Abstract
Background
The low-affinity receptor for IgE, FcεRII (CD23), contributes to allergic inflammation through allergen presentation to T cells, regulation of IgE responses, and enhancement of transepithelial allergen migration.
Objective
We sought to investigate the interaction between CD23, chimeric monoclonal human IgE, and the corresponding birch pollen allergen Bet v 1 at a molecular level.
Methods
We expressed 4 CD23 variants. One variant comprised the full extracellular portion of CD23, including the stalk and head domain; 1 variant was identical with the first, except for an amino acid exchange in the stalk region abolishing the N-linked glycosylation site; and 2 variants represented the head domain, 1 complete and 1 truncated. The 4 CD23 variants were purified as monomeric and structurally folded proteins, as demonstrated by gel filtration and circular dichroism. By using a human IgE mAb, the corresponding allergen Bet v 1, and a panel of antibodies specific for peptides spanning the CD23 surface, both binding and inhibition assays and negative stain electron microscopy were performed.
Results
A hitherto unknown IgE-binding site was mapped on the stalk region of CD23, and the non–N-glycosylated monomeric version of CD23 was superior in IgE binding compared with glycosylated CD23. Furthermore, we demonstrated that a therapeutic anti-IgE antibody, omalizumab, which inhibits IgE binding to FcεRI, also inhibited IgE binding to CD23.
Conclusion
Our results provide a new model for the CD23-IgE interaction. We show that the stalk region of CD23 is crucially involved in IgE binding and that the interaction can be blocked by the therapeutic anti-IgE antibody omalizumab.
Keywords: CD23, allergy, IgE, low-affinity IgE receptor, B cell, allergen
IgE-associated allergy is the most frequent immunologically mediated hypersensitivity disease, affecting more than 25% of the population worldwide.1 IgE is the least abundant immunoglobulin class and therefore exerts its pathologic effects mainly through interaction with 2 cellular receptors: the high-affinity receptor FcεRI and the low-affinity receptor CD23 (FcεRII).2 The molecular and structural interaction between IgE and FcεRI has been investigated in great detail3 and represents an important target for therapeutic strategies for allergy.4–6
The interaction between IgE and the low-affinity receptor CD23 is less well understood than that between IgE and the high-affinity receptor FcεRI, although CD23 plays several important roles in allergic inflammation. CD23 is a key molecule in IgE-facilitated allergen presentation and subsequent activation of allergen-specific T cells,7 a mechanism that can be blocked by allergen-specific IgG after specific immunotherapy.8 Furthermore, CD23 was suggested to be important in the regulation of IgE production,9 and there is growing evidence that CD23 mediates transport of allergens through respiratory and gut epithelial barriers.10,11
Thus far, only parts of the 3-dimensional structure (ie, parts of the head domain) of CD23 have been analyzed by using crystallography12 and nuclear magnetic resonance analysis,13 and results were not in full agreement. By means of site-directed mutagenesis of the head domain and subsequent expression of the mutants in cells, evidence for involvement of the head domain in IgE binding was provided.14 These findings were supported by structural studies that have analyzed the interaction between an incomplete recombinant CD23 head domain expressed in Escherichia coli and a recombinant Cε3-Cε4–comprising subfragment of the Fc part of human IgE.15 One study observed enhanced IgE binding of CD23 variants containing the stalk region compared with CD23 variants representing only the head domain,16 and another study observed enhanced IgE binding of a CD23 variant containing a single amino acid exchange in the stalk region.17 Both attributed the enhanced IgE binding to the fact that the stalk region can contribute to IgE binding through accurate formation of CD23 trimers because the current assumption is that CD23 occurs in trimeric form on the cell surface.
In this study we expressed 4 monomeric folded CD23 variants in insect cells, the first comprising the full extracellular portion of CD23, including the stalk and head domain; the second identical with the first, apart from a single amino acid exchange in the stalk region abolishing the only N-linked glycosylation site of CD23; a third representing the full head domain; and a fourth consisting of a truncated head domain. We were able to directly map hitherto unknown IgE-binding sites on the stalk region of CD23 and could show that a non–N-glycosylated monomeric version of CD23 bound IgE much stronger than glycosylated CD23. Furthermore, we demonstrate that omalizumab, a therapeutic antibody that inhibits IgE binding to FcεRI, also inhibits IgE binding to CD23.
Methods
Construction and recombinant expression of CD23 derivatives in SF9 insect cells
Four constructs of the human low-affinity IgE receptor FcεRII/CD23 (National Center for Biotechnology Information reference sequence NP_001993.2) were expressed in insect cells based on synthetic genes. CD23A represented the full extracellular CD23 protein starting from aspartic acid 48 to the end of the protein (ie, S321). CD23B was identical to CD23A, but asparagine 63 (N63) was substituted by a glutamine residue (N63 → glutamine 63 [Q63]). CD23C represented only the head part of the protein (ie, from methionine 150 to serine 321), and CD23D consisted of a small head part starting at serine 156 (S156) and ending at glutamic acid 298 (E298). Further description of the expression of CD23 derivatives can be found in the Methods section in this article’s Online Repository at www.jacionline.org.
A detailed description of purification and biochemical and biophysical characterization of recombinant CD23 constructs, synthesis of CD23 peptides and generation of CD23 peptide-specific antibodies, and expression and purification of human Bet v 1–specific monoclonal IgE can be found in the Methods section in this article’s Online Repository.
Binding experiments with recombinant CD23 derivatives and statistical analysis
Binding of antibodies specific for CD23 peptides and binding of an anti-CD23 mAb (Isotype M-L233; BD Biosciences, San Jose, Calif) to the CD23 derivatives were tested by using ELISA. A detailed description of the binding experiments can be found in the Methods section in this article’s Online Repository.
Statistical analysis of the ELISA results was done by calculating the Student t test for independent means with the program IBM SPSS Statistics 23.0. Coefficients were considered significant at a P value of less than .05.
A detailed description of binding and inhibition experiments in a cell-based model and the negative stain electron microscopy methods used can be found in the Methods section in this article’s Online Repository.
CD23 structural calculations and predictions
Relative surface exposures were calculated with the MSMS package18 in the USCF Chimera package19 and with Gly-X-Gly tripeptides20 as reference states. Graphic depiction of the CD23 head was rendered with PyMOL (PyMOL Molecular Graphics System, Version 1.7.4; Schrödinger, NY). The structural basis for the calculation and depiction was the PDB model 2H2T.12 The secondary structure prediction was performed with the Psipred server.21,22
Results
Expression and purification of folded and monomeric CD23 derivatives
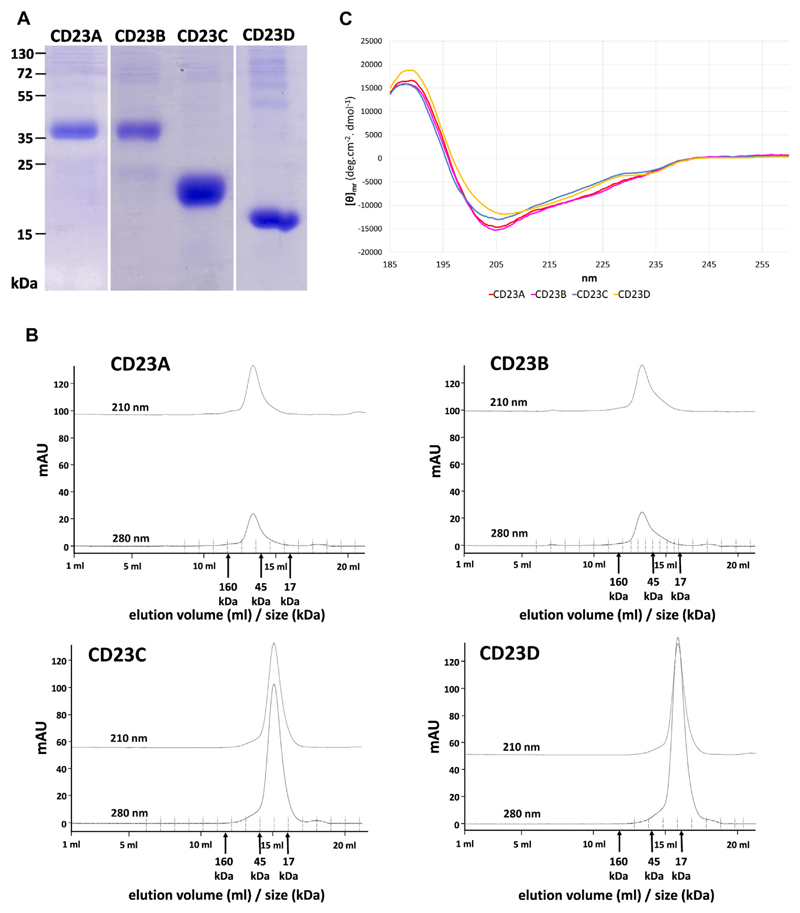
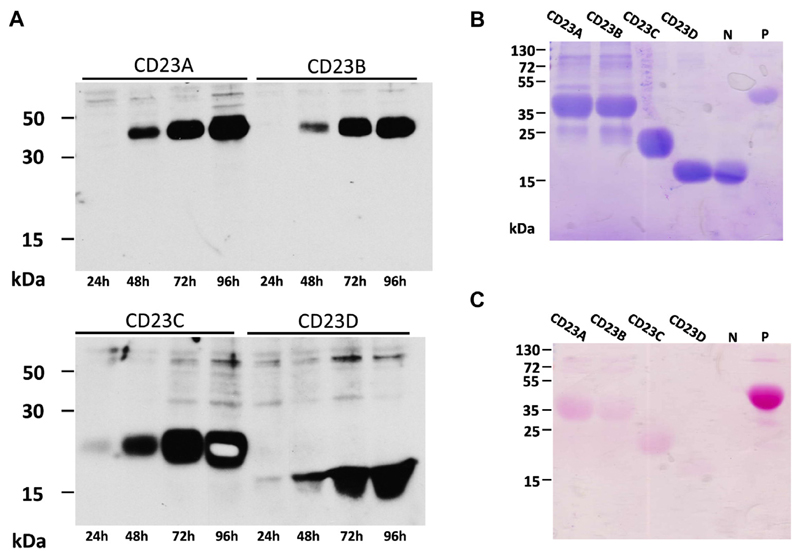
Four different CD23 constructs spanning different regions of CD23 were expressed in SF9 insect cells (Fig 1). Two constructs (ie, CD23A and CD23B) represented the complete stalk and head region of CD23 and differed by a single amino acid exchange (ie, N63 to Q63), which eliminated the only N-linked glycosylation site of CD23 (Fig 1, A and C). The other 2 constructs represented the head domain of CD23, for which the 3-dimensional structures alone (CD23C and CD23D) and in complex with IgE (CD23D) have been reported (Fig 1).12,13,15 CD23D, for which the structure in complex with IgE has been reported, comprises amino acids 156 to 298 and lacks 6 amino acids at the N-terminal end compared with CD23C, as well as 23 amino acids at the C-terminal end (Fig 1). The 4 recombinant CD23 derivatives were expressed with a C-terminal hexahistidine tag in baculovirus-infected insect cells as soluble proteins and could be detected with an anti–his-tag antibody in supernatants from infected cells (see Fig E1, A, in this article’s Online Repository at www.jacionline.org). The molecular masses of the expressed and purified CD23 derivatives observed in SDS-PAGE and after blotting (Fig 2, A; and see Fig E1, A and B) corresponded to those determined by using mass spectrometry (MALDI; CD23A, 33.7 kDa; CD23B, 33.4 kDa; CD23C, 24.7 kDa; and CD23D, 16.8 kDa; data not shown). Only CD23A and CD23B appeared larger (ie, approximately 37 kDa) in SDS-PAGE, which has been reported earlier (see Fig E1, B).23 Compared with a glycosylated control protein containing 9 N-linked glycosylation sites (ie, horseradish peroxidase [HRP]), the CD23 derivatives showed weak reactivity when stained for carbohydrates, with CD23A producing the strongest staining of the 4 constructs, indicating that the mutation of N63 to Q63 has successfully removed the carbohydrates attached to this site. No carbohydrate staining was observed for a nonglycosylated control protein (ie, soybean trypsin inhibitor; see Fig E1, C, lane N).
Fig 1. Overview of CD23 constructs.
A, Amino acid sequence of CD23. Indicated are the constructs CD23A, CD23B, CD23C, and CD23D; peptides (P1-P7) used to raise specific antisera (brown bars); N-linked glycosylation sites (red letters); and secondary structure and surface exposure (blue bars) of amino acids, as derived from the crystal structure PDB:2H2T (Wurzburg et al12). A region in which no data are available (R253-E257) is indicated by a gray bar. Amino acids involved in IgE binding according to Hibbert et al13 and Dhaliwal et al15 are shown in green and blue, respectively. Residues involved in IgE binding according to both publications are shown in orange. Cysteine residues involved in disulfide bond formation are numbered. CD23 domains are shaded (intracellular domain: red; transmembrane domain: yellow; stalk domain: green; head domain: purple). B, Schematic representation of the known (Wurzburg et al12) and predicted 3-dimensional structure of CD23. CD23 peptides are indicated. Data for region R253-E257, as well as for the C-terminal end, are not available. C, Sketch of the recombinant CD23 constructs (CD23A, CD23B, CD23C, and CD23D), as cloned into plasmid pTM1 with a C-terminal hexa-histidine tag (6xHis) and an N-terminal melittin signal sequence. Amino acids at the N- and C-terminal ends of CD23 and one mutated in CD23B are indicated in black.
Fig 2. Biochemical, biophysical, and immunologic characterization of recombinant CD23 constructs.
A, Coomassie-stained gel containing aliquots of purified CD23 constructs. B, Gel filtration profile of the 4 CD23 constructs. Adsorptions at 210 and 280 nm (y-axes, milliabsorbance units [mAU]) recorded during elution (x-axes, elution volumes in milliliters and corresponding protein size ranges in kilodaltons) are shown. C, Circular dichroism analysis of the 4 CD23 constructs. An overlay of the 4 circular dichroism spectra of constructs CD23A to CD23D is shown. CD23A, red; CD23B, pink; CD23C, blue; CD23D, yellow. X-axis, wavelength [in nanometers]; y-axis, [θ]mr (mean-residue molar ellipticity).
All 4 recombinant CD23 derivatives could be purified (Fig 2, A) and, when studied by using size exclusion chromatography, were found to occur in a strictly monomeric form (Fig 2, B). Circular dichroism analyses revealed that all 4 proteins were folded (Fig 2, C) and consisted of a mixture of α-helices (CD23A, 17%; CD23B, 19%; CD23C, 12%; and CD23D, 12%), β-sheets (CD23A, 28%; CD23B, 27%; CD23C, 33%; and CD23D, 34%), turns (CD23A, 24%; CD23B, 24%; CD23C, 25%; and CD23D, 23%), and unordered parts (CD23A, CD23B, and CD23D, 31%; CD23C, 30%; data not shown). Thus we were able to assemble a set of 4 folded and monomeric CD23 derivatives representing different parts of CD23, with and without the N-linked glycosylation site for functional assays.
CD23 derivatives containing the stalk region and in particular the nonglycosylated CD23B variant show superior IgE binding
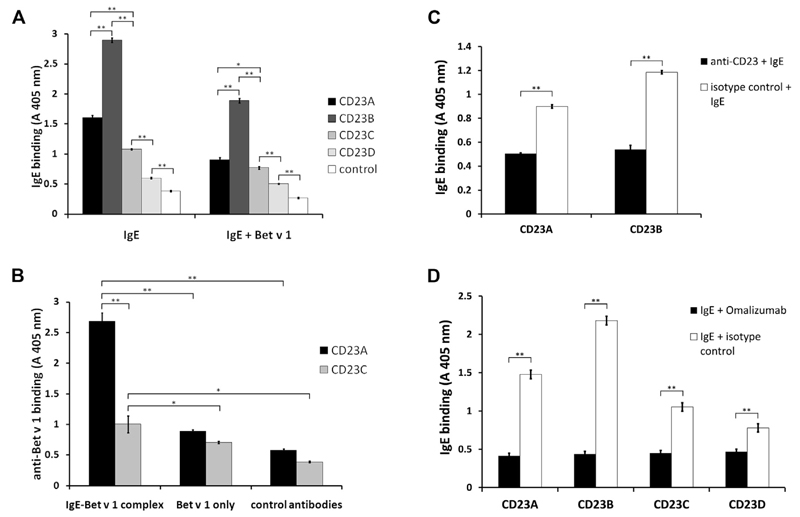
In a first set of experiments, we investigated the binding of IgE and IgE-allergen immune complexes to the CD23 derivatives. For this purpose, the CD23 variants were immobilized to ELISA plates and incubated with a human IgE mAb specific for the major birch pollen allergen Bet v 1 alone or in complex with Bet v 1 (Fig 3). When IgE (Fig 3, A, left panel) or IgE in complex with Bet v 1 (Fig 3, A, right panel) bound to the CD23 derivatives was detected with anti-human IgE antibodies, we observed that the CD23 derivatives containing the stalk region (ie, CD23A and CD23B) bound IgE stronger than the CD23 derivatives without the stalk region (ie, CD23C and CD23D). Interestingly, the strongest IgE binding was obtained for CD23B, which had the amino acid exchange in the N-linked glycosylation site (Fig 3, A). A similar result was obtained when the bound IgE–Bet v 1 complexes were detected with antibodies specific for the allergen. As exemplified for CD23A and CD23C, the CD23 construct containing the stalk region bound the IgE-allergen complexes much better than the construct containing only the head domain (Fig 3, B). The latter experiment also confirmed the correct formation of the IgE-allergen complexes and their binding to the CD23 derivatives because the complex was detected by anti–allergen-specific antibodies.
Fig 3. Binding of IgE to CD23.
A, Varying binding of IgE to the CD23 constructs. ELISA plates were coated with recombinant CD23 constructs A through D or a control protein and incubated with either IgE alone (IgE) or IgE-Bet v 1 complexes (IgE+Bet v 1). Absorbance values (A 405 nm) corresponding to bound IgE are indicated on the y-axis. B, Binding of the IgE–Bet v 1 complexes to CD23A and CD23C. For control purposes, IgE was omitted (Bet v 1 only) or rabbit antibodies without specificity for Bet v 1 (control antibody) were used. Absorbance values (A 405 nm) corresponding to bound Bet v 1 detected with rabbit anti–Bet v 1 antibodies are indicated on the y-axis. Error bars show SDs of triplicates. C and D, Binding of monoclonal IgE to recombinant CD23 constructs can be blocked with α-CD23 (Fig 3, C) or omalizumab (Fig 3, D). In Fig 3, C, ELISA plates with bound CD23A and CD23B were preincubated with α-CD23 (black bars) or with an isotype control (white bars) and incubated with IgE. In Fig 3, D, IgE was preincubated with omalizumab (black bars) or an isotype control (white bars) before incubation with CD23 constructs (x-axis: CD23A, CD23B, CD23C, and CD23D). Absorbance values (A 405 nm) corresponding to bound IgE are indicated on the y-axis. Error bars show SDs of triplicates. *P < .05 and **P < .01, 2-tailed test.
The specificity of IgE binding to the CD23 derivatives was further investigated by using an anti-CD23 mAb that inhibits IgE binding to CD23. We found that this antibody, but not an isotype-matched control antibody, inhibited IgE binding to CD23A and CD23B (Fig 3, C). Next, we investigated whether omalizumab, a registered therapeutic anti-human IgE antibody, is able to block IgE binding to CD23. Using the 4 recombinant CD23 constructs, we were indeed able to show that preincubation of IgE with omalizumab inhibited IgE binding to CD23 compared with an isotype-matched control antibody (Fig 3, D).
Mapping of direct IgE binding sites in the CD23 stalk region with peptide-specific antibodies
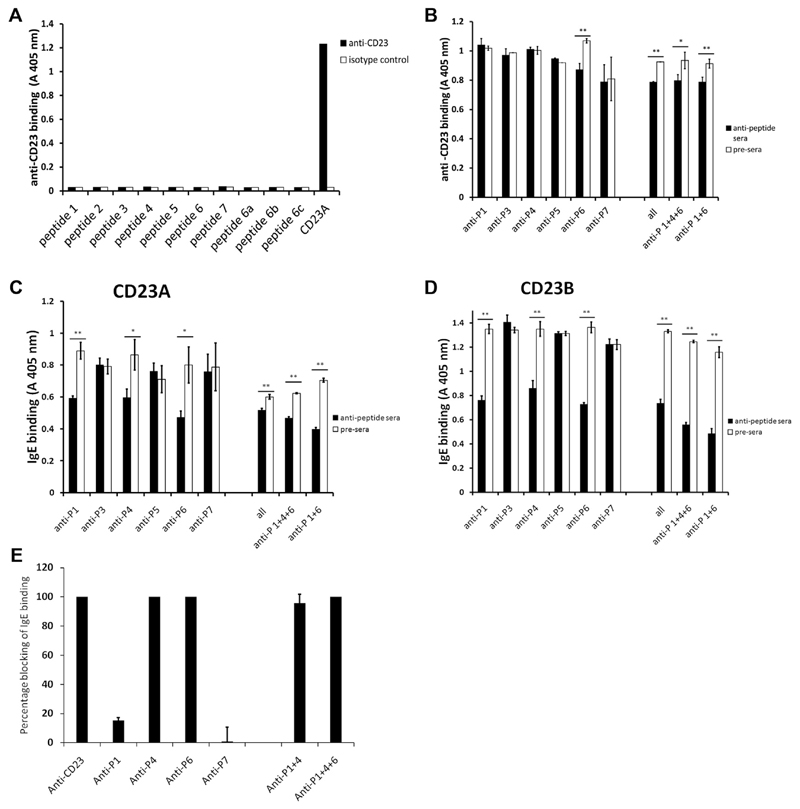
To investigate the involvement of different portions of CD23 in IgE binding, we produced a set of synthetic CD23 peptides (see Table E1 in this article’s Online Repository at www.jacionline.org) derived from different regions in the CD23 molecule. The peptides in the CD23 sequence (Fig 1, A) were located in the transmembrane region (ie, peptide 2) or in the stalk region (ie, peptides 1 and 4), comprised several portions of the head domain (ie, peptides 5, 6 and 7), and included the C-terminus of CD23 (ie, peptide 3). Three shorter versions of peptide 6 (peptides P6a, P6b, and P6c; see Table E1) were synthesized for direct binding experiments with anti-CD23 antibodies. Peptides 1 to 6 were then used to immunize rabbits to generate anti-sera that should be capable of targeting the corresponding sequences in the intact folded CD23 molecule. In a first set of experiments, the resultant antibodies were used as probes to study surface exposure of certain portions of CD23. When we tested the anti-peptide antibodies for reactivity with the folded CD23 derivatives, we found that antibodies raised against peptide 2, primarily including the transmembrane domain, did not bind to the 4 constructs because the peptide was either not present (ie, CD23C and CD23D) or only partially represented (ie, CD23A and CD23B) in the CD23 derivatives (see Fig E2 in this article’s Online Repository at www.jacionline.org). Antibodies raised against peptides 1 and 4 reacted specifically with the constructs containing the stalk region (ie, CD23A and CD23B) but not with the derivatives lacking the stalk region (ie, CD23C and CD23D; see Fig E2). Interestingly, only antibodies raised against peptides 6 and 7 from the head domain, but not peptide 5–specific antibodies, reacted with the CD23 constructs. Because peptide 5–specific antibodies reacted with the isolated peptide 5, lack of reactivity is not due to the failure of obtaining peptide-specific antibodies (data not shown). Anti-peptide 3 antibodies reacted specifically with each of the constructs containing the CD23 C-terminal end (ie, CD23A, CD23B, and CD23C) but not with the construct lacking the C-terminal end (CD23D, see Fig E2). All constructs were recognized by the anti-CD23 mAb, which inhibited IgE binding to CD23 (positive control, see Fig E2).
Next, we investigated whether the anti-CD23 mAb that inhibited IgE binding to CD23 reacts with the synthetic CD23 derived peptides (see Table E1) and found that this antibody only reacted with folded CD23 but not with any of the peptides (Fig 4, A). We then tested whether the peptide-specific antibodies inhibit binding of the anti-CD23 mAb. We observed a weak inhibition with anti–peptide 6 antibodies, as well as with a mix of all anti-peptide antisera or other combinations containing anti–peptide 6 (ie, anti–peptide 1, 4, and 6 antibodies or anti–peptide 1 and 6 antibodies in combination; Fig 4, B). These experiments suggest that the anti-CD23 mAb recognizes an epitope close to the peptide 6 amino acid sequence. Furthermore, it likely binds with higher affinity to CD23 than the peptide-specific antibodies because the inhibition by anti–peptide 6 antibodies was weak (ie, 18.5% inhibition of binding; data not shown).
Fig 4. Blocking ability of CD23 peptide-specific antibodies.
A, Ability of monoclonal anti-CD23 to bind to different anti-CD23 peptides. CD23A was used as a positive control. B, Influence of anti-CD23 peptide antibodies on binding of monoclonal anti-CD23. CD23A was preincubated with anti-peptide antibodies or the corresponding preimmune sera (x-axis) and then incubated with the anti-CD23 antibody. Absorbance values (A 405 nm: y-axes) correspond to bound anti-CD23 antibody. Error bars show SEs of at least triplicates. The experiments were repeated 3 times with similar results. C and D, Influence of anti-CD23 peptide antibodies on the binding of IgE to CD23A (Fig 4, C) and CD23B (Fig 4, D). CD23 constructs were preincubated with anti-peptide antibodies or the corresponding preimmune sera (x-axes) and then incubated with IgE. Absorbance values (A 405 nm: y-axes) correspond to bound IgE. Error bars show SDs of triplicates. E, Influence of antibodies on IgE binding to CD23-expressing B cells. Shown is the percentage blocking of IgE binding (y-axis) to CD23-expressing B cells by an anti-CD23 mAb and by antibodies specific for CD23 peptides (x-axis), as determined by using flow cytometry. Error bars show SDs of triplicates. The experiment was performed twice with similar results. *P < .05 and **P < .01, 2-tailed test.
We then used the peptide-specific antibodies to block IgE binding to CD23 (Fig 4, C and D). We found that the antibodies against peptides 1 and 4 from the stalk region inhibited IgE binding to the CD23A and CD23B variants (Fig 4, C and D). Inhibitions of IgE binding by antibodies specific for peptide 1 were 29% for CD23A and 39% for CD23B. Peptide 4 antibodies were able to inhibit IgE binding to CD23A by 20% and to CD23B by 31% (data not shown). Likewise, antibodies specific for peptide 6 from the head domain partially inhibited IgE binding to both CD23 variants (CD23A, 32%; CD23B, 44%; data not shown), whereas the other peptide antibodies had no effect on IgE binding. Inhibition of IgE binding to CD23A and CD23B was also observed with combinations of anti–peptide 1, 4, and 6 (CD23A, 25%; CD23B, 55%; data not shown) and was even more pronounced when a combination of anti–peptide 1 and 6 antibodies was used (CD23A, 43%; CD23B, 58%; data not shown; Fig 4, C and D). These experiments show that the binding of IgE to monomeric CD23 can be inhibited with antibodies directed against 2 distinct peptides in the CD23 stalk region, which demonstrate a direct involvement of the stalk region in IgE binding. Our experiments also confirm that amino acids in the peptide 6–defined region on the head domain are involved in IgE binding (Fig 1, A). Interestingly, anti–peptide 7 antibodies did not inhibit IgE binding to CD23, although this peptide contained amino acids that were reported to be directly involved in IgE binding (Fig 1, A).13,15 The lack of inhibition of IgE binding by anti–peptide 5 antibodies can be explained by the fact that these antibodies did not react with folded CD23.
Results obtained for the protein interactions could be confirmed in cellular experiments. Antibodies specific for peptides 4 and 6 inhibited IgE binding to B cells expressing CD23 (Fig 4, E). A weak inhibition was also obtained with anti–peptide 1 antibodies but not with anti–peptide 7 antibodies (Fig 4, E).
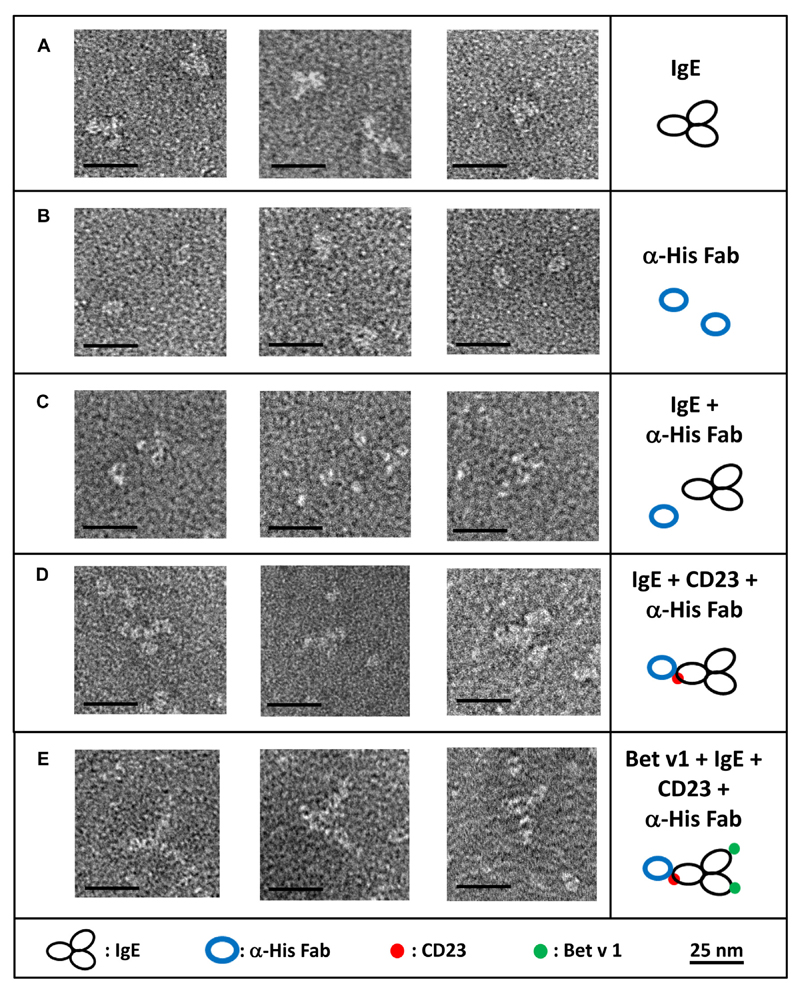
Visualization of the trimolecular complex consisting of CD23A, IgE, and allergen by using negative stain electron microscopy
Negative stain electron microscopy allows us to directly visualize the interaction of molecules.24,25 It has been used successfully to visualize human IgE alone and in complex with FcεRI and an anti-human IgE antibody.26,27 We used CD23A together with a human IgE mAb specific for the major birch pollen allergen Bet v 1 and the Bet v 1 allergen to visualize complexes of the 3 molecules (Fig 5). The challenge regarding visualization of the interaction of CD23 with IgE and the Bet v 1 allergen is that CD23 (33.7 kDa) and Bet v 1 (17 kDa) are very small molecules compared to IgE (190 kDa). Therefore we used an anti-His-Fab, which has a size and shape (ie, donut like shape) distinct from IgE, to investigate the trimolecular interaction. In Fig 5, A, monoclonal Bet v 1–specific IgE alone is shown. The 3 domains (Fc arm and 2 Fab arms) of the molecules can be seen; however, differentiation of Fc from Fab is not possible. Fig 5, B, shows the donut-shaped anti–His-Fab fragment by itself. In Fig 5, C, we added the anti–His-Fab fragment to the IgE preparation. The Fab fragment is not able to bind to the IgE and is therefore separate from IgE. However, when CD23A was added to the preparation, complexes consisting of 1 IgE molecule and 1 CD23 molecule, as indicated by the anti–His-Fab, became visible (Fig 5, D). These complexes were a minority in the preparation; however, they were only seen when CD23 was added compared to control results. A possible explanation for the relatively small number of complexes observed in the mixes could be weak affinity of the IgE-CD23 complex. We then additionally added Bet v 1 allergen to the anti–His-Fab–CD23–IgE complex and observed an increase of the size of the previously observed complexes (Fig 5, E). Interpretation of the samples was challenging because several possible complexes exist in one sample. In addition to the anti–His-Fab–CD23–IgE–Bet v 1 complex, IgE–Bet v 1, IgE–CD23–Bet v 1, anti–His-Fab–CD23–IgE, anti–His-Fab– CD23, and CD23-IgE complexes are also possible, as well as each of the added molecules by themselves. However, again, even though they were a minority, large anti–His-Fab–CD23– IgE–Bet v 1 complexes could be observed repeatedly and only in CD23-containing preparations compared to controls. Their sizes and shapes are best explained by a 1:1 interaction between CD23 and IgE, indicating that CD23 interacts as a monomer with IgE in this experimental setup.
Fig 5. Negative stain electron microscopy of recombinant CD23 in complex with IgE and allergen.
Shown are 3 representative negative stain electron microscopy images and interpretive sketches of IgE alone (A), an anti-His IgG Fab fragment (B), a mix of IgE plus anti-His IgG Fab fragment (C), IgE-CD23 complexes marked with an anti-His IgG Fab fragment (D), and IgE-CD23-allergen complexes marked with an anti-His IgG Fab fragment (E). Black bars represent 25 nm.
Discussion
The IgE-CD23 interaction has regained considerable attention because it contributes to allergic inflammation through IgE-facilitated allergen presentation to T cells and enhancement of transepithelial allergen migration.7,10,11 Our analysis of the interaction between IgE and CD23 has revealed several novel findings that can change current concepts of the interaction of these molecules and in addition might be of relevance for therapeutic approaches for IgE-associated allergy. We expressed and purified a set of folded and monomeric CD23 protein derivatives differing in size and N-linked glycosylation to conduct a series of functional assays investigating the binding of the trimolecular complex between CD23, IgE, and a corresponding allergen.
Direct IgE-binding experiments showed that monomeric CD23 containing the stalk region exhibited superior IgE binding compared with CD23 derivatives without the stalk region, which indicated a direct involvement of the stalk region in IgE binding. This finding was unexpected because thus far, it has been thought that the CD23 stalk region is important for the trimerization of CD23 and that this trimeric state indirectly enhances IgE binding to CD23.16,28 However, our data demonstrate that CD23 derivatives containing the stalk region are monomeric and can bind IgE. According to preliminary plasmon surface resonance (GE Healthcare Biacore, Uppsala, Sweden) experiments, the affinity of the monomeric protein to IgE was similar to that described in earlier studies.13 Further support for a 1:1 binding of monomeric CD23 to IgE and IgE-allergen complexes came from negative stain electron microscopy experiments.
Another interesting observation was that a single amino acid exchange in the only N-linked glycosylation site of CD23 strongly enhanced IgE binding. In fact, it has been reported that single nucleotide polymorphisms in this region were related to the development of asthma, and it has been observed that cells expressing CD23 variants containing an amino acid exchange close to but not within the N-linked glycosylation site (ie, R62-W62) showed better IgE binding.17 The authors who made this observation considered that this exchange might have affected the trimerization of CD23 on the cell surface and thus might have caused increased IgE binding. However, our experiments demonstrate that the exchange directly enhanced the binding of monomeric CD23 to IgE. The enhancement of IgE binding by the mutation in the glycosylation site might be due to the removal of sugar residues hindering IgE binding. Alternatively, it is possible that mutations of certain critical amino acids might improve the affinity of CD23 for IgE. Thus it is quite possible that the binding of IgE to CD23 can be regulated in cells through point mutations through varying glycosylation and/or altered affinity. Hence the reported nucleotide polymorphisms in the DNA encoding the stalk region might directly affect the affinity for IgE binding and thus result in different susceptibilities for asthmatic patients. According to this model, atopic patients expressing less glycosylated CD23 might bind IgE better through CD23 and, consequently, exhibit more severe forms of allergic inflammation because CD23 is important for IgE-facilitated allergen presentation, leading to activation of allergen-specific T-cell and cytokine responses.
Therefore we tested our hypothesis of a critical and direct stalk involvement of CD23 in binding its ligand IgE through inhibition experiments. By using peptide-specific antibodies, binding sites previously identified by structural studies could be confirmed. For example, site-directed mutagenesis in the peptide 6 region in the head domain of CD23 was able to affect IgE binding to the receptor molecule.14 However, we did not observe a blocking effect by anti–peptide 7 antibodies, even though certain amino acids contained in peptide 7 were described to be directly involved in IgE binding.13,15 Peptide 5–specific antibodies did not bind to the CD23 constructs. Because these antibodies reacted with the isolated peptide 5, lack of reactivity is not due to failure of obtaining peptide-specific antibodies but could be a result of a lack of surface exposure of the amino acids contained in peptide 5 on the folded CD23 molecules. In fact, according to the 3-dimensional structure determined for the head domain,12 several amino acids included in peptide 5 are not exposed on the surface of the CD23 molecule. Unlike in peptides 6 and 7, no exposed segment comprising several contiguous amino acids can be found in peptide 5 (Fig 1, A). We also noted that certain amino acids throughout the CD23 head domain (eg, W184, L198, I221, G222, and A279), which have been described as contact residues for the IgE interaction,13,15 were not found to be surface exposed in the 3-dimensional structure determined for the head domain.12
Importantly, antibodies against peptides 1 and 4, which are both located in the stalk region, strongly inhibited IgE binding to CD23 in protein interaction assays and by using cultured cells expressing CD23. Because the CD23 molecules used in the molecular inhibition assays represented monomeric proteins, an indirect involvement of the stalk region by formation of CD23 oligomers with enhanced IgE binding can be excluded. Thus these experiments unambiguously mapped IgE-binding sites on the CD23 stalk region.
Taken together, our experimental data with blocking antibodies identified antibodies against 2 distinct regions in the stalk domain of CD23, which can shield CD23 from IgE binding. However, it is known that cleavage of the head domain from CD23-expressing cells abolishes IgE binding,29 and therefore one must assume that the isolated stalk alone does not bind IgE but rather cooperates with the head in IgE binding. Nevertheless, our observations point to a direct and critical involvement of the CD23 stalk domain in IgE binding and provide a novel view regarding the molecular mode of IgE binding to CD23.
Using the molecular model of the CD23-IgE interaction, we made another observation of high clinical relevance. We found that the therapeutic anti-human IgE antibody omalizumab was also able to inhibit IgE binding to CD23. In fact, it has been hypothesized previously that omalizumab, which inhibits binding of IgE to FcεRI might also affect IgE binding to CD23 and thus could offer additional benefit to patients through an effect that goes beyond inhibition of allergen-induced mast cell and basophil degranulation, such as IgE-facilitated allergen presentation.6,7,30 Recently, the 3-dimensional structure of the omalizumab Fab has been determined by using x-ray crystallography.31 Based on in silico docking experiments, it has been proposed that omalizumab can inhibit IgE binding to CD23,32 but thus far, no definitive experiments have been performed to investigate this possibility. Our experiments now unequivocally demonstrate for the first time in a defined molecular experimental setup that the therapeutic anti-human IgE antibody omalizumab inhibits IgE binding to CD23 and thus has the potential to inhibit CD23-mediated allergic inflammation as well.
In summary, our studies provide a new molecular model for the CD23-IgE-allergen interaction and conceivably might open new doors for the treatment of IgE-associated allergy based on the interference of this interaction.
Methods
Construction and recombinant expression of CD23 derivatives in SF9 insect cells
Each of the 4 constructs contained a C-terminal 6x histidine tag to facilitate purification. Synthetic genes coding for the constructs were codon optimized for insect cell expression (GenScript, Piscataway, NJ) and cloned into the polyhedrin promoter–harboring expression vector pTM1E1 through BamHI and SmaI sites. DNA sequences and their correct insertion were verified by using double-strand sequencing with the primers CGCCCAGGACTCTAGC TATAG (forward) and TGATGATGATGATGATGG (reverse). The melittin sequence of the pTM1 vector ensures export of the protein into the culture supernatant, whereby the export sequence is cleaved off, leaving 2 additional amino acids (aspartic acid and proline) at the N-terminal end of each protein. For generation of recombinant baculovirus harboring CD23 cDNA and subsequent expression of the CD23 proteins in SF9 insect cells, the Bac-to-Bac TOPO expression system (Life Technologies, Carlsbad, Calif) was used, according to the manufacturer’s protocol.E2 The time course of expression was monitored in culture supernatants by using Western blotting. For this purpose, aliquots of the culture supernatants containing CD23 proteins were separated by means of SDS-PAGE and blotted onto a nitrocellulose membrane (Whatman, Little Chalfont, United Kingdom). Membranes were blocked with buffer A (Tris-buffered saline containing 0.05% Tween 20) containing 5% wt/vol milk powder (AppliChem, Darmstadt, Germany) for 1 hour at room temperature, washed 3 times for 10 minutes with buffer A, and incubated with a 1:4000 in buffer A diluted HRP-labeled anti–His-tag antibody (Thermo Scientific, Waltham, Mass) for 1 hour at room temperature. Bound antibodies were visualized by using chemiluminescence with an ECL detection kit (Bio-Rad Laboratories, Hercules, Calif) and exposure of blots to x-ray films (Kodak, Rochester, NY).
Purification and biochemical and biophysical characterization of recombinant CD23 constructs
Protein-containing culture supernatants were concentrated by using an Amicon concentration cell (Millipore, Darmstadt, Germany), and recombinant proteins were bound through the His-tag to a Co2+-containing column (ClonTech, Mountain View, Calif), eluted with 150 mmol/L imidazole, and dialyzed against 10 mmol/LTris (pH 8) containing 100 mmol/L NaCl. Protein aliquots were stored at −80°C. The purity and identity of the recombinant proteins was checked by using SDS-PAGE, followed by Coomassie Blue staining and by immunoblotting with the anti–His-tag antibody or rabbit anti–peptide 6 antibodies (1:10,000, Table E1). Bound rabbit antibodies were detected with HRP-labeled goat anti-rabbit antibodies (GE Healthcare, Chalfont St Giles, United Kingdom) diluted 1:50,000 in buffer and ECL detection. Glycoprotein staining of an SDS-PAGE gel containing CD23 derivatives was performed with the Pierce glycoprotein staining kit (Pierce, Waltham, Mass), according to the manufacturer’s instructions.
Purified CD23 proteins in 10 mmol/L sodium phosphate (pH 7) were used to acquire mass spectra by using MALDI-TOF (Microflex, Bruker, Billerica, Mass). To this end, the samples were mixed at a 1:1 ratio with matrix (sinapinic acid dissolved in 60% acetonitrile and 0.1% trifluoroacetic acid), spotted onto the target, and air-dried.E2 Folding of the recombinant proteins was analyzed by using circular dichroism spectroscopy on a Jasco J-810 spectropolarimeter (Japan Spectroscopic, Tokyo, Japan) with a 1-mm path length cell at room temperature and a wavelength range from 185 to 260 nm at a resolution of 1 nm at 20 nm/min scanning speed and a 1-second response time.E3 Data of 3 measurements were averaged. The final spectra were baseline corrected, and results were expressed as the mean-residue molar ellipticity ([θ]mr) at a given wavelength. The secondary structure content of the CD23 derivatives was calculated by using the secondary structure estimation program CDSSTRE4 and the reference set 4.E5
Recombinant CD23 constructs were checked for possible aggregation by means of gel filtration/size exclusion chromatography.E3 Aliquots of 1 mL containing the different CD23 constructs (250-600 μg/mL in buffer B: 20 mmol/L Tris buffer [pH 7.4] and 150 mmol/L NaCl) were loaded on a Superdex 200 10/300 GL column (GE Healthcare, Uppsala, Sweden) for gel filtration. For gel filtration, buffer B was used at a flow rate of 0.5 mL/min at 4°C. Absorbance was monitored at 210 and 280 nm. Gel filtration standard proteins (Bio-Rad Laboratories: thyroglobulin, 670 kDa; bovine gamma globulin, 158 kDa; chicken ovalbumin, 44 kDa; equine myoglobin, 17 kDa; and vitamin B12, 1.35 kDa) were run under similar conditions to determine the constructs’ size.
Synthesis of CD23 peptides and generation of CD23 peptide-specific antibodies
CD23 peptides (Table E1) were synthesized by using the Fmoc (9 fluorenyl methoxy carbonyl) strategy with HBTU (2-[1H-benzotriazol-1-yl]1,1,3,3 tetramethyluronium hexafluorophosphate) activation (0.1 mmol small-scale cycles) on an Applied Biosystems (Foster City, Calif) Peptide Synthesizer Model 433A, as previously described.E6 Cysteine residues were added to peptides P1, P2, P3, P4, and P6, as indicated in Table E1, whereas peptides P5 and P7 contained cysteine residues. The short peptides P6a, P6b, and P6c were made without modification. Peptides were purified to greater than 90% purity by means of preparative HPLC, and their identity was confirmed by using mass spectrometry (piChem, Graz, Austria). Antibodies against KLH-coupled peptides (P1, P2, P3, P4, and P7; maleimide-activated KLH conjugation kit; Sigma-Aldrich, St Louis, Mo) or Blue Carrier Protein–coupled peptides (peptides P5 and P6; Imject EDC Blue Carrier Protein Spin kit, Thermo Scientific) were raised in rabbits (Charles River, Kisslegg, Germany).E6
Expression and purification of human Bet v 1–specific monoclonal IgE
Human monoclonal IgE specific for the major birch pollen allergen Bet v 1 was expressed, as previously described.E7 High-yield IgE production was obtained in a bioreactor (CELLine CL 350; Sartorius, Göttingen, Germany) with DMEM high-glucose media (Life Technologies) supplied with 5% ultralow IgG FBS (Life Technologies). Bet v 1–specific IgE was purified by using affinity chromatography with an anti-IgE mAb.E8
Binding experiments with recombinant CD23 derivatives and statistical analysis
ELISA plates (Nunc Maxisorp, Roskilde, Denmark) were coated with recombinant CD23 proteins dissolved in coating buffer (100 mmol/L sodium carbonate-bicarbonate [pH 9.3], c = 5 μg/mL; 50 μL/well). Plates were then washed with buffer C (PBS, 0.02% vol/vol Tween 20) and reacted with either the peptide-specific rabbit antisera and the corresponding preimmune sera as a control (diluted 1:1000) or with the anti-CD23 antibody (1:1000) and mouse IgG1 as control overnight at 4°C. Plates were again washed with buffer C. Bound rabbit antibodies were detected with HRP-labeled anti-rabbit IgG antibodies (GE Healthcare) diluted 1:5000 and ABTS (2,2’-azino-bis[3-ethylbenzothiazoline-6-sulphonic acid]) as a substrate. Bound mouse antibodies were detected with an HRP-labeled anti-mouse IgG-ECL antibody (GE Healthcare) diluted 1:5000 and ABTS instead. Results represent means of triplicate determinations ± SD after subtraction of background, as determined with the preimmune sera and isotype control, respectively.
Binding of human IgE and IgE-allergen immune complexes to CD23 derivatives was also investigated by using ELISA experiments. Plates were coated with equimolar amounts of the recombinant CD23 proteins dissolved in coating buffer (100 mmol/L sodium carbonate-bicarbonate [pH 9.3], c = 200 nmol/L; 50 μL/well). Plates were then washed with buffer C (PBS and 0.02% vol/vol Tween 20). Bet v 1–specific IgE (5 μg/mL in buffer C) alone or preincubated with rBet v 1 (5 μg/mL in buffer C; Biomay, Vienna, Austria) for 1 hour at 37°C was then added and incubated overnight at 4°C. After washing with buffer C, bound Bet v 1–specific IgE and IgE-allergen complexes were detected with biotin-labeled anti-human IgE antibodies (KPL, Gaithersburg, Md) at a dilution of 1:2500 in buffer C and incubated overnight at 4°C. After a further washing step, avidin–alkaline phosphatase (Sigma-Aldrich, Saint Louis, Mo) was added at a concentration of 1:5000 for 1 hour at room temperature. Plates were washed with buffer C, 4-nitrophenyl phosphate (Sigma-Aldrich) was added as a substrate, and the color reaction was read at 405 nm with an ELISA reader (Victor3; PerkinElmer, Waltham, Mass). Mean values of triplicate determinations ± SDs were calculated. In another set of experiments, bound IgE–Bet v 1 immune complexes were detected with rabbit anti–Bet v 1 antibodies (1:1000),E9 followed by HRP-labeled anti-rabbit IgG antibodies (GE Healthcare) diluted 1:5000 and ABTS as substrate. For control purposes, experiments were performed only with Bet v 1 without IgE and by using control rabbit antibodies without specificity for Bet v 1 instead of the rabbit anti–Bet v 1 antibodies.
The specificity of IgE binding to the CD23 derivatives was tested in a series of ELISA inhibition experiments. First, inhibition of IgE binding was investigated by preincubating plate-bound CD23 derivatives with the α-CD23 antibody (Isotype M-L233, BD Biosciences) at a concentration of 5 μg/mL or mouse IgG1 as a negative control for 2 hours at room temperature before IgE was added. Second, on another set of experiments, IgE was preincubated with 25 μg/mL omalizumab (Xolair; Novartis, Basel, Switzerland) or an isotype-matched human IgG1 (Sigma-Aldrich) at 378C for 1 hour before addition to plate-bound CD23 derivatives. Third, the ability of the anti-CD23 peptide antibodies to inhibit IgE binding to CD23 derivatives was investigated. For this purpose, plate-bound CD23 derivatives were incubated with the 1:50 diluted peptide-specific antibodies or the corresponding 1:50 diluted preimmune immunoglobulin for 2 hours at room temperature, washed, and then incubated with IgE.
We also tested whether the anti-CD23 mAb reacts with the synthetic CD23 peptides. Peptides were coated onto ELISA plates (5 μg/mL) and reacted with the 1:1000 diluted antibody or isotype control. Furthermore, it was studied whether the peptide-specific antibodies can inhibit binding of the mAb to CD23A. For this purpose, ELISA plate–coated CD23A was preincubated with the peptide-specific antibodies (1:50) or the corresponding preimmunoglobulin, and then binding of the mAb was assessed.
Binding and inhibition experiments in a cell-based model
EBV-transformed B cells (2 × 105 per condition) expressing high levels of CD23E10 were blocked with PBS containing 1% BSA (PAA, Pasching, Austria) and 10% goat serum (Sigma-Aldrich) for 20 minutes on ice. Then cells were washed (PBS with 1% wt/vol BSA) and incubated with peptide-specific rabbit anti-sera or the respective preimmune serum as a control in a 1:2 dilution for 20 minutes on ice. For control purposes, cells were incubated with an anti-CD23 antibody (clone M-L233, BD Biosciences) or the respective isotype control. After washing (PBS and 1% wt/vol BSA), cells were resuspended in PBS and 1% wt/vol BSA, and complexes of monoclonal anti–Bet v 1 IgE (5 μg/ml) and major birch pollen allergen Bet v 1 trimer (5 μg/mL)E3 were added. IgE and Bet v 1 trimers were preincubated for 1 hour at 37°C to ensure complex formation. After another washing step, cells were incubated with fluorescein isothiocyanate–labeled anti-IgE (KPL) and the fixable viability dye eFluor 450 (eBioscience, San Diego, Calif) for 20 minutes on ice. Expression of CD23 was confirmed in each experiment by staining 1 extra sample with a phycoerythrin-labeled anti-CD23 antibody (clone EBVCS2, eBioscience) or the respective isotype control, as well as viability dye. All experiments were performed in duplicates. Flow cytometry was performed with a FACSCanto II (BD Biosciences). Thirty thousand events were acquired per sample and analyzed with FlowJo Software (TreeStar, Ashland, Ore). Mean fluorescence intensity (MFI) was calculated with FlowJo Software, and MFI of cells stained with fluorescein isothiocyanate–labeled anti-IgE in the absence of monoclonal anti–Bet v 1 IgE were subtracted as background. The percentage blocking was calculated according to the following formula:
Negative stain electron microscopy
Negative stain electron microscopy was performed with purified Bet v 12specific IgE,E7,E8 the purified CD23 constructs, and rBet v 1. Furthermore, a His-tag–specific Fab fragment was generated to indicate the His-tagged CD23 constructs. For this purpose, a mouse IgG1 anti-His–specific antibody (37-2900, Life Technologies) was incubated in digestion buffer (100 mmol/L Tris [pH 8], 2 mmol/L EDTA, and 25 mmol/L 2-mercaptoethanol) for 2.5 hours at room temperature. Subsequently, the antibody was incubated with a papain resin (Pierce, Rockford, Ill) overnight at 37°C, and Fab fragments were purified by depleting full antibodies and Fc fragments with protein A (Pierce). Negative stain of the samples was done, as previously described.E11 For this purpose, CD23A was buffered in 10 mmol/LTris (pH 8), 100 mmol/L NaCl, and 2 mmol/L CaCl2. The different molecules (ie, CD23A, IgE, anti–His-Fab, and Bet v 1) were then complexed in borate-buffered saline (10 mmol/L sodium borate and 150 mmol/L NaCl [pH 8.2]) at 4°C overnight (CD23A, 2 μg/mL; IgE, 3.5 μg/mL; anti–His-Fab, 2 μg/ml; and Bet v 1, 2 μg/mL). Samples were floated onto a 2-nm carbon layer, stained with 2% uranyl formate (Polysciences Europe, Eppelheim, Germany), and transferred to a T 600-Cu grid (Ted Pella, Redding, Calif). Samples were then viewed under an electron microscope (EM-900 TEM; Carl Zeiss, Oberkochen, Germany).
Extended Data
Fig E1.
Expression in insect cells and purification. A, Time course of expression (x-axes) of CD23 constructs, as detected in nitrocellulose-blotted insect cell supernatants with an anti2His-tag antibody. Molecular weights are indicated on the y-axes. B and C, Coomassie (Fig E1, B) and glycan (Fig E1, C) staining of purified constructs (CD23A, CD23B, CD23C, and CD23D), as well as of a nonglycosylated (N) and glycosylated (P) control protein.
Fig E2.
Binding of rabbit antisera specific for CD23 peptides to recombinant CD23 constructs: A, CD23A; B, CD23B; C, CD23C; and D, CD23D. CD23 constructs were incubated with anti-CD23 antibodies with different specificities (x-axes: rabbit anti-CD23 peptide antibodies anti-P1, anti-P2, anti-P3, anti-P4, anti-P5, anti-P6, and anti-P7; monoclonal anti-CD23 antibody). Absorbance values (A 405 nm: y-axes) correspond to bound antibodies. Error bars show SDs of triplicates.
Table E1.
Characteristics of synthetic CD23 peptides
| CD23 peptides | Sequence | Theoretical isoelectric point | Amino acid residues | Molecular weight (Da) |
|---|---|---|---|---|
| P1 | LESHHGDQMAQKSQSTQISQELEELRAEQC | 4.68 | 30 | 3441.7 |
| P2 | CLWAGLLTLLLLWHWDTTQSLKQLEE | 4.65 | 26 | 3111.7 |
| P3 | CPASEGSAESMGPDSRPDPDGRLPTPSAPLHS | 4.43 | 32 | 3219.5 |
| P4 | CLSSFKSQELNERNEASDLLERLREEVTKLR | 4.19 | 31 | 3694.1 |
| P5 | PEKWINFQRKCYYFGKGTKQWVHARYAC | 9.71 | 28 | 3509.1 |
| P6 | CSPEEQDFLTKHASHTGSWIGLRNLDLKGEF | 5.39 | 31 | 3516.9 |
| P6a | SPEEQDFLTKHASHT | 5.26 | 15 | 1726.8 |
| P6b | GSWIGLRNLDLKGEF | 6.07 | 15 | 1705.0 |
| P6c | LTKHASHTGSWIGLRN | 11 | 16 | 1778.0 |
| P7 | NWAPGEPTSTSQGEDCVMMRGSGRWNDAFC | 4.32 | 30 | 3345.7 |
Cysteine residues added for carrier coupling are shown in boldface.
Clinical implications.
The new interaction model of CD23 and IgE and its inhibition by omalizumab might open new opportunities for allergy treatment.
Acknowledgments
Supported by the Austrian Science Fund (FWF): SFB 4604, SFB 4605, and SFB 4613.
Abbreviations used
- ABTS
2,2′-Azino-bis(3-ethylbenzothiazoline-6-sulphonic acid)
- HRP
Horseradish peroxidase
- MFI
Mean fluorescence intensity
- N63
Asparagine 63
- Q63
Glutamine 63
Footnotes
Disclosure of potential conflict of interest: R. Selb and J. Eckl-Dorna have received research support from FWF. C. Lupinek has received lecture fees from Thermo Fisher. G. Hofer has received research support from the Austrian Science Fund. M. Focke-Tejkl has received research support from the Austrian Science Fund (FWF). B. Linhart has received research support from the European Union (FP7). H. Breiteneder has received research support from the Austrian Science Fund. W. Keller has received research support from the Austrian Science Fund. R. Valenta has received research support from the Austrian Science Fund (FWF), Biomay AG, Thermo Fisher, and Fresenius Medical Care and has received consultancy fees from Biomay AG, Thermo Fisher, and Fresenius Medical Care. V. Niederberger has received research support from FWF (SFB4613). The rest of the authors declare that they have no conflicts of interest.
References
- 1.Pawankar R, Canonica GW, Holgate ST, Lockey RF. Allergic diseases and asthma: a major global health concern. Curr Opin Allergy Clin Immunol. 2012;12:39–41. doi: 10.1097/ACI.0b013e32834ec13b. [DOI] [PubMed] [Google Scholar]
- 2.Galli SJ, Tsai M. IgE and mast cells in allergic disease. Nat Med. 2012;18:693–704. doi: 10.1038/nm.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garman SC, Wurzburg BA, Tarchevskaya SS, Kinet JP, Jardetzky TS. Structure of the Fc fragment of human IgE bound to its high-affinity receptor Fc epsilonRI alpha. Nature. 2000;406:259–66. doi: 10.1038/35018500. [DOI] [PubMed] [Google Scholar]
- 4.Eggel A, Baravalle G, Hobi G, Kim B, Buschor P, Forrer P, et al. Accelerated dissociation of IgE-FcepsilonRI complexes by disruptive inhibitors actively desensitizes allergic effector cells. J Allergy Clin Immunol. 2014;133:1709–19.e8. doi: 10.1016/j.jaci.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holgate ST, Djukanovic R, Casale T, Bousquet J. Anti-immunoglobulin E treatment with omalizumab in allergic diseases: an update on anti-inflammatory activity and clinical efficacy. Clin Exp Allergy. 2005;35:408–16. doi: 10.1111/j.1365-2222.2005.02191.x. [DOI] [PubMed] [Google Scholar]
- 6.van Neerven RJ, van Roomen CP, Thomas WR, de Boer M, Knol EF, Davis FM. Humanized anti-IgE mAb Hu-901 prevents the activation of allergen-specific T cells. Int Arch Allergy Immunol. 2001;124:400–2. doi: 10.1159/000053770. [DOI] [PubMed] [Google Scholar]
- 7.van der Heijden FL, Joost van Neerven RJ, van Katwijk M, Bos JD, Kapsenberg ML. Serum-IgE-facilitated allergen presentation in atopic disease. J Immunol. 1993;150:3643–50. [PubMed] [Google Scholar]
- 8.van Neerven RJ, Wikborg T, Lund G, Jacobsen B, Brinch-Nielsen A, Arnved J, et al. Blocking antibodies induced by specific allergy vaccination prevent the activation of CD4+ T cells by inhibiting serum-IgE-facilitated allergen presentation. J Immunol. 1999;163:2944–52. [PubMed] [Google Scholar]
- 9.Aubry JP, Pochon S, Graber P, Jansen KU, Bonnefoy JY. CD21 is a ligand for CD23 and regulates IgE production. Nature. 1992;358:505–7. doi: 10.1038/358505a0. [DOI] [PubMed] [Google Scholar]
- 10.Tu Y, Salim S, Bourgeois J, Di Leo V, Irvine EJ, Marshall JK, et al. CD23-mediated IgE transport across human intestinal epithelium: inhibition by blocking sites of translation or binding. Gastroenterology. 2005;129:928–40. doi: 10.1053/j.gastro.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 11.Palaniyandi S, Tomei E, Li Z, Conrad DH, Zhu X. CD23-dependent transcytosis of IgE and immune complex across the polarized human respiratory epithelial cells. J Immunol. 2011;186:3484–96. doi: 10.4049/jimmunol.1002146. [DOI] [PubMed] [Google Scholar]
- 12.Wurzburg BA, Tarchevskaya SS, Jardetzky TS. Structural changes in the lectin domain of CD23, the low-affinity IgE receptor, upon calcium binding. Structure. 2006;14:1049–58. doi: 10.1016/j.str.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 13.Hibbert RG, Teriete P, Grundy GJ, Beavil RL, Reljic R, Holers VM, et al. The structure of human CD23 and its interactions with IgE and CD21. J Exp Med. 2005;202:751–60. doi: 10.1084/jem.20050811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bettler B, Texido G, Raggini S, Ruegg D, Hofstetter H. Immunoglobulin E-binding site in Fc epsilon receptor (Fc epsilon RII/CD23) identified by homolog-scanning mutagenesis. J Biol Chem. 1992;267:185–91. [PubMed] [Google Scholar]
- 15.Dhaliwal B, Yuan D, Pang MO, Henry AJ, Cain K, Oxbrow A, et al. Crystal structure of IgE bound to its B-cell receptor CD23 reveals a mechanism of reciprocal allosteric inhibition with high affinity receptor FcepsilonRI. Proc Natl Acad Sci U S A. 2012;109:12686–91. doi: 10.1073/pnas.1207278109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen BH, Ma C, Caven TH, Chan-Li Y, Beavil A, Beavil R, et al. Necessity of the stalk region for immunoglobulin E interaction with CD23. Immunology. 2002;107:373–81. doi: 10.1046/j.1365-2567.2002.01512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan MA, Gigliotti NM, Aubin BG, Rosenwasser LJ. FCER2 (CD23) asthma-related single nucleotide polymorphisms yields increased IgE binding and Egr-1 expression in human B cells. Am J Respir Cell Mol Biol. 2014;50:263–9. doi: 10.1165/rcmb.2013-0112OC. [DOI] [PubMed] [Google Scholar]
- 18.Sanner MF, Olson AJ, Spehner JC. Reduced surface: an efficient way to compute molecular surfaces. Biopolymers. 1996;38:305–20. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 19.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 20.Bendell CJ, Liu S, Aumentado-Armstrong T, Istrate B, Cernek PT, Khan S, et al. Transient protein-protein interface prediction: datasets, features, algorithms, and the RAD-T predictor. BMC Bioinformatics. 2014;15:82. doi: 10.1186/1471-2105-15-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 2013;41:W349–57. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 23.Graber P, Jansen K, Pochon S, Shields J, Aubonney N, Turcatti G, et al. Purification and characterization of biologically active human recombinant 37 kDa soluble CD23 (sFc epsilon RII) expressed in insect cells. J Immunol Methods. 1992;149:215–26. doi: 10.1016/0022-1759(92)90253-p. [DOI] [PubMed] [Google Scholar]
- 24.Roux KH. Immunoelectron microscopy of idiotype-anti-idiotype complexes. Methods Enzymol. 1989;178:130–44. doi: 10.1016/0076-6879(89)78010-1. [DOI] [PubMed] [Google Scholar]
- 25.Roux KH. Negative-stain immunoelectron-microscopic analysis of small macromolecules of immunologic significance. Methods. 1996;10:247–56. doi: 10.1006/meth.1996.0099. [DOI] [PubMed] [Google Scholar]
- 26.Laffer S, Hogbom E, Roux KH, Sperr WR, Valent P, Bankl HC, et al. A molecular model of type I allergy: identification and characterization of a nonanaphylactic anti-human IgE antibody fragment that blocks the IgE-FcepsilonRI interaction and reacts with receptor-bound IgE. J Allergy Clin Immunol. 2001;108:409–16. doi: 10.1067/mai.2001.117593. [DOI] [PubMed] [Google Scholar]
- 27.Lupinek C, Roux KH, Laffer S, Rauter I, Reginald K, Kneidinger M, et al. Trimolecular complex formation of IgE, Fc epsilon RI, and a recombinant nonanaphylactic single-chain antibody fragment with high affinity for IgE. J Immunol. 2009;182:4817–29. doi: 10.4049/jimmunol.0800726. [DOI] [PubMed] [Google Scholar]
- 28.Kilmon MA, Shelburne AE, Chan-Li Y, Holmes KL, Conrad DH. CD23 trimers are preassociated on the cell surface even in the absence of its ligand, IgE. J Immunol. 2004;172:1065–73. doi: 10.4049/jimmunol.172.2.1065. [DOI] [PubMed] [Google Scholar]
- 29.Schulz O, Sutton BJ, Beavil RL, Shi J, Sewell HF, Gould HJ, et al. Cleavage of the low-affinity receptor for human IgE (CD23) by a mite cysteine protease: nature of the cleaved fragment in relation to the structure and function of CD23. Eur J Immunol. 1997;27:584–8. doi: 10.1002/eji.1830270303. [DOI] [PubMed] [Google Scholar]
- 30.Klunker S, Saggar LR, Seyfert-Margolis V, Asare AL, Casale TB, Durham SR, et al. Combination treatment with omalizumab and rush immunotherapy for ragweed-induced allergic rhinitis: inhibition of IgE-facilitated allergen binding. J Allergy Clin Immunol. 2007;120:688–95. doi: 10.1016/j.jaci.2007.05.034. [DOI] [PubMed] [Google Scholar]
- 31.Jensen RK, Plum M, Tjerrild L, Jakob T, Spillner E, Andersen GR. Structure of the omalizumab Fab. Acta Crystallogr F Struct Biol Commun. 2015;71:419–26. doi: 10.1107/S2053230X15004100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wright JD, Chu HM, Huang CH, Ma C, Wen Chang T, Lim C. Structural and physical basis for anti-IgE therapy. Sci Rep. 2015;5:11581. doi: 10.1038/srep11581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E1.Marlovits TC, Zechmeister T, Schwihla H, Ronacher B, Blaas D. Recombinant soluble low-density lipoprotein receptor fragment inhibits common cold infection. J Mol Recognit. 1998;11:49–51. doi: 10.1002/(SICI)1099-1352(199812)11:1/6<49::AID-JMR388>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- E2.Pahr S, Selb R, Weber M, Focke-Tejkl M, Hofer G, Dordic A, et al. Biochemical, biophysical and IgE-epitope characterization of the wheat food allergen, Tri a 37. PLoS One. 2014;9:e111483. doi: 10.1371/journal.pone.0111483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E3.Campana R, Vrtala S, Maderegger B, Dall’Antonia Y, Zafred D, Blatt K, et al. Altered IgE epitope presentation: a model for hypoallergenic activity revealed for Bet v 1 trimer. Mol Immunol. 2011;48:431–41. doi: 10.1016/j.molimm.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E4.Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–73. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E5.Sreerama N, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal Biochem. 2000;287:252–60. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- E6.Focke M, Mahler V, Ball T, Sperr WR, Majlesi Y, Valent P, et al. Nonanaphylactic synthetic peptides derived from B cell epitopes of the major grass pollen allergen, Phl p 1, for allergy vaccination. FASEB J. 2001;15:2042–4. doi: 10.1096/fj.01-0016fje. [DOI] [PubMed] [Google Scholar]
- E7.Laffer S, Hogbom E, Roux KH, Sperr WR, Valent P, Bankl HC, et al. A molecular model of type I allergy: identification and characterization of a nonanaphylactic anti-human IgE antibody fragment that blocks the IgE-FcepsilonRI interaction and reacts with receptor-bound IgE. J Allergy Clin Immunol. 2001;108:409–16. doi: 10.1067/mai.2001.117593. [DOI] [PubMed] [Google Scholar]
- E8.Lupinek C, Roux KH, Laffer S, Rauter I, Reginald K, Kneidinger M, et al. Trimolecular complex formation of IgE, Fc epsilon RI, and a recombinant nonanaphylactic single-chain antibody fragment with high affinity for IgE. J Immunol. 2009;182:4817–29. doi: 10.4049/jimmunol.0800726. [DOI] [PubMed] [Google Scholar]
- E9.Focke M, Linhart B, Hartl A, Wiedermann U, Sperr WR, Valent P, et al. Non-anaphylactic surface-exposed peptides of the major birch pollen allergen, Bet v 1, for preventive vaccination. Clin Exp Allergy. 2004;34:1525–33. doi: 10.1111/j.1365-2222.2004.02081.x. [DOI] [PubMed] [Google Scholar]
- E10.Reginald K, Eckl-Dorna J, Zafred D, Focke-Tejkl M, Lupinek C, Niederberger V, Keller W, et al. Different modes of IgE binding to CD23 revealed with major birch allergen, Bet v 1-specific monoclonal IgE. Immunol Cell Biol. 2013;91:167–72. doi: 10.1038/icb.2012.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E11.Roux KH. Immunoelectron microscopy of idiotype-anti-idiotype complexes. Methods Enzymol. 1989;178:130–44. doi: 10.1016/0076-6879(89)78010-1. [DOI] [PubMed] [Google Scholar]