Abstract
Animal phyla vary dramatically in species richness (from 1 species to >1.2 million), but the causes of this variation remain largely unknown. Animals have also evolved striking variation in morphology and ecology, including sessile marine taxa lacking heads, eyes, limbs, and complex organs (e.g. sponges), parasitic worms (e.g. nematodes, platyhelminths), and taxa with eyes, skeletons, limbs, and complex organs that dominate terrestrial ecosystems (arthropods, chordates). Relating this remarkable variation in traits to the diversification and richness of animal phyla is a fundamental yet unresolved problem in biology. Here, we test the impacts of 18 traits (including morphology, ecology, reproduction, and development) on diversification and richness of extant animal phyla. Using phylogenetic multiple regression, the best-fitting model includes five traits that explain ~74% of the variation in diversification rates (dioecy, parasitism, eyes/photoreceptors, a skeleton, non-marine habitat). However, a model including just three (skeleton, parasitism, habitat) explains nearly as much variation (~67%). Diversification rates then largely explain richness patterns. Our results also identify many striking traits that have surprisingly little impact on diversification (e.g. head, limbs, and complex circulatory and digestive systems). Overall, our results reveal the key factors that shape large-scale patterns of diversification and richness across >80% of all extant, described species.
Keywords: animals, diversification, habitat, morphology, parasitism, species richness
Introduction
A major goal of ecology and evolutionary biology is to explain patterns of species richness. For example, why do some clades have a single species whereas others (of similar age) have more than a million? A closely related question is: what kinds of traits might be particularly important for explaining these patterns? For example, are the habitats where organisms live as important as their evolutionary innovations in morphology, development, or reproduction? Does parasitism increase the diversification rates of parasitic lineages? If so, is it as important as other traits in explaining diversity patterns?
New time-calibrated phylogenies and phylogenetic comparative methods offer exciting opportunities to address these questions. Here, we use these approaches to analyze patterns of species richness and diversification in one of the largest groups of organisms, the metazoans (i.e. animals, including >80% of all extant, described species; Roskov et al. 2016). We focus on finding the correlates of net diversification rates of clades (speciation – extinction over time), to identify traits that may be particularly important in accelerating speciation and/or buffering lineages from extinction. We focus on diversification rates rather than richness alone, since the latter ignores the ages of clades.
Animal phyla offer a compelling system in which to address these questions. First, animal phyla show dramatic variation in species richness, from a single described species in Placozoa to >1.2 million species in Arthropoda (Zhang 2013). Second, animals have evolved remarkable diversity in their morphology, ecology, development, and reproduction (Nielsen 2001; Hickman et al. 2012) over the past ~800 million years (Fig. 1). Animals range from simple, microscopic, asexual taxa found only in oceans (e.g. placozoa), to sessile, predominantly marine taxa lacking heads, eyes, limbs, and organs (e.g. sponges), to parasitic worms (e.g. many nematodes and platyhelminths), to highly mobile taxa with heads, eyes, skeletons, limbs, and complex organ systems for circulation, digestion, and excretion, that now dominate terrestrial ecosystems in terms of species richness (e.g. arthropods) and body size (e.g. chordates; Nielsen 2001; Hickman et al. 2012). How this incredible variation in traits among animal phyla might be related to their striking differences in richness is a fundamental but unresolved problem in biology.
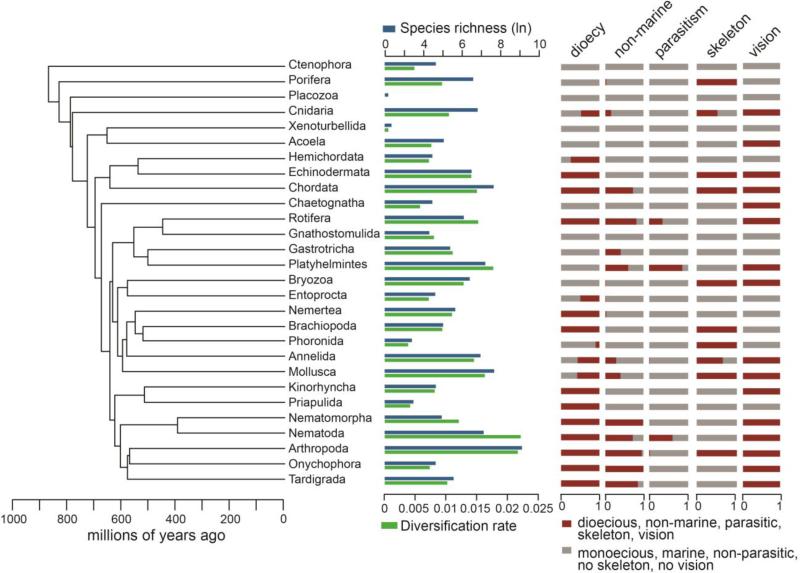
Figure 1.
Summary of phylogeny, richness, diversification rates, and character data among animal phyla. Time-calibrated phylogeny of 28 animal phyla based on topology of Dunn et al. (2014) with a root age at ~836 million years (Tree 2 from Wiens 2015b). For each phylum, species richness and diversification rates (in species per million years) are shown, as well as the distributions of five traits that showed significant relationships with diversification rates: dioecy, occupancy of non-marine habitats, parasitism, presence of a skeleton, and vision. Occupancy of terrestrial habitats is not depicted as it is largely redundant with the occupancy of non-marine habitats (Wiens 2015b). Proportions of character states smaller than ~5% are not clearly visible but exact values are shown in Supplementary File S1.
The question of which traits explain large-scale patterns of animal diversification has remained unresolved for several reasons. First, many traits have been hypothesized to play a role in explaining differences in diversification and richness among animal phyla, but their importance has not been explicitly tested (Heard and Hauser 1995; Mayhew 2007). Second, studies that have explicitly tested relationships between traits and diversification have usually focused on only one trait at a time (e.g. body size [Orme et al. 2002; McClain and Boyer 2009], presence of eyes [de Queiroz 1999], or occurrence in non-marine habitats [Wiens 2015b]). Therefore, the relative importance of different traits (and the potential impact of correlations among traits) remains highly uncertain. Third, many traits that were tested showed weak or no relationship with richness or diversification (e.g. presence of eyes [de Queiroz 1999], body size [Orme et al. 2002]). Similarly, traits that were found to be significant explained only a minority of the variation in diversification rates among animal phyla (e.g. ~33% for non-marine habitat use; Wiens 2015b). Furthermore, some studies have focused on richness instead of diversification (e.g. Orme et al. 2002), without accounting for the fact that some clades may have more species simply because they are older. Finally, some studies have studied potentially relevant traits across animals, but not at the level of phyla (e.g. de Queiroz 1999). Thus, it remains unclear how much variation in diversification rates (or richness) among phyla these traits explain.
In this paper, we test the morphological, ecological, and developmental correlates of diversification among animal phyla (Nielsen 2001; Hickman et al. 2012). We utilize three time-calibrated phylogenies (Wiens 2015b) that include most (28 of 32) widely recognized phyla. We assemble a dataset of 18 diverse ecological, morphological, developmental, and reproductive traits from the literature, each having the potential to impact diversification rates. We perform phylogenetic comparative analyses to identify those traits significantly related to diversification (noting that diversification rates explain ~85–89% of the variation in richness among phyla; Wiens 2015b). We then use multiple regression to identify a model explaining the maximum variation in diversification rates, while including the fewest variables. We identify a set of five traits that together explain ~74% of the variation in diversification rates among animal phyla, and a set of three traits that explain nearly as much (~67%). Our results support the importance of ecology, a few key morphological innovations, and parasitism in explaining diversification and species richness patterns in animals at the largest scales.
Materials and Methods
Traits
We identified 18 traits that were potentially related to diversification rates of animal phyla (Online Appendix A). Most traits involved morphology and development, whereas a few described ecology and reproduction. These traits were previously recognized as potentially promoting diversification of animal phyla (Heard and Hauser 1995; de Queiroz 1999; Orme et al. 2002; Mayhew 2007; Wiens 2015b) or being important in animal macroevolution in general (Dunn et al. 2014). Further, we limited ourselves to those traits for which data were available from the literature for all phyla. Therefore, based on this criterion, we excluded several potential traits, including diet, regenerative ability, generation time, and number of offspring. The final set included external characters (body size and symmetry; presence of eyes, legs, segmentation, and cephalization), internal characters (presence of an excretory system, circulatory system, digestive system, and an endo- or exoskeleton), characters associated with reproduction and development (asexual reproduction, dioecy, metamorphosis), and ecological characters (parasitism, occurrence in marine vs. non-marine and terrestrial vs. aquatic habitats). Note that “non-marine” includes both terrestrial and freshwater habitats, whereas “aquatic” includes both freshwater and marine habitats. These two characters (nonmarine and terrestrial habitat) were previously tested for relationships with diversification of animal phyla by Wiens (2015b). Similarly, Orme et al. (2002) tested for a relationship between body size and richness across animal phyla. Data for these three characters were therefore obtained primarily from Wiens (2015b) and Orme et al. (2002; but see also Online Appendix A). Data for the other 15 characters were assembled from the literature for this study.
Traits were generally scored based on the estimated proportion of species in a phylum exhibiting a given state (ranging from 0 to 1), given that many traits varied among species within one or more phyla. In cases in which a trait showed little variation within phyla, it was treated as a categorical variable. Note that it is appropriate to perform phylogenetic generalized least squares (PGLS) analyses with categorical independent variable as long as the dependent variable is continuous (Martins and Hansen 1997), as is the case here for diversification rates. One trait (body size) was treated as a continuous variable. Data on trait distributions among phyla were often obtained from Hickman et al. (2012), especially for the most obvious traits (e.g. head, limbs). When information was not available in Hickman et al. (2012), we searched the primary literature for additional information. Detailed information on all characters and their coding can be found in Online Appendix A. Distributions of traits among taxa are summarized in Supplementary File S1 (all Supplementary Files have been deposited on Dryad doi:10.5061/dryad.ck52b).
We recognize that some problems could arise when using proportions to associate traits with diversification for large-scale clades. For example, for a given phylum with two subclades A and B, a trait might be present in subclade A, but increased diversification in that phylum might be restricted to subclade B. However, such a pattern would need to be repeated within multiple phyla to give a strong relationship between that trait and diversification rates among phyla, which seems unlikely. Similarly, a trait might be present at very low frequencies within one or more phyla, but could still show a relationship with diversification (although such a rare trait would be highly unlikely to be causally related to diversification across the clade). We confirmed that all traits significantly related to diversification in our results occurred in >50% of the species in two or more phyla (Fig. 1). In general, we recognize that statistical relationships between traits and diversification can support (but not prove) causation.
Phylogeny and Phylum Delimitation
We analyzed relationships between traits and diversification among the 28 animal phyla included in the three time-calibrated phylogenies of Wiens (2015b). These phyla generally correspond to the 34 phyla described in Hickman et al. (2012) with the following exceptions. The phyla Cycliophora, Loricifera, Mesozoa, and Micrognathozoa were not included given that lack of comparable sequence data prevented their inclusion in the tree of Wiens (2015b). Acanthocephala is considered a separate phylum in Hickman et al. (2012) but is included within Rotifera here. Similarly, Sipuncula is considered a separate phylum by Hickman et al. (2012) but here was included within Annelida (for details on delimitation of phyla see Wiens 2015b).
Time-calibrated phylogenies were used to both infer diversification rates of phyla (i.e. clade ages) and account for phylogenetic non-independence of phyla (using phylogenetic comparative methods). We used the three time-calibrated phylogenies from Wiens (2015b). All trees used are given in NEXUS format in Supplementary File S2. These phylogenies were based on 16 genes from 73 metazoan species, using relatively well-supported relationships among phyla as constraints. The first two trees were based on the topology of Dunn et al. (2014), which summarized animal phylogenies from many recent studies. Two sets of fossil calibration points were used, one that estimated the root of the tree at ~1.3 billion years (Tree 1) and the second one that estimated the root at 836 Ma (Tree 2). For the last tree (Tree 3) an alternative tree topology based largely on Philippe et al. (2011) was used, with an estimated root age of 820 Ma. Results were similar using all three trees, and we focused primarily on Tree 2 (which yields a more standard set of divergence dates, along with the relatively well-established topology).
We also performed a series of secondary analyses to address the impacts of clade delimitation on the results. First, some phyla were subdivided into smaller subclades (specifically Annelida, Arthropoda, Bryozoa, Chaetognatha, Chordata, Cnidaria, Echinodermata, Hemichordata, Mollusca, and Porifera; following Wiens 2015b) which yielded 49 higher-level clades. We also independently tested the clades Deuterostomia (i.e. including the higher-level clades within Chordata, Echinodermata, and Hemichordata) and Ecdysozoa (including the six higher-level clades within Arthropoda, and the phyla Kinorhyncha, Nematoda, Nematomorpha, Onychophora, Priapulida, and Tardigrada). However, these were not the primary analyses for our study, given that our main goal was to infer the causes of variation in diversification rates and richness among phyla (rather than simultaneously trying to explain variation both among and within phyla). For example, much of the variation within phyla might be explained by other variables not included here (i.e. that show limited variation among phyla), and traits important for explaining variation among phyla might be less important within them.
Diversification Rates
The net diversification rate for each phylum-level clade was estimated using the method-of-moments estimator for stem group ages (Magallon and Sanderson 2001). Estimated rates are given in Supplementary File S3, along with species richness and ages of clades. The stem-group estimator was used because the phylogenies included too few species for some phyla to infer their crown group age (e.g. some species-poor phyla are represented by single species). Furthermore, use of crown-group ages might give a highly distorted view of a group's net diversification (e.g. a very young crown group age in an ancient group with low richness could suggest a nonsensical high diversification rate). Estimating the diversification rate of a clade using the method-of-moments estimator requires the clade's age, species richness, and an assumed relative extinction fraction (epsilon, or extinction/speciation). Note that epsilon is intended to correct for the failure to sample extinct clades entirely when extinction rates are high (Magallón and Sanderson 2001). It is not an estimate of extinction rates within extant clades. Clade ages were derived from the three time-calibrated trees. Following standard practice, we used three different values of epsilon, two extreme values (0 and 0.9) and an intermediate value (0.5). However, use of different values had relatively minor impact on the relationship between traits and diversification (Supplementary File S4). Species richness estimates based on numbers of described species were obtained from Wiens (2015b). Because actual species numbers are clearly much greater than described species numbers for some phyla, we also performed a set of analyses using diversification rates estimated with projected species numbers (see Online Appendix B for details). Specifically, we used projections of the actual number of extant species in each phylum, not merely the current number of described species.
We note that some authors have claimed that these net diversification rate estimators require that rates of diversification are constant within clades, and should therefore only be used if there is a positive relationship between clade age and richness among clades (e.g. Rabosky et al. 2012; Rabosky and Adams 2012). There are two main problems with these arguments. First, the net diversification rate estimator for stem-group ages is mathematically agnostic with regards to variation in diversification within clades over time. Thus, a young clade with many species will have a relatively fast net diversification rate, regardless of the exact pattern of lineage accumulation over time within that clade (just as an older clade with fewer species will have a slower net rate). Second, recent simulations suggest that the accuracy of the net diversification estimator used here is similar regardless of whether there is a positive or negative relationship between clade age and richness (Kozak and Wiens 2016). In fact, the previous studies (e.g. Rabosky et al. 2012; Rabosky and Adams 2012) did not directly address the accuracy of diversifiction estimators. A simulation study that did address their accuracy suggests that there can be strong relationships between true and estimated rates using these estimators, and that these relationships strengthen dramatically as clade ages increase (Kozak and Wiens 2016). Importantly, the clade ages used here are dramatically older than in that study (i.e. ~15–40 Myr vs. ~500 Myr in this study).
More generally, alternative approaches to estimating diversification rates would generally be problematic given that the phylogenies used here include only a small fraction of the total species richness of these clades. Moreover, the approach used here allows us to test the relationship between traits and richness, and to estimate the variance in diversification rates among phyla that is explained by individual traits and combinations of traits. Such questions would be difficult to address using alternative approaches to diversification that do not estimate an overall diversification rate for each phylum (as done here).
Testing Relationships between Traits and Diversification Rates
The relationships between traits and diversification rates among phyla were tested using phylogenetic generalized least-squares regression (PGLS; Martins and Hansen 1997). This approach accounts for the potential statistical non-independence of clades due to phylogeny. For PGLS, the R package CAPER version 0.5.2 (Orme 2013) was used. Following standard practice, the maximum likelihood transformation of branch lengths optimized for the data (‘lambda = ML’) was used, based on estimated values of lambda (Pagel 1999). Kappa and delta were each fixed at 1.
We first tested the relationship between diversification rates and each of the 18 traits separately. The analysis was repeated using the three phylum-level topologies and the three values of epsilon. Results were largely insensitive to different topologies and epsilon values.
We then conducted multiple regression analyses, including only traits that showed a significant relationship with diversification rates of phyla when tested individually for Tree 2 (which has a relatively standard topology and divergence dates) and epsilon = 0.5 (the intermediate value). For the multiple regression analyses, we first included all traits that were significantly related to diversification in separate analyses in a single multiple regression model. We then repeated the analysis, after excluding one trait at a time (until all n-1 combinations were reached, where n is the number of traits). From these analyses, we selected the analysis with the lowest AIC value. We then excluded one additional trait at a time from this analysis until all possible combinations were tested. We then again selected the analysis with the lowest AIC. We repeated these steps until only a single pair of traits remained. From these analyses, we then selected the model with the lowest AIC value overall, which should simultaneously maximize the variance explained and minimize the number of variables included. We also used the P-values associated with each individual trait in the multiple regression analyses to assess whether each variable significantly contributed to the best-fitting analysis.
We recognize that other approaches to model selection are possible. For example, we could have initially included all traits regardless of their relationship to diversification in pairwise analyses, and sequentially removed them. However, this would create an extremely large number of models to compare (i.e. all the possible combinations of traits from those including 17 traits down to those with only 2), with most combinations almost certainly being unhelpful (since most variables show no significant relationship with diversification). Adding other traits might increase the r2 slightly, but such models would also be penalized for their extra parameters.
The relationships between individual traits and diversification rates were further explored with three additional analyses. First, we repeated the analyses of individual traits for 28 phyla using projected species richness values, instead of numbers of described species (for details see Online Appendix B and Supplementary File S5). Second, we repeated the analyses of individual traits using the 49 higher-level clades, after subdividing some phyla into subclades (for details see Wiens [2015b] and Supplementary File S6). These additional analyses were conducted using Tree 2 and all three values of epsilon. Third, we repeated the analyses individually for Annelida, Arthropoda, and Mollusca (i.e. analyzing variation among the higher-level clades within these phyla), and for all subdivided clades within Ecdysozoa (12 clades total) and Deuterostomia (7 clades). We specifically tested those characters that showed significant relationships with diversification rate in the main analysis of 28 phyla. This latter set of analyses was conducted using the Tree 2 and the intermediate value of epsilon. Results are described in Supplementary File S7.
Results
We first separately tested each of 18 traits (Table 1, Supplementary File S4) for their potential impacts on diversification. Only six traits showed significant relationships with diversification rates across three different phylogenetic trees and different diversification analyses (Table 1, Supplementary File S4). The distribution of these traits across the phylogeny is summarized in Fig. 1, along with patterns of diversification rates and species richness among phyla (note that non-marine and terrestrial habitat use are redundant and not shown separately; see below). The presence of photoreceptors and/or eyes in a phylum (vision for short hereafter) explained 36–39% (P <0.001 to 0.001) of the variation in diversification rates (ranges summarize results across the three trees and three different epsilon values used for estimating diversification rates for each tree). An alternative coding for vision, using the proportion of species with eyes only (i.e. excluding photoreceptors), was also significantly related to diversification (r2 = 0.23–0.32, P = 0.002–0.010) but this coding explained less variation than when eyes and photoreceptors were combined into a single state. Phyla with higher proportions of non-marine species (i.e. freshwater and terrestrial) and higher proportions of terrestrial species had significantly higher diversification rates. Occurrence in non-marine and terrestrial habitats respectively explained 30–37% (P = 0.001–0.002) and 25–28% (P = 0.003–0.006) of the variation in diversification rates, as previously reported (Wiens 2015b). Diversification rates were also significantly related to the presence of dioecy (r2 = 0.15–0.22, P = 0.006–0.046), a skeleton (either internal or external; r2 = 0.15–0.26, P = 0.012–0.050), and the proportion of parasitic species in a clade (r2 = 0.16–0.27; p = 0.005–0.038).
Table 1.
Results of PGLS analyses individually testing relationships between 18 traits and diversification rates of animal phyla. Brief descriptions of each trait and regression results are shown. The results are for Tree 2 and the intermediate relative extinction fraction (epsilon=0.5). Results for other trees and epsilon values are in Supplementary File S4. Correlation coefficients (r2) and P-values are shown with significant values boldfaced.
| Trait | r 2 | P |
|---|---|---|
| Proportion of species exhibiting sexual reproduction | 0.03 | 0.354 |
| Presence or absence of cephalization | 0.06 | 0.209 |
| Presence or absence of a circulatory system | 0.04 | 0.323 |
| Presence or absence of coelom | 0.01 | 0.672 |
| Presence of a digestive system with two openings | 0.09 | 0.111 |
| Proportion of dioecious species | 0.17 | 0.027 |
| Presence or absence of an excretory organ | 0.12 | 0.072 |
| Presence or absence of legs | 0.08 | 0.153 |
| Median biovolume in mm3 (ln transformed) | 0.02 | 0.516 |
| Proportion of species undergoing metamorphosis | 0.04 | 0.300 |
| Proportion of non-marine species | 0.37 | 0.001 |
| Proportion of parasitic species | 0.26 | 0.005 |
| Presence or absence of segmentation | 0.09 | 0.123 |
| Proportion of species that have a skeleton | 0.14 | 0.047 |
| Presence or absence of bilateral symmetry | 0.06 | 0.214 |
| Proportion of terrestrial species | 0.27 | 0.005 |
| Proportion of vagile species | 0.01 | 0.597 |
| Presence or absence of photoreceptors/eyes | 0.38 | <0.001 |
Most other traits were not significantly related to diversification across any trees or epsilon values (Supplementary File S4). However, the presence of a digestive tract with two openings was significantly related to higher diversification rates on one tree (Tree 3, see Materials and Methods), when using lower epsilon values (0, 0.5).
We then conducted multiple regression analyses (Table 2, Supplementary File S8) to identify the combination of traits that best explained patterns of diversification among phyla. For simplicity, we conducted these analyses only on the preferred tree (Tree 2) and an intermediate epsilon value (0.5), given that alternate trees and epsilon values gave similar results in pairwise analyses (Supplementary File S4). We included five of the six traits that were each significantly related to diversification rates (Table 1; Supplementary File S4). We excluded terrestrial habitat since it is largely redundant with non-marine habitat (Wiens 2015b). We compared models including different combinations of traits using the Akaike Information Criterion (AIC).
Table 2.
Results of phylogenetic multiple regression analyses, showing the five models with the best fit based on AIC values (for full results see Supplementary File S8). The results are provided for Tree 2 and an epsilon of 0.5. Correlation coefficients (r2), AIC, and P-values are shown for each model, along with P-values for each trait in each model.
| Traits included in model | r 2 | AIC | P-value | Trait | P-value |
|---|---|---|---|---|---|
| dioecy, non-marine habitat, parasitism, skeleton, vision | 0.740 | 152.50 | <0.000 | dioecy | 0.130 |
| non-marine habitat | 0.144 | ||||
| parasitism | 0.001 | ||||
| skeleton | 0.002 | ||||
| vision | 0.114 | ||||
| non-marine habitat, parasitism, skeleton, vision | 0.710 | 153.48 | <0.000 | non-marine habitat | 0.028 |
| parasitism | 0.003 | ||||
| skeleton | 0.002 | ||||
| vision | 0.097 | ||||
| dioecy, parasitism, skeleton, vision | 0.712 | 153.28 | <0.000 | dioecy | 0.025 |
| parasitism | 0.000 | ||||
| skeleton | 0.003 | ||||
| vision | 0.019 | ||||
| dioecy, non-marine habitat, parasitism, skeleton | 0.708 | 153.75 | <0.000 | dioecy | 0.111 |
| non-marine habitat | 0.024 | ||||
| parasitism | 0.001 | ||||
| skeleton | 0.001 | ||||
| non-marine habitat, parasitism, skeleton | 0.673 | 154.91 | <0.000 | non-marine habitat | 0.001 |
| parasitism | 0.002 | ||||
| skeleton | 0.001 |
The most variation in diversification rates (74%) was explained (and the lowest AIC obtained, 152.5) when all five traits were included (vision, parasitism, skeleton, dioecy, non-marine habitat; Table 2). However, separate analyses that individually excluded vision, dioecy, non-marine habitat, or dioecy and vision each resulted in only a small decrease in explained variation and in similar model fit. In particular, 71% of the variation was explained when either vision (AIC = 153.8), dioecy (AIC = 153.5), or non-marine habitat (AIC = 153.3) was excluded (Table 2). Importantly, 67% of the variation was still explained when both dioecy and vision were excluded (Table 2), with a negligible increase in AIC (154.9). In comparison, excluding parasitism or the skeleton resulted in a model with substantially worse fit and less variation explained (with parasitism excluded: AIC = 164.1, r2 = 0.58; with skeleton excluded: AIC = 163.0, r2 = 0.59). Thus, a model including only non-marine habitat, skeleton, and parasitism seemed to maximize fit and explanatory power with the fewest traits.
In the analysis that included all five traits, only parasitism (P = 0.001) and the presence of a skeleton (P = 0.002) significantly contributed to the model whereas vision (P = 0.11), dioecy (P = 0.13), and non-marine habitat (P = 0.14) did not (Table 2). This pattern presumably occurred because non-marine habitat is significantly related to both vision and dioecy (vision and non-marine: r2 = 0.33, P = 0.001; dioecy and non-marine: r2 = 0.21, P = 0.014; see Supplementary File S9 for all pairwise relationships among these five traits). These relationships between non-marine habitat, vision, and dioecy also explain why the r2 and AIC did not improve substantially when each one of these three traits was removed (Table 2).
We also performed additional analyses to further explore our main results. First, we used projected species numbers (Supplementary File S5) to estimate the diversification rate for each phylum instead of numbers of described species (utilizing Tree 2 and all three epsilon values). This analysis (Supplementary File S5) supported relationships between diversification and five of the six traits supported in the analyses based on described species (vision: r2 = 0.32–33, P = 0.001–0.002; parasitism: r2 = 0.35, P = 0.001; dioecy: r2 = 0.15–17, P = 0.031–0.046; non-marine habitat: r2 = 0.36–0.37, P = 0.001; terrestrial habitat: r2 = 0.26–0.27, P = 0.004–0.006). However, the presence of a skeleton was non-significant across all three epsilon values. Parasitism explained more variation when projected species richness was used than using described richness, presumably because of the high projected richness of parasitic nematodes (Supplementary File S5). The remaining five significant variables explained equal or less variation when projected species richness was used, relative to diversification rates estimated from numbers of described species.
Second, we subdivided many of the more diverse phyla to yield a total of 49 clades (Wiens 2015b). Using Tree 2 and three epsilon values, we found that three traits still showed significant relationships with diversification rates (vision: r2 = 0.21–0.23; P < 0.001; non-marine habitat: r2 = 0.11–0.17; P = 0.001–0.006; terrestrial habitat: r2 = 0.15–0.21; P = 0.001–0.006; Supplementary File S6). The relationship between diversification rates and all three traits is weaker for 49 clades in comparison with 28 clades, as expected given the problem of simultaneously explaining diversification rates both within and between phyla (see above).
Finally, we tested for relationships between traits and diversification rates separately within selected clades (Supplementary File S7). Specifically, we tested the six candidate traits within the phyla Annelida (n = 5 clades), Arthropoda (n = 6), and Mollusca (n = 5) and within the clades Deuterostomia (n = 7 clades, including the subdivided clades of Chordata, Echinodermata, and Hemichordata) and Ecdysozoa (n = 12 clades, including the subdivided clades of Arthropoda, and the phyla Kinorhyncha, Nematoda, Nematomorpha, Onychophora, Priapulida, and Tardigrada). Although many traits that were important among phyla were not significantly related to diversification rates within these clades, we found a significant relationship with the presence of a skeleton within Annelida (r2 = 0.83; P = 0.03). We also found a significant relationship between diversification rates and non-marine habitat within Mollusca ( r2 = 0.91; P = 0.01; see also Wiens 2015b). However, sample sizes of clades were very small within these groups, and some characters were largely invariant within them.
Discussion
In this study we analyzed the morphological, developmental, and ecological traits that potentially underlie patterns of diversification and species richness among clades in the dominant group of living organisms (animals). Specifically, we evaluated 18 traits for their potential to explain variation in diversification rates among animal phyla. We identified five traits that together explain ~74% of the variation in diversification rates among animal phyla (Table 2). These traits include morphological innovations (skeleton, eyes/photoreceptors), ecological characteristics (occurrence in non-marine habitats, parasitism), and a trait associated with reproduction (dioecy). However, a model including just three traits (habitat, parasitism, skeleton) explained nearly as much variation (~67%). To our knowledge, this may be the first study to show the importance of parasitism to patterns of diversification at the largest phylogenetic scales. Perhaps just as importantly, we identified many striking traits that have surprisingly little impact on the diversification rates of animal phyla. These included heads, limbs, body size, vagility, sexuality (vs. asexuality), metamorphosis, and complex organ systems used for circulation, digestion, and excretion (Table 1). Overall, our results show the importance of both ecology and morphological innovations in explaining large-scale patterns of diversity and diversification across >80% of all described species.
Our results also demonstrate the importance of considering multiple traits when explaining diversity patterns. First, by including multiple variables, we were able to explain most (74%) of the variation in diversification rates among animal phyla, with a relatively limited number of variables (five). Each variable separately explained only a limited amount of variation. Further, most of this variation could be explained by three variables alone. In contrast, previous studies of diversification among animal phyla that considered only a single trait (non-marine habitat) were able to explain only 30–37% of the variation in diversification rates among anima phyla (Wiens 2015b). Importantly, our results also suggest that an analysis of a single trait might conclude that a trait is important, when in fact the trait that is more directly influencing diversification is one that is correlated with it. In our analysis, there were five traits with seemingly strong effects on diversification, but three appeared to be overlapping in their effects (dioecy, vision, non-marine habitat). Thus, multiple regression analyses that excluded one of these three traits had only slightly higher AIC scores and explained only slightly less variation relative to a model including all five (Table 2). Pairwise comparisons showed these traits to be significantly related in their distributions among phyla (vision and non-marine: r2 = 0.33, P = 0.001; dioecy and non-marine: r2 = 0.21, P = 0.014; Supplementary File S9). Indeed, vision is believed to have facilitated terrestrial invasion and dioecy is sometimes considered an adaptation to terrestrial life (Little 1983). The former idea is supported by our post-hoc ancestral-state reconstructions of vision and habitat on the tree, which suggest that vision evolved prior to the invasion of non-marine habitats (Supplementary File S10). In contrast, parasitism and the presence of a skeleton seem to be independent of both each other and these other traits, given the low correlations among them (Supplementary File S9). An important goal for future studies should be to tease out the relative effects of these traits on patterns of animal diversification, possibly by analyzing patterns within the more species rich phyla that vary for these traits (e.g. annelids, chordates, molluscs, arthropods). For example, Wiens (2015b) found that non-marine habitats explained >90% of the variation in diversification rates among major clades within molluscs. We found this same pattern, and that the other traits analyzed were not significantly related to large-scale diversification patterns in molluscs. Similarly, we found that the presence of a skeleton explained 83% of the variation in diversification rates among major clades of annelids, whereas the other traits analyzed did not (Supplementary File S7).
We acknowledge that other traits might be important in explaining richness and diversification patterns beyond the ones that we included here. Furthermore, the effects of these other traits might be masked by their correlations with traits that we included here. Nevertheless, we included a large number of dramatic traits that varied among phyla (e.g. heads, limbs, major organ systems), and showed that most were unrelated to diversification. Furthermore, the traits that we did include explained most variation in diversification rates among phyla. This suggests that other traits, if they are unrelated to those we included, would have little additional variation left to explain. However, explaining richness patterns within phyla will likely require including additional traits or might show different traits to be important relative to those that explained diversification rates among phyla. For example, among major vertebrate clades, terrestrial habitat use explains the majority of variation in diversification rates, whereas non-marine habitat use has no significant impact (Wiens 2015a). Similarly, our analyses of 49 subdivided clades and within select phyla supported only some of the traits that were found to be important across animal phyla.
We recognize that some readers may be dismayed that our study is not based on fossil taxa. However, our goal here was to explain patterns of net diversification and extant species richness among animal phyla. Thus, even if patterns of species richness and diversification in fossil taxa were different from those analyzed here (e.g. Alroy 2010), those are not the patterns that we are trying to explain. Furthermore, our primary focus is on explaining extant species richness across phyla, not the diversity of every subclade of every phylum at every slice of time over the last ~800 million years. Of course, analyses within phyla and analyses utilizing fossils may be critically important in further testing (and disentangling) the importance of these factors. We merely assert that our primary question was related to the richness, net diversification, and traits of living taxa. Furthermore, many of the traits analyzed here would be difficult to incorporate using fossil information (e.g. dioecy, parasitism). Similarly, species richness would also be difficult to estimate for many phyla given the bias of the fossil record towards hard-bodied organisms (Foote et al. 2007).
Our study raises many questions for future research. We think that the most important is: how exactly does each of these traits increase diversification? There are several relevant hypotheses in the literature, depending on the trait. First, non-marine environments may offer more effective barriers to dispersal, which may promote speciation (May 1994; Benton 2001; Vermeij and Grosberg 2010; Carrete-Vega and Wiens 2012; Wiens 2015b). Other hypotheses to explain lower marine richness (and diversification) have also been proposed (May 1994; Benton 2001; Vermeij and Grosberg 2010) but are more ambiguous given available data, such as higher terrestrial productivity or the difficulty of moving in water (Carrete-Vega and Wiens 2012; Wiens 2015b). Marine extinction may also be important (Carrete-Vega and Wiens 2012; Wiens 2015b). Vision may facilitate entering new adaptive zones, such as non-marine habitat, and may increase the ability of organisms to localize prey, predators, and conspecific mates (de Queiroz 1999). Dioecy may facilitate internal fertilization and therefore transitions to terrestrial environments, where external fertilization is problematic (Little 1983). Further, dioecy may increase the evolutionary potential of species by increasing their levels of heterozygosity and genetic diversity through genetic recombination (Lloyd 1980). The evolution of the skeleton has been traditionally associated with the Cambrian explosion, as it might have provided body support and defense from predators (Bengtson and Zhao 1992). Consequently, the presence of a skeleton is believed to have promoted rapid diversification of many animal phyla during the Cambrian (Thomas et al. 2000; Erwin et al. 2011). There has been some discussion of the idea that parasitism and similar types of species interactions might promote diversification (e.g. Yoder and Nuismer 2010; Althoff et al. 2014; Hembry et al. 2014), but strong empirical support at broad scales has been elusive (e.g. Wiegmann et al. 1993). To our knowledge, ours is the first study to support this hypothesis at such a deep phylogenetic scale. One potential mechanism by which parasitism might promote diversification is through co-speciation of hosts and their parasites (either strict or relatively loose), especially if niche partitioning within a host enables coexistence of multiple parasite species in a single host (Feder and Forbes 2010). Another potential mechanism is host switching (e.g. Ricklefs et al. 2004, 2014). We also note that our definition of “parasitism” was quite restrictive (i.e. an obligatory relationship between two heterospecific organisms during which the parasite is metabolically dependent on the host; Appendix A). More inclusive definitions might show even greater impact of parasitism on diversification (e.g. encompassing herbivorous insects; Ehrlich and Raven 1964; Mitter et al. 1988; Futuyma and Agrawal 2009; Wiens et al. 2015). This process also raises a methodological issue, in that increased diversification in phylogenetically disparate host and parasite lineages might be coupled rather than independent. For example, the high overall diversification rate of nematodes might be causally related to the high diversification rate of the arthropods that many nematodes parasitize (especially when using our projected richness values), rather than there being fully independent increases in each clade. Finally, another interesting topic for future research is how these traits might interact with each other and with large-scale historical factors, such as mass extinction events, climatic changes, and the rise of angiosperms.
Our results support the conclusions of some previous studies on higher-level animal diversification but not others. Our results agree with those of Wiens (2015b) on the importance of habitat (but with the caveat that the effects of habitat, dioecy, and vision are difficult to distinguish). Our results also agree with those of Orme et al. (2002) in showing that body size is not important. Orme et al. (2002) found no significant relationship between their index of body size for animal phyla (median biovolume among species) and the species richness of these phyla. In contrast, McClain and Boyer (2009) found a significant relationship between extreme body sizes of species within phyla (minimum and maximum among species) and the species richness of phyla. However, if body sizes evolved randomly across a tree, one would expect clades with more species to have both larger and smaller species (due to chance alone), even if there was no causal relationship between size and diversification. Therefore, we did not perform the same tests as McClain and Boyer (2009), to avoid this potential artifact. Our results disagree with those of de Queiroz (1999), who found no significant relationship between the presence of eyes and diversification. Our results support significant relationships between diversification rate and vision (but again with the important caveat that the effects of vision and habitat are difficult to parse). de Queiroz (1999) did not look at phyla but compared pairs of lower-level sister clades. He speculated that eyes might have been important at the level of phyla (as in our study), but not more recently, within phyla. Also, de Queiroz (1999) assessed only the impact of image-forming eyes on diversification, whereas we included both eyes and photoreceptors. We found that including both eyes and photoreceptors revealed a stronger relationship with diversification.
In conclusion, we found that a limited number of traits can explain most variation in diversification rates among animal phyla. These traits include morphological innovations (i.e. skeleton), as well as ecology (i.e. non-marine habitat). Our results also support parasitism as a key process promoting large-scale patterns of animal diversification. This latter result further supports the idea that local-scale species interactions help drive large-scale patterns of clade diversification over hundreds of million years (e.g. Wiens et al. 2015). We also show that numerous dramatic evolutionary innovations fail to drive diversification patterns among animal phyla (including the evolution of a head, limbs, motility, sexuality, and complex organs systems for circulation, digestion, and excretion). Future research should address the specific processes by which the traits supported here increase diversification, research that will presumably occur at smaller phylogenetic scales. Importantly, our results demonstrate which traits do (and do not) “scale up” to explain these large-scale diversity patterns, regardless of patterns at smaller scales.
Supplementary Material
Acknowledgements
T.J. was supported by a PERT postdoctoral fellowship (5K12GM000708-13). We thank Y. Michalakis, S. Heard, and two anonymous reviewers for many helpful comments that improved the manuscript.
Appendix A. Trait selection and character coding
Overview
We identified a suite of potentially relevant traits and tested whether they were significantly related to the diversification rates of animal phyla, both separately and together. We used traits for which character states are known for all phyla, based on data in the literature. We compiled information for 18 characters, including 17 binary characters and one continuous variable. Binary characters were coded as presence or absence (n=7), or as a proportion of species in a phylum exhibiting a given state (n=10). Characters were coded as presence or absence if they showed no (or little) variation within phyla. Proportions were used when character states varied among species within one or more phyla. One character (vision) was coded both ways, that is, as presence or absence as well as a proportion of species in a phylum exhibiting vision. Data on character states of phyla were primarily obtained from Hickman et al. (2012), which provides an extensive summary of anatomical knowledge on animal phyla. When relevant information was absent in this book, we searched for additional literature using Google Scholar.
For our analyses, we used 28 phyla (for all analyses) and 49 subdivided phyla (for some analyses) as described in Wiens (2015b). The names of phyla and taxa included within each phylum were generally congruent between Wiens (2015b) and Hickman et al. (2012). In a few cases, the phyla or subdivided phyla did not match perfectly between the two sources. However the available information could be applied to the classification used by Wiens (2015b). For example, Acanthocephala are considered as a separate phylum in Hickman et al. (2012), but are nested within Rotifera in Wiens (2015b). We therefore combined information on character states for Rotifera and Acanthocephala, assuming that Acanthocephala represent 1197 species out of 3246 species of Acanthocephala and Rotifera combined (Zhang 2013; Wiens 2015b).
Below, we describe in detail all characters and the justification for coding for each phylum, emphasizing information obtained from sources other than Hickman et al. (2012). That is, if the character state for a phylum is explicitly stated in Hickman et al. (2012), we do not justify the coding here. Character data for each phylum are listed in Supplementary File S1. Note that in the descriptions below, describing states as “0” or “1” does not imply primitive or derived states, and character state polarity is not relevant for the PGLS analyses conducted here.
1. Asexual reproduction
Classification: proportion of species that exhibit asexual reproduction (ranges from 0 to 1)
Character states: (0) no asexual reproduction; (1) asexual reproduction present.
We assessed the proportion of species in each phylum that exhibit asexual reproduction. As asexual reproduction, we considered parthenogenesis (e.g. in Gastrotricha, Rotifera), as well as reproduction via regeneration (e.g. in Echinodermata), or budding (e.g. in Cnidaria; Nielsen 2001; Hickman et al. 2012). Note that the vast majority of animal species exhibiting asexual reproduction also exhibit sexual reproduction (Hickman et al. 2012).
We considered asexual reproduction to be present in all species of Cnidaria and Ctenophora (Hinde 1998), Porifera, Placozoa, Hemichordata, and Rotifera (Nielsen 2001; Hickman et al. 2012), and Ectoprocta (Thomsen and Hakansson 1995). In Tardigrada, parthenogenesis is present but it is unclear whether it occurs in all the species (Bertolani 2001). We tentatively considered parthenogenesis to be present in 100% of Tardigrada. In five phyla asexual reproduction is present in only some species. For these phyla, proportion of species with asexual reproduction were derived as follows:
Echinodermata: Regeneration has been observed in some species of sea stars but the number of species capable of asexual reproduction is not known (Nielsen 2001; Hickman et al. 2012). We tentatively considered 25% of sea stars (Clade Asteroidea) to be capable of asexual reproduction, based on the number of species in genera (WoRMS Editorial Board 2015) in which asexual reproduction has been documented, as reported by Edmondson (1935). Twenty five percent of sea stars correspond to 10% of Echinodermata (Wiens 2015b).
Gastrotricha: many species in this phylum are parthenogenetic (Hickman et al. 2012) but the exact number of species capable of asexual reproduction is unclear. Given this uncertainty, we tentatively considered 50% of Gastrotricha species to exhibit asexual reproduction.
Nematoda: parthenogenesis is present in this phylum but it is quite rare (Nielsen 2001; Hickman et al. 2012). We assumed that it is present in less than 1% of species and we therefore considered Nematoda to not undergo asexual reproduction overall (value set to 0).
Nemertea: some species of this phylum reproduce asexually (Nielsen 2001; Hickman et al. 2012). The highly regenerative species belongs to the genus Lineus which comprise ~100 species (Coe 1930) out of ~1358 species of Nemertea (Wiens 2015b). We therefore considered the percentage of asexual species to be 7%.
Platyhelminthes: asexual reproduction is found mainly in turbellarians although it is unclear whether it is present in all species. We tentatively considered the number of asexually reproducing species to be equal to the number of turbellarians, which is 15% of Platyhelminthes (Ruppert et al. 2004).
2. Cephalization
Classification: presence or absence
Character states: (0) cephalization absent; (1) cephalization present.
Cephalization is an evolutionary trend wherein nervous tissue becomes concentrated towards the anterior end of an organism. Cephalization is typical for triploblastic animals (Hickman et al. 2012). Following Hickman et al. (2012), we therefore considered cephalization to be absent in all diploblastic phyla, that is, Cnidaria, Ctenophora, Placozoa, and Porifera. However, we also considered cephalization to be absent in Acoela and Echinodermata. Acoela is the basal triploblastic phylum but the members of this phylum exhibit diffuse set of nerves rather than anterior ganglia typical for cephalization (Hickman et al. 2012). Echinodermata also exhibit no cephalization (possibly secondarily), and the nervous system is represented by a circumoral ring (Nielsen 2001; Hickman et al. 2012).
3. Circulatory system
Classification: presence or absence
Character states: (0) circulatory system absent; (1) circulatory system present.
The circulatory system is an organ system that permits blood or hemolymph to circulate and transport nutrients, oxygen, carbon dioxide, and hormones to and from the cells in the body (Nielsen 2001; Hickman et al. 2012). Circulatory systems include open circulatory systems, wherein blood or hemolymph is not contained within blood vessels, as well as closed circulatory systems, wherein blood moves to and from tissues within blood vessels (Nielsen 2001; Hickman et al. 2012). Circulatory systems were considered absent in those phyla that exhibit direct diffusion of nutrients to cells from the digestive system (Nielsen 2001; Hickman et al. 2012). Data on the distribution of circulatory systems among phyla were obtained from Hickman et al. (2012).
4. Coelom
Classification: presence or absence
Character states: (0) acoelomate or pseudocoelomate; (1) coelomate
The presence and origin of the body cavity that surrounds and contains the digestive tract and other organs can be used to divide animal phyla into acoelomates (body cavity is absent), pseudocelomates (fluid-filled body cavity is present but is not derived from mesoderm), and coelomates (fluid-filled body cavity is present and is derived from the mesoderm; Hickman et al. 2012). The three character states are well established in the literature and known for most phyla (Nielsen 2001; Hickman et al. 2012; Nakano et al. 2013). Coelomates represent the most derived form, in which organs are attached to each other in a particular order while still being able to move freely within the cavity (Hickman et al. 2012). We therefore distinguished between phyla that are coelomates from those that exhibit the more primitive states of being acoelomate or pseudocoelomate, wherein organs are held in place and are not as well organized as in coelomates. To our knowledge, none of the phyla exhibit a mixture of different character states.
5. Digestive system
Classification: presence or absence
Character states: (0) digestive system absent or blind; (1) digestive system with two openings.
Digestion may occur either via diffusion, or through the presence of blind digestive system (i.e. presence of a gastrovascular cavity where the same opening is used for feeding and excretion), or through a digestive system with two openings, a mouth and an anus (Hickman et al. 2012). A digestive system with two openings is considered a potentially advantageous state, as it allows for continuous food intake and more efficient absorption of nutrients (Hickman et al. 2012). Therefore, we distinguished between taxa with a digestive system with two openings (state 1) and those with digestion via diffusion or a blind digestive system (state 0). Data for all phyla were obtained from Nielsen (2001) and Hickman et al. (2012). All phyla exhibit only one character state with the exception of Rotifera. In particular, Acanthocephala ((~34% of rotiferan species; Wiens 2015b) have no digestive track whereas other Rotifera have a linear digestive track. Using binary coding, we coded Rotifera with the state of the majority species (66%).
6. Dioecy
Classification: proportion of species that are dioecious (ranges from 0 to 1)
Character states: (0) monoecious; (1) dioecious
We assessed the proportion of species in each phylum that are dioecious (having male and female reproductive organs in separate individuals) as opposed to monoecious (having male and female reproductive organs in the same individual; Hickman et al. 2012). The following phyla were considered exclusively dioecious: Arthropoda, Brachiopoda, Chordata, Echinodermata, Kinorhyncha, Nematoda, Nematomorpha, Nemertea, Onychophora, Priapulida, Rotifera, (Hickman et al. 2012), and Tardigrada (Bertolani 2001). Bryozoa are mostly hermaphroditic (Thomsen and Hakansson 1995), so we considered them to be 0% dioecious. The reproduction of Xenoturbellida is poorly understood but they are likely monoecious (Nakano et al. 2013) so we also considered them to be 0% dioecious.
Six phyla exhibit both monoecy and dioecy. Their relative proportions were estimated as follows.
Annelida. In this phylum, Sedentaria and Sipuncula are dioecious, whereas Chaetopteridae, Errantia, and Myzostomida are monoecious (Hickman et al. 2012). A few species of Errantia are dioecious but we considered the number to be negligible and considered Errantia to be 100% monoecious. Sedentaria and Sipuncula together comprise 10,272 species out of 18,114 species of Annelida (Wiens 2015b) so 0.57 (10,272/18,114) of species in this phylum were considered dioecious.
Cnidaria. Anthozoa are mostly monoecious whereas Hydrozoa and Scyphozoa are mostly dioecious (Hickman et al. 2012). Anthozoa comprise 6,972 out of 13323 Cnidaria species (Wiens 2015b) so this phylum was considered 48% dioecious.
Entoprocta. This phylum is both monoecious and dioecious (Hickman et al. 2012), but the percentage of each character state is unknown. We therefore arbitrarily considered this phylum to be 50% dioecious.
Hemichordata. The families Ptychoderidae, Spengelidae_Torquaratoridae are dioecious whereas the family Harrimaniidae and the group Pterobranchia can be monoecious or dioecious (Hickman et al. 2012). We considered 50% of species in the clade that comprises Harrimaniidae and Pterobranchia (62 species) to be dioecious, which corresponds to 25% of hemichordate species (130 species total; Wiens 2015b).
Mollusca. In this phylum, Bivalvia, Cephalopoda, Monoplacophora, Polyplacophora, and Scaphopoda are dioecious (Hickman et al. 2012). Aplacophora (404 species; Wiens 2015b) exhibit both character states (Hickman et al. 2012) but at unknown frequencies. Therefore, we arbitrarily considered this group to be 50% dioecious. Gastropoda (61,105 species; Wiens 2015b) can be monoecious or dioecious. The monoecious Gastropoda belong predominantly to Pulmonata, which represents ~50% species of Gastropoda (Wade et al. 2001). Therefore, 50% of Gastropoda were considered dioecious. Given the total of 73,006 species of Mollusca, 50% of Aplacophora and 50% of Gastropoda being monoecious, we considered this phylum to be 58% dioecious (1-(404/2+61,105/2)/73,006). For the dataset of 49 higher-level clades, Aplacophora were considered 50% dioecious and the clade that comprises Gastropoda and Scaphopoda (61,676 species; Wiens 2015b) was considered 50% dioecious.
Phoronida. The majority of species in this phylum are considered monoecious (Nielsen 2001). The exact number of dioecious species is unknown so we arbitrarily considered 10% of the species in this phylum to be dioecious.
7. Excretory system
Classification: presence or absence
Character states: (0) excretory organ absent, (1) excretory organ present.
Excretory organs were considered to include glands cells, protonephridia, metanephridia, nephridia, malpighian glands, and kidneys (Nielsen 2001; Hickman et al. 2012). Organisms that lack these organs exhibit excretion via diffusion (Hickman et al. 2012). Data for all phyla with the exception of Gnathostomulida were obtained from Nielsen (2001) and Hickman et al. (2012). The presence of protonephridia in Gnathostomulida was based on Lammert (1985). Platyhelminthes generally have protonephridia with the exception of a few species of Turbellaria (Hickman et al. 2012). We considered this phylum to have excretory organs.
8. Legs
Classification: proportion of species with legs (ranges from 0 to 1)
Character states: (0) legs absent, (1) legs present
Legs (appendages used for locomotion) are considered generally present in Arthropoda, Onychophora, and Tardigrada (Hickman et al. 2012). Echinodermata possess appendages used for locomotion (Hickman et al. 2012) with the exception of Holothuroidea. Holothuroidea comprise 1,716 species of the 7,288 species of Echinodermata (Wiens 2015b). Therefore, the proportion of Echinodermata with legs is 0.76. Holothuroidea are nested within the higher-level clade Echinoidea, which comprises a total of 4,795 species (Wiens 2015b). Therefore, the proportion of Echinoidea with legs is 0.64 (i.e. 3,079/4,795). In Chordata, legs are generally present in tetrapods which represent 48% of chordate species (33,122 out of 69255 of Chordata; Wiens 2015b). Legs are considered absent in other phyla. Mollusca is a problematic phylum as some members possess an organ called a muscular foot (e.g. gastropods and bivalves) which has further evolved into tentacles in cephalopods (Shigeno et al. 2008). We did not consider a muscular foot to characterize presence of legs as the muscular foot is not specifically used for locomotion (i.e. some groups use it to attach themselves to a hard surface or to burrow into sediment). However, we did consider tentacles in cephalopods as legs as they are generally used for locomotion (Hickman et al. 2012). Cephalopoda consists of 811 species (Wiens 2015b) out of 73,006 species of Mollusca (Wiens 2015b), so the proportion of Mollusca with legs is 0.01.
9. Metamorphosis
Classification: proportion of species undergoing metamorphosis (ranges from 0 to 1)
Character states: (0) direct development; (1) metamorphosis.
Metamorphosis is defined as a process of transformation from an immature form to an adult form during which the individual undergoes drastic morphological or physiological changes (Wald 1981). However, identifying metamorphosis is not straightforward, since species vary greatly in the amount of change that they undergo during their transformation from a juvenile to an adult form. Herein, we generally follow Wald (1981), who provided a general overview of all groups undergoing metamorphosis (i.e. those having a larva) versus those undergoing direct development. Additional literature was accessed for phyla not included in Wald (1981), as described below.
All species in the phyla Cnidaria, Ctenophora, Nematomorpha, and Porifera undergo metamorphosis (Wald 1981), as do Bryozoa (Nielsen and Worsaae 2010). For Platyhelminthes, most undergo metamorphosis, although a few turbellarian species seemingly undergo direct development (Wald 1981). It is unclear how prevalent direct development is in this phylum. Therefore, metamorphosis was tenatively considered to be present in all species in this phylum.
The phyla Acoela, Gastrotricha, Gnathostomulida, Onychophora, Placozoa, Tardigrada, and Xenoturbellida are considered to undergo direct development (Wald 1981; Hickman et al. 2012; Nakano et al. 2013). The presence of metamorphosis in Nematoda is difficult to assess, as the juveniles resemble parents more than what is usually expected under metamorphosis (Wald 1981). We therefore tentatively consider Nematoda to undergo direct development. Similarly, juvenile forms of Chaetognatha resemble the adult forms although they have a different lifestyle (free swimming; Wald 1981). We tentatively consider this phylum to undergo direct development. The following phyla contain both species undergoing metamorphosis as well as those with direct development.
In Annelida, metamorphosis is present in the clade Sedentaria, which represents ~65% of annelid species (Wiens 2015b). Therefore, metamorphosis was considered to be present in 65% of species in this phylum.
In Arthropoda, the clades Pycnogonida, Xiphosura, Vericrustacea, and Hexapoda undergo metamorphosis, whereas Arachnida and Myriapoda undergo direct development (Gilbert 1981; Wald 1981; Hickman et al. 2012). Out of 1,257,040 species of Arthropoda, Arachnida and Myriapoda comprise 112,434 and 11,999 of species, respectively (Wiens 2015b). Therefore, 90% or Arthropoda were considered to undergo metamorphosis.
In Chordata, all species in Tunicata undergo metamorphosis, as do some species of Craniata, in particular some amphibians (Wald 1981). Wiens (2015a) estimated, that 70% of amphibians (corresponding to 5,082 out of 7,260 species) are aquatic for part of their lives. We tentatively used this proportion to estimate the number undergoing metamorphosis. Therefore, we estimate that in Craniata, comprising 66,199 species (Wiens 2015b), 7% of species (5,082/66,199) undergo metamorphosis. For the phylum Chordata (69,255 species; Wiens 2015b), metamorphosis was considered present in 3,206 species of tunicates and 5,082 species of amphibians (Wiens 2015b) which corresponds to 12% of Chordata. Note that we did not consider fish to undergo metamorphosis as the number of species that unquestionably meet the criteria of metamorphosis (i.e. lampreys; Wald 1981) is negligible (i.e. there are 62 species of lampreys; Wiens 2015b) with respect to the number of Chordates.
Hemichordata exhibits both direct development and development via metamorphosis but the ratio between the two character states is not clear (Wald 1981). We therefore arbitrarily considered 50% of species in this phylum to undergo metamorphosis.
In Mollusca, metamorphosis is present in Aplacophora, Bivalvia, Monoplacophora, Polyplacophora, Scaphopoda, and marine Gastropoda (Wald 1981; Hickman et al. 2012). Direct development occurs in terrestrial Gastropoda (Pulmonata), and in Cephalopoda (Wade et al. 2001). The group Pulmonata represents ~50% of 61,105 species of Gastropoda (Wade et al. 2001; Wiens 2015b) and Cephalopoda consists of 811 species (Wiens 2015b). Given 73,006 species of Mollusca (Wiens 2015b), we considered metamorphosis to occur in 57% of species in this phylum.
In Nemertea, metamorphosis is present in 1/3 (~33%) of species (Maslakova 2010).
10. Non-marine habitat
Classification: proportion of species inhabiting non-marine environment (ranges from 0 to 1)
Character states: (0) marine; (1) non-marine
Data were obtained from Wiens (2015b). Note that non-marine habitat includes both terrestrial and freshwater environments.
11. Parasitism
Classification: proportion of species that are parasitic (ranges from 0 to 1)
Character states: (0) not parasitic; (1) parasitic
Parasitism is defined as an obligatory relationship between two heterospecific organisms during which the parasite is metabolically dependent on the host (Cheng 1986g). The relationship may be permanent (e.g. tapeworm), or temporary (e.g. mosquitoes, leeches, and ticks; Cheng 1986g). Parasitism is present in five phyla for which the proportions of parasitic species were estimated as follows.
In Arthropoda, exclusive parasites can be found in Siphonaptera (2,500 species), Mallophaga (3,000 species), and Anoplura (500 species; (Cheng 1986d). Most species in Acarina/Metastigmata (ticks) are parasitic (Cheng 1986e; Cheng 1986d) so we considered parasitism to be present in 50% of the 48,200 species of Acarina (corresponding to the number of Metastigmata species within Acarina; (Chapman 2009). A small number of Diptera and Hymenoptera are also parasitic (Cheng 1986e; Cheng 1986d). We were unable to find the exact number of parasitic Diptera, and Hymenoptera so we tentatively considered parasitism to be present in 5% of the 153,000 species of Diptera (Chapman 2009), and 5% of the 115,000 species of Hymenoptera (Chapman 2009). Overall, we considered 37,756 out of 1,257,040 species of Arthropoda (Wiens 2015b) to be parasitic, which corresponds to 3%. For the dataset of 49 higher-level clades, the number of parasitic species corresponded to 13% in Arachnida (24,100 out of 112,434 species; Wiens 2015b) and 1% in Hexapoda (13,567 out of the 1,063,532 species; Wiens 2015b). Note that we did not consider herbivorous insects to be parasitic (following the review on arthropod parasitism cited here; Cheng 1986d). If we did, this would increase the proportion of parasitic arthropods even further, and potentially show an even stronger relationship between parasitism and diversification among animal phyla.
In Annelida, all species in Myzostomida are parasitic and a negligible number of species in Hirudinea are parasitic (Cheng 1986f). We considered Myzostomida to be parasitic, which represent ~1% of annelid species (Wiens 2015b).
We considered 60% of nematode species to be parasitic (Lambert and Bekal 2002).
In Platyhelminthes, all species in Trematoda, Monogenea, and Cestoda are parasitic (Cheng 1986c; Cheng 1986b). These three groups contain ~85% of all species of Platyhelminthes (Ruppert et al. 2004).
In Rotifera, all species in Acanthocephala were considered parasitic (Cheng 1986a). Acanthocephalans comprise ~34% of rotiferan species (Wiens 2015b),
The following phyla have negligible (<1%) numbers of parasite species: Porifera, Ctenophora, Mollusca, Rotifera, and Chordata (Cheng 1986f). These and all other phyla were therefore considered non-parasitic.
12. Segmentation
Classification: presence or absence
Character states: (0) segmentation absent, (1) segmentation present
Body segmentation represents division of the body into a series of repetitive segments that are often grouped into larger functional units (Hickman et al. 2012). Body segmentation evolved independently several times (e.g. in protostomes and deuterostomes; (Nielsen 2001). Segmentation is associated with the presence of a coelom (Hickman et al. 2012) and true segmentation (also called coelomic segmentation), occurs in Annelida, Arthropoda, Chordata, Onychophora, and Tardigrada (Tautz 2004; Hickman et al. 2012). We followed this classification of true segmentation and did not consider weak, non-coelomic segmentation, as occurring in Kinorhyncha, Platyhelminthes or Rotifera. Also, the secondary loss of segmentation in some Echiura (within Annelida) was not considered as less than 1% of annelid species lack segmentation (Hickman et al. 2012). Otherwise, no phylum exhibited a mixture of both character states.
13. Size
Classification: continuous
Unit: median biovolume of the phylum in mm3
Data on biovolume values for most phyla (median value among sampled species within a phylum) were taken directly from Orme et al. (2002). Biovolume for a given species was calculated as the product of the length, width, and height of the maximum recorded dimensions for that species (Orme et al. 2002). The median biovolume was calculated for each phylum to represent a central tendency among sampled species (Orme et al. 2002). The values for Porifera, Ctenophora, and Cnidaria were not available in Orme et al. (2002) and we therefore calculated values for each of these phyla as an average between the biovolume of the smallest and the largest species in each phylum. The latter data were taken from McClain and Boyer (2009). We did not include this trait in the analysis of our dataset of 49 higher-level clades, as we were unable to obtain values for some subclades.
14. Skeleton
Classification: proportion of species that possess a skeleton (ranges from 0 to 1)
Character states: (0) skeleton absent; (1) skeleton.
We identified the proportion of species in each phylum possessing a skeleton, defined as an internal or external framework of rigid material supporting, containing, or protecting the body of the animal. We considered skeletons to include an exoskeleton (external skeleton secreted by ectoderm or epidermis that is usually made of chitin or collagen) and endoskeleton (skeleton that develops within the skin or in the deeper body tissues; Hickman et al. 2012).
Following Hickman et al. (2012), we considered a skeleton to be present in all species of Arthropoda (exoskeleton), Brachiopoda (exoskeleton), Bryozoa (exoskeleton), Chordata (endoskeleton), Echinodermata (endoskeleton), Phoronida (exoskeleton), and Porifera (exoskeleton). The following three phyla exhibit a mixture of species that possess a skeleton and species that do not possess a skeleton.
In Annelida, an exoskeleton is present in Sedentaria, Myzostomida, Chaetopteridae, and Sipuncula (Hickman et al. 2012). These groups represent ~65% of Annelida species (Wiens 2015b). Therefore the value for this character was set to 0.65.
In Cnidaria, an exoskeleton is present in Anthozoa (Hickman et al. 2012), which represents ~52% of cnidarian species (Wiens 2015b).
In Mollusca, an exoskeleton is present in Bivalvia, Monoplacophora, Polypacophora, and Scaphopoda (Hickman et al. 2012). In Gastropoda, some species have secondarily lost their shells (e.g. slugs) but remnants of a shell are usually still present (Hickman et al. 2012). Therefore, we considered all Gastropoda to have an exoskeleton. In Cephalopoda, only one species (Nautilus) possesses an exoskeleton and a few species possess an endoskeleton (e.g. cuttlefish). However, the majority of Cephalopoda do not possess a skeleton (Hickman et al. 2012). We therefore considered Cephalopoda to be without a skeleton. Given that cephalopods comprise ~1% of molluscan species (Wiens 2015b), we considered the proportion of Mollusca possessing a skeleton to be 0.99.
15. Symmetry
Classification: Proportion of species in a phylum with bilateral symmetry.
Character states: (0) asymmetrical or with radial symmetry; (1) bilateral symmetry
Body symmetry is the distribution of matching body parts within an organism, and its distribution among animal phyla is well established in the literature (Hickman et al. 2012). All but one phyla exhibit only one type of symmetry. Porifera have a disparate arrangement of body parts and therefore exhibit an asymmetrical body plan. The phyla Cnidaria, Ctenophora, and Placozoa have a radial symmetry, a form of symmetry wherein identical body parts are arranged in a circular fashion around a central axis. These four phyla exhibiting asymmetry or radial symmetry (Porifera, Cnidaria, Ctenophora, and Placozoa) were coded as “0”. All other phyla (with the exception of Echinodermata) exhibit bilateral symmetry, as they have an equal arrangement of parts along an axis running from head to tail. These phyla were coded “1”.
Echinodermata is the only phylum that exhibits both types of symmetry. Echinodermata exhibit radial symmetry with the exception of Holothuroidea, which are bilateral (Hickman et al. 2012). Holothuroidea comprise 1,716 species of the 7,288 species of Echinodermata (Wiens 2015b). Therefore, the proportion of bilateral Echinodermata is 0.24. For our dataset of 49 higher-level clades, Holothuroidea are nested within the clade Echinoidea, which comprises a total of 4,795 species (Wiens 2015b). Therefore, the proportion of bilateral Echinoidea is 0.36 (i.e. 1,716/4,795).
16. Terrestrial habitat
Classification: proportion of species inhabiting terrestrial environment (ranges from 0 to 1)
Character states: (0) aquatic; (1) terrestrial
Data were obtained from Wiens (2015b). Note that aquatic habitats here include both freshwater and marine environments.
17. Vagility
Classification: proportion of species that are vagile (range from 0–1)
Character states: (0) sessile; (1) vagile.
Species coded as vagile were those in which the adult form is free-moving. Sessile species were those whose adult form is attached to a substrate and stays in one place for its entire adult life (Webb 1969; Nielsen 2001; Hickman et al. 2012). Parasitic species were generally considered vagile, since they often move inside the host (Cheng 1986g). However, we recognize that at least some species of parasites (e.g. tapeworms) could be classified as sessile also. Vagility is the most common state among animal phyla, although some phyla are exclusively sessile and some exhibit a mixture of vagility and sessility (Webb 1969). Following Webb (1969), we considered as exclusively sessile the phyla Brachiopoda, Bryozoa, Entoprocta, Phoronida, and Porifera. The proportion of vagile species in these phyla was therefore set to 0. Phyla with both vagile and sessile species include Annelida, Cnidaria, Hemichordata, and Rotifera. The remaining phyla were considered exclusively vagile. For phyla with mixed vagility, the proportions were derived as follows,
Annelida: Vagile species belong to the clade Errantia which represents 35% (6,250 out of 18,114) of all species of Annelida (Wiens 2015b). Therefore, the proportion of vagility in Annelida was considered 0.35. For our dataset of 49 higher-level clades, Chaetopteridae, Myzostomida, Sedentaria, and Sipuncula were considered exclusively sessile whereas Errantia was considered exclusively vagile.
Cnidaria: Sessile cnidarians belong to the clade Anthozoa, which includes 6,972 out of 13,323 species of Cnidaria (Wiens 2015b). Therefore, the proportion of vagile species was set to 0.48. For our dataset of 49 higher-level clades, the clade Anthozoa was considered exclusively sessile, whereas the clade comprising Medusozoa and Myxozoa was considered exclusively vagile.
Hemichordata: Sessile hemichordates belong to the clade Pterobranchia, which includes ~23 species (WoRMS Editorial Board 2015) of a total of 130 species of Hemichordata (Wiens 2015b). Therefore, the proportion of vagile species of Hemichordata was estimated to be 0.82. For our dataset of 49 higher-level clades, Pterobranchia are grouped with Harrimaniidae, with the latter being a vagile group (Deland et al. 2010). The total number of species in the group that comprises Pterobranchia plus Harrimaniidae is 62 (Wiens 2015b; WoRMS Editorial Board 2015), therefore the proportion of vagile species in this higher-level clade was set to 0.63 (23/62).
Rotifera: All species in the families Collothecidae, Conochilidae, and Flosculariidae, are sessile (Wallace 1980). The total, combined number of species in these three families is 200, based on the Rotifer World Catalog (accessed at http://rotifera.hausdernatur.at/ on the 1st Dec 2015). The total number of rotifers is 3,246 (Zhang 2013; Wiens 2015b), therefore the proportion of vagile rotifers was estimated to be 0.94.
18a. Vision
Classification: presence or absence
Character states: (0) no photoreceptor and no vision; (1) photoreceptor or vision
Each phylum for which at least some members possess either eyes or photoreceptors were coded as 1, whereas phyla in which neither eyes nor photoreceptors were found were coded as 0. We used presence-absence coding instead of estimating the percentage of species in a phylum that possess photoreceptors or vision because photoreceptors often remain undetected for taxa that have not been closely examined (Autrum et al. 1979). Therefore it would be very difficult to determine the proportion of species in a given phylum that do or do not have photoreceptors.
Herein, we follow the characterization of phyla from Autrum et al. (1979) and Ali (1982), augmented from other sources as needed. Note that we reference individual chapters from Autrum et al. (1979) and Ali (1982) as these two books represent an assemblage of independent manuscripts written by different authors.
We did not find any reports of photoreceptors or eyes in the following phyla: Brachiopoda, Ctenophora, Entoprocta, Gastotricha, Gnathostomulida, Hemichordata, Nemertea, Phoronida, Placozoa, Porifera, Priapulida, and Xenoturbellida. For the dataset of 49 higher-level clades, neither eyes nor photoreceptors were documented in Mezostomida within the Annelida (Purschke 2005) nor in Aplacophora within Mollusca (Boyle 1969; Autrum et al. 1979).
Photoreception has been reported as present in at least some members of the phyla Acoela (Yoshida 1979), Cnidaria (Burr 1982), Echinodermata (Menzel 1979; Burr 1982), , Kinorhyncha (Hickman et al. 2012), Nematoda (Burr 1982), Nematomorpha (Hickman et al. 2012), and Platyhelminthes (Fournier 1984). Photoreception seems to be present in all members of the phyla Bryozoa (Burr 1982), Chaetognatha (Goto and Yoshida 1982), Onychophora (Hickman et al. 2012), and Tardigrada (Greven 2007). Combinations of photoreception and eyes are present in Annelida, Chordata, and Mollusca and eyes are exclusively present in Arthropoda (Autrum et al. 1979; Nielsen 2001; Purschke 2005; Hickman et al. 2012).
18b. Vision (alternative coding)
Classification: proportion of species with vision (ranges from 0 to 1)
Character states: (0) eyes absent; (1) eyes present.
We also explored an alternative coding for vision, wherein we coded the proportion of species in each phylum that possess eyes. Possession of eyes, however, is not straightforward. Eyes evolved independently several times from simpler photoreceptors, with no clear distinction between eyes and photoreceptors (Autrum et al. 1979; Ali 1982). Lens-bearing eyes and compound eyes (Ali 1982) were considered as present in the following four phyla.
Within Annelida, photoreception and vision with eyes are present exclusively in Sedentaria (Autrum et al. 1979; Purschke 2005). The complexity of eyes in Sedentaria increases from simple photoreceptors to well-developed eyes with no clear distinction between photoreceptors and vision (Purschke 2005). Therefore, we arbitrarily considered vision present in 50% of Sedentaria. Given 10,125 species of Sedentaria and 18,114 species of Annelida, this yields a proportion of 0.3 annelid species with vision. For our dataset of 49 higher-level clades, the value was set to 0.5 for Sedentaria whereas all other Annelida clades were set at 0.
In Arthropoda, vision was considered present in all species (Hickman et al. 2012).
Within Chordata, vision is present in Craniata but absent in Tunicata and Cephalochordata (Hickman et al. 2012). Tunicata and Cephalochordata together comprise 4% of chordate species (Wiens 2015b), so the proportion of species with eyes was estimated to be 0.96 for this phylum.
In Mollusca, all members of Cephalopoda (811 species; Wiens 2015b) possess eyes (Ali 1982). In Bivalvia, eyes are found only in scallops of the family Pectinidae (Boyle 1969; Autrum et al. 1979), with ~516 species (WoRMS Editorial Board 2015). In Gastropoda (61,105 species; Wiens 2015b), there is a continuum from simple photoreceptors to complex eyes (Burr 1982) so the exact number of species with each state is unclear. Therefore, we arbitrarily considered 50% of gastropod species to have eyes. Overall, given a total number of 73,006 molluscan species (Wiens 2015b), the proportion of species with vision was estimated to be 0.44 ((811+516+61,105/2)/73,006). For the dataset of 49 higher-level clades, Cephalopoda and Monoplacophora are grouped together, and the former group has eyes whereas the latter group does not. Given 31 species of Monoplacophora and 811 species of Cephalopoda (Wiens 2015b), the proportion of species with vision in the clade Cephalopoda + Monoplacophora group was estimated at 0.96. For Bivalvia, the family Pectinidae includes 6% of the total number of bivalve species (Boyle 1969; Autrum et al. 1979), and so the value was estimated to be 0.06. For the higher-level clade Gastropoda + Scaphopoda, Scaphopoda do not have eyes whereas 50% of Gastropoda were considered as having eyes (see above). Considering 571 species of Scaphopoda and a total of 61,676 species in this higher-level clade (Wiens 2015b), the proportion of species with vision for this clade was estimated to be 0.5 ((571+61,105/2)/61,676).
Appendix B. Methodology for estimating projected species richness
In addition to using numbers of described species, we also tested the relationship between traits and diversification rates among phyla using the projected species richness values for each phylum, given that the number of currently described species may underestimate actual species richness for many phyla. These projected species richness values were then used to calculate net diversification rates (Magallón and Sanderson 2001), as described in the main text.
The projected number of species for exclusively marine phyla were obtained from estimates by Appeltans et al. (2012). When available, we used the “total estimated” value from Table 2 in Appeltans et al. (2012). When unavailable, the number of projected species was calculated based on the “% known” value in the same table. For several phyla, a range of values was given (e.g. total estimated number of Echinodermata is 7,288–9,617 species) and in these cases we used the midpoint of these ranges (e.g. 8453 projected species of Echinodermata). For Cnidaria, the estimated number of species was not provided directly by Appeltans et al. (2012) but could be calculated from their estimated numbers for individual subclades. For Priapulida and Xenoturbellida, no estimated species numbers were provided by Appeltans et al. (2012) so we simply used the number of described species (note that these two phyla only have 19 and 2 described species, respectively; Wiens 2015b). For Brachiopoda, the estimated number of species provided by Appeltans et al. (2012) is 388 which is less than the 392 described species used by Wiens (2015b). We therefore used 392 for Brachiopoda.
The projected number of species for most phyla that are not exclusively marine (with the exception of Arthorpoda and Nematoda, see below) were obtained from Chapman (2009). We used the “Estimate World” values provided by Chapman (2009) for most phyla. For Tardigrada, Gastotricha, and Rotifera, Chapman (2009) did not provide a number of projected species. Therefore, we used the estimate number of marine species for these phyla from Appeltans et al. (2012) and extrapolated this estimate to all species in each of the three phyla. For example, Appeltans et al. (2012) estimated that 13–19% of marine Gastotricha are described. We used the same percentage (that is, 13–19%) for all Gastotricha and calculated the projected number of Gastotricha species to be 5260 (((812*100/13)+(812*100/19))/2=5,260), given that the total number of described species is 812 (Wiens 2015b). We also used the extrapolation of estimated numbers of marine species from Appeltans et al. (2012) for Entoprocta and Bryozoa because the projected number of species provided by Chapman (2009) is less than the number of described species provided by Wiens (2015b).
Most arthropods are insects, and the actual number of living insect species is generally agreed to be much higher than the number of described species (~1 million; Roskov et al. 2016). However, the magnitude of undescribed diversity has been relatively uncertain, with some estimates ranging as high as ~30 million species (e.g. Erwin 1982). Nevertheless, in the past ~15 years, many estimates appear to have converged on a similar value of ~5 million species (Novotny et al. 2002; Basset et al. 2012; Stork et al. 2015). Furthermore, Stork et al. (2015) recently estimated total terrestrial arthropod richness to be 5.9–7.4 million, with a mean among estimates of 6.8 million. We use this mean estimate here, but with the important caveat that it might underestimate arthropod richness given the many species of mites that occur on insects and other terrestrial arthropods (Walter and Proctor 2013). Furthermore, this estimate does not include cryptic species revealed by molecular methods. However, we add to this number (6.8 million) to the approximate number of marine arthropods (maximum ~370,000; Appeltans et al. 2012), to yield a total of 7.2 million arthropods, of which 6% are marine and 94% are terrestrial.
Species richness of Nematoda may be greatly underestimated given the numerous nematode species that occur on arthropods, especially insects. We tentatively estimate that there is (on average) at least one nematode species per insect species. This estimate is based on studies of nematode diversity in the most diverse insect orders (Coleoptera, Diptera, Hymenoptera, Lepidoptera). For example Grucmanová and Holuša (2013) summarized the nematode fauna of beetles in the genus Ips in Central Europe. Each beetle species hosted an average of 3.2 unique nematode species. Diptera have also been shown to host many nematode species. As one example, simulid flies can host several genera of mermithid nematodes (Poinar 1977). In Hymenoptera, nematodes are known to infect both wasps (e.g. Powers et al. 2009; Saito-Morooka 2014), and ants (Poinar 2012). Powers et al. (2009) suggested that each fig species they examined in a site in Costa Rica (La Selva) contained two unique nematode species in their fruits, with different fig wasp species associated with each fig species. In ants, many genera and families of nematode can infect a single host species or genus (the ant genus Lasius may host species in the nematode families Mermithidae, Diplogratridae, and Rhabditidae; Poinar 2012). Furthermore, some nematode families have species that can be specific to a single ant host species (e.g. Mermithidae; Poinar 2012).
We know of few studies of nematodes in Lepidoptera. However, nematodes are known in other insect orders. In Orthoptera (grasshoppers, crickets, and relatives), it is clear that each host species can harbor many nematode species, and that these can show some host specificity within the order. For example, Camino and Achinelly (2011) described nematodes in two sympatric orthopteran species from two families. They found 15 and 8 unique nematode species per host species. In Isoptera (termites) there are estimated to be 0.8 unique endoparasitic nematode species per termite species in a site in Costa Rica (Powers et al. 2009). These authors also summarized data from previous studies suggesting this ratio may be widespread for termite species in temperate and tropical sites (Powers et al. 2009).
Overall, we tentatively estimate that there is (on average) at least one nematode species per terrestrial arthropod species. Therefore, we estimate that there are 6.8 million nematode species. We did not include nematodes parasitizing other organisms besides arthropods or free-living nematodes. Poulin and Morand (2004) summarized the species richness and host specificity of nematodes parasitizing various major vertebrate clades. Almost half of vertebrate species are actinopterygian fish, yet each hosts an average of only 0.145 unique nematode species each, and other vertebrate groups have ~1 nematode species per host species (or fewer). Therefore, we assumed that nematodes parasitizing other animals would not significantly impact our estimate of 6.8 million nematodes overall. Similarly, free-living nematodes can be highly diverse, especially in soils and possibly in deep marine environments. However, estimates of >1 million free-living nematodes have been challenged by the authors of some of these same estimates (Lambshead and Boucher 2003). Furthermore, Appeltans et al. (2012) estimated only 64,000 marine nematodes (both free-living and parasitic). Given this, we tentatively assumed that free-living nematodes would not significantly impact our (already large) estimate of 6.8 million nematode species. We assumed that this huge overall number of undescribed nematodes are predominantly terrestrial and predominantly parasitic.
References from the Appendices
- Ali MA. Photoreception and vision in invertebrates. University of Montreal; Montreal, Quebec, Canada: 1982. [Google Scholar]
- Appeltans W, Ahyong ST, Anderson G, Angel MV, Artois T, Bailly N, Bamber R, et al. The Magnitude of Global Marine Species Diversity. Current Biology. 2012;22:2189–2202. doi: 10.1016/j.cub.2012.09.036. [DOI] [PubMed] [Google Scholar]
- Autrum H, Bennett MF, Diehn B, Hamdorf K, Heisenberg M, Järvilehto M, Kunze P, et al. Comparative Physiology and Evolution. Vision in Invertebrates A: Invertebrate Photoreceptors. Springer-Verlag; Berlin, Heidelberg, New York: 1979. [Google Scholar]
- Basset Y, Cizek L, Cuénoud P, Didham RK, Guilhaumon F, Missa O, Novotny V, et al. Arthropod diversity in a tropical forest. Science. 2012;338:1481–1484. doi: 10.1126/science.1226727. [DOI] [PubMed] [Google Scholar]
- Bertolani R. Evolution of the reproductive mechanisms in tardigrades - A review. Zoologischer Anzeiger. 2001;240:247–252. [Google Scholar]
- Boyle PR. Fine structure of eyes of Onithochiton neglectus (Mollusca - Polyplacophora). Zeitschrift Fur Zellforschung Und Mikroskopische Anatomie. 1969;102:313–332. doi: 10.1007/BF00335443. [DOI] [PubMed] [Google Scholar]
- Burnham KP, Anderson DR. Multimodel inference - understanding AIC and BIC in model selection. Sociological Methods & Research. 2004;33:261–304. [Google Scholar]
- Burr AH. Evolution of eyes and photoreceptor organelles in lower phyla. In: Ali MA, editor. Photoreception and vision in invertebrates. University of Montreal; Montreal, Quebec, Canada: 1982. [Google Scholar]
- Camino N, Achinelly M. Biodiversity of insect-parasitic nematodes in soil pest insect (Orthoptera, Gryllidae and Gryllotalpidae) in wheat fields of Buenos Aires, Argentina. Anales de Biología. 2011;33:15–21. [Google Scholar]
- Chapman AD. Numbers of living species in Australia and the World. 2nd edition 2009. pp. 1–84. [Google Scholar]
- Cheng TC. Acanthocephala: The spiny-headed worms. In: Cheng TC, editor. General Parasitology. Second Edition Academic Press; San Diego: 1986a. pp. 445–464. [Google Scholar]
- Cheng TC. Cestoidea: The tapeworms Cestodaria: The unsegmented tapeworms. In: Cheng TC, editor. General Parasitology. Second Edition Academic Press; San Diego: 1986b. pp. 378–386. [Google Scholar]
- Cheng TC. Eucestoda: The true tapeworms. In: Cheng TC, editor. General Parasitology. Second Edition Academic Press; San Diego: 1986c. pp. 387–444. [Google Scholar]
- Cheng TC. Introduction to the parasitic arthropods. In: Cheng TC, editor. General Parasitology. Second Edition Academic Press; San Diego: 1986d. pp. 572–591. [Google Scholar]
- Cheng TC. The mites. In: Cheng TC, editor. General Parasitology. Second Edition Academic Press; San Diego: 1986e. pp. 592–613. [Google Scholar]
- Cheng TC. Other zooparasites. In: Cheng TC, editor. General Parasitology. Second Edition Academic Press; San Diego: 1986f. pp. 706–787. [Google Scholar]
- Cheng TC. Parasitism and symbiosis. In: Cheng TC, editor. General Parasitology. Second Edition Academic Press; San Diego: 1986g. pp. 1–34. [Google Scholar]
- Coe WR. Asexual reproduction in nemerteans. Physiological Zoology. 1930;3:297–308. [Google Scholar]
- Deland C, Cameron CB, Rao KP, Ritter WE, Bullock TH. A taxonomic revision of the family Harrimaniidae (Hemichordata: Enteropneusta) with descriptions of seven species from the Eastern Pacific. Zootaxa; 2010. pp. 1–30. [Google Scholar]
- Edmondson CH. Autotomy and regeneration in Hawaiian star-fishes. Occasional Papers of the Bishop Museum Honolulu. 1935;11:1–29. [Google Scholar]
- Erwin TL. Tropical forests their richness in Coleoptera and other arthropod species. Coleopterists Bulletin. 1982;36:74–75. [Google Scholar]
- Fournier A. Photoreceptors and Photosensitivity in Platyhelminthes. In: Ali MA, editor. Photoreception and Vision in Invertebrates. NATO ASI Series. Springer; US: 1984. pp. 217–239. [Google Scholar]
- Gilbert LI. Metamorphosis: a problem in developlental biology. 2nd edition Plenum Press; New York: 1981. [Google Scholar]
- Goto T, Yoshida M. Photoreception in Chaetognatha. In: Ali MA, editor. Photoreception and vision in invertebrates. University of Montreal; Montreal, Quebec, Canada: 1982. [Google Scholar]
- Greven H. Comments on the eyes of tardigrades. Arthropod Structure & Development. 2007;36:401–407. doi: 10.1016/j.asd.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Grucmanová Š, Holuša J. Nematodes associated with bark beetles, with focus on the genus Ips (Coleoptera: Scolytinae) in Central Europe. Acta Zoologica Bulgarica. 2013;65:547–556. [Google Scholar]
- Hinde RT. The Cnidaria and Ctenophora. In: Anderson DT, editor. Invertebrate Zoology. Oxford University Press; Oxford: 1998. pp. 28–57. [Google Scholar]
- Lambert K, Bekal S. Introduction to plant-parasitic nematodes. The Plant Health Instructor; 2002. DOI:10.1094/PHI-I-2002-1218-01. [Google Scholar]
- Lambshead PJD, Boucher G. Marine nematode deep-sea biodiversity - hyperdiverse or hype? Journal of Biogeography. 2003;30:475–485. [Google Scholar]
- Lammert V. The fine structure of protonephridia in Gnathostomulida and their comparison within Bilateria. Zoomorphology. 1985;105:308–316. [Google Scholar]
- Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. Version 2.75. 2011 Available at: http://mesquiteproject.org.
- Maslakova SA. Development to metamorphosis of the nemertean pilidium larva. Frontiers in Zoology. 2010;7:1–18. doi: 10.1186/1742-9994-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel R. Spectral sensitivity and color vision in invertebrates. In: Autrum H, editor. Comparative Physiology and Evolution of Vision in Invertebrates A: Invertebrate Photoreceptors. Springer-Verlag Berlin; Heidelberg, New York: 1979. [Google Scholar]
- Nakano H, Lundin K, Bourlat SJ, Telford MJ, Funch P, Nyengaard JR, Obst M, et al. Xenoturbella bocki exhibits direct development with similarities to Acoelomorpha. Nature Communications. 2013;4:1–6. doi: 10.1038/ncomms2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen C, Worsaae K. Structure and occurrence of Cyphonautes larvae (Bryozoa, Ectoprocta). Journal of Morphology. 2010;271:1094–1109. doi: 10.1002/jmor.10856. [DOI] [PubMed] [Google Scholar]
- Novotny V, Basset Y, Miller SE, Weiblen GD, Bremer B, Cizek L, Drozd P. Low host specificity of herbivorous insects in a tropical forest. Nature. 2002;416:841–844. doi: 10.1038/416841a. [DOI] [PubMed] [Google Scholar]
- Poinar G., Jr. Nematode parasites and associates of ants: past and present. Psyche (Cambridge) 2012;192017:1–13. [Google Scholar]
- Poinar GO., Jr. A synopsis of the nematodes occurring in blackflies (Diptera: Simuliidae). Bulletin of the World Health Organization. 1977;55:509–515. [PMC free article] [PubMed] [Google Scholar]
- Poulin R, Morand S. Parasite Biodiversity. Smithsonian; Washington, DC.: 2004. [Google Scholar]
- Powers TO, Neher DA, Mullin P, Esquivel A, Giblin-Davis RM, Kanzaki N, Stock SP, et al. Tropical nematode diversity: vertical stratification of nematode communities in a Costa Rican humid lowland rainforest. Molecular Ecology. 2009;18:985–996. doi: 10.1111/j.1365-294X.2008.04075.x. [DOI] [PubMed] [Google Scholar]
- Purschke G. Sense organs in polychaetes (Annelida). In: Bartolomaeus T, Purschke G, editors. Morphology, molecules, evolution and phylogeny in Polychaeta and related taxa. Developments in Hydrobiology; Springer Netherlands; 2005. pp. 53–78. [Google Scholar]
- Ruppert EE, Fox RS, Barnes RD. Invertebrate Zoology: A Functional Evolutionary Approach. Seventh Edition Thomson, Brooks/Cole; New York: 2004. [Google Scholar]
- Saito-Morooka F. The prevalence of the parasitic nematode Sphaerularia sp in the overwintering gynes of Parapolybia spp. (Hymenoptera, Polistinae). Journal of Hymenoptera Research. 2014;38:37–43. [Google Scholar]
- Shigeno S, Sasaki T, Moritaki T, Kasugai T, Vecchione M, Agata K. Evolution of the cephalopod head complex by assembly of multiple molluscan body parts: Evidence from Nautilus embryonic development. Journal of Morphology. 2008;269:1–17. doi: 10.1002/jmor.10564. [DOI] [PubMed] [Google Scholar]
- Stork NE, McBroom J, Gely C, Hamilton AJ. New approaches narrow global species estimates for beetles, insects, and terrestrial arthropods. Proceedings of the National Academy of Sciences, USA. 2015;112:7519–7523. doi: 10.1073/pnas.1502408112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D. Segmentation. Developmental Cell. 2004;7:301–312. doi: 10.1016/j.devcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Thomsen E, Hakansson E. Sexual versus asexual dispersal in clonal animals: Examples from cheilostome bryozoans. Paleobiology. 1995;21:496–508. [Google Scholar]
- Wade CM, Mordan PB, Clarke B. A phylogeny of the land snails (Gastropoda: Pulmonata). Proceedings of the Royal Society of London B: Biological Sciences. 2001;268:413–422. doi: 10.1098/rspb.2000.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wald G. In: Metamorphosis: An Overview. Metamorphosis: a problem in developmental biology. Gilbert L, Frieden E, editors. Plenum Press; New York and London: 1981. pp. 1–39. [Google Scholar]
- Wallace RL. Ecology of sessile rotifers. Hydrobiologia. 1980;73:181–193. [Google Scholar]
- Walter DE, Proctor HC. Mites: ecology, evolution, and behavior. Life at a microscale. Second edition Springer; 2013. [Google Scholar]
- Webb M. An evolutionary concept of some sessile and tubicolous animals. Sarsia. 1969;38:1–8. [Google Scholar]
- WoRMS Editorial Board [2015-12-01];World Register of Marine Species. 2015 Available from http://www.marinespecies.org at VLIZ.
- Yoshida M. Extraocular photoreception. In: Autrum H, editor. Comparative Physiology and Evolution of Vision in Invertebrates A: Invertebarte Photoreceptors. Springer-Verlag Berlin; Heidelberg, New York: 1979. [Google Scholar]
Literature Cited
- Alroy J. The shifting balance of diversity among major marine animal groups. Science. 2010;329:1191–1194. doi: 10.1126/science.1189910. [DOI] [PubMed] [Google Scholar]
- Althoff DM, Segraves KA, Johnson MTJ. Testing for coevolutionary diversification: linking pattern with process. Trends in Ecology and Evolution. 2014;29:82–89. doi: 10.1016/j.tree.2013.11.003. [DOI] [PubMed] [Google Scholar]
- Bengtson S, Zhao Y. Predatorial borings in late precambrian mineralized exoskeletons. Science. 1992;257:367–369. doi: 10.1126/science.257.5068.367. [DOI] [PubMed] [Google Scholar]
- Benton MJ. Biodiversity on land and in the sea. Geological Journal. 2001;36:211–230. [Google Scholar]
- Carrete-Vega G, Wiens JJ. Why are there so few fish in the sea? Proceedings of the Royal Society of London, Series B: Biological Sciences. 2012;279:2323–2329. doi: 10.1098/rspb.2012.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Queiroz A. Do image-forming eyes promote evolutionary diversification? Evolution. 1999;53:1654–1664. doi: 10.1111/j.1558-5646.1999.tb04551.x. [DOI] [PubMed] [Google Scholar]
- Dunn CW, Giribet G, Edgecombe GD, Hejnol A. Animal phylogeny and its evolutionary implications. Annual Review of Ecology, Evolution, and Systematics. 2014;45:371–395. [Google Scholar]
- Ehrlich PR, Raven PH. Butterflies and plants: a study in coevolution. Evolution. 1964;18:586–608. [Google Scholar]
- Erwin DH, Laflamme M, Tweedt SM, Sperling EA, Pisani D, Peterson KJ. The Cambrian conundrum: Early divergence and later ecological success in the early history of animals. Science. 2011;334:1091–1097. doi: 10.1126/science.1206375. [DOI] [PubMed] [Google Scholar]
- Feder JL, Forbes AA. Sequential speciation and the diversity of parasitic insects. Ecological Entomology. 2010;35:67–76. [Google Scholar]
- Foote M, Miller AI, Raup DM, Stanley SM. Principles of Paleontology. Macmillan; New York: 2007. [Google Scholar]
- Futuyma DJ, Agrawal AA. Macroevolution and the biological diversity of plants and herbivores. Proc. Natl Acad. Sci. USA. 2009;106:18054–18061. doi: 10.1073/pnas.0904106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard SB, Hauser DL. Key evolutionary innovations and their ecological mechanisms. Historical Biology. 1995;10:151–173. [Google Scholar]
- Hembry DH, Yoder JB, Roesch Goodman K. Coevolution and the diversification of life. American Naturalist. 2014;184:425–438. doi: 10.1086/677928. [DOI] [PubMed] [Google Scholar]
- Hickman C, Roberts L, Keen S, Larson A, Elsenhour D. Animal Diversity. 6th edn. McGraw-Hill Higher Education; NY: 2012. [Google Scholar]
- Jezkova T, Wiens JJ. Data from: What explains patterns of diversification and richness among animal phyla? Dryad Digital Repository. 2016 doi: 10.1086/690194. ( http://dx.doi.org/10.5061/dryad.2kd28) [DOI] [PMC free article] [PubMed]
- Kozak KH, Wiens JJ. Testing the relationships between diversification, species richness, and trait evolution. Systematic Biology. 2016 doi: 10.1093/sysbio/syw029. (published online, swy029) [DOI] [PubMed] [Google Scholar]
- Little C. The colonisation of land: origins and adaptations of terrestrial animals. Cambridge University Press; Cambridge: 1983. [Google Scholar]
- Lloyd DG. Benefits and handicaps of sexual reproduction. Evolutionary Biology. 1980;13:69–111. [Google Scholar]
- Magallón S, Sanderson MJ. Absolute diversification rates in angiosperm clades. Evolution. 2001;55:1762–1780. doi: 10.1111/j.0014-3820.2001.tb00826.x. [DOI] [PubMed] [Google Scholar]
- Martins EP, Hansen TF. Phylogenies and the comparative method: A general approach to incorporating phylogenetic information into the analysis of interspecific data. American Naturalist. 1997;149:646–667. [Google Scholar]
- May RM. Biological diversity: Differences between land and sea. Philosophical Transactions of the Royal Society of London, Series B: Biological Sciences. 1994;343:105–111. [Google Scholar]
- Mayhew PJ. Why are there so many insect species? Perspectives from fossils and phylogenies. Biological Reviews of the Cambridge Philosophical Society. 2007;82:425–454. doi: 10.1111/j.1469-185X.2007.00018.x. [DOI] [PubMed] [Google Scholar]
- McClain CR, Boyer AG. Biodiversity and body size are linked across metazoans. Proceedings of the Royal Society of London, Series B: Biological Sciences. 2009;276:2209–2215. doi: 10.1098/rspb.2009.0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitter C, Farrell B, Wiegmann B. The phylogenetic study of adaptive zones: Has phytophagy promoted insect diversification? American Naturalist. 1988;132:107–128. [Google Scholar]
- Nielsen C. Animal evolution: Interrelationships of Living Phyla. Oxford University Press; Oxford: 2001. [Google Scholar]
- Orme CDL, Quicke DLJ, Cook JM, Purvis A. Body size does not predict species richness among the metazoan phyla. Journal of Evolutionary Biology. 2002;15:235–247. [Google Scholar]
- Orme D. The caper package: comparative analysis of phylogenetics and evolution in R. R package version 0.5.2. 2013.
- Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999;401:877–884. doi: 10.1038/44766. [DOI] [PubMed] [Google Scholar]
- Philippe H, Brinkmann H, Lavrov DV, Littlewood DTJ, Manuel M, Woerheide G, Baurain D. Resolving difficult phylogenetic questions: Why more sequences are not enough. PloS Biology. 2011;9:e1000602. doi: 10.1371/journal.pbio.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabosky DL, Slater GJ, Alfaro ME. Clade age and species richness are decoupled across the eukaryotic tree of life. PLoS Biology. 2012;10:e1001381. doi: 10.1371/journal.pbio.1001381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabosky DL, Adams DC. Rates of morphological evolution are correlated with species richness in salamanders. Evolution. 2012;66:1807–1818. doi: 10.1111/j.1558-5646.2011.01557.x. [DOI] [PubMed] [Google Scholar]
- Ricklefs RE, Fallon SM, Bermingham E. Evolutionary relationships, cospeciation, and host switching in avian malaria parasites. Systematic Biology. 2004;53:111–119. doi: 10.1080/10635150490264987. [DOI] [PubMed] [Google Scholar]
- Ricklefs RE, Outlaw DC, Svensson-Coelho M, Medeiros MCI, Ellis VA, Latta S. Species formation by host shifting in avian malaria parasites. Proceedings of the National Academy of Sciences, U.S.A. 2014;111:14816–14821. doi: 10.1073/pnas.1416356111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskov Y, Abucay L, Orrell T, Nicolson D, Flann C, Bailly N, Kirk P, et al. 2016 Species 2000 & ITIS Catalogue of Life, 27th February 2016. Digital resource at www.catalogueoflife.org/col. Species 2000, Naturalis, Leiden, the Netherlands.
- Thomas RDK, Shearman RM, Stewart CW. Evolutionary exploitation of design options by the first animals with hard skeletons. Science. 2000;288:1239–1242. doi: 10.1126/science.288.5469.1239. [DOI] [PubMed] [Google Scholar]
- Vermeij GJ, Grosberg RK. The great divergence: When did diversity on land exceed that in the sea? Integrative and Comparative Biology. 2010;50:675–682. doi: 10.1093/icb/icq078. [DOI] [PubMed] [Google Scholar]
- Wiegmann BM, Mitter C, Farrell B. Diversification of carnivorous parasitic insects: extraordinary radiation or specialized dead end? American Naturalist. 1993;142:737–754. [Google Scholar]
- Wiens JJ. Explaining large-scale patterns of vertebrate diversity. Biology Letters. 2015a;11:20150506. doi: 10.1098/rsbl.2015.0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiens JJ. Faster diversification on land than sea helps explain global biodiversity patterns among habitats and animal phyla. Ecology Letters. 2015b;18:1234–1241. doi: 10.1111/ele.12503. [DOI] [PubMed] [Google Scholar]
- Wiens JJ, Lapoint RT, Whiteman NK. Herbivory increases diversification across insect clades. Nature Communications. 2015;6:8370. doi: 10.1038/ncomms9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JB, Nuismer SL. When does coevolution promote diversification? American Naturalist. 2010;176:802–817. doi: 10.1086/657048. [DOI] [PubMed] [Google Scholar]
- Zhang Z-Q. Animal biodiversity: An update of classification and diversity in 2013. Zootaxa. 2013;3703:5–11. doi: 10.11646/zootaxa.3703.1.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.