Abstract
Background
Associating a patient’s profile with the memories of prototypical patients built through previous repeat clinical experience is a key process in clinical judgment. We hypothesized that a similar process using a cognitive computing tool would be well suited for learning and recalling multidimensional attributes of speckle tracking echocardiography (STE) data sets derived from patients with known constrictive pericarditis (CP) and restrictive cardiomyopathy (RCM).
Methods and Results
Clinical and echocardiographic data of 50 patients with CP and 44 with RCM were used for developing an associative memory classifier (AMC) based machine learning algorithm. The STE data was normalized in reference to 47 controls with no structural heart disease, and the diagnostic area under the receiver operating characteristic curve (AUC) of the AMC was evaluated for differentiating CP from RCM. Using only STE variables, AMC achieved a diagnostic AUC of 89·2%, which improved to 96·2% with addition of 4 echocardiographic variables. In comparison, the AUC of early diastolic mitral annular velocity and left ventricular longitudinal strain were 82.1% and 63·7%, respectively. Furthermore, AMC demonstrated greater accuracy and shorter learning curves than other machine learning approaches with accuracy asymptotically approaching 90% after a training fraction of 0·3 and remaining flat at higher training fractions.
Conclusions
This study demonstrates feasibility of a cognitive machine learning approach for learning and recalling patterns observed during echocardiographic evaluations. Incorporation of machine learning algorithms in cardiac imaging may aid standardized assessments and support the quality of interpretations, particularly for novice readers with limited experience.
Keywords: pericardial disease, cardiomyopathy, cardiovascular imaging, speckle tracking echocardiography, database, machine learning, cognitive tools
Echocardiography is the most widely used cardiac imaging modality and is indispensable in the management of most patients with a suspected or known cardiac illness. However, echocardiography is highly operator dependent, and requires considerable expertise.1–4 This is especially relevant in the contemporary clinical environments that demand higher precision in diagnosis while the required high-level diagnostic expertise remains in short supply. Automated techniques using novel machine learning approaches may potentially help transforming the interpretation process and render clinical imaging much smarter, efficient and cost effective.
The new generation of so called “Big Data” machine learning techniques has potential applications for non-parametric analysis of cardiac imaging data during routine clinical assessments. However, using traditional statistical model-based or logic/rule-based tools5–7 would differ from the working of the human brain, which draws relevant inferences by recognizing patterns stored in memories built through previous and repeated experiences in assessing cardiac structure and function.8 Associative memory can be thought of as conceptually similar to “clinical judgment” in the medical setting, where the brain of a trained doctor intuitively attempts to ‘connect the dots’ in search for the ‘best fit’ or ‘associations’ for understanding a pattern of medical abnormality. Associative memory-based ‘brain-like’ machine learning algorithms have been recently applied successfully in the operational risk intelligence areas of national security and defense, however, their application in clinical medicine or cardiac imaging has not been hitherto reported.9
In this investigation we hypothesized that a cognitive machine learning approach would be well suited for integrating clinical and echocardiographic data for differentiating complex patterns of cardiac structural and functional abnormalities in different cardiac pathologies.10–12 This may be particularly relevant for integrating novel echocardiographic techniques like speckle tracking echocardiography (STE) or features tracking of cardiac magnetic resonance images where a large volume of the spatially and temporally diverse data are generated and clinical interpretations currently are time-constrained by graph-based assessments of spatially averaged functional data over a cardiac cycle.13 We utilized constrictive pericarditis (CP) and restrictive cardiomyopathy (RCM) as an initial clinical model to test the feasibility and effectiveness of employing an associative memory-based machine learning approach for differentiating the disparate patterns of cardiac tissue motion abnormalities.
METHODS
We analyzed the echocardiography data of patients pooled from two previously described databases10, 14, with a total of 54 patients with CP and 49 patients with RCM. Appropriate institutional review board approval was previously obtained for each database 10, 14. Four patients with CP and five patients with RCM were excluded due to errors in tracking the stored echocardiography images. Thus, 94 patients [50 with CP, mean age 57·8±13·4 years, 32 (64·0%) males; 44 with RCM, mean age 64·0±11·8 years, 30 (68·2%) males] were included in this investigation. The etiology and the diagnostic criteria for CP and RCM is presented in Table 1.10, 14 Briefly, these patients had presented with heart failure with preserved left ventricular (LV) ejection fraction and the initial echocardiogram had suggested a possibility of CP or RCM. CP was surgically confirmed at pericardiectomy in 41 (82%) cases. In the remaining 9 patients who did not undergo pericardiectomy the diagnosis of CP was confirmed by multimodality imaging (echo and CT or CMR) and cardiac catheterization. CP in the later cases was diagnosed by the presence of at least one of the following four additional criteria: (1) cardiac catheterization findings consistent with CP; (2) evidence of thickened pericardium (thickness >4 mm by CMR); or (3) evidence of increased LV-right ventricular (RV) coupling (septal shift with respiration) by both echocardiography and CMR. The catheterization criteria included presence of ≥2 of the following: (1) a difference between LV end-diastolic pressure and RV end-diastolic pressure of ≤5 mm Hg; (2) pulmonary arterial systolic pressure <55 mm Hg; (3) a ratio of RV end-diastolic pressure to RV systolic pressure of >1/3; (4) inspiratory decrease in pulmonary capillary wedge pressure/LV end-diastolic pressure difference of >5 mm Hg; and (5) the ratio of the right ventricular to left ventricular systolic pressure-time area during inspiration versus expiration (systolic area index) >1·1.10, 14 Restrictive cardiomyopathy was defined as myocardial disease that was characterized by restrictive physiology demonstrated by Doppler transmitral diastolic flow velocity, reduced diastolic LV volumes, and preserved LV ejection fraction. The underlying etiology of RCM was biopsy proven in 33 (75%) and based upon delayed enhanced CMR demonstration of cardiac infiltrative disease in 11 (25%) cases. Biopsy confirmed cardiac amyloidosis in 26 (59%) and was consistent with idiopathic RCM in remaining 7 (16%) patients.
Table 1.
Diagnostic criteria and etiology of constrictive pericarditis and restrictive cardiomyopathy
| Constrictive Pericarditis | ||
|---|---|---|
| Final Diagnostic Criteria | Surgical pericardiectomy | 41 (82%) |
| Multimodality (CT/CMR, Catheterization) | 9 (18%) | |
| Etiology | Idiopathic | 20 (40%) |
| Previous Cardiac Surgery | 11 (22%) | |
| Radiotherapy | 9 (18%) | |
| Previous Known Pericarditis | 10 (20%) | |
|
| ||
| Restrictive Cardiomyopathy | ||
|
| ||
| Final Diagnostic Criteria | Biopsy | 33 (75%) |
| Echocardiogram and Late Gadolinium enhanced CMR | 11 (25%) | |
| Etiology | Cardiac amyloidosis | 26 (59%) |
| Idiopathic RCM | 7 (16%) | |
| Unknown (CMR based evidence of myocardial infiltrative cardiomyopathy) | 11 (25%) | |
Study Parameters
For the purpose of the present study, clinical parameters and conventional echocardiographic and STE data were collected for all of the patients (Table 2). STE was performed offline using the vendor-customized two-dimensional Cardiac Performance Analysis software for echocardiography (2D-CPA, version 1·1·3, TomTec Imaging Systems GmbH, Unterschleissheim, Germany). Gray scale images, obtained from the apical four chamber and the mid-ventricular short-axis views and stored at a frame rate of 25–40 frames/sec, were used for this purpose. A total of 20 measurements (including velocity, displacement, area, strain, and strain rate) were derived from these two views.
Table 2.
Clinical and echocardiographic parameters in the two groups
| CP (n=50) | RCM (n=44) | P-value | |
|---|---|---|---|
| Clinical variables | |||
| Age, years | 57·8±13·4 | 64·1±11·8 | 0·018 |
| Male gender, n (%) | 32 (64·0) | 30 (68·2) | 0·669 |
| Body surface area, m2 | 1·94±0·27 | 1·88±0·22 | 0·271 |
| Systolic blood pressure, mmHg | 113±16·7 | 115±18.0 | 0·556 |
| Diastolic blood pressure, mmHg | 68·5±8·2 | 69·6±11·1 | 0·616 |
| Heart rate, beats/min | 80·1±17·7 | 74·9±13·2 | 0·105 |
| Conventional echocardiographic variables | |||
| End-diastolic ventricular septal thickness, mm | 8·8±1·6 | 13·4±2·7 | <0·0001 |
| End-diastolic LV posterior wall thickness, mm | 9·0±1·7 | 13·6±2·6 | <0·0001 |
| End-diastolic LV cavity size, mm | 43±6·1 | 41·1±5·6 | 0·121 |
| End-systolic LV cavity size, mm | 28±5·4 | 26·9±5·7 | 0·364 |
| LV ejection fraction, % | 60±8·9 | 59·6±7·6 | 0·795 |
| Left atrial volume index, ml/m2 | 40·2±18·6 | 44±14·3 | 0·277 |
| Mitral inflow E velocity, cm/sec | 88.2±33.0 | 95.5±36.3 | 0·318 |
| Mitral inflow A velocity, cm/sec | 49.9±17.5 | 64.5±32.5 | 0·017 |
| Mitral inflow E/A ratio | 1.80±0·69 | 1·75±0·98 | 0·775 |
| Mitral inflow E wave deceleration time, msec | 166±40·8 | 172.6±64.6 | 0·566 |
| Mitral E wave respiratory variation >25%, n (%) | 27 (54%) | - | - |
| Medial mitral annular e′ velocity, cm/sec | 11·1±4·5 | 5·02±1·6 | <0·0001 |
| Mitral E/e′ ratio | 10±7·4 | 19·9±8·5 | <0·0001 |
| Speckle tracking echocardiographic variables | |||
| Longitudinal strain, % | −14·2±5·47 | −12·1±4·44 | 0·047 |
| Circumferential strain, % | −22·6±8.2 | −22·5±6·91 | 0·925 |
| Radial strain, % | 21·7±20·5 | 24·4±16·7 | 0·479 |
A, late diastolic mitral inflow velocity; CP, constrictive pericarditis; E, early diastolic mitral inflow velocity; e′, early diastolic mitral annular velocity; LV, left ventricular; RCM, restrictive cardiomyopathy
Associative memory machine learning algorithm
The development of an associative memory-based machine learning algorithm was a multi-step process. The algorithm was implemented and executed using Classification Application Programming Interface in Saffron’s Natural Intelligence Platform, version 10·0·0, a commercially available cognitive software solution (Saffron Technology, NC).
STE data normalization and binning
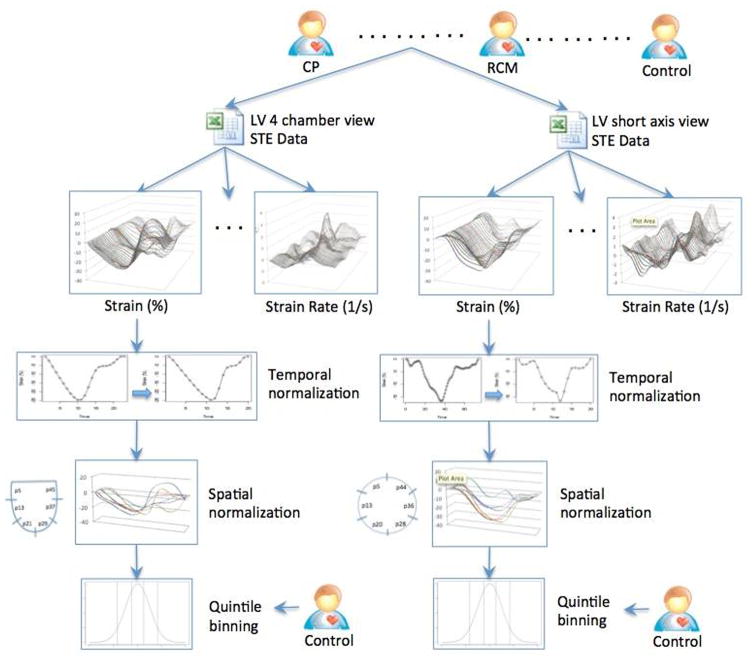
STE measurements were performed by a single individual and the STE data was stored in text and excel files. During image analysis, the STE software measures each parameter at multiple spatial locations within the myocardium, and at multiple time-points within the cardiac cycle (Supplemental Figure 1). To allow for proper comparisons, the data was transformed using spatial and temporal normalization techniques.15 For temporal normalization, the entire cardiac cycle was divided into 20 time intervals (t1 – t20), each corresponding to a 5% increment. All measurements for a specific STE variable were then binned into these intervals using interpolation of a spline function. The myocardial 4-chamber or short-axis views were further divided into six segments (s1–s6) and measurements were obtained for locations within each myocardial segment. AMC can handle multiple data types and integrate with approaches like (autoregressive integrated moving average) for analyzing real-time, time series data streams. For the present study quintiles of the raw data from the echo images were used and thresholding-based categorization used to improve the speed and scalability. Thus all continuous data were categorized or binned into quintiles (Figure 1). For quintile binning, we used a comparison cohort of 47 control subjects (54±14 years, 29 males) with no structural heart disease from the previously described databases.10, 14 Break points of quintile binning were produced for each variable to categorize continuous values in one of the five bins numbered from 1 to 5, where bin-1 represents the lowest value and bin-5 the highest. The process of data normalization and discretization yielded approximately 1800 STE data points for each patient as compared to roughly 20000 variables available originally (for further details, please refer to ‘Data Discretization’ sections in Supplemental Methods).
Figure 1.
Data normalization for speckle tracking echocardiography based data. Each measurement was subjected first to temporal normalization followed by spatial normalization. All normalized variables were then binned in to quintiles based on similar data derived from normal subjects.
CP, constrictive pericarditis; LV, left ventricular; RCM, restrictive cardiomyopathy; STE, speckle tracking echocardiography
Variable Selection
From the available 1800 STE data points and 17 clinical and conventional echocardiographic variables for each patient, those with the highest predictive accuracy were selected for inclusion in the associative memory algorithm. For this purpose, we used the wrapper method for feature reduction and prioritization16, which relies on a machine learning approach to assess the usefulness of subsets of variables. AMC was used as the wrapper in this study. All variables were ranked according to their L1-distance value, which estimates the discriminatory ability of a single variable by measuring the non-overlapping area between the two resultant probability distributions for the two outcomes (in this case, CP and RCM) (for formal definition and properties of L1-distance, please see ‘L1-distance’ section in Supplemental Methods). The variables were arranged in the descending order of their L1-distance ranks and the best-ranked variables were then entered one-by-one into the set of selected variables to determine the gain in diagnostic accuracy with each addition. Variable selection was assumed to be complete when there was no further increase in the accuracy with the addition of more variables. The accuracy at each step of variable selection was assessed by constructing receiver operating characteristic (ROC) curves and computing the area under the curve (AUC).17 By default, the accuracy was averaged over rounds of cross-validation tests with random splits, typically at 50/50 or 90/10 ratio, between training and test data. This process yielded a final subset that contained the top four conventional echocardiographic variables (end-diastolic septal and posterior wall thickness, e′ and the ratio of mitral E to e′) and the top 15 STE variables (Table 3). We also tested the validity of the analysis by performing a more conservative 50–50 hold out validation in which randomly held out 50% of the patients from each class to reduce the total number of patients to 44 with 22 CP and 22 RCM patients respectively (for further details, please refer to the ‘Variable Selection’, ‘Accuracy Assessment’ sections in Supplemental Methods).
Table 3.
L1-distance ranking of speckle tracking echocardiography variables included in the final dataset
| Variable* | L1-Distance | Data Set |
|---|---|---|
| Segmental volume_s1_t12 | 0·828 | STE (apical four-chamber) |
| Longitudinal strain rate_s5_t12 | 0·728 | STE (apical four-chamber) |
| Minimum displacement_t12 | 0·707 | STE (apical four-chamber) |
| Longitudinal strain rate _s2_t3 | 0·678 | STE (apical four-chamber) |
| Transverse strain_s4_t12 | 0·653 | STE (short-axis) |
| Transverse velocity_s6_t3 | 0·637 | STE (apical four-chamber) |
| Longitudinal strain_s5_t4 | 0·629 | STE (apical four-chamber) |
| Longitudinal velocity_s6_t3 | 0·626 | STE (apical four-chamber) |
| Longitudinal strain rate_s5_t4 | 0·617 | STE (apical four-chamber) |
| Longitudinal strain rate_s1_t4 | 0·610 | STE (apical four-chamber) |
| Segmental volume_s4_t3 | 0·578 | STE (apical four-chamber) |
| Transverse strain rate_s4_t4 | 0·573 | STE (apical four-chamber) |
| Maximum displacement_t12 | 0·570 | STE (apical four-chamber) |
| Heart rate# | 0·563 | Clinical/conventional echocardiographic |
| Circumferential strain rate_s1_t12 | 0·557 | STE (short axis) |
| Longitudinal strain rate_s2_t12 | 0·556 | STE (apical four-chamber) |
sN in the name of a variable refers to a specific spatial segment location within the myocardium and tN refers to a specific time interval within the cardiac cycle (please refer to the text for further details);
Heart rate was used a probe to remove unwarranted STE variables from the initial dataset.
STE, speckle tracking echocardiography.
Associative Memory Classifier
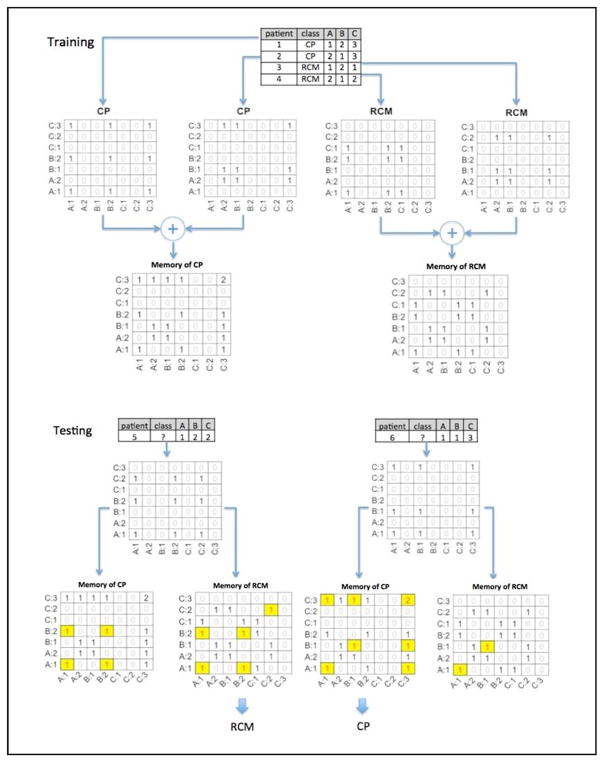
AMC is a cognitive computing machine learning algorithmic approach used for making predictions based on sets of matrices, called associative memories, developed by observing co-occurrences of predictors under outcomes.9 Outcomes are the conditions that the AMC is being trained to predict.
To classify CP and RCM, AMC constructed two classes of matrices: a CP matrix and an RCM matrix. During the training phase, variables of each patient in the training set were transformed into a set of predictors. AMC observed the co-occurrence of the different predictors in the corresponding class matrix and counted the number of times any two predictors were observed together in the training data for the class. Predictors were paired to form the associations and matched against the two classes of matrices. The ratio between the numbers of matches between the two classes of matrices provided the likelihood of prediction for one class over the other (Figure 2) (for formal treatment of algorithm and scoring of AMC, please see the ‘Associative Memory Classifier’ section in Supplemental Methods). The bin numbers were treated like integers and could be organized in sorted order under the variable such as A:1, A:2, B:1, B2, C:1, C:2, and C:3 (Figure 2). If a test patient happened to have a variable instance, say A:3, which had not been observed in the training dataset, a windowing function was applied with a radius centered around A:3 to find matches. If the radius was equal to one, A:2 would be matched with much less weight comparing to a direct match of A:3 (weight=1.0). The weight was computed by a window function (e−αd2), decreasing sharply with the increase of distance (d) to the center and the weight becomes zero when the distance is larger than the radius. This windowing function allowed recognition of the individual quintiles treated along an ordinal scale.
Figure 2.
Classification training and testing of associative memories. The figure illustrates the concept of training data of four patients with variables A, B, and C (Patient 1 and 2 represent examples of CP patients and, patients 3 and 4 are examples of RCM). The patient 1 (CP) has there variables (A, B, C) where their magnitude is falling into respective quintiles 1, 2, 3 and hence the labels A: 1, B: 2 and C: 3. The fully connected associations are displayed in the CP matrix: (A:1, A:1), (A:1, B:2), (B:2, A:1), (B:2, B:2), (A:1, C:3), (C:3, A1), (C:3, C:3), (B:2, C:3), and (C:3, B2) with co-occurrence count 1. The same process when next repeated for patient-2, one observes that the co-occurrence count (C:3, C:3) became 2 in matrix CP because C:3 was shared by patient 1 and 2. This process was repeated for all CP patients to develop a memory of associations and co-occurrences that represent CP in the training phase. Similarly, matrix RCM was trained using Patient 3 and 4. During the testing phase an unknown patient (patient 5) is shown with variable A:1, B:2, and C:2. This unknown association was matched against the memory of two trained classes of matrices CP and RCM. The ratio between the numbers of matches (highlighted cells) between the two classes of matrices provided the likelihood of prediction of the unknown patient as CP or RCM.
CP, constrictive pericarditis; RCM, restrictive cardiomyopathy
Comparative assessment using general machine learning algorithms
To evaluate classification performance and generalizability of the selection, we also evaluated diagnostic accuracies of four other machine learning algorithms. Specific R packages (‘randomForest’ for Random Forest, ‘class’ for k-Nearest Neighbor, ‘e1071’ for Support Vector Machine) were used for this purpose. Learning curves for all the different machine learning algorithms, including AMC, were constructed by plotting respective diagnostic accuracies at different training fractions and were compared with each other.
Statistical analysis
The clinical and echocardiographic data were managed on Microsoft Excel spreadsheet (version 2007, Microsoft Corp, Seattle, Washington) and analyzed using SPSS for Windows (Release 15·0, IBM, Armonk, NY, USA). All values were expressed as mean (±standard deviation) or as percentages. The comparisons between CP and RCM groups were performed using the chi-square test for categorical variables and Student’s independent sample t-test for continuous variables. As outlined above, the diagnostic accuracies of the different classifiers were assessed by constructing ROC curves under 10-fold cross-validation tests and using different bootstrap samples.18 AUCs were calculated for each classifier approach and used as the basis of comparison (pROC package for R).19 A p-value <0·05 was considered statistically significant.
The sponsor of the study had no role in study design, data collection or data interpretation. The primary author had full access to all the data, had final responsibility for the decision to submit for publication and is guarantor.
RESULTS
Clinical and conventional echocardiography variables in the two groups are presented in Table 2. Although the RCM patients were older (64.0±11·8 years vs 57·8±13·4 years, P = 0·018), there were no other significant differences between the two groups in the clinical parameters.
The basic echocardiographic parameters in the two groups were consistent with their respective hemodynamic profiles. As compared to the patients with CP, those with RCM had greater LV wall thickness (septum 8·8±1·6 mm vs 13·4±2·7 mm, P <0·0001; posterior wall 9·0±1·7 mm vs 13·6±2·56 mm, P <0·0001), reduced medial mitral e′ velocity (11·1±4·5 cm/s vs. 5·02±1·6 cm/s, P <0·0001) and significantly elevated mitral E/e′ ratios (10±7·5 vs 19·9±8·5, P <0·0001).
On STE, RCM patients had reduced average longitudinal strain (−12·1±4·44% vs −14·2±5·47, P 0·047) as compared to those with CP. The radial and circumferential strain values, however, were not different between the two groups.
Classification accuracy of AMC
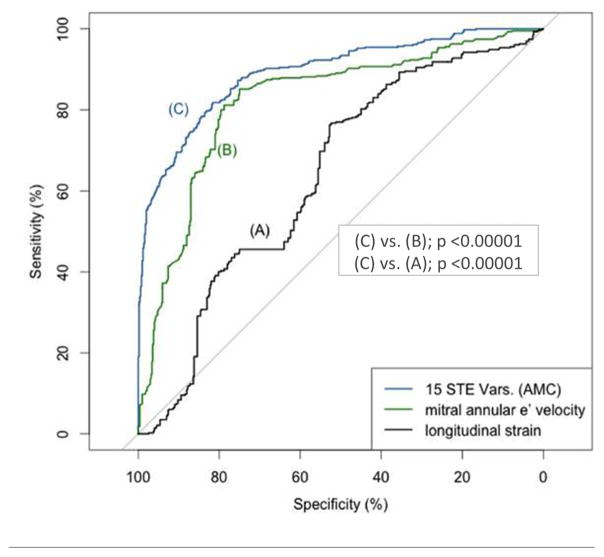
Under 10-fold cross-validation tests, AMC achieved an average AUC of 89·2% using the top 15 STE variables (Figure 3). In comparison, the AUC of e′ was 82·1%, and that of longitudinal strain was 63·7% (Figure 3, p<0.0001 for both). Four echocardiographic variables (e′, E/e′, septal and posterior wall thickness) had an AUC of 94.2%, and on combining with the top 15 STE variables, the AUC of the resultant AMC increased marginally from 94·2% to 96·2% (Table 4). The incremental diagnostic value of AMC using top 15 STE variables over e′ and 4 combined echocardiography variables (e′, E/e′, septal and posterior wall thickness) was also confirmed using a more conservative holdout validation testing (Table 5).
Figure 3.
Diagnostic accuracy of different variables assessed using receiver operating characteristics curves under 10-fold cross-validation tests. (A) medial mitral annular early diastolic velocity, (B) average left ventricular longitudinal strain, derived from the apical four-chamber view, (C) top 15 speckle tracking echocardiography variables as included in the associative memory classifier, and (D) all the above three compared together.
AMC, associative memory classifier; AUC, area-under-the-curve; e′, medial mitral annular early diastolic velocity; STE, speckle tracking echocardiography
Table 4.
Results from 10-fold cross validation
| Variables | AUCLGT | AUCAMC | AUCRF | AUCSVM | AUCKNN |
|---|---|---|---|---|---|
| e′ | 82.1% | - | - | - | - |
| 4 variables* | 94.2% | - | - | - | - |
| 15 STE | - | 89.2% | 84.7% | 78.0% | 71.3% |
| e′ + 15 STE | - | 93.8% | 92.7% | 78.0% | 78.7% |
| 4 variables + 15 STE | - | 96.2% | 94.2% | 92.2% | 87.0% |
| ROC #1 | ROC #2 | p-value AMC | p-value RF | p-value SVM | p-value kNN |
|---|---|---|---|---|---|
| e′ | e′ + 15 STE | < 0.0001 | < 0.0001 | 0.04 | 0.10 |
| 15 STE | e′ + 15 STE | < 0.0001 | < 0.0001 | 0.89 | < 0.0001 |
| e′ +15 STE | 4 variables + 15 STE | < 0.0001 | 0.09 | < 0.0001 | < 0.0001 |
| 4 variables | 4 variables + 15 STE | 0.01 | 0.98 | 0.03 | < 0.0001 |
AUC – area under the curve
4 variables included e′, E/e′, IVS and PW thickness of the LV.
Table 5.
Results of 50-50 holdout validation
| Variables | AUC | AUCAMC | AUCRF | AUCSVM | AUCKNN |
|---|---|---|---|---|---|
| e′ | 73.2% | - | - | - | - |
| 4 variables* | 89.6% | - | - | - | - |
| 15 STE | - | 68.2% | 68.1% | 68.3% | 61.3% |
| e′ + 15 STE | - | 78.9% | 85.2% | 70.0% | 70.5% |
| 4 variables + 15 STE | - | 91.0% | 94.6% | 85.7% | 79.5% |
| ROC #1 | ROC #2 | p-value AMC | p-value RF | p-value SVM | p-value kNN |
|---|---|---|---|---|---|
| e′ | e′ + 15 STE | <0.0001 | < 0.0001 | 0.025 | 0.04 |
| 15 STE | e′ + 15 STE | <0.0001 | <0.0001 | 0.0001 | <0.0001 |
| e′ +15 STE | 4 variables + 15 STE | < 0.0001 | < 0.0001 | < 0.0001 | < 0.0001 |
| 4 variables | 4 variables + 15 STE | 0.04 | < 0.0001 | < 0.0001 | < 0.0001 |
4 variables included e′, E/e′, IVS and PW thickness of the LV.
Learning curve of AMC
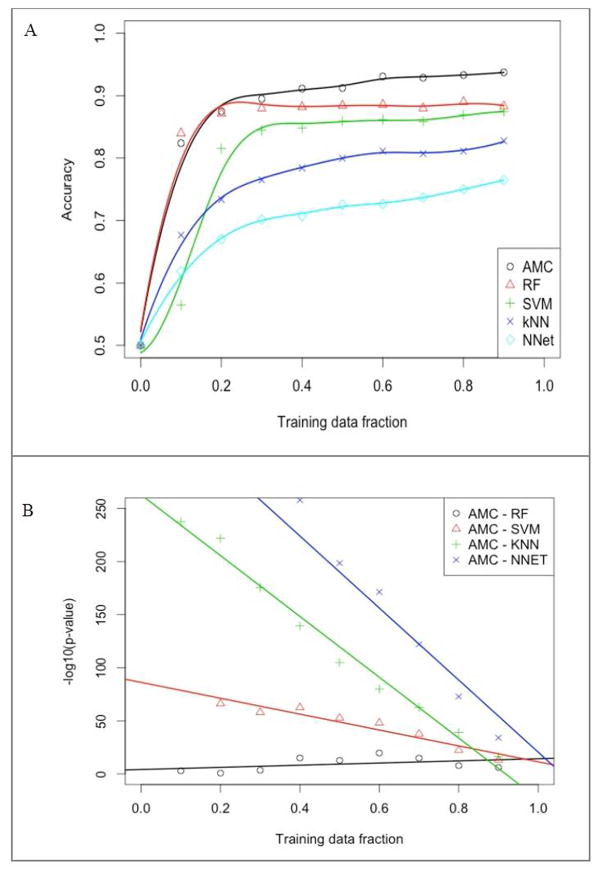
On plotting the prediction accuracies achieved with increasing sizes of training data, significant differences were noted in learning curves for different machine learning approaches (Figure 4A, 4B). Among all machine learning algorithms evaluated, AMC performed the best [averaged classification accuracy (ACC) 93·7%, AUC 96·2%], with Random Forest (ACC 88·3%, AUC 94·2%) and Support Vector Machines (ACC 87·4%, AUC 92·2%) ranking second and third, respectively. For AMC, the accuracy at a high training fraction was relatively flat and asymptotically approached 90% after a training fraction of 0·3. This indicated that at least 30 trials were needed for AMC to be sufficiently trained, but only small gains occurred with more data (Figure 4A). However, at all training fractions, the diagnostic accuracy of AMC remained superior as compared to the other machine learning algorithms [all p-values <0·05 (−log10 p-value >1·3)] (Figure 4B). This remained true asymptotically as the differences became smaller with increasing training fractions. Random Forest showed the second best performance and the p-values for its comparison with AMC are listed in Table 6.
Figure 4.
(A) Learning curves for various machine learning algorithms using the top four clinical and conventional echocardiographic variables and the top 15 speckle tracking echocardiography variables; (B) Log10 (p-value) for differences in the area-under-the-curve for associative memory classifier and each of the other four machine learning algorithms.
AMC, associative memory classifier; kNN, k-Nearest Neighbor; NN, Neural Networks; RF, Random Forests; SVM, Support Vector Machines.
Table 6.
P-values for differences in diagnostic accuracy of Associate Memory Classifier and Random Forest algorithms at different fractions of training data
| Training Fraction | P-value for AUC difference |
|---|---|
| 0·1 | 0·001 |
| 0·2 | 0·156 |
| 0·3 | 2·97E-04 |
| 0·4 | 8·21E-16 |
| 0·5 | 1·43E-13 |
| 0·6 | 1·57E-20 |
| 0·7 | 1·24E-15 |
| 0·8 | 1·19E-08 |
| 0·9 | 7·31E-07 |
AUC, area-under-the-curve.
DISCUSSION
To the best of our knowledge, this is the first report describing the development and validation of a cognitive computing machine learning approach for interpreting large, high-dimensional data from cardiac imaging. The entire clinical, echocardiography and STE data with ≈2000 motion and deformation features obtained from a single patient were used to train and develop an associative memory-based algorithm. Total 15 STE and the 4 echo variables were automatically selected by the AMC. The addition of 15 STE to 4 echo variables (e′, E/e′, septal and posterior LV wall thickness) showed incremental diagnostic value in distinguishing CP from RCM. The approach presented here does not contest the value of traditional imaging and interpretation; however, illustrates a technique for automation and acceleration of diagnostic decisions. The application of machine-learning algorithms to automated STE could enable development of real-time decision support system for differentiating disease phenotype directly from just grey scale 2D echo images. Further addition of parameters (like tissue Doppler, clinical variables or manual extracted data-points) could help enrich the diagnostic yield. The ability to handle large volumes of data using this approach and ability to integrate it with clinical echo variables is an unmet need and has a potential for standardizing and improving workflow in busy echocardiography labs.
The need for machine-based automation
Cardiac imaging techniques like two-dimensional echocardiography generate several thousand data points during each examination and these numbers are likely to grow further with advent of multidimensional cardiac imaging techniques like STE. However, it is difficult for clinicians to fully assimilate and interpret such large and complex data sets, and therefore, only a fraction of the available potentially useful information is fully utilized for diagnostic interpretations and clinical decision making. Interestingly, recent standardization efforts20 have made surprising revelation that STE derived measurements are more reproducible than conventional 2D and Doppler measurements. Furthermore, automated approaches in STE have been recently developed and shown to improve efficiency and reduce inter- and intra-observer variability21. While echo-Doppler measurements are pivotal for differentiating CP from RCM, the clinical validity of some of the older variables have been challenged. For example, a recent study of 130 surgically confirmed CP from Mayo Clinic showed little diagnostic value of respiratory variations in mitral inflow E velocity for differentiate CP from RCM22. Investigators have recently described unique patterns of speckle tracking derived abnormalities in CP14, 23, 24. Since complex pattern recognition in big data is better performed using machine approaches, a potential solution to meet this challenge is to develop computer-based cognitive tools for automated analysis. Such cognitive computing tools have been successfully applied in various fields including national security, defense, and manufacturing and are beginning to be employed in medical field.9, 25–29 However, their feasibility and effectiveness in automated analysis and interpretation of echocardiography images have not been explored previously.
To meet this objective, we compared various machine learning approaches, including a cognitive computing approach, to the problem of differentiating CP and RCM. Although rare, the differentiation of RCM and CP is perhaps one of the most challenging and complex echocardiographic conundrums, because of the inherent similarities in the clinical and echocardiographic profiles of these two diseases.1
In our study, using a set of selected STE variables, the AMC achieved AUC of 89·2%. This result was superior to the performance of commonly used echocardiographic variables such as early diastolic mitral annular longitudinal velocity and global longitudinal strain for differentiating CP from RCM. The observed diagnostic values on 10-fold cross validation was higher than holdout validation. This is expected since holdout sets (different splits) result in smaller training samples with heterogeneous results. Additionally, depending on the size of available data, one can underestimate of the generalization capability. On the other hand, 10-fold cross validation allows a better use of the available dataset to obtain a better aggregate measure of classification accuracy.
The AMC algorithm was able to automatically use multiple data points, learning to filter, store and then recall those predictors that had the highest yield in differentiating the two conditions. Interestingly, two other machine learning algorithms- Random Forest and Support Vector Machines- also achieved similar high level of performance (AUC >90%) under the same variable selection but differed in the extent of training required. These results indicate that machine learning algorithms can perform well for the diagnosis of cardiac pathologies, and the reliability of diagnosis can be increased with an ensemble of algorithms.
The choice of machine learning approach
Learning curves are an important consideration in the choice of a machine learning algorithm, as the amount of data required for each algorithm would vary accordingly. For rare diseases, data is often scarce and therefore it would be important to ascertain whether an algorithm could be as effective with less data. In this regard AMC not only had much higher accuracy, it had a shorter learning curve as well.
Given the same variable subset, differences in classification accuracy likely resulted from how the variables were modeled by the different machine learning algorithms. By default, most algorithms follow the logic of decision-tree based approaches and use a linear combination of the predictors to model the training data. In contrast, AMC relies on recognizing patterns by observing pair-wise as well as triangular associations of variables to make diagnostic predictions. In this manner, the functioning of AMC is analogous to aspects of human cognition. Our minds do not incorporate the slow, sequential, logical ways of thinking that has been a characteristic of earlier algorithms. Instead, our thinking is fast and mostly based on our memory of learned associations built through repeated previous experiences. Using statistical modeling, AMC explores data sets to discover the most predictive candidate variables and the relationships between these variables to address a complex-use case question. The hypothesis-free data representation, a key characteristic of AMC, helps accelerate the identification of the most relevant connections between variables. This ability to recognize the unknown nonlinear entity relationships is perhaps the most valuable feature of a cognitive computing approach such as AMC.
Limitations and Future Directions
First, the selection and ranking of variables and selection are data- and algorithm-dependent. Thus, the variables selected by our algorithm should be viewed as one of the many possible selections and not the only possible selection. Interestingly, however, the currently selected STE variables were seen to cluster at early systole or end-systole, which is consistent with previous description of pre-ejection and end-ejection variables for differentiating CP from RCM.10, 30 Although STE had incremental value over conventional echocardiography variables like e′, the diagnostic gain was marginal when STE parameters were added to 4 echocardiographic variables for differentiating CP from RCM. Despite these limitations, the study illustrated the superiority of machine learning techniques to analyze STE data. The traditional STE analysis yielded differences in Longitudinal strain with AUC of 63.7%. There were no differences in radial and circumferential strain. Whereas, AUC of AMC for integrating the STE data using 15 variables was 89.2%. The ability to automate and handle large volumes of data using this approach and ability to integrate it with clinical echo variables is an unmet need and has a potential for imparting automation during reporting. The incremental value of machine-learning algorithm for STE in other disease states requires systematic exploration. Secondly, although our sample size can be considered modest because of the rarity of the disease1, given the large number of available STE variables, the analysis may be susceptible to over-fitting. This approach for high dimensionality (several variables tested) for small sample size (<100 patients) mirrors microarray-related studies that often involve a very large number of genes (100–1000 variables) for small sample size 31, 32. Interestingly a previous method paper showed that such morphophenotypic classifications could be achieved with 10–20 training samples with “reasonably accurate” extrapolation requiring only 30–40 training samples in studies that have small sample size data 32. Moreover, we applied a combination of techniques such as cross-validation, bootstrapping, plotting learning curves, and favoring the selection with fewer variables to detect and prevent such over-fitting. The data showed consistency using both 10-fold cross-validation and the 50–50 hold out analysis, with k-fold validation providing higher accuracy numbers because of the larger training data set. Our learning curves illustrated that the accuracies from rounds of bootstraps were asymptotically stable under different algorithms as more training data were made available, indicating that the prediction for selected variables was likely well generalized. The technique to use learning curve post-hoc to understand the stability of classification accuracy provides additional validity where the sample size is small 33. Finally, it should be noted that for various machine-learning algorithms other than AMC, we used the default settings of the relevant software packages as listed above. Hence, the relative performance of these different machine-learning algorithms demonstrated in our study is applicable only to those settings and should not be generalized beyond the conditions tested. The performance also may vary for the etiologies of CP and RCM which was not separately assessed due to the small sample size in the subgroups. The direct head-to-head comparison of AMC with good clinical judgement or expert echo interpretation was not attempted and would require further investigation. It would be important to recognize that the machine learning algorithms do not contest the value of ‘traditional parameters’ and manual analysis; however, provides oppurtunities for automation and acceleration of diagnostic decisions. For example, we have shown that acquisition of STE can be automated and data can be extracted automatically within ‘8-seconds’ with ‘zero’ intra-observer and ‘inter-observer’ variability 34. Acceleration and automation may represent the logical next steps in the presence complex high-dimensional data in echocardiography and requires systematic investigations in the future.
CONCLUSIONS
This investigation used a complex chronic disease model of CP and RCM for demonstrating the feasibility and effectiveness of a cognitive computing machine learning approach for automated interpretations of STE data. Incorporation of a cognitive computing machine learning algorithm in cardiac imaging may aid standardized assessments and support the quality of interpretations, particularly for novice readers with limited experience.
Supplementary Material
Clinical Perspective.
The currently available cardiac imaging techniques have the capabilities to generate vast amount of cardiac structural and functional data during routine cardiac imaging. However, due to the inability of a clinician to fully assimilate and interpret such large and complex data sets, only a fraction of the available potentially useful information is fully utilized for diagnostic interpretations and clinical decision making. A potential solution to meet this challenge is to develop computer-based cognitive tools for automated analysis of big functional data sets. Although such cognitive computing tools have been successfully applied in various fields, their application in the medical field has been very limited with no previous study describing their feasibility and effectiveness in analysis and interpretation of echocardiography images. We investigated the feasibility and diagnostic accuracy of an associative memory-based machine-learning algorithm in automated analysis and interpretation of speckle tracking echocardiography data sets derived from patients with constrictive pericarditis and restrictive cardiomyopathy. The associative memory classifier showed a short learning curve achieving over 90% of asymptotic accuracy with only 30% of the data trained, and achieved a diagnostic area under the curve of 89.2%, which was superior to that of conventional echocardiographic variables like early diastolic mitral annular velocity and longitudinal strain (82.1% and.63.7% respectively) used for differentiating the two conditions. These findings suggest that incorporation of an automated cognitive machine learning algorithm in cardiac imaging is feasible and may aid standardized assessments and support the quality of interpretations, particularly for novice readers with limited experience.
Acknowledgments
We sincerely thank Dr. Paul Hofmann for his guidance and inspiring discussions on this project. We also thank Deb Taylor for her editing of the final version of the manuscript. KS and JTD would like to acknowledge the following grant from National Institutes of Health (NIH): National Center for Advancing Translational Sciences (NCATS, UL1TR000067) Clinical and Translational Science Award (CTSA).
Footnotes
AUTHOR CONTRIBUTIONS
Partho P Sengupta: Hypothesis generation, study design, literature search, figures, method design, data analysis, data interpretation, writing, contributing equally as the first author.
Yen-Min Huang: Literature search, figures, method design, data analysis, data interpretation, writing, contributing equally as the first author.
Manish Bansal: data interpretation, editing, writing
Ali Ashrafi: methodology, data analysis
Matt Fisher: data analysis, data interpretation, editing
Shameer Khader: data interpretation, editing
Walt Gall: study design, data interpretation, editing, writing
Joel T Dudley: data interpretation, editing, writing
DISCLOSURES
Dr Partho P Sengupta is an advisor for Saffron Technology, TeleHealthRobotics Inc., Heart Test Labs, and consultant for Edward Lifesciences.
Dr. Yen-Min Huang is employee of Saffron Technology with the role of Chief Scientist.
Dr. Ali Ashrafi is an employee of Saffron Technology with the role Senior Data Scientist.
Dr. Matt Fisher is an employee of Saffron Technology with the role Senior Data Scientist.
Dr. Watt Gall is an employee of Saffron Technology with the role of Vice President, Healthcare & Strategic Partnerships.
Dr Manish Bansal, Dr Shameer Khader and Dr Joel T Dudley have no competing interests to declare.
Contributor Information
Partho P. Sengupta, Zena and Michael A. Wiener Cardiovascular Institute and the Marie-Josée and Henry R. Kravis Center for Cardiovascular Health, Mount Sinai School of Medicine, One Gustave L. Levy Place, New York, NY.
Yen-Min Huang, Saffron Technology, Inc., Cary, North Carolina.
Manish Bansal, Zena and Michael A. Wiener Cardiovascular Institute and the Marie-Josée and Henry R. Kravis Center for Cardiovascular Health, Mount Sinai School of Medicine, New York, NY.
Ali Ashrafi, Saffron Technology, Inc., Cary, North Carolina.
Matt Fisher, Saffron Technology, Inc., Cary, North Carolina.
Khader Shameer, Icahn Institute for Genomics and Multiscale Biology, Icahn School of Medicine at Mount Sinai, New York, NY.
Walt Gall, Saffron Technology, Inc., Cary, North Carolina.
Joel T Dudley, Icahn Institute for Genomics and Multiscale Biology, Icahn School of Medicine at Mount Sinai, New York, NY.
References
- 1.Dal-Bianco JP, Sengupta PP, Mookadam F, Chandrasekaran K, Tajik AJ, Khandheria BK. Role of echocardiography in the diagnosis of constrictive pericarditis. J Am Soc Echocardiogr. 2009;22:24–33. doi: 10.1016/j.echo.2008.11.004. quiz 103–104. [DOI] [PubMed] [Google Scholar]
- 2.Picano E, Lattanzi F, Orlandini A, Marini C, L’Abbate A. Stress echocardiography and the human factor: The importance of being expert. J Am Coll Cardiol. 1991;17:666–669. doi: 10.1016/s0735-1097(10)80182-2. [DOI] [PubMed] [Google Scholar]
- 3.Varga A, Picano E, Dodi C, Barbieri A, Pratali L, Gaddi O. Madness and method in stress echo reading. Eur Heart J. 1999;20:1271–1275. doi: 10.1053/euhj.1999.1541. [DOI] [PubMed] [Google Scholar]
- 4.Nagueh SF, Appleton CP, Gillebert TC, Marino PN, Oh JK, Smiseth OA, Waggoner AD, Flachskampf FA, Pellikka PA, Evangelista A. Recommendations for the evaluation of left ventricular diastolic function by echocardiography. J Am Soc Echocardiogr. 2009;22:107–133. doi: 10.1016/j.echo.2008.11.023. [DOI] [PubMed] [Google Scholar]
- 5.Altman NS. An introduction to kernel and nearest-neighbor nonparametric regression. The American Statistician. 1992;46:175–185. [Google Scholar]
- 6.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 7.Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20:273–297. [Google Scholar]
- 8.Hawkins J, Blakeslee S. On intelligence. New York: Time Books; 2004. [Google Scholar]
- 9.Manuel A. [Accessed august 25, 2015];Your brain is cognitive, not a database. Http://www.Saffrontech.Com/wp-content/uploads/sites/8/2014/06/brain-is-cognitive.Pdf.
- 10.Amaki M, Savino J, Ain DL, Sanz J, Pedrizzetti G, Kulkarni H, Narula J, Sengupta PP. Diagnostic concordance of echocardiography and cardiac magnetic resonance-based tissue tracking for differentiating constrictive pericarditis from restrictive cardiomyopathy. Circ Cardiovasc Imaging. 2014;7:819–827. doi: 10.1161/CIRCIMAGING.114.002103. [DOI] [PubMed] [Google Scholar]
- 11.Geyer H, Caracciolo G, Abe H, Wilansky S, Carerj S, Gentile F, Nesser HJ, Khandheria B, Narula J, Sengupta PP. Assessment of myocardial mechanics using speckle tracking echocardiography: Fundamentals and clinical applications. J Am Soc Echocardiogr. 2010;23:351–369. doi: 10.1016/j.echo.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 12.Mor-Avi V, Lang RM, Badano LP, Belohlavek M, Cardim NM, Derumeaux G, Galderisi M, Marwick T, Nagueh SF, Sengupta PP, Sicari R, Smiseth OA, Smulevitz B, Takeuchi M, Thomas JD, Vannan M, Voigt JU, Zamorano JL. Current and evolving echocardiographic techniques for the quantitative evaluation of cardiac mechanics: ASE/eae consensus statement on methodology and indications endorsed by the japanese society of echocardiography. J Am Soc Echocardiogr. 2011;24:277–313. doi: 10.1016/j.echo.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 13.Sengupta PP. Intelligent platforms for disease assessment: Novel approaches in functional echocardiography. JACC Cardiovasc Imaging. 2013;6:1206–1211. doi: 10.1016/j.jcmg.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 14.Sengupta PP, Krishnamoorthy VK, Abhayaratna WP, Korinek J, Belohlavek M, Sundt TM, 3rd, Chandrasekaran K, Mookadam F, Seward JB, Tajik AJ, Khandheria BK. Disparate patterns of left ventricular mechanics differentiate constrictive pericarditis from restrictive cardiomyopathy. JACC Cardiovasc Imaging. 2008;1:29–38. doi: 10.1016/j.jcmg.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 15.Mitsa T. Temporal data mining. Florida: Chapman & Hall/CRC Press; 2010. [Google Scholar]
- 16.Kohavi R, John GH. Wrappers for feature subset selection. Artificial Intelligence. 1997;97:273–324. [Google Scholar]
- 17.Fawcett T. An introduction to roc analysis. Pattern Recognition Letters. 2006;27:861–874. [Google Scholar]
- 18.Kohavi R. A study of cross-validation and bootstrap for accuracy estimation and model selection. Proceedings of the 14th international joint conference on Artificial intelligence - Volume 2; 1995; pp. 1137–1143. [Google Scholar]
- 19.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 20.Farsalinos KE, Daraban AM, Unlu S, Thomas JD, Badano LP, Voigt JU. Head-to-head comparison of global longitudinal strain measurements among nine different vendors: The eacvi/ASE inter-vendor comparison study. J Am Soc Echocardiogr. 2015;28:1171–1181. e1172. doi: 10.1016/j.echo.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 21.Knackstedt C, Bekkers SC, Schummers G, Schreckenberg M, Muraru D, Badano LP, Franke A, Bavishi C, Omar AM, Sengupta PP. Fully automated versus standard tracking of left ventricular ejection fraction and longitudinal strain: The fast-efs multicenter study. J Am Coll Cardiol. 2015;66:1456–1466. doi: 10.1016/j.jacc.2015.07.052. [DOI] [PubMed] [Google Scholar]
- 22.Welch TD, Ling LH, Espinosa RE, Anavekar NS, Wiste HJ, Lahr BD, Schaff HV, Oh JK. Echocardiographic diagnosis of constrictive pericarditis: Mayo clinic criteria. Circ Cardiovasc Imaging. 2014;7:526–534. doi: 10.1161/CIRCIMAGING.113.001613. [DOI] [PubMed] [Google Scholar]
- 23.Kusunose K, Dahiya A, Popovic ZB, Motoki H, Alraies MC, Zurick AO, Bolen MA, Kwon DH, Flamm SD, Klein AL. Biventricular mechanics in constrictive pericarditis comparison with restrictive cardiomyopathy and impact of pericardiectomy. Circ Cardiovasc Imaging. 2013;6:399–406. doi: 10.1161/CIRCIMAGING.112.000078. [DOI] [PubMed] [Google Scholar]
- 24.Negishi K, Popovic ZB, Negishi T, Motoki H, Alraies MC, Chirakarnjanakorn S, Dahiya A, Klein AL. Pericardiectomy is associated with improvement in longitudinal displacement of left ventricular free wall due to increased counterclockwise septal-to-lateral rotational displacement. J Am Soc Echocardiogr. 2015;28:1204–1213. e1202. doi: 10.1016/j.echo.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 25.Tantimongcolwat T, Naenna T, Isarankura-Na-Ayudhya C, Embrechts MJ, Prachayasittikul V. Identification of ischemic heart disease via machine learning analysis on magnetocardiograms. Comput Biol Med. 2008;38:817–825. doi: 10.1016/j.compbiomed.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 26.Krizmaric M, Verlic M, Stiglic G, Grmec S, Kokol P. Intelligent analysis in predicting outcome of out-of-hospital cardiac arrest. Comput Methods Programs Biomed. 2009;95:S22–32. doi: 10.1016/j.cmpb.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 27.Afshin M, Ben Ayed I, Islam A, Goela A, Peters TM, Li S. Global assessment of cardiac function using image statistics in mri. Med Image Comput Comput Assist Interv. 2012;15:535–543. doi: 10.1007/978-3-642-33418-4_66. [DOI] [PubMed] [Google Scholar]
- 28.Xiong G, Kola D, Heo R, Elmore K, Cho I, Min JK. Myocardial perfusion analysis in cardiac computed tomography angiographic images at rest. Med Image Anal. 2015;24:77–89. doi: 10.1016/j.media.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Syed Z, Stultz CM, Scirica BM, Guttag JV. Computationally generated cardiac biomarkers for risk stratification after acute coronary syndrome. Science translational medicine. 2011;3:102ra195. doi: 10.1126/scitranslmed.3002557. [DOI] [PubMed] [Google Scholar]
- 30.Sengupta PP, Krishnamoorthy VK, Abhayaratna WP, Korinek J, Belohlavek M, Sundt TM, 3rd, Chandrasekaran K, Seward JB, Tajik AJ, Khandheria BK. Comparison of usefulness of tissue doppler imaging versus brain natriuretic peptide for differentiation of constrictive pericardial disease from restrictive cardiomyopathy. Am J Cardiol. 2008;102:357–362. doi: 10.1016/j.amjcard.2008.03.068. [DOI] [PubMed] [Google Scholar]
- 31.Molinaro AM, Simon R, Pfeiffer RM. Prediction error estimation: a comparison of resampling methods. Bioinformatics. 2005;21:3301–7. doi: 10.1093/bioinformatics/bti499. [DOI] [PubMed] [Google Scholar]
- 32.Mukherjee S, Tamayo P, Rogers S, Rifkin R, Engle A, Campbell C, Golub TR, Mesirov JP. Estimating dataset size requirements for classifying DNA microarray data. J Comput Biol. 2003;10:119–42. doi: 10.1089/106652703321825928. [DOI] [PubMed] [Google Scholar]
- 33.Wang LY, Lee WC. One-step extrapolation of the prediction performance of a gene signature derived from a small study. BMJ Open. 2015 Apr 17;5(4):e007170. doi: 10.1136/bmjopen-2014-007170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knackstedt C, Bekkers SC, Schummers G, Schreckenberg M, Muraru D, Badano LP, Franke A, Bavishi C, Omar AM, Sengupta PP. Fully Automated Versus Standard Tracking of Left Ventricular Ejection Fraction and Longitudinal Strain: The FAST-EFs Multicenter Study. J Am Coll Cardiol. 2015;66:1456–66. doi: 10.1016/j.jacc.2015.07.052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.