 32.5 | 132.1 | 245.3 | 721.8 | 679.5 | 721.8 | |
| 5 | 0 | 53.8 | 130.3 | 275.0 | 482.9 | 442.3 | 482.9 | |
| 6 | 0 | 23.9 | 90.3 | 233.9 | 583.7 | 519.5 | 583.7 | |
| 7 | 0 | 34.9 | 132.3 | 243.6 | 669.7 | 585.1 | 669.7 | |
| 8 | 0 | 29.5 | 101.1 | 244.4 | 676.4 | 569.8 | 676.4 |
Supplementary Material
Supplementary Information
Raw scan data in obj format and text files containing individual and species-averaged landmarks are available from the Natural History Museum Data Portal here: http://dx.doi.org/10.5519/0005413.
AvianPhylogenies.zip: this archive contains all alternative genus level phylogenies used in our analyses.
Excel file (PC_scores_all_genera.csv): this file contains scores for the first eight non-phylogenetic PC’s for all species (n = 2028) in our data.
Excel file (PrumMerge_CRC.xlsx): this file details the mapping of Jetz et al. clades to the Prum et al. backbone phylogeny. The table shows the nodes used to attach patch clades from the Jetz et al. stage 2 Hackett tree to the Prum et al. backbone phylogeny.
PrumMerge.zip: this archive contains data files and an R script to combine the backbone (approximately family level) phylogeny of Prum et al. with the species level resolution of the Jetz et al avian phylogeny.
Acknowledgements
We thank Mark Adams, Hein van Grouw, and Robert Prys-Jones from the Bird Group at the NHM, Tring and Henry McGhie at the Manchester Museum for providing access to and expertise in the collections; Shai Meiri of Tel Aviv University for providing a sample of study skins; Simon Stone of MechInovation Ltd. for providing training and advice on 3D scanning; Matthew Groves, Jamie McLaughlin, Mike Pidd of HRI Digital for the construction of www.markmybird.org; Andrew Beckerman for advice on analysing P matrices; Emily Rayfield, Alex Pigot, Arne Mooers, and Alex White for providing valuable comments on pre-submission drafts of the manuscript. Finally, we are indebted to the wonderful volunteer citizen scientists at www.markmybird.org for generously giving up their time to help build the database of bird bill shape and contribute to our understanding of avian evolution. This work was funded by the European Research Council (grant number 615709 Project ‘ToLERates’) and by a Royal Society University Research Fellowship to GHT (UF120016).
Footnotes
Contributions
Christopher R. Cooney, Jen A. Bright, and Gavin H. Thomas conceived of the study, designed analytical protocols, analysed the data and wrote the manuscript. All authors collected and processed data and provided editorial input into the manuscript.
Competing financial interests. None.
References
- 1.Bright JA, Marugan-Lobon J, Cobb SN, Rayfield EJ. The shapes of bird beaks are highly controlled by nondietary factors. Proc Natl Acad Sci U S A. 2016;113:5352–5357. doi: 10.1073/pnas.1602683113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamichhaney S, et al. Evolution of Darwin's finches and their beaks revealed by genome sequencing. Nature. 2015;518:371–375. doi: 10.1038/nature14181. [DOI] [PubMed] [Google Scholar]
- 3.Phillimore AB, Price TD. Density-dependent cladogenesis in birds. PLoS Biol. 2008;6:e71. doi: 10.1371/journal.pbio.0060071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simpson GG. Tempo and mode in evolution. Columbia University Press; 1944. [Google Scholar]
- 5.Ezard TH, Purvis A. Environmental changes define ecological limits to species richness and reveal the mode of macroevolutionary competition. Ecol Lett. 2016 doi: 10.1111/ele.12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Price T. Speciation in birds. 1st edn. Roberts and Co; 2008. [Google Scholar]
- 7.Price TD, et al. Niche filling slows the diversification of Himalayan songbirds. Nature. 2014;509:222–225. doi: 10.1038/nature13272. [DOI] [PubMed] [Google Scholar]
- 8.Losos JB, Mahler DL. In: Evolution Since Darwin: The First 150 Years. Bell MA, Futuyma DJ, Eanes WF, Levinton JS, editors. Sinauer Associates; 2010. pp. 381–420. [Google Scholar]
- 9.Reznick DN, Ricklefs RE. Darwin's bridge between microevolution and macroevolution. Nature. 2009;457:837–842. doi: 10.1038/nature07894. [DOI] [PubMed] [Google Scholar]
- 10.Alfaro ME, et al. Nine exceptional radiations plus high turnover explain species diversity in jawed vertebrates. Proc Natl Acad Sci U S A. 2009;106:13410–13414. doi: 10.1073/pnas.0811087106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jetz W, Thomas GH, Joy JB, Hartmann K, Mooers AO. The global diversity of birds in space and time. Nature. 2012;491:444–448. doi: 10.1038/nature11631. [DOI] [PubMed] [Google Scholar]
- 12.Hedges SB, Marin J, Suleski M, Paymer M, Kumar S. Tree of life reveals clock-like speciation and diversification. Mol Biol Evol. 2015;32:835–845. doi: 10.1093/molbev/msv037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Venditti C, Meade A, Pagel M. Multiple routes to mammalian diversity. Nature. 2011;479:393–396. doi: 10.1038/nature10516. [DOI] [PubMed] [Google Scholar]
- 14.Etienne RS, et al. Diversity-dependence brings molecular phylogenies closer to agreement with the fossil record. Proc Biol Sci. 2012;279:1300–1309. doi: 10.1098/rspb.2011.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rabosky DL, Glor RE. Equilibrium speciation dynamics in a model adaptive radiation of island lizards. P Natl Acad Sci USA. 2010;107:22178–22183. doi: 10.1073/pnas.1007606107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jonsson KA, et al. Ecological and evolutionary determinants for the adaptive radiation of the Madagascan vangas. Proc Natl Acad Sci U S A. 2012;109:6620–6625. doi: 10.1073/pnas.1115835109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rabosky DL, et al. Rates of speciation and morphological evolution are correlated across the largest vertebrate radiation. Nat Commun. 2013;4:1958. doi: 10.1038/ncomms2958. [DOI] [PubMed] [Google Scholar]
- 18.Grant PR. Ecology and evolution of Darwin's finches. 2nd edn. Princeton University Press; 1999. [Google Scholar]
- 19.Pigot AL, Trisos CH, Tobias JA. Functional traits reveal the expansion and packing of ecological niche space underlying an elevational diversity gradient in passerine birds. Proc Biol Sci. 2016;283 doi: 10.1098/rspb.2015.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lovette IJ, Bermingham E, Ricklefs RE. Clade-specific morphological diversification and adaptive radiation in Hawaiian songbirds. Proc Biol Sci. 2002;269:37–42. doi: 10.1098/rspb.2001.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kraft NJB, Valencia R, Ackerly DD. Functional traits and niche-based tree community assembly in an amazonian forest. Science. 2008;322:580–582. doi: 10.1126/science.1160662. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell JS. Extant-only comparative methods fail to recover the disparity preserved in the bird fossil record. Evolution. 2015;69:2414–2424. doi: 10.1111/evo.12738. [DOI] [PubMed] [Google Scholar]
- 23.Finarelli JA, Goswami A. Potential Pitfalls of Reconstructing Deep Time Evolutionary History with Only Extant Data, a Case Study Using the Canidae (Mammalia, Carnivora) Evolution. 2013;67:3678–3685. doi: 10.1111/evo.12222. [DOI] [PubMed] [Google Scholar]
- 24.Slater GJ. Iterative adaptive radiations of fossil canids show no evidence for diversity-dependent trait evolution. P Natl Acad Sci USA. 2015;112:4897–4902. doi: 10.1073/pnas.1403666111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ricklefs RE. Small clades at the periphery of passerine morphological space. Am Nat. 2005;165:651–659. doi: 10.1086/429676. [DOI] [PubMed] [Google Scholar]
- 26.Robinson MR, Beckerman AP. Quantifying multivariate plasticity: genetic variation in resource acquisition drives plasticity in resource allocation to components of life history. Ecol Lett. 2013;16:281–290. doi: 10.1111/ele.12047. [DOI] [PubMed] [Google Scholar]
- 27.Hadfield JD. MCMC Methods for Multi-Response Generalized Linear Mixed Models: The MCMCglmm R Package. J Stat Softw. 2010;33:1–22. [Google Scholar]
- 28.Hughes M, Gerber S, Wills MA. Clades reach highest morphological disparity early in their evolution. Proc Natl Acad Sci U S A. 2013;110:13875–13879. doi: 10.1073/pnas.1302642110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchell JS, Makovicky PJ. Low ecological disparity in Early Cretaceous birds. P Roy Soc B-Biol Sci. 2014;281 doi: 10.1098/rspb.2014.0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brusatte SL, O'Connor JK, Jarvis ED. The Origin and Diversification of Birds. Curr Biol. 2015;25:R888–898. doi: 10.1016/j.cub.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 31.Klingenberg CP. Visualizations in geometric morphometrics: how to read and how to make graphs showing shape changes. Hystrix. 2013;24:15–24. [Google Scholar]
- 32.Zelditch M. Geometric morphometrics for biologists : a primer. Elsevier Academic Press; 2004. [Google Scholar]
- 33.Fruciano C. Measurement error in geometric morphometrics. Dev Genes Evol. 2016;226:139–158. doi: 10.1007/s00427-016-0537-4. [DOI] [PubMed] [Google Scholar]
- 34.Adams DC, Otarola-Castillo E. geomorph: an r package for the collection and analysis of geometric morphometric shape data. Methods Ecol Evol. 2013;4:393–399. [Google Scholar]
- 35.Gunz P, Mitteroecker P, Bookstein FL. In: Modern Moprhometrics in Physical Anthropology. Slide DE, editor. Kluwer Academic/Plenum Publishers; 2004. pp. 73–98. [Google Scholar]
- 36.tpsSmall, testing amount of shape variation, version 1.30. Department of Ecology and Evolution, State University of New York at Stony Brook; 2014. [Google Scholar]
- 37.Revell LJ. Size-Correction and Principal Components for Interspecific Comparative Studies. Evolution. 2009;63:3258–3268. doi: 10.1111/j.1558-5646.2009.00804.x. [DOI] [PubMed] [Google Scholar]
- 38.Revell LJ. phytools: an R package for phylogenetic comparative biology (and other things) Methods Ecol Evol. 2012;3:217–223. [Google Scholar]
- 39.Uyeda JC, Caetano DS, Pennell MW. Comparative Analysis of Principal Components Can be Misleading. Systematic Biology. 2015;64:677–689. doi: 10.1093/sysbio/syv019. [DOI] [PubMed] [Google Scholar]
- 40.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prum RO, et al. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature. 2015;526:569–573. doi: 10.1038/nature15697. [DOI] [PubMed] [Google Scholar]
- 42.Thomas GH, Freckleton RP. MOTMOT: models of trait macroevolution on trees. Methods Ecol Evol. 2012;3:145–151. doi: 10.1111/j.2041-210X.2011.00132.x. [DOI] [Google Scholar]
- 43.Pennell MW, et al. geiger v2.0: an expanded suite of methods for fitting macroevolutionary models to phylogenetic trees. Bioinformatics. 2014;30:2216–2218. doi: 10.1093/bioinformatics/btu181. [DOI] [PubMed] [Google Scholar]
- 44.Rabosky DL. Automatic Detection of Key Innovations, Rate Shifts, and Diversity-Dependence on Phylogenetic Trees. Plos One. 2014;9 doi: 10.1371/journal.pone.0089543. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
AvianPhylogenies.zip: this archive contains all alternative genus level phylogenies used in our analyses.
Excel file (PC_scores_all_genera.csv): this file contains scores for the first eight non-phylogenetic PC’s for all species (n = 2028) in our data.
Excel file (PrumMerge_CRC.xlsx): this file details the mapping of Jetz et al. clades to the Prum et al. backbone phylogeny. The table shows the nodes used to attach patch clades from the Jetz et al. stage 2 Hackett tree to the Prum et al. backbone phylogeny.
PrumMerge.zip: this archive contains data files and an R script to combine the backbone (approximately family level) phylogeny of Prum et al. with the species level resolution of the Jetz et al avian phylogeny.
Data Availability Statement
Scan and landmark data that support the findings of this study have been deposited in the NHM Data Portal with the identifier doi:10.5519/0005413. All other data analysed during this study are included as source and supplementary information files.