Summary
The Neolithic transition was a dynamic time in European prehistory of cultural, social, and technological change. Although this period has been well explored in central Europe using ancient nuclear DNA [1, 2], its genetic impact on northern and eastern parts of this continent has not been as extensively studied. To broaden our understanding of the Neolithic transition across Europe, we analyzed eight ancient genomes: six samples (four to ∼1- to 4-fold coverage) from a 3,500 year temporal transect (∼8,300–4,800 calibrated years before present) through the Baltic region dating from the Mesolithic to the Late Neolithic and two samples spanning the Mesolithic-Neolithic boundary from the Dnieper Rapids region of Ukraine. We find evidence that some hunter-gatherer ancestry persisted across the Neolithic transition in both regions. However, we also find signals consistent with influxes of non-local people, most likely from northern Eurasia and the Pontic Steppe. During the Late Neolithic, this Steppe-related impact coincides with the proposed emergence of Indo-European languages in the Baltic region [3, 4]. These influences are distinct from the early farmer admixture that transformed the genetic landscape of central Europe, suggesting that changes associated with the Neolithic package in the Baltic were not driven by the same Anatolian-sourced genetic exchange.
Keywords: ancient DNA, Neolithic transition, genomics, population genetics, Baltic, Ukraine
Highlights
-
•
A degree of genetic continuity from the Mesolithic to the Neolithic in the Baltic
-
•
Steppe-related genetic influences found in the Baltic during the Neolithic
-
•
No Anatolian farmer-related genetic admixture in Neolithic Baltic samples
-
•
Steppe ancestry in Latvia at the time of the emergence of Balto-Slavic languages
Jones et al. present genome-wide data spanning the Mesolithic-Neolithic transition in Latvia and Ukraine that show that massive migration of Anatolian farmers was not a universal driver for the spread of Neolithic lifeways and possibly Indo-European languages throughout Europe.
Results
Sample Context and Genomic Data
In Europe, the Neolithic transition marked the beginning of a period of innovations that saw communities shift from a mobile lifestyle, dependent on hunting and gathering for survival, to a more sedentary way of life based on food production. This new lifeway, which originated in the Near East ∼11,500 calibrated years before present (cal BP) [5, 6], had arrived in southeast Europe by ∼8,500 cal BP [7], from where it spread quickly across the continental interior of Europe and introduced animal husbandry, cultivated cereals, pottery, and ground stone tools to the region. There is a long-standing debate among archaeologists whether this spread was due to the dispersal of farmers into new lands (i.e., demic diffusion) or horizontal cultural transmission [8]. Genetic evidence suggests that these cultural and technological changes were accompanied by profound genomic transformation, consistent with the migration of people of most likely Anatolian origin [9, 10, 11, 12]. In contrast to central Europe, the adoption of agriculture in northern and eastern parts of this continent, in the areas which encompass modern-day Latvia and Ukraine, was slow and relatively recent [13, 14, 15, 16]. Although some features of the Neolithic package, such as ceramics, appeared as early as 8,500–7,500 cal BP [17, 18], agriculture was not adopted as a primary subsistence economy until the Late Neolithic/Bronze Age [13, 14, 15, 16, 19].
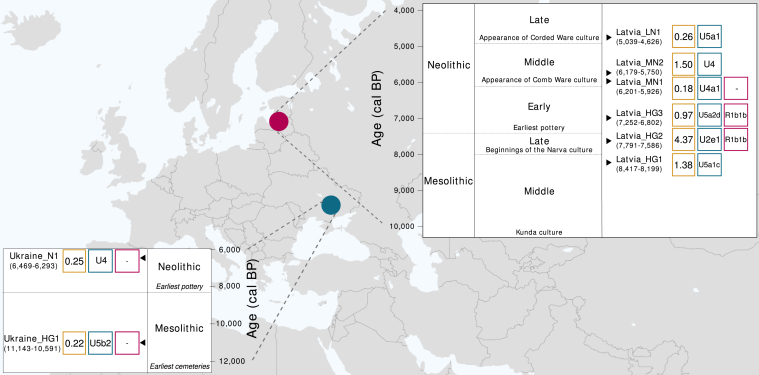
The Neolithic transition in the Baltic and Ukraine thus had a different tempo to that of central Europe, and it is unclear how this may have shaped the genetic composition of these regions. To investigate this, we sampled three Mesolithic and three Neolithic individuals from the archaeological site of Zvejnieki (Latvia), which is one of the richest Stone Age cemeteries in Northern Europe for number of inhumations, as well as duration of use [20, 21] (see the Supplemental Experimental Procedures for site details). We also sampled a Mesolithic and a Neolithic individual from cemeteries found along the Dnieper River in Ukraine (Vasilyevka 3 and Vovnigi 2, respectively). DNA was extracted from the petrous portion of the temporal bone (see the Experimental Procedures), which yielded between 4.30% and 55.99% endogenous DNA for all samples. Samples were shotgun sequenced using Illumina sequencing technology to between 0.22- and 4.37-fold coverage (Figure 1). The authenticity of the data was assessed in silico by examining the data for signatures of post-mortem DNA damage and evaluating the mitochondrial contamination rate in all samples along with the X chromosome contamination rate in males (see the Supplemental Experimental Procedures). All samples had degradation patterns typical of ancient DNA (Figure S1) and low contamination estimates of ∼1% or less (Table 1).
Figure 1.
Geographic Location and Chronologies for Latvian and Ukrainian Sites
Radiocarbon dates (in cal BP) are shown under the sample name. Mean genome coverage is shown in yellow squares, mitochondrial haplogroups in blue squares, and Y chromosome haplogroups for male samples (where discernible) in magenta squares. The chronology of the Latvian site of Zvejnieki is adapted from [21]. The Ukrainian chronology is taken from [18, 22, 23].
Table 1.
Alignment and Contamination Results for Latvian and Ukrainian Ancient Samples
| Sample | Site | Context | Burial | Aligned Reads | Aligned Reads (%) | MT Coverage | MT Contamination (c+md/c−md) | X Contamination (Test 1/Test 2) |
|---|---|---|---|---|---|---|---|---|
| Latvia_HG1 | Zvejnieki | Mesolithic; Kunda culture | 313 | 54,784,565 | 49.26 | 47.83 | 0.68/0.04 | − |
| Latvia_HG2 | Zvejnieki | Mesolithic; Narva culture | 93 | 172,707,718 | 55.99 | 114.97 | 0.94/0.19 | 0.92 ± 0.08/0.88 ± 0.17 |
| Latvia_HG3 | Zvejnieki | Mesolithic/Early Neolithic | 121 | 37,749,963 | 45.51 | 40.29 | 0.77/0.10 | 0.99 ± 0.26/0.72 ± 0.37 |
| Latvia_MN1 | Zvejnieki | Middle Neolithic | 124 | 6,648,453 | 5.22 | 8.14 | 0.97/0.50 | − |
| Latvia_MN2 | Zvejnieki | Middle Neolithic; Comb Ware culture | 221 | 59,800,396 | 37.51 | 48.54 | 0.69/0.10 | − |
| Latvia_LN1 | Zvejnieki | Late Neolithic; Corded Ware culture | 137 | 9,222,060 | 7.48 | 9.58 | 1.09/0.00 | − |
| Ukraine_HG1 | Vasilyevka | Mesolithic | 37 | 9,528,908 | 4.30 | 5.49 | 0.29/0.29 | − |
| Ukraine_N1 | Vovnigi | Neolithic; Dnieper-Donets culture | 2 | 10,741,415 | 12.04 | 6.06 | 1.06 0.28 | − |
MT, mitochondria; c+md, percentage contamination including sites with potentially damaged bases; c−md, percentage contamination excluding sites with potentially damaged bases. For X chromosome contamination estimation, two tests were performed as described by Rasmussen et al. [24]. Test 1 used all high-quality reads provided per sample, whereas only a single read was sampled per site for test 2, thereby removing the assumption of independent error rates. The associated p values calculated using Fisher’s exact test [24] were ≤0.05 for all tests. See also Table S1 for imputed genotypes probabilities for selected loci.
Episodes of Continuity and Change during the Mesolithic and Neolithic in the Baltic
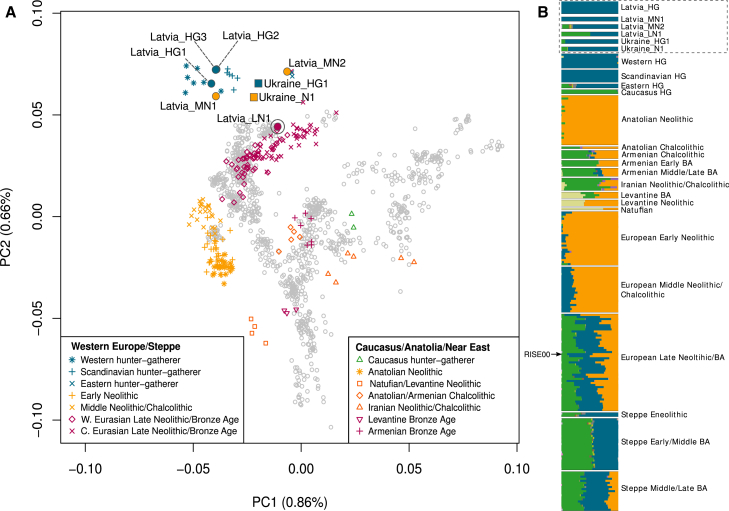
The two earliest samples in our Baltic time series, Latvia_HG1 (8,417–8,199 cal BP), associated with the Kunda culture, and Latvia_HG2 (7,791–7,586 cal BP), associated with the Narva culture, derive from the Late Mesolithic period [17, 21]. A third sample, Latvia_HG3 (7,252–6,802 cal BP), dates to the Late Mesolithic/Early Neolithic period, with the burial showing no major departures from the preceding Mesolithic traditions [21]. Principal component analysis (PCA) with ancient samples projected onto modern Eurasian genetic variation (see the Supplemental Experimental Procedures) shows that these three hunter-gatherer samples group together in a PCA plot (first two components, Figures 2A and S1A). In keeping with their geographical origins, they are in an intermediate position between Western European hunter-gatherer samples (WHG; from Luxembourg, Hungary, Italy, France, and Switzerland) and Eastern European hunter-gatherer samples (EHG; from Russia). They are composed of the same (blue) major component as these other hunter-gatherer groups in an ancestry coefficient decomposition analysis performed using ADMIXTURE [25] (Figure 2B), suggesting a close relationship between these groups. We found that although the Latvian Mesolithic samples share closer affinity to WHG than to EHG, the Latvian Mesolithic samples do not belong entirely to either hunter-gatherer group (tested using D statistics [27], which offer a formal test of admixture; Table 2). This suggests that they may be a previously unsampled component of a hunter-gatherer meta-population that stretched across Northern Europe during the early Holocene.
Figure 2.
PCA and ADMIXTURE Analysis for Ancient Latvian and Ukrainian Samples
(A) Ancient data presented in this study as well as published ancient data (see Data S1 for sample details) were projected onto the first two principal components defined by selected modern Eurasians from the Human Origins dataset (see the Supplemental Experimental Procedures). Our Latvian Mesolithic samples cluster tightly together between western and eastern hunter-gatherers in PCA space, whereas the Latvian Neolithic samples are more variable in their position, suggesting impacts from exogenous populations. The Ukrainian Mesolithic and Neolithic samples fall close together between western and eastern hunter-gatherers, suggesting a degree of continuity across the Mesolithic-Neolithic boundary in this region.
(B) ADMIXTURE ancestry components (K = 17) [25] for ancient samples showing that the Latvian Neolithic samples do not have the yellow component that dominates in Anatolian and early European farmers. The Latvian and Ukrainian samples presented in this study are displayed in a gray box and at twice the height of the other ancient samples for ease of visualization. The arrow shows an Estonian Bronze Age sample (RISE00) [26] that has a yellow component, suggesting that an early European farmer genetic influence had arrived in the Baltic by the Bronze Age.
HG, hunter-gatherer; BA, Bronze Age; W, western; C, Central. See also Figures S1–S4.
Table 2.
Key D Statistics of the Form D(A,B; X,Y) for Latvian Samples
| A | B | X | Y | D | Z Score | Loci |
|---|---|---|---|---|---|---|
| Latvian Mesolithic Samplesa | ||||||
| Mbuti | Latvia_HG | EHG | WHG | 0.0787 | 13.064 | 103,420 |
| Mbuti | EHG | Latvia_HG | WHG | −0.0281 | −4.797 | 103,420 |
| Mbuti | WHG | Latvia_HG | EHG | −0.1065 | −17.418 | 103,420 |
| Mbuti | Latvia_HG | Karelia (EHG) | Bichon (WHG) | 0.0801 | 11.725 | 3,188,010 |
| Mbuti | Latvia_HG | Karelia (EHG) | Loschbour (WHG) | 0.0928 | 13.521 | 3,204,237 |
| Mbuti | Karelia (EHG) | Latvia_HG | Bichon (WHG) | −0.0432 | −6.494 | 3,188,010 |
| Mbuti | Karelia (EHG) | Latvia_HG | Loschbour (WHG) | −0.0354 | −5.367 | 3,204,237 |
| Mbuti | Bichon (WHG) | Karelia (EHG) | Latvia_HG | 0.1228 | 17.680 | 3,188,010 |
| Mbuti | Loschbour (WHG) | Karelia (EHG) | Latvia_HG | 0.1277 | 18.643 | 3,204,237 |
| Latvia_MN1b | ||||||
| Mbuti_AF | Iran_LN | Latvia_HG | Latvia_MN1 | 0.0427 | 1.459 | 7,261 |
| Mbuti_AF | Anatolia_ChL | Latvia_HG | Latvia_MN1 | 0.0249 | 0.907 | 8,289 |
| Mbuti_AF | Kennewick | Latvia_HG | Latvia_MN1 | 0.0181 | 0.685 | 9,026 |
| Latvia_MN2c | ||||||
| Mbuti | Zapotec | Latvia_HG | Latvia_MN2 | 0.0295 | 4.171 | 74,461 |
| Mbuti | Guarani | Latvia_HG | Latvia_MN2 | 0.0303 | 4.027 | 74,461 |
| Mbuti | Aymara | Latvia_HG | Latvia_MN2 | 0.0282 | 3.926 | 74,461 |
| Mbuti | EHG | Latvia_HG | Latvia_MN2 | 0.0346 | 3.734 | 70,000 |
| Mbuti | AfontovaGora3 | Latvia_HG | Latvia_MN2 | 0.0658 | 3.519 | 19,541 |
| Mbuti | MA1 | Latvia_HG | Latvia_MN2 | 0.0381 | 3.441 | 53,389 |
| Mbuti | Karelia (EHG) | Latvia_HG | Latvia_MN2 | 0.0386 | 5.742 | 2,165,498 |
| Mbuti | MA1 | Latvia_HG | Latvia_MN2 | 0.0553 | 7.355 | 1,799,638 |
| Mbuti | Karitiana | Latvia_HG | Latvia_MN2 | 0.0366 | 5.307 | 2,693,045 |
| Latvia_LN1d | ||||||
| Mbuti | CHG | Latvia_HG | Latvia_LN1 | 0.0312 | 2.081 | 20,994 |
| Mbuti | Iran_N | Latvia_HG | Latvia_LN1 | 0.0275 | 1.483 | 16,000 |
| Mbuti | Iran_ChL | Latvia_HG | Latvia_LN1 | 0.0203 | 1.457 | 19,027 |
| Mbuti | Iran_LN | Latvia_HG | Latvia_LN1 | 0.0319 | 1.320 | 9,481 |
| Mbuti | Iranian_Jew_WA | Latvia_HG | Latvia_LN1 | 0.0106 | 1.091 | 20,998 |
| Mbuti | Kotias (CHG) | Latvia_HG | Latvia_LN1 | 0.0182 | 2.138 | 715,061 |
| Latvian Neolithic Samplese | ||||||
| Mbuti | Anatolia_N | Latvia_HG | Latvia_MN1 | −0.0013 | −0.108 | 16,255 |
| Mbuti | Europe_EN | Latvia_HG | Latvia_MN1 | −0.0055 | −0.476 | 16,272 |
| Mbuti | Anatolia_N | Latvia_HG | Latvia_MN2 | −0.0246 | −3.607 | 74,355 |
| Mbuti | Europe_EN | Latvia_HG | Latvia_MN2 | −0.0335 | −5.055 | 74,460 |
| Mbuti | Anatolia_N | Latvia_HG | Latvia_LN1 | −0.0221 | −2.136 | 20,969 |
| Mbuti | Europe_EN | Latvia_HG | Latvia_LN1 | −0.0308 | −2.952 | 20,998 |
| Mbuti | Stuttgart | Latvia_HG | Latvia_MN1 | 0.0168 | 1.804 | 581,303 |
| Mbuti | NE1 | Latvia_HG | Latvia_MN1 | 0.0161 | 1.704 | 581,758 |
| Mbuti | Stuttgart | Latvia_HG | Latvia_MN2 | −0.0300 | −4.527 | 2,722,280 |
| Mbuti | NE1 | Latvia_HG | Latvia_MN2 | −0.0272 | −3.871 | 2,724,548 |
| Mbuti | Stuttgart | Latvia_HG | Latvia_LN1 | −0.0246 | −2.955 | 715,090 |
| Mbuti | NE1 | Latvia_HG | Latvia_LN1 | −0.0215 | −2.373 | 715618 |
Latvia_HG, Latvian hunter-gatherers; WHG, western hunter-gatherers; EHG, eastern hunter-gatherers; Iran_LN, Iranian Late Neolithic; Anatolia_ChL, Anatolian Chalcolithic; CHG, Caucasus hunter-gatherers; Iran_N, Iranian Neolithic; Iran_ChL, Iranian Chalcolithic; Anatolian_N, Anatolian Neolithic; Europe_EN, European Early Neolithic. Tests performed using the whole genome panel are italicized; otherwise, tests were performed using the Human Origin transversion SNP panel. Ancient samples are shown in bold. Samples include in each ancient group can be found in Data S1. See Table S1 for key D statistics for Ukrainian samples.
The Latvian Mesolithic samples share more affinity to WHG than to EHG, but they do not belong entirely to either group.
There is no evidence for admixture in our Latvian Neolithic sample, Latvia_MN1 (the three largest positive statistics are shown).
There is an eastern influence in the Latvian Middle Neolithic sample, Latvia_MN2, as compared to the Latvian Mesolithic samples (the three most significantly positive results with ancient and modern populations/individuals from the Human Origins SNP panel dataset are shown).
Largest positive results for the test D(Mbuti, X; Latvia_HG, Latvia_LN1). ADMIXTURE results suggest that there may have been a CHG-related influence in Latvia during the Late Neolithic period; however, although D statistics to test this are positive, they do not reach significance.
We do not find evidence for early European/Anatolian farmer admixture in our Latvian Neolithic samples.
Next we sampled two Middle Neolithic individuals, Latvia_MN1 (6,201–5,926 cal BP), from an isolated grave located among burials from earlier periods, and Latvia_MN2 (6,179–5,750 cal BP), who was interred in a collective burial with five other individuals. During the Middle Neolithic at Zvejnieki, mortuary practices from the preceding periods were partially maintained, but some new features appeared, including collective burials and votive deposits, which are associated with the Comb Ware culture or its influences in the Baltic [21]. Despite having been roughly contemporaneous, these Middle Neolithic samples cluster in different regions of our PCA plot (Figure 2A) and have distinct profiles in ADMIXTURE analysis (Figure 2B). In both analyses, Latvia_MN1 groups with the Mesolithic Latvian samples, suggesting a degree of continuity across the Mesolithic-Neolithic transition in this region and consistent with suggestions that the eastern Baltic was a genetic refugium for hunter-gatherer populations during the Neolithic period [28]. The persistence of hunter-gatherer ancestry in the Baltic until at least the Middle Neolithic also provides a possible source for the resurgence of hunter-gatherer ancestry that is proposed to have occurred in central Europe from 7,000–5,000 cal BP [1]. In contrast, Latvia_MN2 is placed toward EHG in PCA space and has several components in ADMIXTURE analysis that are found in Native Americans, Siberians, and hunter-gatherer samples from the Caucasus. In keeping with these results, we found that there has been a northern Eurasian influence in the Baltic region since the Mesolithic period, as suggested by significantly positive statistics for the test D(Mbuti, X; Latvia Mesolithic, Latvia_MN2) when X was an EHG, modern and ancient Siberian (including the Upper Palaeolithic Mal’ta genome [29]), or Native American (Table 2). This influence is supported archaeologically by the appearance of copper rings and amber jewelry in Middle Neolithic collective burials that bear similarities to artifacts found in Estonia, Finland, and northwestern Russia [21, 30].
The latest Neolithic sample in our Baltic time series, Latvia_LN1 (5,039–4,626 cal BP), which was found in a crouched burial of the type associated with the Late Neolithic Corded Ware culture [21], falls near other Late Neolithic and Bronze Age European and Steppe samples in PCA analysis (Figure 2A). In ADMIXTURE analysis, it is composed of the blue component (Figure 2B), which is predominant in all of the older Latvian samples, but also a green component, which is maximized in hunter-gatherer samples from the Caucasus. A Caucasus-related influence in this sample is also suggested by positive results (although without formal significance, Z > 2) for tests of the form D(Mbuti, Caucasus hunter-gatherer; Latvian Mesolithic, Latvia_LN1). Ancestry related to hunter-gatherers from the Caucasus has previously been postulated to have arrived in Europe through herders from the Pontic Steppe [1, 31], and these migrations could potentially be the source of this ancestry in our sample. Interestingly, this individual lived around the time of later date estimates (∼4,500–7,000 cal BP) proposed for the split of Proto-Balto-Slavic from other Indo-European languages [3, 4]. There are two major theories to explain the distribution of Indo-European languages that constitute the most widely spoken language family in the world: (1) they have an Anatolian origin and were spread by Neolithic agriculturalists [32, 33] and (2) they developed in the Pontic Steppe and proliferated through Late Neolithic/Bronze Age migrations [1, 3, 34]. The presence of a Steppe-related component in Latvia_LN1 in the absence of an Anatolian farmer-related genetic input supports a Steppe rather than an Anatolian origin for the Balto-Slavic branch of the Indo-European language family.
It is striking that we did not find evidence for early European or Anatolian farmer admixture in any of our Latvian Neolithic samples using both D statistics (Table 2) and ADMIXTURE (Figure 2A). This lack of admixture is also supported by the mitochondrial haplogroup of the Latvian Neolithic samples (all belong to U; Figure 1), which is prevalent in European hunter-gatherers [1, 35], including our Latvian Mesolithic samples, but not in early farmers. It is interesting that among the grave goods found in the burial of Latvia_LN1 was a chisel made from the bone of a domesticated goat or sheep [17, 21]. The presence of this tool made from a domesticate as well as dietary isotope data (δ15N and δ13C), which show greater reliance on terrestrial resources than in previous periods [17], is consistent with either the adoption of farming without early European farmer-related genetic admixture or the existence of trade networks with farming communities that were largely independent of genomic exchange. Although we find no genetic input from Anatolian or early European farmers in our time series, ADMIXTURE analysis of an Estonian Corded Ware sample [26] (Figure 2B) has suggested that this farmer genetic influence, which is present in contemporary Northern European populations (Figure S2), had arrived in the Baltic by at least the Bronze Age.
The Neolithic Transition in Ukraine
The Ukrainian Mesolithic and Neolithic male samples (Ukraine_HG1 [11,143–10,591 cal BP] and Ukraine_N1 [6,469–6,293 cal BP], respectively) cluster tightly together between WHG and EHG samples in PCA analysis (Figure 2A). They form a clade with respect to other modern and ancient samples when tested using genome-wide D statistics (D(Mbuti, X; Ukraine_HG1, Ukraine_N1); Table S1), and their mitochondria belong to the U haplogroup, which has been found in ∼80% of European hunter-gatherer samples [1, 35]. These results suggest a degree of continuity across 4,000 years from the Mesolithic to the Neolithic period in the Dnieper Rapids. In ADMIXTURE analysis (Figure 2B), both Ukrainian samples are composed almost entirely of the European hunter-gatherer (blue) component, with a smaller green component that is also found in EHG. This green component is slightly larger in the Neolithic sample than in the Mesolithic sample, which is in keeping with D statistics that suggest increased affinity with ancient northern Eurasians from the Mesolithic to the Neolithic in Ukraine (Table S1). It is intriguing that we find an increased affinity to northern Eurasian samples in both Ukraine and Latvia during the Neolithic period. This could be the result of increased connectivity in Europe at this time. More extensive sampling will reveal whether this is a feature of the Neolithic across Northern and Eastern Europe.
Relationship of Ancient Samples to Modern Populations
The ancient Latvian and Ukrainian samples fall close to modern Northern and Eastern European populations in PCA analysis (Figures 2A and S1A), suggesting a degree of continuity in both regions since the Mesolithic period. Outgroup f3 statistics, which measure shared genetic drift between populations, further support this as they show that these ancient samples share most affinity with modern populations from Northern and Eastern Europe (Figure S3). Further, the Y chromosomes of two of our Latvian Mesolithic samples were assigned to haplogroup R1b (the maximum-likelihood sub-haplogroup is R1b1b), which is the most common haplogroup found in modern Western Europeans [36]. This haplogroup has been found at low frequencies before the Late Neolithic in Western Europe [1, 35] but at higher frequencies in Russia and is suggested to have spread into Europe from the East after 5,000 cal BP [1]. The presence of this haplogroup in Mesolithic Latvia points to a more westward ancestral range. We found that the three Mesolithic Latvian samples are predicted to have had the derived variant (rs12913832) of the HERC2 gene associated with blue eye color (Figure S3B and Table S2). Blue eye color is found at high frequencies in Northern Europe today, and these results suggest that this phenotype was already present in the Baltic by the Mesolithic period. We also found tentative evidence for progressive skin depigmentation in Latvia based on mutations in the SLC24A5 and SLC45A2 genes (rs1426654 and rs16891982, respectively; Figure S3B and Table S2).
Discussion
The Neolithic transitions in the Baltic and Dnieper Rapids region of Ukraine show very different archaeological and genetic dynamics to those observed in Central and Western Europe. Although in central Europe pottery and agriculture arrive as a package, in the Baltic and Dnieper Rapids the onset of the Neolithic is characterized by the appearance of ceramics, with a definitive shift to an agro-pastoralist economy only occurring during the Late Neolithic/Bronze Age [13, 14, 15, 16, 19]. Although the prolonged and piecemeal uptake of Neolithic characteristics in these regions makes it challenging to attribute a definitive shift in ideology or lifestyle, it does, along with evidence for continuities in material culture and settlement patterns, suggest that Neolithic features were predominantly adopted by indigenous hunter-gatherers in this region [13, 14, 15, 16, 37]. We find genetic evidence in support of this in the affinity of the Latvian and Ukrainian Neolithic samples, Latvian_MN1 and Ukrainian_N1, to earlier Mesolithic samples from the same respective regions. However, we also find indications of genetic impact from exogenous populations during the Neolithic, most likely from northern Eurasia and the Pontic Steppe. These influences are distinct from the Anatolian-farmer-related gene flow found in central Europe during this period. It is interesting to note that even in outlying areas of Europe, such as Sweden and Ireland [38, 39], an Anatolian-farmer-related genetic signature is present by the Middle to Late Neolithic period (∼5,300–4,700 cal BP). We conclude that the gradual appearance of features associated with the Neolithic package in the Baltic and Dnieper Rapids was not tied to the same major genetic changes as in other regions of Europe. The emergence of Neolithic features in the absence of immigration by Anatolian farmers highlights the roles of horizontal cultural transmission and potentially independent innovation during the Neolithic transition.
Experimental Procedures
DNA was extracted from a petrous bone of our samples in dedicated ancient DNA facilities (see the Supplemental Experimental Procedures). Libraries were prepared and sequenced using 50–100 bp Illumina single-end sequencing, and reads were aligned to the GRCh37 build of the human genome with the mitochondrial sequence replaced by the revised Cambridge reference sequence. The authenticity of the data was assessed by looking for short average sequence length and patterns of molecular damage and estimating the mitochondrial and X chromosome contamination rates (see the Supplemental Experimental Procedures). Sex was determined by examining the ratio of Y chromosome reads to reads aligning to both sex chromosomes [40]. Pseudo-diploid genotypes were called in our sample at positions that overlapped autosomal genotypes in the Human Origins dataset (110,507 positions) and merged with publicly available ancient genotypes (see the Supplemental Experimental Procedures). The relationship between our ancient samples and other modern and ancient populations was assessed using PCA, ADMIXTURE, D statistics, and f3 statistics (see the Supplemental Experimental Procedures). Mitochondrial and Y chromosome haplogroups were determined using HAPLOFIND and Yfitter, respectively. Genotypes associated with particular phenotypes were examined using observed and imputed genotypes as in [2] (see the Supplemental Experimental Procedures, Figure S3, and Table S2).
Author Contributions
D.G.B., R.P., and A.M. supervised the study. E.R.J., D.G.B., and A.M. analyzed genetic data. R.P., G.Z., V.M., and P.R.N. provided samples and/or input about archaeological context. E.L. helped interpret isotope analyses. E.R.J., A.M., P.R.N., and D.G.B. wrote the manuscript with input from all co-authors.
Acknowledgments
This research was supported by a European Research Council (ERC) Starting Grant (ERC-2010-StG 263441) and Irish Research Council Advanced Research Project Grant to R.P. D.G.B. and A.M. were also supported by the ERC (295729-CodeX and 647787-LocalAdaptation, respectively). P.R.N. was supported by a FP7MC Career Integration Grant (NEMO-ADAP; no. 322261). We would like to thank Veronika Siska, Cristina Gamba, Rui Martiniano, Russell McLaughlin, Lara Cassidy, and Olesia Kononenko for their support.
Published: February 2, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, two tables, and one data file and can be found with this article online at http://dx.doi.org/10.1016/j.cub.2016.12.060.
Contributor Information
Eppie R. Jones, Email: erj35@cam.ac.uk.
Andrea Manica, Email: am315@cam.ac.uk.
Ron Pinhasi, Email: ron.pinhasi@ucd.ie.
Daniel G. Bradley, Email: dbradley@tcd.ie.
Accession Numbers
The accession number for the sequence data reported in this paper is European Nucleotide Archive (ENA): PRJEB18067.
Supplemental Information
References
- 1.Haak W., Lazaridis I., Patterson N., Rohland N., Mallick S., Llamas B., Brandt G., Nordenfelt S., Harney E., Stewardson K. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature. 2015;522:207–211. doi: 10.1038/nature14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gamba C., Jones E.R., Teasdale M.D., McLaughlin R.L., Gonzalez-Fortes G., Mattiangeli V., Domboróczki L., Kővári I., Pap I., Anders A. Genome flux and stasis in a five millennium transect of European prehistory. Nat. Commun. 2014;5:5257. doi: 10.1038/ncomms6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anthony D.W. Princeton University Press; 2010. The Horse, the Wheel, and Language: How Bronze-Age Riders from the Eurasian Steppes Shaped the Modern World. [Google Scholar]
- 4.Bouckaert R., Lemey P., Dunn M., Greenhill S.J., Alekseyenko A.V., Drummond A.J., Gray R.D., Suchard M.A., Atkinson Q.D. Mapping the origins and expansion of the Indo-European language family. Science. 2012;337:957–960. doi: 10.1126/science.1219669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bar-Yosef O., Meadow R.H. The origins of agriculture in the Near East. In: Price T.D., Gebauer A.B., editors. Last Hunters, First Farmers: New Perspectives on the Prehistoric Transition to Agriculture. School of American Research Press; 1995. pp. 39–94. [Google Scholar]
- 6.Zeder M.A. Domestication and early agriculture in the Mediterranean Basin: Origins, diffusion, and impact. Proc. Natl. Acad. Sci. USA. 2008;105:11597–11604. doi: 10.1073/pnas.0801317105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Özdoğan M. A new look at the introduction of the Neolithic way of life in Southeastern Europe. Changing paradigms of the expansion of the Neolithic way of life. Doc. Praehistorica. 2014;41:33. [Google Scholar]
- 8.Robb J. Material culture, landscapes of action, and emergent causation: a new model for the origins of the European Neolithic. Curr. Anthropol. 2013;54:657–683. [Google Scholar]
- 9.Mathieson I., Lazaridis I., Rohland N., Mallick S., Patterson N., Roodenberg S.A., Harney E., Stewardson K., Fernandes D., Novak M. Genome-wide patterns of selection in 230 ancient Eurasians. Nature. 2015;528:499–503. doi: 10.1038/nature16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hofmanová Z., Kreutzer S., Hellenthal G., Sell C., Diekmann Y., Díez-Del-Molino D., van Dorp L., López S., Kousathanas A., Link V. Early farmers from across Europe directly descended from Neolithic Aegeans. Proc. Natl. Acad. Sci. USA. 2016;113:6886–6891. doi: 10.1073/pnas.1523951113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Omrak A., Günther T., Valdiosera C., Svensson E.M., Malmström H., Kiesewetter H., Aylward W., Storå J., Jakobsson M., Götherström A. Genomic Evidence Establishes Anatolia as the Source of the European Neolithic Gene Pool. Curr. Biol. 2016;26:270–275. doi: 10.1016/j.cub.2015.12.019. [DOI] [PubMed] [Google Scholar]
- 12.Lazaridis I., Nadel D., Rollefson G., Merrett D.C., Rohland N., Mallick S., Fernandes D., Novak M., Gamarra B., Sirak K. Genomic insights into the origin of farming in the ancient Near East. Nature. 2016;536:419–424. doi: 10.1038/nature19310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zvelebil M., Lillie M. Transition to agriculture in eastern Europe. In: Price T.D., editor. Europe’s First Farmers. Cambridge University Press; 2000. pp. 57–92. [Google Scholar]
- 14.Zvelebil M., Domanska L., Dennell R. Bloomsbury Academic; 1998. Harvesting the Sea, Farming the Forest: The Emergence of Neolithic Societies in the Baltic Region. [Google Scholar]
- 15.Antanaitis I. Concerning the transition to farming in the East Baltic. Doc. Praehistorica. 1999;26:89–100. [Google Scholar]
- 16.Zvelebil M., Dolukhanov P. The transition to farming in Eastern and Northern Europe. J. World Prehist. 1991;5:233–278. [Google Scholar]
- 17.Meadows J., Bērziņš V., Legzdiņa D., Lübke H., Schmölcke U., Zagorska I., Zariņa G. Stone-age subsistence strategies at Lake Burtnieks, Latvia. J. Archaeol. Sci. Rep. 2016 Published online April 22, 2016. [Google Scholar]
- 18.Matuzevičiūtė G.M. Neolithic Ukraine: a review of theoretical and chronological interpretations. Archaeol. Baltica. 2014;20:136–149. [Google Scholar]
- 19.Lillie M., Budd C. The Mesolithic-Neolithic transition in Eastern Europe: integrating stable isotope studies of diet with palaeopathology to identify subsistence strategies and economy. In: Stock J.T., Pinhasi R., editors. Human Bioarchaeology of the Transition to Agriculture. John Wiley & Sons; 2011. pp. 43–62. [Google Scholar]
- 20.Zagorskis F.A. Archaeopress; 2004. Zvejnieki (Northern Latvia) Stone Age Cemetery. [Google Scholar]
- 21.Larsson L., Zagorska I., editors. Back to the Origin: New Research in the Mesolithic-Neolithic Zvejnieki Cemetery and Environment, North Latvia. Almqvist & Wiksell International; 2006. [Google Scholar]
- 22.Lillie M., Richards M.P., Jacobs K. Stable isotope analysis of 21 individuals from the Epipalaeolithic cemetery of Vasilyevka III, Dnieper Rapids region, Ukraine. J. Archaeol. Sci. 2003;30:743–752. [Google Scholar]
- 23.Kośko A., Domańska L., Jacobs K. The Ukrainian Steppe as a region of intercultural contacts between Atlantic and Mediterranean zones of European Mesolithic. Baltic-Pontic-Studies. 1998;5:102–119. [Google Scholar]
- 24.Rasmussen M., Guo X., Wang Y., Lohmueller K.E., Rasmussen S., Albrechtsen A., Skotte L., Lindgreen S., Metspalu M., Jombart T. An Aboriginal Australian genome reveals separate human dispersals into Asia. Science. 2011;334:94–98. doi: 10.1126/science.1211177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allentoft M.E., Sikora M., Sjögren K.-G., Rasmussen S., Rasmussen M., Stenderup J., Damgaard P.B., Schroeder H., Ahlström T., Vinner L. Population genomics of Bronze Age Eurasia. Nature. 2015;522:167–172. doi: 10.1038/nature14507. [DOI] [PubMed] [Google Scholar]
- 27.Green R.E., Krause J., Briggs A.W., Maricic T., Stenzel U., Kircher M., Patterson N., Li H., Zhai W., Fritz M.H.-Y. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malmström H., Gilbert M.T.P., Thomas M.G., Brandström M., Storå J., Molnar P., Andersen P.K., Bendixen C., Holmlund G., Götherström A., Willerslev E. Ancient DNA reveals lack of continuity between neolithic hunter-gatherers and contemporary Scandinavians. Curr. Biol. 2009;19:1758–1762. doi: 10.1016/j.cub.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 29.Raghavan M., Skoglund P., Graf K.E., Metspalu M., Albrechtsen A., Moltke I., Rasmussen S., Stafford T.W., Jr., Orlando L., Metspalu E. Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature. 2014;505:87–91. doi: 10.1038/nature12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zagorska I. Amber graves of Zvejnieki burial ground. Acta Academiae Artium Vilnensis. 2001;22:109–124. [Google Scholar]
- 31.Jones E.R., Gonzalez-Fortes G., Connell S., Siska V., Eriksson A., Martiniano R., McLaughlin R.L., Gallego Llorente M., Cassidy L.M., Gamba C. Upper Palaeolithic genomes reveal deep roots of modern Eurasians. Nat. Commun. 2015;6:8912. doi: 10.1038/ncomms9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Renfrew C. Cambridge University Press; 1990. Archaeology and Language. [Google Scholar]
- 33.Gray R.D., Atkinson Q.D. Language-tree divergence times support the Anatolian theory of Indo-European origin. Nature. 2003;426:435–439. doi: 10.1038/nature02029. [DOI] [PubMed] [Google Scholar]
- 34.Mallory J.P., Adams D.Q. Fitzroy Dearborn; 1997. Encyclopedia of Indo-European Culture. [Google Scholar]
- 35.Fu Q., Posth C., Hajdinjak M., Petr M., Mallick S., Fernandes D., Furtwängler A., Haak W., Meyer M., Mittnik A. The genetic history of Ice Age Europe. Nature. 2016;534:200–205. doi: 10.1038/nature17993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Myres N.M., Rootsi S., Lin A.A., Järve M., King R.J., Kutuev I., Cabrera V.M., Khusnutdinova E.K., Pshenichnov A., Yunusbayev B. A major Y-chromosome haplogroup R1b Holocene era founder effect in Central and Western Europe. Eur. J. Hum. Genet. 2011;19:95–101. doi: 10.1038/ejhg.2010.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rigaud S., d’Errico F., Vanhaeren M. Ornaments reveal resistance of North European cultures to the spread of farming. PLoS ONE. 2015;10:e0121166. doi: 10.1371/journal.pone.0121166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skoglund P., Malmström H., Omrak A., Raghavan M., Valdiosera C., Günther T., Hall P., Tambets K., Parik J., Sjögren K.-G. Genomic diversity and admixture differs for Stone-Age Scandinavian foragers and farmers. Science. 2014;344:747–750. doi: 10.1126/science.1253448. [DOI] [PubMed] [Google Scholar]
- 39.Cassidy L.M., Martiniano R., Murphy E.M., Teasdale M.D., Mallory J., Hartwell B., Bradley D.G. Neolithic and Bronze Age migration to Ireland and establishment of the insular Atlantic genome. Proc. Natl. Acad. Sci. USA. 2016;113:368–373. doi: 10.1073/pnas.1518445113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skoglund P., Storå J., Götherström A., Jakobsson M. Accurate sex identification of ancient human remains using DNA shotgun sequencing. J. Archaeol. Sci. 2013;40:4477–4482. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.