Abstract
Background
Increasing evidence suggests that the low-affinity receptor for IgE, CD23, plays an important role in controlling the activity of allergen-specific T cells through IgE-facilitated allergen presentation.
Objective
We sought to determine the number of CD23 molecules on immune cells in allergic patients and to investigate whether the number of CD23 molecules on antigen-presenting cells is associated with IgE levels and influences allergen uptake and allergen-specific T-cell activation.
Methods
Numbers of CD23 molecules on immune cells of allergic patients were quantified by using flow cytometry with QuantiBRITE beads and compared with total and allergen-specific IgE levels, as well as with allergen-induced immediate skin reactivity. Allergen uptake and allergen-specific T-cell activation in relation to CD23 surface density were determined by using flow cytometry in combination with confocal microscopy and T cells transfected with the T-cell receptor specific for the birch pollen allergen Bet v 1, respectively. Defined IgE-allergen immune complexes were formed with human monoclonal allergen-specific IgE and Bet v 1.
Results
In allergic patients the vast majority of CD23 molecules were expressed on naive IgD+ B cells. The density of CD23 molecules on B cells but not the number of CD23+ cells correlated with total IgE levels (RS = 0.53, P = .03) and allergen-induced skin reactions (RS = 0.63, P = .008). Uptake of allergen-IgE complexes into B cells and activation of allergen-specific T cells depended on IgE binding to CD23 and were associated with CD23 surface density. Addition of monoclonal IgE to cultured PBMCs significantly (P = .04) increased CD23 expression on B cells.
Conclusion
CD23 surface density on B cells of allergic patients is correlated with allergen-specific IgE levels and determines allergen uptake and subsequent activation of T cells.
Keywords: CD23, allergy, IgE, low-affinity IgE receptor, B cell, allergen
IgE is known to have 2 different receptors, the high-affinity receptor FcεRI and the low-affinity receptor CD23 (FcεRII). Cross-linking of FcεRI-bound IgE by allergens mediates degranulation of mast cells and basophils and thus leads to immediate symptoms of allergic disease.1 In addition, FcεRI is expressed on eosinophils2 and antigen-presenting cells (APCs; eg, monocytes and dendritic cells) and was shown to be involved in IgE-facilitated allergen presentation to T cells.3,4 Interestingly, expression of FcεRI on mast cells, basophils, and even APCs is upregulated by increasing IgE levels,5,6 and it was found that omalizumab, an anti-IgE antibody, prevents IgE binding to FcεRI and thereby also downregulates FcεRI expression.7
Expression of the low-affinity receptor for IgE (CD23), a 45-kDa calcium-binding protein belonging to the family of C-type lectins on various cell types, has been investigated mainly by using cells cultured under various conditions. These studies have shown that CD23 is expressed on B cells,8 monocytes,9 T cells,10 dendritic cells,11 platelets,12 and neutrophils.13 However, expression of the numbers of CD23 molecules on these cell types has not been studied in detail by using ex vivo isolated cells from allergic patients. CD23 has an important function in IgE-facilitated allergen presentation to T cells.14,15 In fact, IgE-facilitated antigen presentation strongly activates allergen-specific T cells and secretion of proinflammatory and TH2-driving cytokines.14–17 It has been shown that facilitated antigen presentation can be inhibited with a therapeutic anti-CD23 antibody18 and by allergen-specific IgG antibodies induced by allergen-specific immunotherapy.19 An association between improvement of symptoms after specific immunotherapy with a reduction of allergen-IgE binding to CD23 (facilitated antigen binding) on B cells by enhanced levels of blocking IgG antibodies has been demonstrated by using facilitated antigen-binding assays.20,21
Despite the importance of CD23 in activating allergen-specific T cells, several aspects of its biology have not been investigated as meticulously as for FcεRI. For example, there are no studies that have investigated the density of the expression of CD23 molecules on ex vivo isolated cells from allergic patients. Studies investigating CD23 mainly focused on the relative number and percentage of cells expressing CD23.22–29 Therefore it has also not been studied whether the number of CD23 molecules on the cells is associated with total and allergen-specific IgE levels. Furthermore, there are no systematic studies in defined experimental human model systems that have analyzed whether and how the number of CD23 molecules on APCs has an effect on the magnitude of IgE-facilitated allergen presentation and subsequent T-cell activation.
In the present study we established a new technique for measurement of CD23 receptor molecule numbers on the surfaces of immune cells. We investigated the distribution frequency of CD23 on immune cells in allergic patients and whether and how this parameter is correlated with IgE levels. We also studied whether addition of IgE to PBMC cultures has effects on CD23 expression on B cells. Furthermore, we used CD23 cell lines expressing different numbers of CD23 molecules on their surfaces to study whether and how the density of CD23 molecules on APCs influences IgE-facilitated allergen uptake and allergen-specific T-cell activation.
Methods
Patients
Blood samples from 17 study participants with a positive history suggestive of grass pollen allergy and a positive skin prick test reaction with grass pollen extract were analyzed. Apart from their allergy, none of the subjects had a history of a chronic or current acute disease. Subjects were included in the study during the grass pollen season (ie, during the months of June/July in Vienna). The presence of symptoms of grass pollen allergy (rhinitis, conjunctivitis, and asthma) was recorded at that time. Furthermore, a history of other allergies was obtained. No patients were analyzed who had a contraindication against skin prick testing or were receiving long-term treatment with systemic corticosteroids, immunosuppressive drugs, tranquilizers, or psychoactive drugs. Before the study, patients were not allowed to use oral antihistamines for 3 days and local (in the skin test area) and systemic corticosteroids for 14 days. Blood samples were analyzed in an anonymized manner with approval of the Ethics Committee of the Medical University of Vienna (EK508/2011) after written informed consent was obtained from the patients.
Skin prick tests
Skin test solutions (positive control, timothy grass pollen extract; negative control solution, codeine phosphate; Stallergenes, Antony, France) were applied to the lower arms of patients and were pricked with commercial prick lancets (Allergopharma, Reinbek, Germany). After 20 minutes, the wheal reaction was surrounded with a felt pen and transferred to paper by using adhesive tape. The size of the wheal reactions was measured by using planimetry, as previously described.30
Blood samples and total and allergen-specific IgE measurements
Immediately after venipuncture from the antecubital vein, cells from heparinized blood samples were assessed for CD23 expression. Serum was obtained from clotted blood samples by means of centrifugation and stored at −20°C until use. Total IgE and timothy grass pollen–specific IgE levels were measured with the Phadia CAP system (Thermo Fisher, Uppsala, Sweden).
Data analysis
All clinical data (patient history and skin prick test results) were obtained by a clinical investigator and deposited in a database. Measurement of total and specific IgE levels was performed by an independent external laboratory, which was unaware of clinical data and CD23 measurements. Measurement of CD23 levels on different cell types was performed by another independent investigator who was not in contact with the study participants and who was blinded regarding total and specific IgE levels and clinical data (ie, results from skin prick tests and clinical symptoms). After all 3 independent data sets (ie, IgE levels, clinical data, and CD23 measurements) were completed, they were submitted to a database, and correlations were analyzed.
Blood sample preparation and flow cytometry
Red blood cell lysis solution (155 mmol/L ammonium chloride, 10 mmol/L potassium bicarbonate, and 12 mmol/L EDTA) was applied to heparinized blood samples from patients. For flow cytometry, the following surface markers of cells were stained: T cells (positive with anti-CD3, clone OKT3), natural killer (NK) cells (negative with anti-CD3 and positive with anti-CD56, clone TULY56), B cells (positive with anti-CD19, clone HIB19), monocytes (positive with anti-CD14, clone 61D3), platelets (positive with anti-CD61, clone VI-PL2, and with anti-CD41a, clone HIP8), basophils (positive with anti-CD123, clone 6H6, and anti-CCR3, clone 5E8-G9-B4), neutrophils (granulocytes negative with anti-CD49d, clone HP2/1), eosinophils (granulocytes positive with anti-CD49d, clone HP2/1, and negative with anti-CD19), dendritic cells (lineage cocktail negative, positive with anti-CD11c, clone 3.9), naive B cells (positive with anti-CD19, clone J3-119, and anti-IgD, clone 11-26), memory B cells (positive with anti-CD19, clone J3-119, and positive with anti-CD27, clone O323). All cells were additionally stained with anti-CD23 (clone EBVCS2). Matching nonbinding isotype antibodies were used as controls. All antibodies were obtained from eBioscience (San Diego, Calif), except for anti-CD49d, anti-CD19 for naïve/memory staining (Beckman Coulter, Brea, Calif), and lineage cocktail lin1 (BD Biosciences, Franklin Lakes, NJ). Aliquots of 1.5 × 106 cells were used for each staining. Before staining, cells were blocked with 10% vol/vol mouse serum (Life Technologies, Carlsbad, Calif). Dead cells were excluded from the analysis with eFluor 780 Fixable Viability Dye (eBioscience). Flow cytometric analysis was performed on a Beckman Coulter FC 500 flow cytometer (Beckman Coulter). Depending on the cell type, 3 × 105 (T cells, B cells, NK cells, and monocytes), 1 × 106 (basophils), or 5 × 105 (all other cell types; ie, neutrophils, eosinophils, dendritic cells, and platelets) events were recorded. FlowJo Software 7.5 (TreeStar, Ashland, Ore) was used for data analysis. Gates were set according to the matching nonbinding isotype control of each antibody for each cell type.
Measurement and calculation of CD23 surface density
Quantification of CD23 expression was performed with BD QuantiBRITE PE beads (BD Biosciences), according to the manufacturer’s instructions. Briefly, beads with different intensity levels in the phycoerythrin (PE) channel FL-2 and defined numbers of surface PE molecules were used as a standard in flow cytometry and used for back-calculation of CD23 stained with a PE-labeled anti-CD23 antibody. FlowJo Software 7.5 (TreeStar) was used for data analysis. Molecule density on cells was calculated only when more than 20 cells of the assessed cell type were positive for CD23.
Measurement of soluble CD23 in sera
Measurement of soluble CD23 in patients’ sera was performed with the Novex CD23 (soluble) Human Direct ELISA Kit (Life Technologies), according to the manufacturer’s instructions. Nanogram per milliliter values of soluble CD23 were calculated with standard measurement of the 25-kDa form of soluble CD23.
Recombinant allergens and uptake of IgE-allergen complexes by CD23-expressing B-cell lines
Recombinant Bet v 1 and Art v 1 were obtained from Biomay AG (Vienna, Austria). A recombinant trimer of Bet v 131 was used for experiments in which high fluorescence intensity was required. Three EBV-transformed, CD23-expressing B-cell lines (high, medium, and low CD23 expression) were generated, as previously described.32 For each sample of 1 to 2 × 105 EBV-transformed B cells, 300 ng of a Bet v 1–specific recombinant mAb with a human IgE heavy chain constant region33 were incubated with 300 ng of Bet v 1 trimer fluorescently labeled with the Pierce DyLight 488 Labeling Kit (Thermo Scientific, Rockford, Ill), according to the manufacturer’s instructions. Alternatively, Bet v 1 trimer31 was fluorescently labeled with pHrodo Green STP Ester (Life Technologies), according to the manufacturer’s instructions. Cells were first blocked with 10% human serum for 20 minutes at 4°C. Subsequently, the IgE-allergen complex or allergen alone was added to the cells and incubated for 20 minutes at 4°C. Cells were washed with PBS and incubated for 3.5 or 5 hours in RPMI (Gibco, Life Technologies) at 37°C and 5% CO2. For DyLight 488 labeling, surface fluorescence was removed by means of acid washing immediately after the 20-minute incubation at 4°C, as well as after 3.5 and 5 hours of incubation at 37°C. For this purpose, cells were exposed to acid wash I (130 mmol/L NaCl, 0.5 mmol/L KCl, and 10 mmol/L lactic acid, pH 3.9)34 3 times for 5 minutes and additionally to acid wash II (150 mmol/L NaCl and 20 mmol/L HCl, pH 1.7)35 2 times for 1 minute at room temperature. Surface CD23 was stained by using anti-CD23–PE antibody or matching isotype control. Binding of fluorescently labeled allergen binding (0 hours) or uptake (3.5 and 5 hours) by the cells was measured by means of flow cytometry, as stated above.
CD23 dependency of binding and uptake of IgE-allergen complexes was shown with an anti-CD23 antibody (clone M-L233, BD Biosciences). Anti-CD23 or a matching isotype control (mouse IgG1) was incubated with the cells at a concentration of 15 μg/mL for 20 minutes on ice before incubation with the allergen-IgE complexes.
Uptake of fluorescently (DyLight 488) labeled allergen in confocal microscopy was visualized by using a Zeiss LSM 510 (Zeiss, Oberkochen, Germany) with an oil immersion ×60 lens. For this purpose, silhouettes of cells were stained with anti-CD19 (Alexa Fluor 647, clone HIB19; BioLegend, San Diego, Calif) after incubation with allergen-IgE complexes and after acid washing. Cells were then added to adhesion slides (Paul Marienfeld, Lauda-Koenigshofen, Germany), fixed with acetone, and washed twice with PBS, and slides were covered with Fluormount (Sigma-Aldrich, St Louis, Mo). A representative cell sample for each cell line was chosen for depiction.
Cell sorting and T-cell activation assay
An EBV-immortalized B-cell line (HLA-DRB1:0701) was cytometrically sorted for CD23 expression (FACSAria, Becton Dickinson). Briefly, EBV-immortalized B cells were stained with α-human CD23–fluorescein isothiocyanate (TU1; Caltag, Invitrogen–Fisher Scientific) for 30 minutes at 4°C, washed once with 1× PBS supplemented with 0.5% wt/vol BSA, and incubated with α-human CD11a (efalizumab, 3 × 106 EBV cells, 5-10 µg/mL) for 30 minutes at 4°C to prevent homotypic aggregation of the EBV-immortalized B cells.36 Cells were washed once with 1× PBS/0.5% wt/vol BSA and subsequently sorted for high (mean fluorescence intensity [MFI], 325), intermediate (MFI, 138) and negative/low (MFI, 11) CD23-expressing EBV-immortalized B cells (FACSAria).
After 48 hours of recovery, sorted EBV-immortalized B cells (5 × 104 per well) were incubated with titrated concentrations of allergen/anti–Bet v 1 specific IgE complexes in V-bottom plates at 37°C for 3 hours. The dependency of IgE binding on binding to CD23 was investigated by adding an anti-CD23 antibody (clone M-L233, BD Biosciences) or a matching isotype control (mouse IgG1) to the cells. After washing with 1 × Iscove modified Dulbecco medium, 1 × 105 Bet v 1142–156–specific Jurkat T cells37 stably transfected with an IL-2–luciferase reporter were added to each well and co-cultured for 6 hours. Phorbol 12-myristate 13-acetate (10−7 mol/L) plus PHA (12.5 μg/mL) served as a positive control and medium alone served as a negative control, respectively. After the coincubation period, cells were lysed, and luciferase activity was determined (Promega, Madison, Wis) on a Luminoskan Ascent Luminometer (Thermo Scientific), as previously described.38
Flow cytometric measurement of MHC and costimulatory molecules
Immunophenotyping of sorted EBV-immortalized B cells was performed with directly conjugated mAbs directed against HLA class II (HLA-DR, L243, and fluorescein isothiocyanate), CD80 (2D10 and PE), CD86 (IT2.2 and allophycocyanin), and CD19 (HIB19 and peridinin-chlorophyll-protein complex), as well as nonbinding isotype control antibodies (BioLegend). Four-color data acquisition was performed on a fluorescence-activated cell sorter FACSCalibur flow cytometer (Becton Dickinson) and analyzed with CellQuest software (Becton Dickinson). MFI data were calculated from the geometric mean of the fluorescent intensity for all positive cells (above isotype control) in the flow cytometric channel used. Only data acquired in the same experiment with the exact same settings were used to compare MFIs.
Stimulation of PBMCs with IgE
PBMCs from blood donors (5 allergic patients with total IgE levels between 200-312 kU/L and 3 nonallergic subjects with total IgE levels <100 kU/L) were isolated by means of Ficoll gradient centrifugation (GE Healthcare, Fairfield, Conn). Aliquots of 1 × 106 cells/well were cultivated in 1× RPMI medium (Life Technologies, Grand Island, NY) in 12-well plates together with 1 μg/mL of a purified recombinant human IgE mAb33 or PBS alone (Life Technologies). All experiments were done in duplicates. CD23 expression on B cells was assessed by means of flow cytometry after 6 days of culture. To this end, 5 × 105 total cells were assessed, and B cells were stained with an anti-CD19 antibody (clone HIB19) and an anti-CD23 antibody (clone EBVCS2). Quantification with QuantiBRITE beads was performed, as described above, and results represent means of duplicates with an error of less than 5%.
Statistical analysis
To calculate correlations, we used the Pearson correlation coefficient (R) or the Spearman rank correlation coefficient (Rs), depending on data distribution. Therefore Rs was used for correlation of the patient’s CD23 levels with IgE and skin sensitivity values because of the skewed distribution of the data. For correlation in in vitro B-cell uptake and T-cell activation experiments, R was used because of the normal data distribution. Coefficients were considered significant at a P value of less than .05.
A paired t test was performed to test the null hypothesis, stating that the mean relative increase of CD23 expression after addition of IgE to PBMCs is zero. Analyses were performed with the software package R, version 3.1.1.
Results
Characterization of patients with grass pollen allergy
Blood samples from 17 patients (9 male and 8 female) with a history of grass pollen allergy (ie, rhinitis, conjunctivitis, and/or asthma during the grass pollen season [May-July]) were analyzed. Clinical and demographic data of the patients are displayed in Table I. All patients had current symptoms of grass pollen allergy (Table I) when skin prick tests were performed and blood samples for flow cytometry and other experiments were obtained. All but 2 patients had a history of other allergies. Total IgE levels ranged between 33 and 10,856 kU/L (median, 462 kU/L), whereas IgE levels specific for timothy grass pollen ranged between 1.1 and 1,204 kU of antigen/L (median, 33 kU of antigen/L). All patients had positive skin prick test reactions to grass pollen extract (12.6-238.7 mm2; median, 61 mm2).
Table I.
Demographic and clinical data of patients with grass pollen allergy
| Patient no. | Sex | Age (y) | Current grass pollen symptoms* | Other allergen sources† | Total IgE (kU/L) | Grass pollen IgE (kUA/L) | SPT (mm2) |
|---|---|---|---|---|---|---|---|
| 1 | F | 28 | r | a, h, t | 77 | 3.3 | 72.4 |
| 2 | M | 34 | c, r | a, f, m, t | 94 | 33.1 | 14.8 |
| 3 | M | 26 | c, r, a | a, f, h, t, w | 462 | 22.5 | 101.7 |
| 4 | M | 30 | c, r | — | 33 | 14.6 | 20.6 |
| 5 | F | 29 | c, r | t, w | 395 | 82.6 | 17.4 |
| 6 | M | 44 | c, r, d | — | 82 | 5.9 | 61.2 |
| 7 | M | 34 | c, r, a, d | a, f, t | 680 | 19.7 | 34.2 |
| 8 | M | 22 | c, r, a, d | a | 740 | 167.0 | 55.7 |
| 9 | M | 30 | c, r, d | a, h, t | 10856 | 1204.0 | 238.7 |
| 10 | F | 31 | c, r, a, d | f, h, t | 4967 | 338.0 | 169.5 |
| 11 | F | 18 | c, r, d | a, h, t, w | 1249 | 92.3 | 61.9 |
| 12 | F | 32 | c, r, a, d | a | 695 | 31.9 | 20.4 |
| 13 | F | 29 | c, r, a, d | a, t | 110 | 19.7 | 58.4 |
| 14 | M | 41 | a, d | a, f, t, w | 649 | 93.8 | 119.7 |
| 15 | F | 24 | r, a, d | a, t, f | 346 | 33.8 | 159.3 |
| 16 | M | 40 | r, d | a, h, t | 9113 | 222.0 | 78.5 |
| 17 | F | 28 | c, r | h, t | 277 | 1.1 | 12.6 |
| Median | 462 | 33 | 61 |
Shown are sex, age, current symptoms to grass pollen, sensitization to other allergen sources, total IgE levels, timothy grass pollen–specific IgE levels, and sizes of wheal reactions to grass pollen mix.
F, Female; kUA, kilounits of antigen; M, male; SPT, skin prick test.
a, Asthma; c, conjunctivitis; d, dermatitis; r, rhinitis.
—, No other allergen source; a, animal dander; f, food; h, house dust mite; m, mold; t, tree pollen; w, weed pollen.
CD23 was expressed mainly on naive IgD+ B cells in patients with grass pollen allergy
When CD23 expression was determined on blood cells from patients with symptoms of grass pollen allergy, we found that the receptor was expressed almost exclusively on B cells. Table II shows that 49.8% of patients’ B cells on average expressed CD23 molecules, whereas in most patients only a few monocytes (mean, 1.38%), NK cells (mean, 0.57%), or dendritic cells (mean, 0.43%) were CD23+ (Table II). Next, we investigated the CD23 receptor density on the CD23+ cell populations (Table III). We found a high density of CD23 molecules expressed on B cells (2168-6719 molecules per cell; mean, 3968 molecules per cell), whereas it was possible to calculate the number of CD23 molecules on monocytes in only 6 of the 17 patients, and the density was lower than on B cells (ie, 388-1465 molecules per cell; mean, 1089 molecules per cell).
Table II.
Percentages of CD23+ cells among different cell types
| Mean (SD), n = 17 | Range, n = 17 | |
|---|---|---|
| T cells | 0.02 (0.02) | 0-0.07 |
| NK cells | 0.57 (0.97) | 0-3.35 |
| Monocytes | 1.38 (1.89) | 0-7.88 |
| Neutrophils | 0.00 (0.00) | 0.00 |
| Platelets | 0.09 (0.13) | 0-0.54 |
| Dendritic cells | 0.43 (0.97) | 0-4 |
| Eosinophils | 0.04 (0.07) | 0-0.23 |
| Basophils | 0.00 (0.00) | 0.00 |
| B cells | 49.81 (21.47) | 3.3-82.33 |
CD23+ cells in blood samples from patients with grass pollen allergy (Table I, n = 17). Shown are the mean percentages and SDs of CD23+ cells within the investigated cell types and ranges.
Table III.
Numbers of CD23 molecules per cell and concentrations of soluble CD23 in serum
| Patient no. | CD23 molecules per B cell | CD23 molecules per monocyte | Soluble CD23 (ng/mL) |
|---|---|---|---|
| 1 | 4869 | ND | 1.2 |
| 2 | 2168 | ND | 0.9 |
| 3 | 3183 | ND | 1.8 |
| 4 | 3088 | ND | 1.6 |
| 5 | 2497 | 1230 | 1.0 |
| 6 | 2758 | 1228 | 2.3 |
| 7 | 2327 | 1465 | 1.1 |
| 8 | 3552 | ND | 1.3 |
| 9 | 6539 | ND | 0.9 |
| 10 | 6719 | 388 | 1.1 |
| 11 | 5749 | ND | 1.5 |
| 12 | 2930 | 1399 | 1.9 |
| 13 | 3278 | 826 | 1.5 |
| 14 | 3989 | ND | 1.0 |
| 15 | 3125 | ND | 1.3 |
| 16 | 6530 | ND | 2.1 |
| 17 | 4150 | ND | 1.3 |
| Mean | 3968 | 1089 | 1.4 |
Average numbers of CD23 molecules per CD23+ B cell and monocyte and concentrations of soluble CD23 in patients’ sera (in nanograms per milliliter) were measured in the blood of the 17 patients with grass pollen allergy (Table I).
ND, Numbers of CD23 molecules could not be calculated when less than 20 of the monocytes among the analyzed cells were positive for CD23.
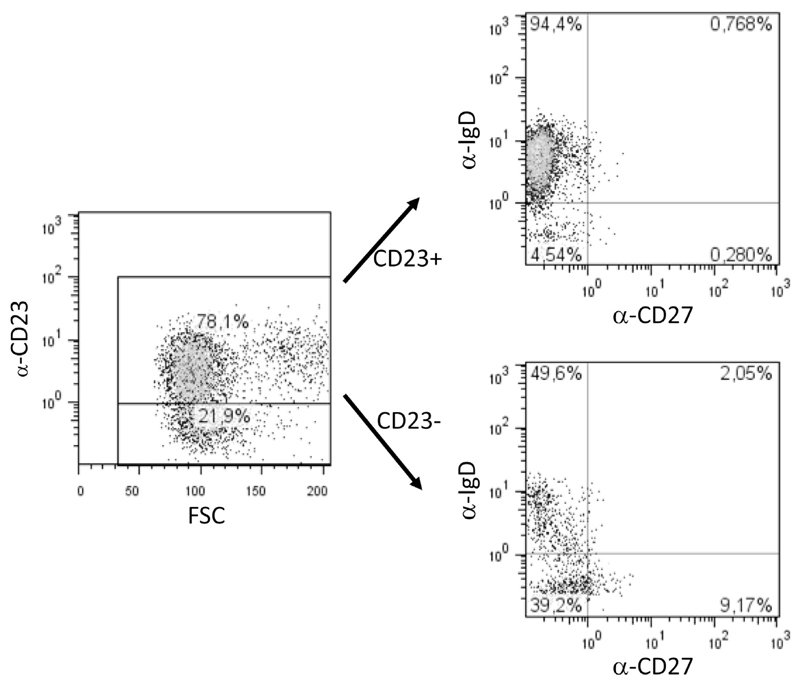
To characterize the CD23+ and CD23− populations of B cells, we stained blood cells of 3 patients (patients 8, 11, and 17) with antibodies specific for CD19, CD23, IgD, and CD27 (see Table E1 in this article’s Online Repository at www.jacionline.org). On average, 65.1% of B cells were CD23+ in these patients. The majority of the CD23+ cells were also positive for IgD and negative for the memory marker CD27 (naive B cells: mean, 89.2%; see Table E1). Only 0.5% of the CD23+ B cells were IgD+CD27+ preswitch memory B cells, 1.1% were CD27+IgD− postswitch memory cells, and 9.2% were IgD−CD27− resting memory B cells.
Within the CD23− B-cell population (ie, 34.9% of all B cells), 42.9% were naive IgD+CD27− B cells, 47.1% were IgD−CD27− resting memory B cells, 8.9% were CD27+IgD− postswitch memory cells, and 1.1% of the CD23− B cells were preswitch memory B cells (IgD+CD27+ cells, see Table E1). The flow cytometric charts of 1 representative patient (patient 11, Table I) are displayed in fig E1 in this article’s Online Repository at www.jacionline.org.
Total IgE levels are correlated with numbers of CD23 molecules on B cells
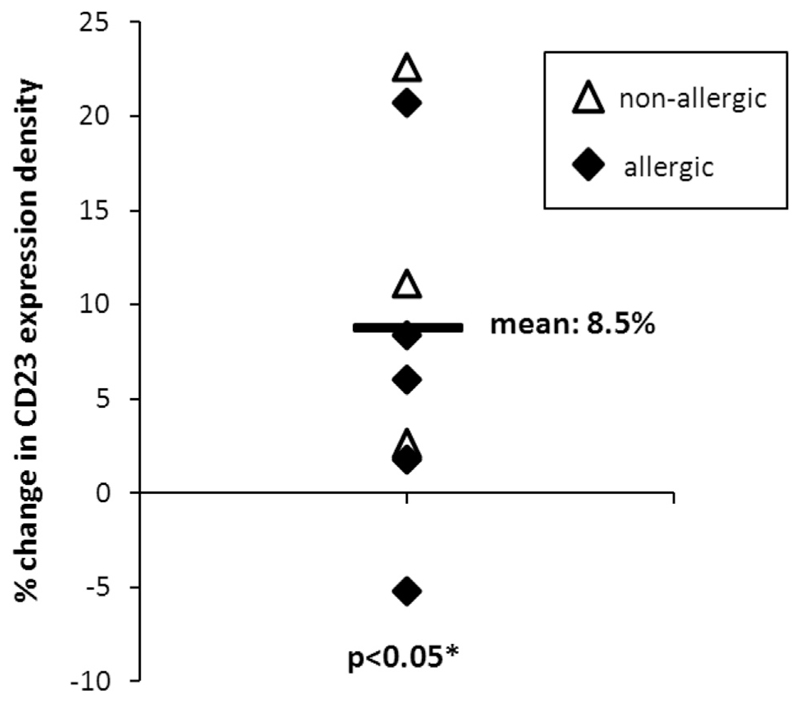
In our group of 17 patients with grass pollen allergy, we found a significant correlation between the density of CD23 on CD23+ B cells (Table III) and total serum IgE levels (RS = 0.53, P = .03; Fig 1, A). A pilot experiment showed that addition of purified monoclonal human IgE to cultured PBMCs resulted in a significant increase (mean, 8.5%; P = .04) of CD23 expression on B cells, suggesting that IgE can upregulate CD23 expression (see Fig E2 in this article’s Online Repository at www.jacionline.org).
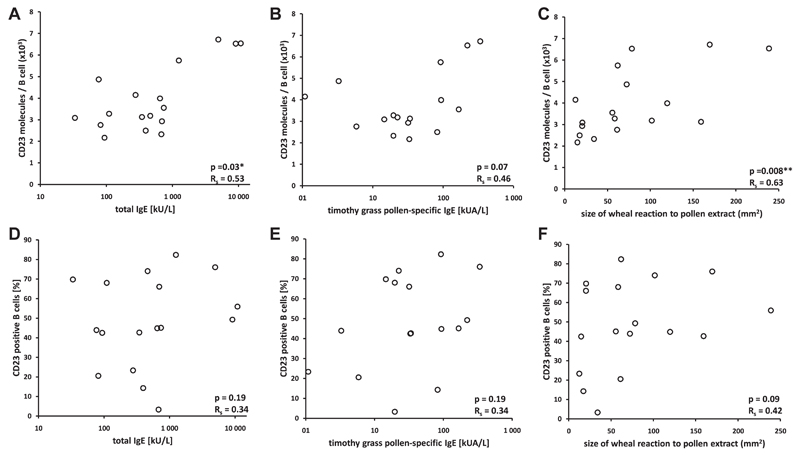
Fig 1.
Associations of numbers of CD23 molecules per B cell and percentages of CD23+ B cells with skin sensitivity to grass pollen allergens and total and grass pollen–specific IgE levels. Numbers of CD23 molecules per CD23+ B cells (A-C) or percentages of CD23+ B cells (D-F) are displayed on the y-axis. Total IgE levels (Fig 1, A and D), timothy grass pollen–specific IgE levels (Fig 1, B and E), or sizes of grass pollen–induced wheal reactions (Fig 1, C and F) are plotted on the x-axis. *P values of less than .05. **P values of less than .01.
Furthermore, grass pollen–specific serum IgE levels were also linked to the number of CD23 molecules on B cells (RS = 0.46, P =.07; Fig 1, B). We also found a significant correlation between the molecule density of CD23 on B cells (Table III) and the size of skin prick test reactions to grass pollen extract (RS = 0.63, P = .008; Fig 1, C). Additionally, skin prick test reaction size correlated significantly with grass pollen–specific serum IgE levels (RS = 0.54, P = .03, data not shown). Because only 6 patients showed substantial CD23 expression on monocytes, we did not correlate the number of CD23 molecules on monocytes with respective IgE and skin prick test data.
Neither CD23+ B-cell numbers nor soluble CD23 levels are correlated with total and specific IgE levels or skin sensitivity
Fig 1 shows correlations of percentages of CD23+ B cells with total and allergen-specific IgE levels, as well as with grass pollen–specific wheal reactions. In fact, neither total serum IgE levels (RS = 0.34, P = .19; Fig 1, D) nor grass pollen–specific serum IgE levels (RS = 0.34, P = .19; Fig 1, E) correlated with the number of CD23+ B cells. We also did not find a statistically significant correlation between the percentage of CD23+ B cells with skin prick test reactivity to grass pollen (RS = 0.42, P = .09; Fig 1, F).
Also, we analyzed levels of soluble CD23 in sera of the 17 patients (Table III). However, soluble CD23 serum levels did not correlate with patients’ total IgE levels (RS = −0.12, P =.66), grass pollen–specific IgE levels (RS = −0.33, P = .19), or grass pollen–specific wheal reactions (RS = −0.06, P = .81, data not shown).
IgE-facilitated allergen uptake is dependent on CD23 density on B lymphocytes
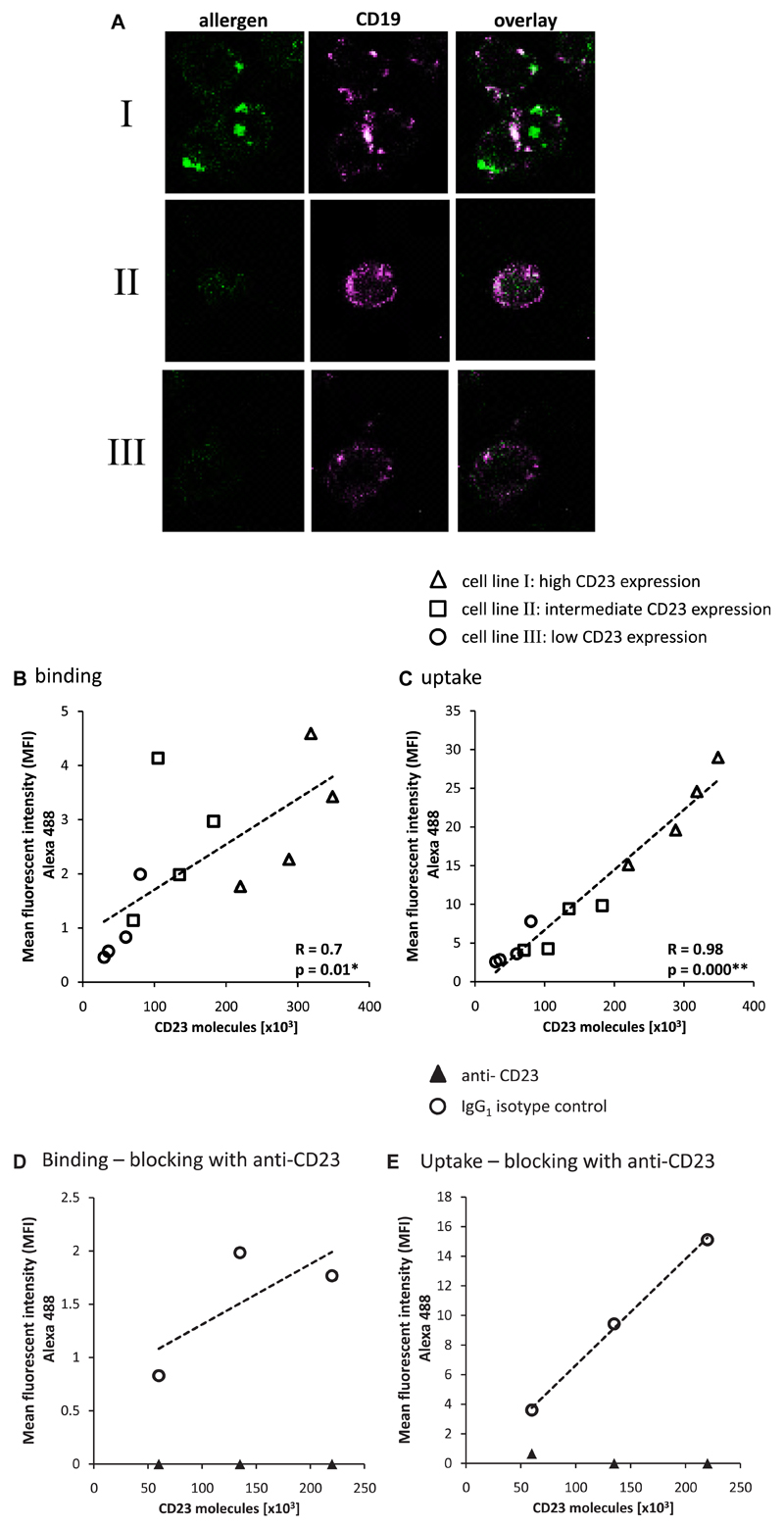
We used 3 different CD23-expressing EBV-transformed B-cell clones to investigate the possible dependence of IgE-facilitated allergen uptake into B cells on CD23 density. Cell line I had an average number of 3 × 105 CD23 molecules per cell, cell line II expressed 1.2 × 105 CD23 molecules per cell on average, and cell line III expressed 5 × 104 molecules per cell on average (Fig 2). In a first set of experiments, we measured and compared uptake of the complexes into the different cell lines after removal of bound surface molecules by means of acid washing by incubating the cells with fluorescently labeled allergen-IgE complexes. Allergen uptake into different cell lines was studied by using confocal microscopy, and CD19+ cells were stained (Fig 2, A). We found that staining of allergen-IgE immune complexes was more intensive on the cell lines expressing high levels of CD23 molecules (ie, cell lines I and II) compared with the cell line expressing fewer CD23 molecules (ie, cell line III; Fig 2, A). Cells had been stripped with an acidic wash after incubation with the fluorescently labeled IgE-allergen complexes, followed by anti-CD19 staining. Under these conditions, CD19 stained the silhouette of the sections, whereas labeled allergen appeared intracellularly, indicating that the allergen had been taken up. No allergen staining was observed with any of the 3 CD23-expressing cell lines when allergen-specific IgE was omitted (data not shown).
Fig 2.
Dependence of allergen uptake on CD23 cell-surface density. A, Confocal microscopy of 3 EBV-transformed B-cell lines expressing different numbers of CD23 molecules on their surfaces (I, 3.5 × 105; II, 1 × 105; and III, 5 × 104 CD23 molecules per cell). Cells stained with anti-CD19 are shown in pink, whereas allergen (Bet v 1 trimer) was labeled in green with DyLight 488, preincubated with allergen-specific IgE, and then exposed to the cells. The pictures marked overlay show an overlay of anti-CD19 and allergen staining. B and C, Binding (Fig 2, B) and uptake (Fig 2, C) of a fluorescent allergen-IgE complex by CD23-expressing EBV-transformed B-cell lines (I, II, and III) shown by using flow cytometry. CD23 expression was measured before and after 3.5 hours of incubation with IgE-allergen complexes. Different symbols (circles, I; squares, II: and triangles, III) represent the 3 different B-cell lines. Four independent experiments performed on 4 different study days are displayed. Each data point represents the mean of triplicate experiments done on the same study day. Background fluorescence measured after incubation with allergen alone was subtracted. Dotted lines, Trend lines of correlations. D and E, Binding (Fig 2, D) and uptake (Fig 2, E) of allergen-IgE complexes was blocked by an anti-CD23 antibody but not by the matching isotype control (IgG1). Solid triangles represent blocking by anti-CD23, and open circles show cells incubated with the isotype control.
The findings of the confocal staining experiments were corroborated by a second set of experiments in which allergen-IgE binding to the cell surface (Fig 2, B) and uptake into cells (Fig 2, C) were studied by using flow cytometry. In these experiments we found that there was a significant correlation (R = 0.7, P = .01) between the number of CD23 molecules on the cells and binding of the allergen-IgE complexes after 20 minutes of incubation at 4°C (Fig 2, B). An almost linear and striking correlation (R = 0.98, P = .000; Fig 2, C) was found between the uptake of allergen after 3.5 hours of incubation at 37°C and the surface density of CD23 molecules on these cells. Both binding and uptake of allergen-IgE complexes could be blocked by an anti-CD23 antibody (Fig 2, D and E), and both complex binding and uptake were unaffected if a matching isotype control was used instead (Fig 2, D and E). Our experiments demonstrate the causal dependency of allergen uptake and T-cell activation on IgE-facilitated allergen presentation through CD23. In these experiments the extent of IgE-facilitated allergen uptake was correlated with the surface density of CD23.
To further confirm that allergen-specific signals measured by using flow cytometry were derived only from intracellular compartments and did not represent remaining allergen residing on the cell surface, we repeated the uptake experiment using allergen labeled with the pH-sensitive pHrodo Green, which only emits fluorescence after uptake into endolysosomal compartments because of acidic pH. Again, the measured uptake of allergen significantly correlated with the CD23 surface density (R = 0.97, P = .000, data not shown).
Density of CD23 on the surfaces of B cells determines activation of allergen-specific T cells by IgE-facilitated allergen presentation
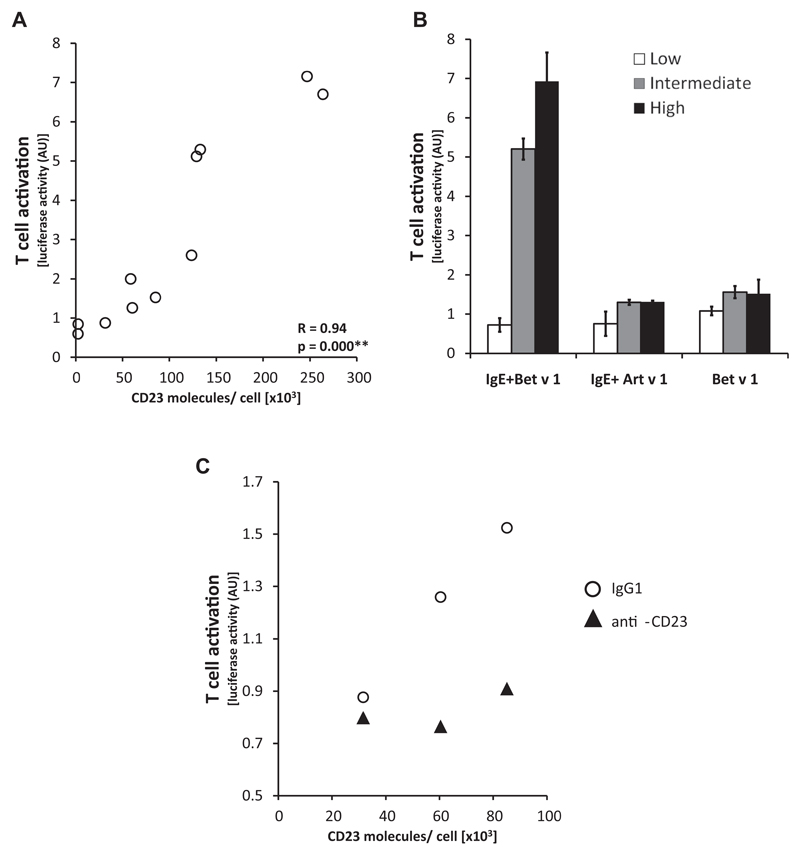
Next, we measured whether the density of CD23 on antigen-presenting B cells that had been loaded with IgE-allergen complexes influences the activation of MHC-matched Bet v 1–specific T cells, which expressed a single Bet v 1–specific T-cell receptor. For this purpose, we sorted EBV-transformed B cells into 3 different subpopulations according to their CD23 expression levels (high, intermediate, or low expression), incubated them with equal amounts of Bet v 1–IgE complexes, and measured Bet v 1–specific T-cell activation (Fig 3). Our experiments showed that T-cell activation was directly dependent on the CD23 surface density of cocultured B cells when incubated together with allergen-IgE complexes (Fig 3, A and B). Initial CD23 expression on B cells was correlated with subsequent T-cell activation (R = 0.94, P = .000; Fig 3, A). No relevant T-cell activation was observed when an unrelated allergen (Art v 1) or Bet v 1 without Bet v 1–specific IgE was used for stimulation (Fig 3, B), and IgE-facilitated T-cell activation could be blocked by an anti-CD23 antibody (Fig 3, C). In addition to CD23, we also measured MHC class II, CD80, and CD86 expression on sorted B cells. We found some differences in expression of the costimulatory molecules CD80 and CD86. However, there was no difference regarding MHC class II expression in CD23-high and CD23-intermediate cell populations, although T-cell activation was different (see Table E2 in this article’s Online Repository at www.jacionline.org).
Fig 3.
IgE-facilitated T-cell activation is dependent on CD23 surface density. EBV-immortalized B cells were sorted according to their CD23 expression. A, Correlation between numbers of CD23 molecules on B cells (x-axis) and T-cell activation (y-axis, luciferase activity in arbitrary units) induced by IgE–Bet v 1 immune complexes. Four independent experiments on 4 different study days are displayed. B, Activation of Bet v 1–specific T cells (y-axis) after incubation of sorted EBV-immortalized B cells (white, low expression; gray, intermediate expression; and black, high expression) with IgE–Bet v 1 immune complexes, Bet v 1–specific IgE and the unrelated allergens Art v 1 or Bet v 1 alone, respectively. Data from 2 independent experiments showed similar results and were summarized. C, IgE-facilitated T-cell activation was blocked by preincubation of B cells with an anti-CD23 antibody (anti-CD23) but not by an isotype control antibody (IgG1).
Discussion
Several studies performed in patients during allergen-specific immunotherapy have provided evidence that CD23, the low-affinity receptor for IgE, has an important role in controlling allergen-specific T-cell activation through IgE-facilitated allergen presentation.19–21,39 To investigate the number of CD23 molecules on immune cells of allergic patients, we developed a flow cytometry–based assay using QuantiBRITE beads. Using this assay, we found that CD23 is expressed primarily on naive IgD+ B cells in the blood of allergic patients and only in negligible numbers on other immune cells.
In fact, CD23 was present on other APCs, such as dendritic cells or monocytes; however, either only a few of these cells were CD23+, or expression levels were very low, or both (Table II). Therefore it was not possible to calculate reliable CD23 expression on their cell surfaces. We must thus assume that mainly B cells play a role in IgE-facilitated allergen presentation through CD23. Interestingly, the number of CD23 molecules per cell and not the percentage of CD23+ cells was correlated with total IgE levels and allergen-induced immediate type skin reactions and was associated with allergen-specific IgE levels. Pilot experiments even suggested that addition of highly purified human monoclonal IgE might upregulate CD23 expression on B cells in cultured PBMCs from allergic and nonallergic subjects. Thus the surface expression of CD23 seems to follow the rules that guide the expression of the high-affinity receptor for IgE (FcεRI), which was shown to depend on IgE levels5 and to be regulated directly by IgE binding.40 In fact, it has also been shown that an anti-IgE antibody (omalizumab), which prevents binding of IgE to FcεRI by shielding IgE binding to the receptor, can downregulate FcεRI expression on basophils, mast cells, and APCs.7
Next, we were interested in whether the surface density of CD23 on B cells might have an influence on IgE-facilitated allergen uptake and subsequent allergen-specific T-cell proliferation. To study this question, we used defined model systems. We used the purified major birch allergen Bet v 1, a human Bet v 1–specific IgE mAb, T cells transfected with a Bet v 1–specific T-cell receptor, and MHC-matched, EBV-transformed B cells, which were characterized regarding the numbers of CD23 molecules on their surface. The experiments performed showed that allergen uptake by IgE-facilitated allergen presentation was significantly correlated with the numbers of CD23 molecules on the surfaces of APCs and depended on the presence of allergen-specific IgE. It was causally related to the ability of IgE to bind to CD23 because it could be blocked with an anti-CD23 antibody. Furthermore, the strongest T-cell activation was observed when APCs expressing high CD23 levels were used for IgE-facilitated allergen presentation, and we found a significant correlation (P = .000) between CD23 levels on B cells and T-cell activation. Taken together, our findings suggest that high levels of allergen-specific IgE upregulate the surface density expression of CD23 on B cells in allergic patients, which might then lead to enhanced IgE-facilitated allergen presentation and activation of allergen-specific T cells. This possible interplay between IgE levels, CD23 surface density on B cells as APCs, and allergen-specific T-cell activation might open possibilities to control T cell–mediated allergic inflammation caused by IgE-facilitated allergen presentation by several nondirectly T cell– or cytokine-targeting approaches and nonpharmacologic strategies. For example, it can be considered to block the CD23-binding site on IgE with anti-IgE antibodies, as has been suggested earlier,17,41 to suppress allergen-specific T-cell activation. In this context it was found that omalizumab indeed suppressed markers of T-cell activation when administered to patients with atopic dermatitis.42 Another possibility would be to block binding of IgE to CD23, such as with therapeutic anti-CD23 antibodies or CD23 variants.43 In this context it was found that lumiliximab, an anti-CD23 antibody, suppressed allergen-specific T-cell activation in vitro.18 Also, other “humoral” approaches, which primarily focus on IgE production,44 and interference of the IgE-allergen interaction (eg, passive immunization with blocking allergen-specific IgG45) might have an influence on T cell–mediated allergic inflammation.
In summary, our study suggests that IgE might play a role in the upregulation of CD23 on B cells and demonstrates that the surface density of CD23 on B cells determines allergen uptake and subsequent T-cell activation. These findings conceivably have implications for the treatment of allergic diseases.
Extended Data
Fig E1.
CD23+ B cells mainly have a naive IgD+ phenotype. Data from 1 representative patient are shown. Of the 78.1% CD23+ B cells, 94.4% were IgD+ naive cells. The population of CD23− B cells consisted of 21.9% of the cells, of which 49.6% were IgD+ naive cells and 9.17% were CD27+IgD− memory B cells.
Fig E2.
Changes of CD23 surface density on B cells after addition of IgE. Shown are percentage changes of CD23 density on B cells in PBMCs from allergic (black) and nonallergic (white) subjects cultured after addition of an IgE mAb (1 µg/mL IgE) for 6 days compared with untreated cells.
Table E1.
Expression of IgD and CD27 on CD23+ and CD23− B cells
| CD23+ (range) | CD23− (range) | |
|---|---|---|
| Total B cells | ||
| CD19+ | 65.1 (55.8-77.9) | 34.9 (22.1-44.2) |
| Naive B cells | ||
| IgD+CD27− | 89.2 (81.8-94.5) | 42.9 (31.9-42.9) |
| Postswitch memory B cells | ||
| IgD−CD27+ | 1.1 (0.25-1.72) | 8.9 (6.9-10.2) |
| Resting memory B cells | ||
| IgD−CD27− | 9.2 (4.8-15.9) | 47.1 (41.2-57.3) |
| Preswitch memory B cells | ||
| IgD+CD27+ | 0.5 (0.5-0.6) | 1.1 (0.6-1.5) |
The mean percentage of CD23+ and CD23− cells was investigated in total B cells, as well as among naive (CD27−IgD+), postswitch memory (CD27+IgD−), resting memory (IgD−CD27−), and preswitch memory (IgD+CD27+) B cells.
Table E2.
Expression of surface markers on sorted EBV-transformed B cells
| MFI |
|||
|---|---|---|---|
| High | Intermediate | Low | |
| CD23 | 325 | 138 | 11 |
| CD19 | 68 | 48 | 17 |
| HLA class II | 271 | 271 | 175 |
| CD80 | 231 | 139 | 34 |
| CD86 | 793 | 540 | 204 |
B cells sorted according to their CD23 expression (high, intermediate, and low) were stained for expression of CD19, HLA class II (HLA-DR), and the costimulatory molecules CD80 and CD86.
Acknowledgments
Supported by the Austrian Science Fund (FWF): SFB 4605, SFB 4613, and SFB 4609.
R. Selb and J. Eckl-Dorna were paid by grants from the Austrian Science Fund. K. Marth has received payment for lectures from Thermo Fisher. W. F. Pickl has received a grant from the Austrian Science Fund (SFB F4609) and has stock options in Biomay AG. R. Valenta has received grants from the Austrian Science Fund, Biomay, Thermo Fisher, and Fresenius Medical Care and has consultant arrangements with Biomay, Thermo Fisher, and Fresenius Medical Care. V. Niederberger has received a grant from the Austrian Science Fund (SFB4613).
Abbreviations used
- APC
Antigen-presenting cell
- MFI
Mean fluorescence intensity
- NK
Natural killer
- PE
Phycoerythrin
- R
Pearson correlation coefficient
- Rs
Spearman rank correlation coefficient
Footnotes
Disclosure of potential conflict of interest: The rest of the authors declare that they have no relevant conflicts of interest.
References
- 1.Bischoff SC. Role of mast cells in allergic and non-allergic immune responses: comparison of human and murine data. Nat Rev Immunol. 2007;7:93–104. doi: 10.1038/nri2018. [DOI] [PubMed] [Google Scholar]
- 2.Dombrowicz D, Woerly G, Capron M. IgE receptors on human eosinophils. Chem Immunol. 2000;76:63–76. doi: 10.1159/000058781. [DOI] [PubMed] [Google Scholar]
- 3.Stingl G, Maurer D. IgE-mediated allergen presentation via Fc epsilon RI on antigen-presenting cells. Int Arch Allergy Immunol. 1997;113:24–9. doi: 10.1159/000237499. [DOI] [PubMed] [Google Scholar]
- 4.Novak N, Kraft S, Bieber T. Unraveling the mission of FcepsilonRI on antigen-presenting cells. J Allergy Clin Immunol. 2003;111:38–44. doi: 10.1067/mai.2003.2. [DOI] [PubMed] [Google Scholar]
- 5.Malveaux FJ, Conroy MC, Adkinson NF, Jr, Lichtenstein LM. IgE receptors on human basophils. Relationship to serum IgE concentration. J Clin Invest. 1978;62:176–81. doi: 10.1172/JCI109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sihra BS, Kon OM, Grant JA, Kay AB. Expression of high-affinity IgE receptors (Fc epsilon RI) on peripheral blood basophils, monocytes, and eosinophils in atopic and nonatopic subjects: relationship to total serum IgE concentrations. J Allergy Clin Immunol. 1997;99:699–706. doi: 10.1016/s0091-6749(97)70033-2. [DOI] [PubMed] [Google Scholar]
- 7.Beck LA, Marcotte GV, MacGlashan D, Togias A, Saini S. Omalizumab-induced reductions in mast cell Fc epsilon RI expression and function. J Allergy Clin Immunol. 2004;114:527–30. doi: 10.1016/j.jaci.2004.06.032. [DOI] [PubMed] [Google Scholar]
- 8.Yokota A, Kikutani H, Tanaka T, Sato R, Barsumian EL, Suemura M, et al. Two species of human Fc epsilon receptor II (Fc epsilon RII/CD23): tissue-specific and IL-4-specific regulation of gene expression. Cell. 1988;55:611–8. doi: 10.1016/0092-8674(88)90219-x. [DOI] [PubMed] [Google Scholar]
- 9.Vercelli D, Jabara HH, Lee BW, Woodland N, Geha RS, Leung DY. Human recombinant interleukin 4 induces Fc epsilon R2/CD23 on normal human monocytes. J Exp Med. 1988;167:1406–16. doi: 10.1084/jem.167.4.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Armitage RJ, Goff LK, Beverley PC. Expression and functional role of CD23 on T cells. Eur J Immunol. 1989;19:31–5. doi: 10.1002/eji.1830190106. [DOI] [PubMed] [Google Scholar]
- 11.Krauss S, Mayer E, Rank G, Rieber EP. Induction of the low affinity receptor for IgE (Fc epsilon RII/CD23) on human blood dendritic cells by interleukin-4. Adv Exp Med Biol. 1993;329:231–6. doi: 10.1007/978-1-4615-2930-9_39. [DOI] [PubMed] [Google Scholar]
- 12.Klouche M, Klinger MH, Kuhnel W, Wilhelm D. Endocytosis, storage, and release of IgE by human platelets: differences in patients with type I allergy and nonatopic subjects. J Allergy Clin Immunol. 1997;100:235–41. doi: 10.1016/s0091-6749(97)70230-6. [DOI] [PubMed] [Google Scholar]
- 13.Yamaoka KA, Arock M, Issaly F, Dugas N, Le Goff L, Kolb JP. Granulocyte macrophage colony stimulating factor induces Fc epsilon RII/CD23 expression on normal human polymorphonuclear neutrophils. Int Immunol. 1996;8:479–90. doi: 10.1093/intimm/8.4.479. [DOI] [PubMed] [Google Scholar]
- 14.Kehry MR, Yamashita LC. Low-affinity IgE receptor (CD23) function on mouse B cells: role in IgE-dependent antigen focusing. Proc Natl Acad Sci U S A. 1989;86:7556–60. doi: 10.1073/pnas.86.19.7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van der Heijden FL, Joost van Neerven RJ, van Katwijk M, Bos JD, Kapsenberg ML. Serum-IgE-facilitated allergen presentation in atopic disease. J Immunol. 1993;150:3643–50. [PubMed] [Google Scholar]
- 16.van der Heijden FL, van Neerven RJ, Kapsenberg ML. Relationship between facilitated allergen presentation and the presence of allergen-specific IgE in serum of atopic patients. Clin Exp Immunol. 1995;99:289–93. doi: 10.1111/j.1365-2249.1995.tb05547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Neerven RJ, Knol EF, Ejrnaes A, Wurtzen PA. IgE-mediated allergen presentation and blocking antibodies: regulation of T-cell activation in allergy. Int Arch Allergy Immunol. 2006;141:119–29. doi: 10.1159/000094714. [DOI] [PubMed] [Google Scholar]
- 18.Poole JA, Meng J, Reff M, Spellman MC, Rosenwasser LJ. Anti-CD23 monoclonal antibody, lumiliximab, inhibited allergen-induced responses in antigen-presenting cells and T cells from atopic subjects. J Allergy Clin Immunol. 2005;116:780–8. doi: 10.1016/j.jaci.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 19.van Neerven RJ, Wikborg T, Lund G, Jacobsen B, Brinch-Nielsen A, Arnved J, et al. Blocking antibodies induced by specific allergy vaccination prevent the activation of CD4+ T cells by inhibiting serum-IgE-facilitated allergen presentation. J Immunol. 1999;163:2944–52. [PubMed] [Google Scholar]
- 20.Wachholz PA, Soni NK, Till SJ, Durham SR. Inhibition of allergen-IgE binding to B cells by IgG antibodies after grass pollen immunotherapy. J Allergy Clin Immunol. 2003;112:915–22. doi: 10.1016/s0091-6749(03)02022-0. [DOI] [PubMed] [Google Scholar]
- 21.Shamji MH, Wilcock LK, Wachholz PA, Dearman RJ, Kimber I, Wurtzen PA, et al. The IgE-facilitated allergen binding (FAB) assay: validation of a novel flow-cytometric based method for the detection of inhibitory antibody responses. J Immunol Methods. 2006;317:71–9. doi: 10.1016/j.jim.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung CM, Prinz JC, Rieber EP, Ring J. A reduction in allergen-induced Fc epsilon R2/CD23 expression on peripheral B cells correlates with successful hyposensitization in grass pollinosis. J Allergy Clin Immunol. 1995;95:77–87. doi: 10.1016/s0091-6749(95)70155-9. [DOI] [PubMed] [Google Scholar]
- 23.Roever AC, Heine G, Zuberbier T, Worm M. Allergen-mediated modulation of CD23 expression is interferon-gamma and interleukin-10 dependent in allergic and non-allergic individuals. Clin Exp Allergy. 2003;33:1568–75. doi: 10.1046/j.1365-2222.2003.01797.x. [DOI] [PubMed] [Google Scholar]
- 24.Vecchiarelli A, Siracusa A, Monari C, Pietrella D, Retini C, Severini C. Cytokine regulation of low-affinity IgE receptor (CD23) on monocytes from asthmatic subjects. Clin Exp Immunol. 1994;97:248–53. doi: 10.1111/j.1365-2249.1994.tb06076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hakansson L, Heinrich C, Rak S, Venge P. Activation of B-lymphocytes during pollen season. Effect of immunotherapy. Clin Exp Allergy. 1998;28:791–8. doi: 10.1046/j.1365-2222.1998.00295.x. [DOI] [PubMed] [Google Scholar]
- 26.Nakamura K, Okubo Y, Minami M, Furue M, Ishibashi Y. Phenotypic analysis of CD23+ peripheral blood mononuclear cells in atopic dermatitis. Br J Dermatol. 1991;125:543–7. doi: 10.1111/j.1365-2133.1991.tb14791.x. [DOI] [PubMed] [Google Scholar]
- 27.Corominas M, Mestre M, Bas J, Buendia E. Distinct modulation by interferon-gamma (IFN-gamma) of CD23 expression on B and T lymphocytes of atopic subjects. Clin Exp Immunol. 1998;112:276–80. doi: 10.1046/j.1365-2249.1998.00570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corominas M, Mestre M, Bas J, Verdaguer J, Valls A, Romeu A, et al. CD23 expression on B-lymphocytes and its modulation by cytokines in allergic patients. Clin Exp Allergy. 1993;23:612–7. doi: 10.1111/j.1365-2222.1993.tb00902.x. [DOI] [PubMed] [Google Scholar]
- 29.Dehlink E, Baker AH, Yen E, Nurko S, Fiebiger E. Relationships between levels of serum IgE, cell-bound IgE, and IgE-receptors on peripheral blood cells in a pediatric population. PLoS One. 2010;5:e12204. doi: 10.1371/journal.pone.0012204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niederberger V, Marth K, Eckl-Dorna J, Focke-Tejkl M, Weber M, Hemmer W, et al. Skin test evaluation of a novel peptide carrier-based vaccine, BM32, in grass pollen-allergic patients. J Allergy Clin Immunol. 2015;136:1101–3.e8. doi: 10.1016/j.jaci.2015.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vrtala S, Hirtenlehner K, Susani M, Akdis M, Kussebi F, Akdis CA, et al. Genetic engineering of a hypoallergenic trimer of the major birch pollen allergen Bet v 1. FASEB J. 2001;15:2045–7. doi: 10.1096/fj.00-0767fje. [DOI] [PubMed] [Google Scholar]
- 32.Sugden B, Mark W. Clonal transformation of adult human leukocytes by Epstein-Barr virus. J Virol. 1977;23:503–8. doi: 10.1128/jvi.23.3.503-508.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laffer S, Vangelista L, Steinberger P, Kraft D, Pastore A, Valenta R. Molecular characterization of Bip 1, a monoclonal antibody that modulates IgE binding to birch pollen allergen, Bet v 1. J Immunol. 1996;157:4953–62. [PubMed] [Google Scholar]
- 34.Pruzansky JJ, Grammer LC, Patterson R, Roberts M. Dissociation of IgE from receptors on human basophils. I. Enhanced passive sensitization for histamine release. J Immunol. 1983;131:1949–53. [PubMed] [Google Scholar]
- 35.Karagiannis SN, Warrack JK, Jennings KH, Murdock PR, Christie G, Moulder K, et al. Endocytosis and recycling of the complex between CD23 and HLA-DR in human B cells. Immunology. 2001;103:319–31. doi: 10.1046/j.1365-2567.2001.01238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang SC, Kanner SB, Ledbetter JA, Gupta S, Kumar G, Nel AE. Evidence for LFA-1/ICAM-1 dependent stimulation of protein tyrosine phosphorylation in human B lymphoid cell lines during homotypic adhesion. J Leukoc Biol. 1995;57:343–51. doi: 10.1002/jlb.57.2.343. [DOI] [PubMed] [Google Scholar]
- 37.Neunkirchner A, Leb-Reichl VM, Schmetterer KG, Mutschlechner S, Kueng HJ, Haiderer D, et al. Human TCR transgenic Bet v 1-specific Th1 cells suppress the effector function of Bet v 1-specific Th2 cells. J Immunol. 2011;187:4077–87. doi: 10.4049/jimmunol.1003220. [DOI] [PubMed] [Google Scholar]
- 38.Leb VM, Jahn-Schmid B, Schmetterer KG, Kueng HJ, Haiderer D, Neunkirchner A, et al. Molecular and functional analysis of the antigen receptor of Art v 1-specific helper T lymphocytes. J Allergy Clin Immunol. 2008;121:64–71. doi: 10.1016/j.jaci.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 39.Larche M, Akdis CA, Valenta R. Immunological mechanisms of allergen-specific immunotherapy. Nat Rev Immunol. 2006;6:761–71. doi: 10.1038/nri1934. [DOI] [PubMed] [Google Scholar]
- 40.Gomez G, Jogie-Brahim S, Shima M, Schwartz LB. Omalizumab reverses the phenotypic and functional effects of IgE-enhanced Fc epsilonRI on human skin mast cells. J Immunol. 2007;179:1353–61. doi: 10.4049/jimmunol.179.2.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Neerven RJ, van Roomen CP, Thomas WR, de Boer M, Knol EF, Davis FM. Humanized anti-IgE mAb Hu-901 prevents the activation of allergen-specific T cells. Int Arch Allergy Immunol. 2001;124:400–2. doi: 10.1159/000053770. [DOI] [PubMed] [Google Scholar]
- 42.Iyengar SR, Hoyte EG, Loza A, Bonaccorso S, Chiang D, Umetsu DT, et al. Immunologic effects of omalizumab in children with severe refractory atopic dermatitis: a randomized, placebo-controlled clinical trial. Int Arch Allergy Immunol. 2013;162:89–93. doi: 10.1159/000350486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Griffith Q, Liang Y, Whitworth P, Rodriguez-Russo C, Gul A, Siddiqui AA, et al. Immuno-evasive tactics by schistosomes identify an effective allergy preventative. Exp Parasitol. 2015;153:139–50. doi: 10.1016/j.exppara.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gauvreau GM, Harris JM, Boulet LP, Scheerens H, Fitzgerald JM, Putnam WS, et al. Targeting membrane-expressed IgE B cell receptor with an antibody to the M1 prime epitope reduces IgE production. Sci Transl Med. 2014;6:243ra85. doi: 10.1126/scitranslmed.3008961. [DOI] [PubMed] [Google Scholar]
- 45.Flicker S, Gadermaier E, Madritsch C, Valenta R. Passive immunization with allergen-specific antibodies. Curr Top Microbiol Immunol. 2011;352:141–59. doi: 10.1007/82_2011_143. [DOI] [PubMed] [Google Scholar]