Abstract
Whole-brain fMRI signals are a subject of intense interest: variance in the global fMRI signal (the spatial mean of all signals in the brain) indexes subject arousal, and psychiatric conditions such as schizophrenia and autism have been characterized by differences in the global fMRI signal. Further, vigorous debates exist on whether global signals ought to be removed from fMRI data. However, surprisingly little research has focused on the empirical properties of whole-brain fMRI signals. Here we map the spatial and temporal properties of the global signal, individually, in 1000+ fMRI scans. Variance in the global fMRI signal is strongly linked to head motion, to hardware artifacts, and to respiratory patterns and their attendant physiologic changes. Many techniques used to prepare fMRI data for analysis fail to remove these uninteresting kinds of global signal fluctuations. Thus, many studies include, at the time of analysis, prominent global effects of yawns, breathing changes, and head motion, among other signals. Such artifacts will mimic dynamic neural activity and will spuriously alter signal covariance throughout the brain. Methods capable of isolating and removing global artifactual variance while preserving putative “neural” variance are needed; this paper adopts no position on the topic of global signal regression.
Introduction
Cognitive and clinical correlates of global fMRI signals (the average signal of all gray matter signals or of all in-brain signals, see below) have gained much attention in recent years. For example, more attentive or vigilant (or caffeinated) human subjects exhibit reduced variance in the global fMRI signal (Wong et al., 2016; Wong et al., 2012, 2013). And psychiatric conditions have been characterized by differences in global fMRI signals, for example by altered variance in subjects with schizophrenia (Hahamy et al., 2014; Yang et al., 2014), or by an altered spatial distribution of the global signal in patients with autism spectrum disorder (Gotts et al., 2013; Gotts et al., 2012). Such findings, coupled with a report of an electrophysiological basis for global fMRI signals (Scholvinck et al., 2010), have generated intense interest in global fMRI signals.
Additionally, debates on the practice of removing brain-wide signals are among the most controversial, but consequential, issues in human neuroimaging. Although debates on removal of brain-wide signals date to the 1980s (for PET imaging, (Fox and Raichle, 1984; Horwitz et al., 1984)) and the 1990s (for task fMRI, (Aguirre et al., 1998)), the current debates concern studies of resting state functional connectivity MRI, a widely used technique for assessing brain organization (see (Power et al., 2014b) for review). In most human functional connectivity studies, fMRI data are obtained from subjects lying quietly in the scanner, and covariance between signals is the feature of interest. Several groups have argued that removing global fMRI signals from datasets prior to or during functional connectivity analyses renders the data (nearly) uninterpretable, on the presumption that the global fMRI signal is substantially composed of a mixture of non-global signals (Gotts et al., 2013; Murphy et al., 2009; Saad et al., 2012). These arguments gain force to the extent that a true global signal does not exist, and also gain (or lose) force to the extent that the signals in a scan exhibit low (or high) dimensionality. A related argument is that global neural signals may discriminate conditions or populations and thus ought not be removed. These arguments have been accepted by much of the field, and many groups now avoid removing global signal fluctuations in functional connectivity analyses.
In light of these considerations, it is perhaps surprising how little is firmly established about the empirical properties of global fMRI signals. For example, although the spatial distribution of the global fMRI signal has been used to characterize clinical status and plays an important role in the above-mentioned debates, the spatial distribution of the global fMRI signal in typical adults changes across studies that explicitly examine it: some studies find a rather uniform distribution of the global signal across gray and white matter (Aguirre et al., 1997; Zarahn et al., 1997), whereas others report that the global signal is found primarily in gray matter and not in white matter (Fox et al., 2009) (Figure S1). The reasons for the discrepancy are unclear.
The spatial and temporal characteristics of certain causes of brain-wide signals are partially known. Relatively recently, it was reported that motion can cause transient, brain-wide changes in signals that are typically signal decreases (Satterthwaite et al., 2013) (Figure S1). And a long series of studies have demonstrated that signals across much of the brain can be modulated by breath-holding maneuvers or forced alterations in blood gases that cause hypoxia or hypercapnia (Kastrup et al., 1998; Poulin et al., 1996; Stillman et al., 1995) (Figure S1). Extensions of those studies have linked breathing patterns at rest to fMRI signal changes across much of the brain, with Wise and colleagues reporting modulation in gray and white matter (Wise et al., 2004) and more recent studies emphasizing gray matter changes (Birn et al., 2006; Birn et al., 2008; Chang and Glover, 2009). It is thus known that motion and respiration (among other potential factors, see (Murphy et al., 2013)) can cause potent, brain-wide modulation of fMRI signals. There are some similarities, but also dissimilarities, of the reported spatial distribution of the global fMRI signal to the reported spatial effects of motion and respiration (Figure S1).
In sum, our present knowledge about global fMRI signals is fragmentary. On the one hand, based on the findings of Scholvinck and colleagues, the global fMRI signal might represent a global neural signal. On the other hand, effects of respiration and motion can cause global fMRI signal changes, and are probably present in many datasets, so the global fMRI signal is likely to include signals due to such influences. Although global neural signals would be of considerable interest, global effects of motion or respiration are typically considered artifact that need to be removed: they degrade statistical power in task fMRI modeling and will spuriously alter signal covariance in functional connectivity analyses. But if one decides to remove the global signal, one encounters the argument that the global signal must to some extent represent a mixture of non-global signals and thus should not be removed (in the worst case scenario, there is no true global signal at all, only a mixture of non-global signals). Faced with these considerations, an investigator would like to know the extent to which global artifacts, to the extent that they exist, are identified and removed during typical processing. However, there is almost no information in the literature on the composition of the global signal in typical data, much less the extent to which the global signal can be broken into constituent signals worth preserving or discarding.
This paper aims to help address such issues, which are fundamental to the conduct and interpretation of fMRI studies. We present and analyze whole-brain fMRI signals in over 1000 scans collected from 8 distinct sites, representing over 700 subjects. The scans represent a variety of subject populations, scanners, and scanner sequences, including newer multi-band sequences with relatively high temporal and spatial resolution. These scans thus provide a representation of the quality and kind of data obtained in many human functional neuroimaging studies. All scans are examined at the individual level and group levels. We have found single-subject presentations so useful for understanding global signals that we have prepared individual versions of every figure for each scan included in the paper, which can be viewed immediately as videos on YouTube and/or downloaded at the authors’ website1. A few minutes spent with these videos can familiarize a reader with effects present over hundreds of individuals.
Several clarifications will help define the scope and intent of this paper. First, by the “global signal” we mean the average fMRI signal over the entire brain, as it is typically and historically used (Aguirre et al., 1997; Fox et al., 2005; Fox et al., 2009; Greicius et al., 2003; Murphy et al., 2009; Zarahn et al., 1997). This signal is nearly identical to the mean signal of the gray matter for reasons that will shortly become apparent (the average correlation is r = 0.98 over all subjects of this paper), and is thus nearly synonymous with “mean gray matter timeseries”. Because multiple influences can modulate fMRI signals globally (e.g., motion, respiration, etc.), it is understood that the global signal represents not a single phenomenon, but just the observed average brain signal. When we begin to speak of multiple kinds of global signal, it will be understood that we are referring to multiple but distinct processes with signals sufficiently widespread (or of adequate amplitude) to affect the average whole-brain fMRI signal.
For brevity, we will sometimes refer to all kinds of unwanted signals as artifacts (or noise), to contrast them with the neurally-caused “signal” of interest. While respiratory or other physiological sources of variance constitute true fMRI signals, in most contexts these sources of variance are considered nuisance signals that need to be separated from signal of interest. We will also sometimes refer to “neural” fMRI signals; by this we mean signals contingent on changes in blood oxygenation principally caused by neural activity of interest.
Methods
Six large fMRI datasets are examined, all of which have been published previously. The datasets will be referred to as the Washington University (WU), the National Institute of Health (NIH), the Autism Brain Imaging Data Exchange (ABIDE), the Brain Genomics Superstruct (GSP), the Russ Poldrack (RP), and the Human Connectome Project (HCP) cohorts. Each subject’s data included a high-resolution T1-weighted (MP-RAGE) scan and one or more resting state T2*-weighted (BOLD-weighted) scans. Pertinent details of the functional data are given in Table 1, and more details can be found in the following references: WU: (Power et al., 2014a); NIH: (Gotts et al., 2012); ABIDE2; GSP: (Holmes et al., 2015); RP: (Poldrack et al., 2015); HCP: (Glasser et al., 2013).
Table 1.
Properties of the 6 fMRI datasets
| Site | Location | Scanner equipment | Voxel size at acquisition | TR (TE) | Slice order | Length | Population(s) | Further details of dataset |
|---|---|---|---|---|---|---|---|---|
| WU N=120 | Saint Louis, MO | Siemens Trio 3T + 12-channel head coil | 4 × 4 × 4 mm | 2500 (30) ms | alt+z2 | 14.4 min | Adults Typical only | Power et al., 2014 |
| NIH N=91 | Bethesda, MD | GE Signa 3T + 8-channel head coil | 1.7 × 1.7 × 3 mm | 3500 (27) ms | alt+z | 8.2 min | Adults, adolescents Typical and ASD | Gotts et al., 2012 |
| ABIDE N=164 | New York, NY (NYU) Los Angeles, CA (UCLA) Salt Lake City (USM) |
Siemens Allegra 3T Siemens Trio 3T Siemens Trio 3T |
3 × 3 × 3 mm 3 × 3 × 4 mm 3.4 × 3.4 × 4 mm |
2000 (15) ms 3000 (28) ms 2000 (28) ms |
alt+z alt+z2 alt+z2 |
6 min 6 min 8 min |
Children to adults Typical and ASD | http://fcon_1000.projects.nitrc.org/indi/abide/ |
| GSP N=231 | Boston, MA | Siemens Trio 3T + 12-channel head coil | 3 × 3 × 3 mm | 3000 (30) ms | alt+z | 6.2 min | Adults Typical only | Holmes et al., 2015 |
| RP N=1 (84 scans) | Austin, TX | Siemens Skyra 3T + 32-channel head coil | 2.4 × 2.4 × 2.4 mm | 1016 (30) ms | MB=4 | 10 min | Adults Typical only | Poldrack et al., 2015 |
| HCP N=38 | Saint Louis, MO | Siemens Skyra 3T + 32-channel head coil | 2.0 × 2.0 × 2.0 mm | 720 (33) ms | MB=8 | 14.3 min | Adults Typical only | Glasser et al., 2013 |
Idiosyncratic details of the datasets are mentioned here. Unlike other datasets, the WU dataset contains scans of varying length, ranging from 5.4–30 minutes (14.4 min on average), and variable numbers of runs per subject, due to collating scans from several studies. The ABIDE dataset contains many sites; the 3 sites examined here were chosen simply because they were on local servers. The GSP dataset contains 1000+ subjects; the first 235 were selected simply to conserve disk space. The HCP dataset presently includes 900 subjects; we use only the “40 Unrelated Subjects” download of this dataset (which actually contains only 38 subjects), simply to conserve disk space.
Five of these datasets are examined in detail (WU, NIH, ABIDE, GSP, RP) and processing for these datasets is described below. The HCP dataset was processed per the HCP processing pipelines (Glasser et al., 2013) and did not receive any additional processing here (other than constructing brain compartment masks, described below). The HCP data have undergone an independent components analysis (ICA) denoising procedure designed and administered by experts in that technique (Salimi-Khorshidi et al., 2014), which is the reason this dataset is of interest. The datasets used are the “Pre-processed” and “FIX-ICA Extended” atlas-registered scans, reflecting data before and after ICA denoising.
Spatial masks of brain compartments
All T1-weighted images in the WU, NIH, ABIDE, and GSP cohorts underwent automated segmentation by FreeSurfer version 5.3. FreeSurfer segmentations for the RP and HCP data were already available.
FreeSurfer segmentation yielded high-resolution (1 mm isotropic) masks of brain compartments. Lower resolution masks to match the fMRI data (3 or 2 mm isotropic as appropriate) were created using nearest-neighbor resampling. Separate gray matter masks of the cortical ribbon, cerebellum, and subcortical nuclei were created. Masks of the white matter underwent 0–4 erosion cycles, and ventricle masks underwent 0–2 erosion cycles at 1 mm resolution prior to resampling to fMRI resolution.
To study the properties of signals with varying proximity to gray matter, fMRI-resolution masks were subtracted from each other to identify voxels in superficial, deeper, and deepest masks (in white matter: ero0-ero2, ero2-ero4, and ero4 masks). In the ventricles, which are much smaller, only superficial and deeper masks (ero0-2 and ero2) were possible. In Figures, color darkness corresponds to mask depth (i.e., light green are superficial voxels, dark green are deepest voxels).
Two whole-brain masks are used. The all-brain mask contains all voxels inside the brain based on FreeSurfer segmentation, and is used to calculate the global signal. Because the spatial coverage of the structural image is often larger than that of the fMRI data (e.g., fMRI acquisitions often crop out portions of the cerebellum), another whole-brain mask including only voxels with mean fMRI values over 20% of the modal value was combined with all the above-mentioned masks to exclude any non-scanned parts of the brain.
Video 1 shows all masks in all subjects at both MP-RAGE and fMRI resolutions.
fMRI data processing
The RP cohort was processed exactly as described in (Laumann et al., 2015), and the HCP data was obtained from the HCP website in already-processed form. These datasets underwent no other processing by the authors. For all other cohorts, all fMRI scans underwent identical processing as described in (Power, submittedA), summarized below.
The first 4 volumes from all fMRI scans were ignored to ensure magnetization equilibrium. All scans were slice-time corrected using the AFNI command “3dTshift”. Scan realignment was calculated using AFNI’s “3dVolReg”.
To register data to an atlas space, the subject’s scan-space MP-RAGE was registered using a 12-parameter affine transform to the target atlas space (via AFNI’s “@auto_tlrc” command), the first used volume of the first fMRI scan was registered to the scan-space MP-RAGE, and all fMRI scans were registered to the first volume of the first fMRI scan in the realignment step. These registrations were concatenated into a single transform and the scan-space fMRI data were then transformed to atlas space and resampled to 3 mm isotropic voxels in a single step. The target was the “TT_N27” atlas in AFNI. Note that this procedure enforces cross-run alignment of multiple BOLD runs via the realignment step.
Each functional scan of each subject was visually checked for goodness of spatial registration and for complete brain coverage in the scan. If coverage or spatial registration was deemed inadequate, the scan (and, if necessary, the subject) was removed from further analysis. This selection based on spatial coverage and registration was the only exclusion criterion for the study and was performed prior to any examination of other data characteristics. This exclusion criterion resulted in the loss of 3/120 WU subjects, 0/91 NIH subjects, 3/164 ABIDE subjects, and 27/231 GSP subjects.
fMRI signals are always presented in mode 1000 notation (the modal value is 1000, i.e., 1% signal change is 10 units) unless otherwise noted (occasionally z-scores are used instead).
Spatial smoothing
fMRI data are not smoothed during processing, other than the smoothing implicit in resampling the data during realignment and registration. Explicit smoothing was avoided because we are interested in the distinct signals of various adjacent tissues.
However, smoothing is used for cosmetic purposes to create the 2D gray-scale plots seen in some Figures (Video 2 shows pre- and post-blurred versions, the Gaussian kernel is 6 mm FWHM). This blurring is performed using AFNI’s “3dBlurInMask” command, such that the blurring only occurs within-compartment and not across compartments (i.e., a voxel in the gray matter cortical mask will only be blurred with other voxels in the same gray matter mask, likewise for all masks). This blurring is performed because near-white (Rician) noise is prominent in datasets with small voxels (the NIH, RP, and HCP data) and obscures the presence of widespread signal changes in the gray-scale plots. In other words, gray-scale plots without smoothing in high-resolution datasets tend only to show a wall of static, despite the presence of structured signals “underneath” the noise, which become visible after smoothing.
Mean compartment signals are unchanged by this blurring procedure. The entire paper utilizes only unsmoothed data for computations, with the exception that the nuisance regressions in Figure 7 are performed on the smoothed data; performing the computations on unsmoothed data does not appreciably change the results (data not shown).
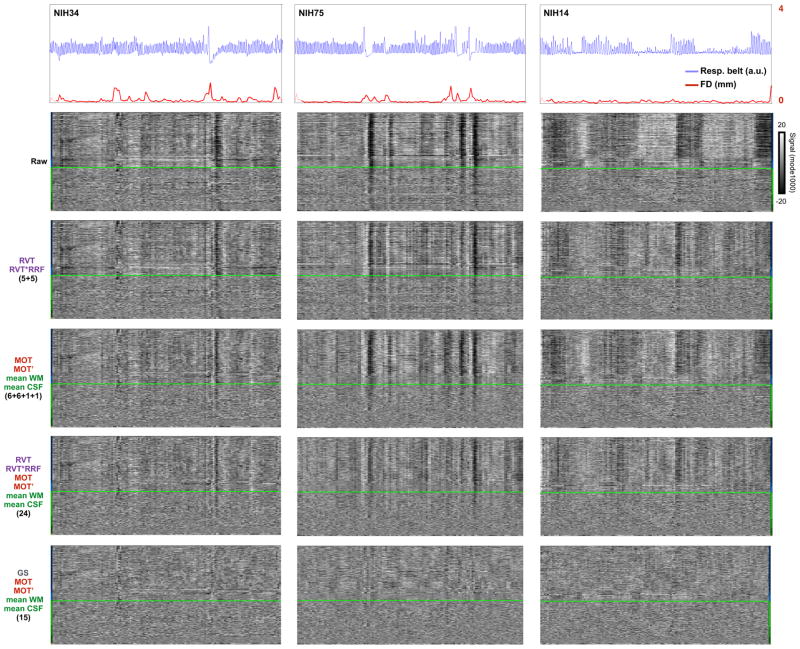
Figure 7. Removal of respiratory-related variance during denoising is often incomplete.
Data for 3 subjects from Figure 6 are shown. At top, respiratory belt and head motion traces. The five panels below show data after various combinations of denoising. The labels at left denote the kinds and number of regressors used.
Data traces
Motion estimates (MOT): head position was estimated using 3dVolReg in unprocessed data for the WU, NIH, ABIDE, and GSP data (since motion estimates after slice time correction under-represent motion (Power, submittedA)). Motion estimates were provided for RP and HCP data.
Framewise displacement (FD): framewise displacement indexes head motion. FD is calculated as the sum of the absolute values of the differentiated (in time) motion estimates, after converting rotational positions to arc length displacements at a radius of 50 mm, as in (Power et al., 2012). By convention, FD = 0 for the first volume.
fMRI signals: signals from various tissue compartments are presented as average signals within brain masks. Nuisance signals and the global signal are derived using the already-described masks.
fMRI signal heatmaps: all voxel signals within one or more brain compartments will often be shown as gray-scale heatmaps, with time on the X axis and voxels on the Y axis. These heatmaps are always scaled −20 to 20 in mode 1000 units. Brain compartments are usually denoted by colored bars beside the heat map (e.g., light green for superficial white matter voxels).
Mapping the spatial distribution of the global fMRI signal: GSCORR
To show how each voxel signal relates to the global signal, the global signal is calculated from the mean signal of the all-brain mask and is correlated with the signal at every voxel using a simple linear Pearson correlation in each subject. Voxelwise mean and linear trend terms are removed within each run prior to global signal computation. For any regression (or correlation) calculated in this article, mean and trend terms, per run, are removed from all involved signals (and for multiple linear regressions, all regressors are standardized).
We call this particular map GSCORR (see (Aguirre et al., 1997; Fox et al., 2009) for other examples of the same map, reproduced in Figure S1). When these maps (or any correlation measure) are combined across subjects, Fisher-z transforms are used prior to computations, and inverse transforms are used to derive r values for reporting purposes.
Physiologic traces
The NIH dataset includes pulse oximeter traces (in arbitrary units) and respiratory belt traces (in arbitrary units) for each scan, both sampled at 50 Hz. The respiratory belt traces are presented without further processing. The pulse oximeter traces underwent automated peak detection to detect systole, and instantaneous heart rate was calculated from peak-to-peak intervals. Pulse oximeter traces contain many artifacts, and suspicious portions of signals were identified using methods such as wavelet transforms of the raw signal and sensible checks on changes in heart rate (e.g., >40 Δbpm in a single beat). All signals and detected peaks were manually checked in their entirety to exclude spurious peaks and it is believed that the resulting heart rate traces are (nearly) completely accurate for all subjects. Peak amplitude was calculated as the difference between an accepted peak and the signal found 0.14 sec (7 samples) prior to the peak. Video 3 shows all traces and derived measures at fine and coarse temporal resolution.
fMRI signal denoising
The ability of several common denoising procedures, in various combinations, to remove global artifacts is evaluated. All voxel signals undergoing regression have per-run mean and linear trend terms removed, as do all regressors, which were also thereafter standardized. Regressors include the signals listed in Table 2, all of which might plausibly capture brain-wide variance. PCA of gray matter signals was not performed because it is already known that the global signal correlates with the 1st principal component of gray matter voxels at r = 0.95–0.99 (Carbonell et al., 2011). Independent component analysis (ICA) of gray matter signals was not performed in the five main cohorts but was instead examined in the HCP dataset.
Table 2.
Nuisance regressors used to denoise fMRI data
| Procedure/regressor | # regressors | Comment |
|---|---|---|
| Motion parameters | ||
| Realignment estimates (MOT) | 6 | |
| Realignment estimate derivatives (MOT′) | 6 | |
| Nuisance signals | ||
| Mean white matter signal (eroded 4x) (WM) | 1 | |
| Mean ventricle signal (eroded 2x) (CSF) | 1 | |
| PCA of all eroded white matter and ventricle signals | 5 | |
| Local white matter | Not performed | Mean and PCA signals should better reflect global signals |
| Physiological models | ||
| RETROICOR (Glover et al., 2000) | 8 | |
| RVT (Birn et al., 2006) | 5 | |
| RVT*RRF (Birn et al., 2008) | 5 | |
| RV (Chang et al., 2009) | 3 | |
| RV*RRF (Chang et al., 2009) | 3 | |
| Gray matter signals | ||
| ICA of all gray matter signals | Not performed | Studied in the HCP dataset, which has undergone FIX-ICA, see Video 5 |
| Global signal (GS) | 1 | |
| PCA of all gray matter signals | Not performed | Correlates with global signal at nearly r = 1 (Carbonell et al., 2011) |
| Combinations of Figure 7 | ||
| MOT+MOT′+WM+CSF (+RVT+RVT*RRF) (+GS) | ||
Several models of physiology-related fMRI signal variance were applied to the NIH dataset (of the main datasets, it alone has the necessary physiological records). These models include RETROICOR (Glover et al., 2000), RVT (Birn et al., 2006), RVT*RRF (Birn et al., 2008), and RV and RVT*RRF models (Chang et al., 2009). RETROICOR was applied using the AFNI command ‘3dretroicor’. Both Respiration Volume per Time (RVT) and Respiratory Variation (RV) measures were calculated, as well as elaborations of these models that convolve those regressors with “respiratory response functions”. RVT was calculated via “retroTS.m” in AFNI, and RV was calculated as the standard deviation of respiratory belt traces over a 6 second window. An empirical respiratory response function (RRF) was derived by measuring the average global signal change in the 42 seconds following isolated deep breaths apparent in several subjects from the NIH cohort (Figure 6). This derived RRF matches the RRFs published in (Birn et al., 2008) and (Chang and Glover, 2009) reasonably well (Figure S6). The 5 RVT regressors (the “default” approach of one RVT regressor with 4 added lags) were convolved with the RRF to yield 5 additional regressors intended to remove delayed effects of respiration (5+5 regressors). A similar convolution procedure was followed with the RV regressor, which was also lagged 7 seconds backward and forward (3+3 regressors). Many other versions of physiological signal modeling exist (e.g., (Shmueli et al., 2007), adding lagged heart rate regressors, or (Chang et al., 2013), adding regressors related to heart rate variability). We chose the RETROICOR, RVT, RVT*RRF, RV, and RV*RRF models because they are among the most commonly used models and because there are compelling reasons to focus on respiratory effects. Because there are collinearities between the derived physiologic regressors used in various models (e.g., between heart rate variability and respiratory volume), we were reluctant to combine large numbers of models without carefully examining the regressors, a procedure that becomes difficult with large numbers of tested models. Our goal is not to test all possible models, but to illustrate the efficacy of frequently used models.
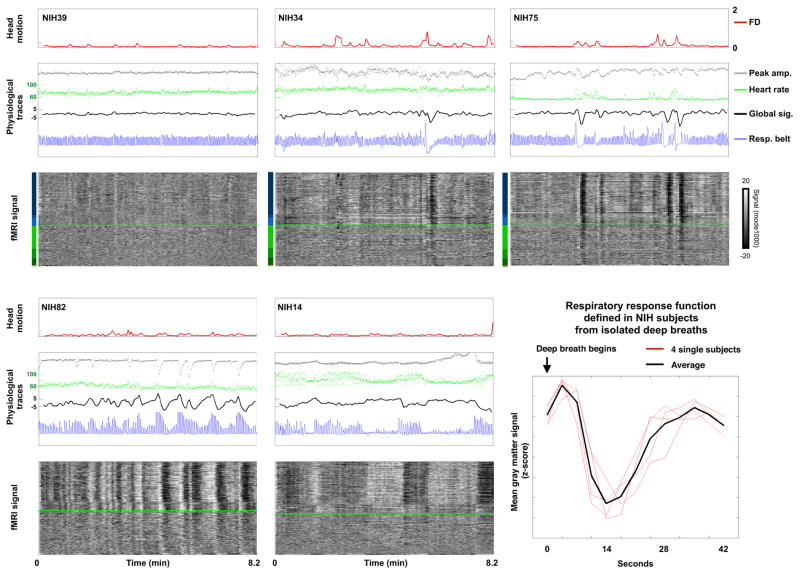
Figure 6. Respiratory traces denote many global signal changes.
Red traces of FD (mm) show head motion. The middle panels show heart rate (green, beats per minute) and peak amplitude (gray, zscore) data derived from a pulse-oximeter trace. The unprocessed respiratory belt record is shown in blue (arbitrary units). At bottom all fMRI signals in the brain are shown. When green heart rate traces appear to be noisy, for example in the bottom middle subject, this variance is not actually random noise, but rather cyclical modulation of heart rate by respiratory cycle (see Video 3a and 3b to resolve individual cycles). Similar statements apply to the gray peak amplitude trace at upper middle. At bottom right, the respiratory response function defined in the NIH data (see Figure S7 for more details).
Video 4 shows many combinations of these regression models in each subject of each cohort.
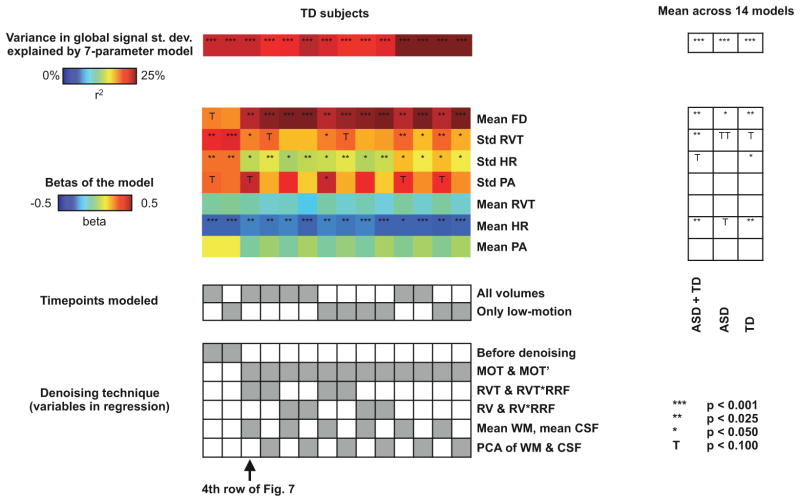
Modeling variance in global signals
In Table 3 and Figures 8–10 we model variance in the global signal, or in GSCORR, as a function of one or more variables. The independent variables are mean FD, mean and standard deviation of the heart rate, mean and standard deviation of a respiratory variable (RVT or RV), and mean and standard deviation of the pulse amplitude. In scatter plots, variance in the global signal is the dependent variable. In the brain slices, GSCORR is the dependent variable. For the permutation tests shown in Figures 9 and 10, dependent variables (variance in the global signal or the GSCORR maps) were shuffled across subjects 10,000 times while preserving the independent variables at each subject. The histogram in Figure 10 reflects the permutation ranks of all voxels in the model of GSCORR as a function of RVT variability, after the denoising shown in the 4th row of Figure 7 (MOT MOT’ WM CSF RVT RVT*RRF regressors).
Table 3.
7-parameter model of global signal variance
| Regressor | All timepoints | Low-motion timepoints | ||
|---|---|---|---|---|
| RVT model | RV model | RVT model | RV model | |
| Mean FD | 0.52 | 0.52 | 0.11 | 0.11 |
| Std. RVT/RV | 0.25 | 0.21 | 0.55 | 0.40 |
| Std. HR | 0.35 | 0.36 | 0.24 | 0.28 |
| Std. PA | 0.09 | 0.10 | 0.11 | 0.14 |
| Mean RVT/RV | −0.02 | 0.02 | −0.10 | 0.05 |
| Mean HR | −0.32 | −0.31 | −0.27 | −0.28 |
| Mean PA | 0.03 | 0.02 | 0.11 | 0.07 |
| % global signal std explained | 0.27 | 0.25 | 0.19 | 0.08 |
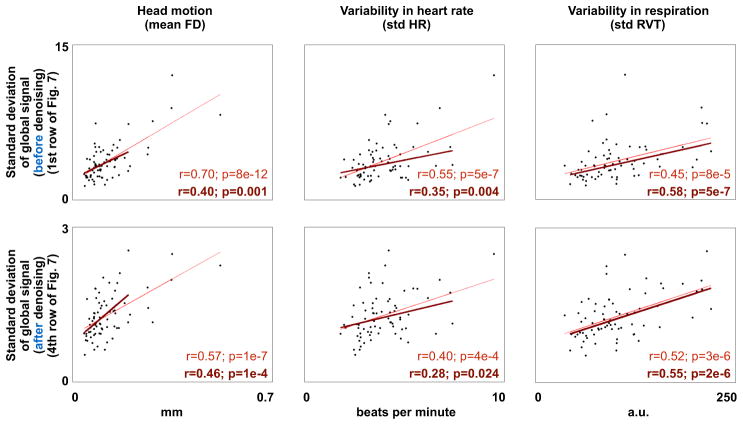
Figure 8. Global signals after denoising continue to reflect unwanted influences.
The standard deviation of the global signal is correlated, across subjects, with motion, heart rate variability, and respiratory variability. The thin red line fits all the data, the thicker dark line fits only subjects with mean FD < 0.2 mm. The top row reflect undenoised data, the bottom row reflects data after the denoising procedure shown in the 4th row of Figure 7.
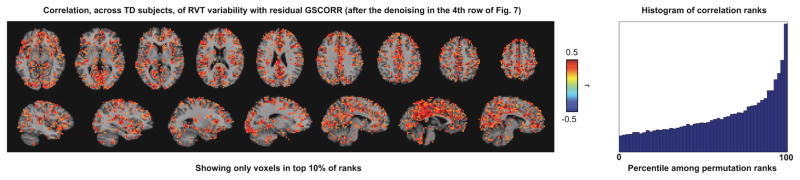
Figure 10. GSCORR after denoising continue to reflect unwanted influences.
GSCORR calculated after denoising is correlated with RVT variability, across subjects, at each voxel. The top 10% of correlations determined by 10,000 permutations are shown in slices, the ranks of all voxels in the image are shown at right.
Figure 9. Global signals after denoising continue to reflect unwanted influences across a variety of denoising strategies.
Global signal variance was modeled, across subjects, as a function of 7 parameters, as in Table 3. The heat maps and tables at left show the percent variance explained and beta values for “typical” (TD) subjects, the timepoints entering the model, and the various denoising strategies tested. At right, mean values across the 14 columns are shown for all NIH subjects, ASD subjects only, and TD subjects only. TT is placed in the mean ASD column to indicate that significant relationships exist in some strategies and in raw data, but the average is a permutation rank of 88%.
Results
Global signal fluctuations are evident in most fMRI scans, and are often artifact
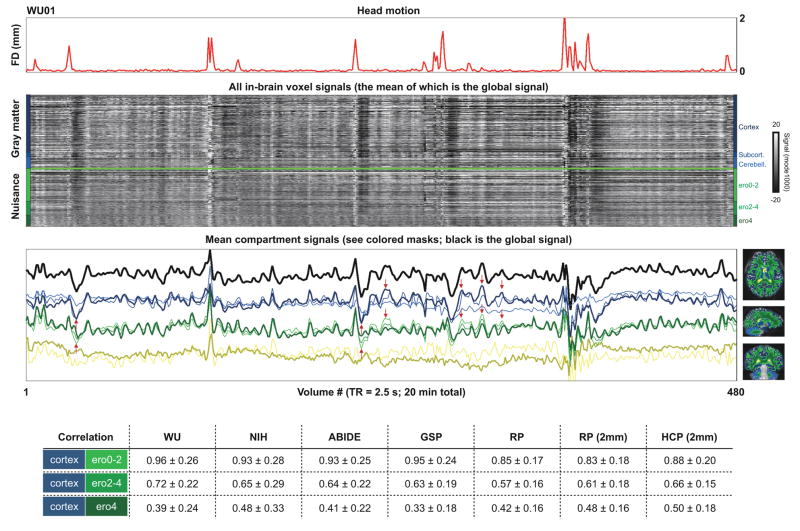
Signals present across most or all of the brain are routine features of fMRI scans. Figure 1 shows all in-brain signals in a single subject of the WU cohort. The brain next to the traces shows tissue compartments defined by segmentation of high-resolution structural images. In the heat map, the vertical black and white bands that span most or all brain compartments illustrate signals that prominently enter the global fMRI signal. Most of the global fluctuations in the gray matter signals are by definition artifact since they are also present throughout white matter and often the cerebrospinal fluid. In the third panel the black trace is the global signal and the colored traces represent the mean signals in each compartment. At top, the red trace shows head motion. Head motion coincides with (or marks the beginning of) many of the most obvious global fluctuations. Such signals are typical of most subjects. Readers may view structural brain segmentations for all subjects in Video 1, and versions of Figure 1 for all subjects in Video 2.
Figure 1. All voxel signals in a single subject.
At top a head motion (FD) trace is shown to establish when artifactual signals are expected. At bottom right are compartment masks. In the middle panels, signals from all in-brain voxels are shown, organized by mask. The traces are z-scored so that differences in signal magnitudes do not obscure signal (dis)similarities. Over half of all white matter voxels are in the ero0-2 erosion mask (lightest green) and signals at these white matter voxels are highly correlated with cortical gray matter signals (see red arrows), especially when compared with the signals deeper in the white matter (ero4, dark green). At bottom, the table shows correlations in each cohort of mean cortical ribbon signals and mean signals found in the various white matter masks.
Signals in the white matter and ventricles are widely considered to be nuisance signals with little to no representation of neural activity. However, nuisance signals in the white matter and ventricles vary depending on how well-isolated they are from gray matter signals. The resolution of the data under study is typical of fMRI data from the last decade: 3 mm isotropic voxels at analysis, derived from coarser or finer spatial resolutions at acquisition. At such resolution, voxels will often include multiple kinds of tissue (e.g., both gray and white matter). Such mixing of signals due to spatial sampling is called a partial volume effect. Nuisance voxels near gray matter tend to have signals highly correlated with the gray matter, but these correlations decrease in the deeper and deepest white matter voxels (medium and dark green masks, see Figure 1). This phenomenon is visible in the grayscale heat maps in many Figures (global gray matter signals often fade with depth in white matter). The effect is also visible in the traces of Figure 1 (see the red arrows pointing out similarities in the dark blue cortical gray matter trace and the light green superficial white matter trace). A similar effect is found in the ventricles, which in places abut subcortical gray matter. For example, mean subcortical gray matter signals (medium blue) correlate with superficial (light yellow) versus deeper (dark yellow) ventricle signals at r = 0.40, 0.29, 0.27, 0.06, and 0.23 versus 0.12, 0.18, 0.04, −0.13, and 0.18 in the WU, NIH, ABIDE, GSP, and RP datasets. In datasets that permit higher resolution analysis, such as the RP and HCP datasets, analyzing 2 mm isotropic voxels makes little difference in these numbers. In the remainder of the paper we will only use the dark green and dark yellow signals as nuisance signals, in all cohorts.
Various kinds of global signal are commonly encountered and can be recognized
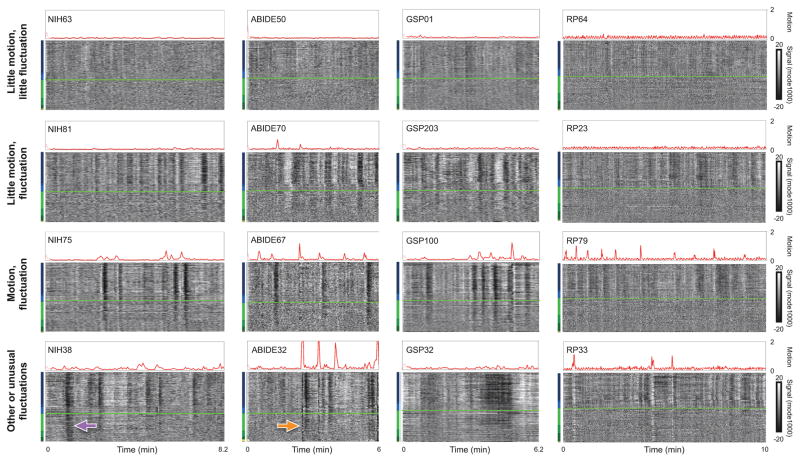
Several patterns emerge from viewing these plots in hundreds of subjects, summarized in Figure 2. First, in a minority of subjects (or scans), there is little evidence of global signal fluctuations. Such subjects always exhibit almost no motion (top row). Second, most subjects (or scans) do exhibit prominent global signal fluctuations of a particular kind. These fluctuations often occur without obvious motion (second row) but also often occur during and after motion (third row). These fluctuations are present throughout gray matter and superficial white matter, but they become less prominent (or absent) in deeper white matter voxels. Note that these fluctuations are slow – the x-axis shows 6–10 minutes of data in each of these subjects. Across hundreds of subjects, most global signal fluctuations exhibit the characteristics just described: slow changes present mainly in the gray matter and superficial white matter but not in deep white matter. We will refer to these fluctuations as “typical” global signals, and will identify their source shortly. The fourth row of Figure 2 shows some less usual but still common and recognizable kinds of global signals: at bottom left a global signal prominent throughout the entire brain without evidence of motion, at bottom middle a motion artifact that affects the entire brain, and at bottom right some unusually protracted changes (of the usual kind shown in rows 2 and 3). The RP scan at bottom right shows a motion artifact (white band) affecting the entire brain. The quality of the data shown in this figure is representative of all datasets, and, if anything, represent the higher end of data quality (see Video 2).
Figure 2. Kinds of global signals.
The top row shows subjects with very little motion and unobtrusive global signal fluctuations. The second row shows a common signal pattern: prominent global signal fluctuations mostly in the absence of motion. The third row shows a common signal pattern: motion followed by a prominent signal fluctuation. The fourth row shows other signals or less common versions of signals. The signal in NIH38 (purple arrow) is likely due to a head coil malfunction. The signal in ABIDE32 (orange arrow) is due to a large motion. The very low frequency signal modulations in GSP32 are likely due to respiratory artifact. The white bands in RP33 are associated with large motions.
To quantify the qualitative observation that there is little variance in the global signal in scans with little motion (row 1), we correlated head motion with the standard deviation in the global signal, across subjects. This correlation is r = 0.35, 0.78, 0.51, 0.54, and 0.58 in the WU, NIH, ABIDE, GSP, and RP cohorts, all highly significant correlations (Figure S2). If timepoints containing motion (e.g., FD > 0.5 mm) are censored to remove most frank motion artifact from standard deviation calculations, the relationships are reduced but remain significant (r = 0.32, 0.45, 0.36, 0.51, 0.38), consistent with the observation that motion often precedes the “typical” fluctuations (e.g., row 3 of Figure 2).
The spatial distributions of the global signal reflect specific artifacts
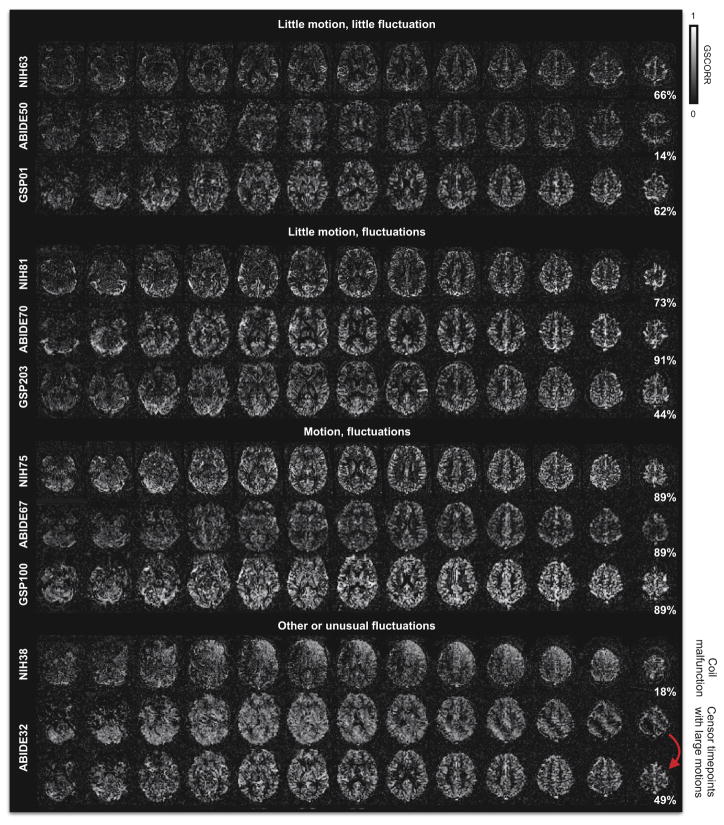
Figure 3 shows the spatial distribution of the global signal for most scans in Figure 2 (RP data are shown in Figure S4). The spatial distribution is illustrated as a heat map of the correlation of the global signal to each voxel’s signal. We will refer to this correlation as GSCORR. In the first 9 GSCORR maps (drawn from the first 3 rows of Figure 2), it is evident that the global signal is most strongly represented in the gray matter and that is only weakly present in the white matter and ventricles. These maps are typical of most subjects (GSCORR maps are presented individually for all subjects in Video 2). The 10th map (for NIH38) illustrates one of the unusual signals from Figure 2, and is an unusual spatial map, reflecting a malfunctioning coil in the scanning apparatus (a known historical problem with this particular site (Jo et al., 2010)). The 11th map (for ABIDE32) is shown first including all timepoints in the scan, resulting in an unusual diagonal banding pattern in the GSCORR map, and then excluding several timepoints with very large motions (FD > 1 mm) from signal calculations. Note that the banding pattern is removed and a more typical pattern emerges when those timepoints are censored.
Figure 3. Spatial distribution of global signals.
For several scans of Figure 2, maps are shown of each voxel’s correlation with the global signal (GSCORR). The first 3 subjects exhibit minimal fluctuations, the next 3 exhibit obvious fluctuations, the next 3 exhibit obvious fluctuations and head motion, and the final maps show examples of global signal distributions that reflect artifact.
Atypical GSCORR maps often reflect an identifiable artifact, and particular kinds of atypical maps are often associated with a particular scanning site (Figure S3 shows 12 examples). The WU site contains many GSCORR maps that are uniformly high across the brain (Figure S3), likely a result of a scanner-related artifact (note the many white bands in Figure 1 present uniformly throughout the brain; such bands are less frequent at other sites). The NIH site contains many instances of the asymmetric GSCORR map, reflecting coil artifacts (Figure S3). The diagonal banding artifact is present at all sites in subjects with large motions (Figure S3). Several other atypical and obviously artifactual GSCORR maps exist in the ABIDE dataset (Figure S3). In the RP dataset most scans conform to the “typical” pattern (Figure S4, see also Video 2), though there are also several atypical maps (Figure S4 and Video 2).
To quantify the qualitative observations of “typical” and “atypical” maps, an all-subject GSCORR map was created from the median value at each voxel across all 583 subjects of the WU, NIH, ABIDE, and GSP datasets (which are all in register (RP and HCP data are in different atlas spaces)). Each individual GSCORR map was then correlated with the all-subject map (Figure S5). The percentiles next to the maps in Figures 3, S3, and S4 indicate the rank of the map’s similarity, within the map’s site, to the all-subject map (100% is the most similar). The maps shown in Figures thus far were selected based on qualitative assessments of GSCORR maps (prior to quantification of the “typicality” of the maps), but the quantitative ranks of the chosen maps reflect these assessments: the presented “typical” maps have much higher rankings than the “atypical” maps (68 ± 27 vs 12 ± 14; p = 7.8e–8).
A suggested relation of the “typical” global signal to respiratory effects
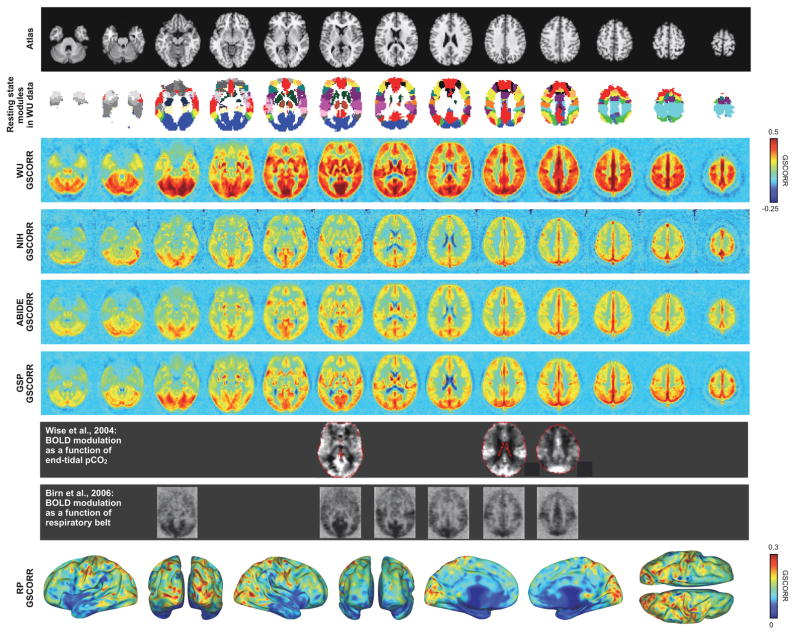
To capture the central tendency of GSCORR, median GSCORR maps for each cohort are presented in Figure 4. These maps reflect the “typical” maps seen in Figure 3. Though the maps differ in magnitude (see Discussion), the spatial pattern of GSCORR is similar across maps: all in-register maps correlate at over r = 0.7 (correlations with the NIH data are the lowest at r = 0.72, 0.79, and 0.72, whereas the other maps correlate at r = 0.82, 0.84, and 0.86). These correlations are all highly significant. GSCORR is near zero in deep white matter, is slightly negative in the ventricles, and is high in gray matter. GSCORR is relatively low in frontal cortex compared to posterior cortex. GSCORR is routinely high in much of the midline and in much of occipital cortex, and is also elevated in portions of the insula and near the Rolandic fissure.
Figure 4. Spatial distribution of global signals in each cohort.
At top, an atlas image and the WU modules reported in [Power, 2011]. At middle, the median GSCORR map across each cohort. In the gray rows, published images are paired to the approximately corresponding slices of GSCORR. At bottom, GSCORR in the RP datasets (6.5 hours of data, see Figure S4). The RP data, from a single individual, are easier to view on a surface because the central tendency of dozens of scans is less spatially blurred than in other cohorts and thus contains complex folding patterns. See Figure S6 for further illustrations of RP and WU data, and surface visualizations of modules in these datasets.
The GSCORR maps bear resemblance to published maps of BOLD signal dependence on pCO2. In Figure 4, the map of Wise et al, 2004 shows how end-tidal pCO2 measured by nasal cannula correlates with BOLD signal changes, and the map of Birn et al., 2006 shows how respiratory belt measures correlate with BOLD signal changes. This correspondence suggests that the “typical” GSCORR maps may in part reflect the influence of pCO2, and, correspondingly, that the “typical” global signals illustrated in Figure 2 may, in part, reflect effects of respiration.
It is useful to contextualize these maps in terms of resting state organization. The modular organizations of the WU and RP data have been extensively characterized in (Gordon et al., 2016; Laumann et al., 2015; Power et al., 2011; Power et al., 2013). Published modules for these datasets are shown next to GSCORR maps in Figures 4 and S6. GSCORR is notably high in visual and auditory and somato-motor modules, as are respiratory effects. Elevations in GSCORR are not limited to these locations, but are also high in lateral occipital, superior temporal, and superior parietal locations. It seems noteworthy that some of these locations are also proximal to major draining veins (e.g., the Labbe, Trolard, and superficial Sylvian veins). In some prior work, respiratory effects were interpreted as being prominent in default regions of the posterior midline. But In comparing the various maps of Figure 4 it appears that respiratory effects may not be particularly prominent in default regions, but rather in neighboring occipital and parietal regions. Relatedly, in the RP data, which has no cross-subject blurring, GSCORR is not nearly as high in the posterior default regions as it is in neighboring occipital and parietal regions (Figure S6).
Measuring respiration and its consequences
Most fMRI datasets are acquired without any accompanying physiological information, and this is true of the WU, ABIDE, GSP, and RP datasets. However, the NIH datasets have pulse oximeter and respiratory belt records from all scans, allowing us to document the extent to which respiration and other physiological changes can account for global signal changes. Respiratory belts (strapped about the abdomen) provide a record of expansion and contraction during breath cycles, and pulse oximeter traces (here, from a finger) track opacity of tissue to light. Because blood volume in the fingertip changes during the heartbeat and dynamically impedes light transmission, pulse oximeters can be used to derive multiple physiologic indices including heart rate and proxies of arterial pulse pressure. Pulse pressure (the difference between systolic and diastolic blood pressure) has only relatively recently been described as a measure that can be derived via peak amplitudes from pulse oximeter traces (Cannesson et al., 2005), and little is known about its correlates in fMRI signals.
Because pulse oximeter records are often contaminated by artifact, after algorithmic detection of peaks corresponding to heart beats, all traces were manually checked in their entirety to accept or omit the detected peaks (see Video 3a for fine-grained traces illustrating artifacts and quality control decisions in all subjects; see Video 3b for scan-summary versions of all traces in all subjects). The regularity of cardiac cycles makes distinction between signal and artifact relatively unambiguous in pulse oximeter traces. Instantaneous heart rate was calculated from peak-to-peak intervals, and peak amplitude was calculated for accepted peaks as the difference between peak signal and signal 0.14 seconds prior the peak (see Video 3a). Respiratory patterns may be regular and rhythmic, but they can also be quite irregular (see examples in Figures 5 and 6), making it much more difficult to distinguish artifacts from an irregular signal. We therefore did not intervene in respiratory belt traces to try to identify artifact.
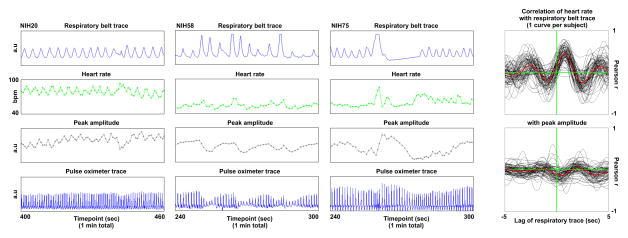
Figure 5. Relationships between respiratory cycles, estimated heart rate, and pulse pressures.
For 1 minute of data from 3 subjects, top and bottom panels show respiratory belt and pulse oximeter traces (both 50 Hz signals). Instantaneous heart rate is calculated from the peak-to-peak interval and plotted in green at the peak time, as is peak amplitude. Cyclical influences of respiration on heart rate and peak amplitude are evident. Such traces can be seen for all subjects in Videos 3a and 3b. The black traces at right show the correlation between respiratory belt traces and heart rate and peak amplitude, with -5 to 5 seconds of lag applied to the respiratory trace.
Our goal is to link global fMRI signal changes to changes in physiologic records, with a focus on respiration. However, prior to examining those links, it is prudent to examine the links between the physiologic records themselves, since respiratory cycles can modulate both heart rate (due to intrathoracic pressure changes) and pulse pressure (mainly in dehydrated individuals) (Cannesson et al., 2007; Cannesson et al., 2005).
Much of the variance in heart rate can be attributed to respiratory cycles (Figure 5, see also subjects 2, 4, 5, and others in Video 3a). The peak correlation between respiratory traces and heart rate traces is 0.47 ± 0.22 at a 0.8 second lag of respiratory traces, a highly significant correlation. Aside from cyclic heart rate changes due to respiration, another common phenomenon is intermittent and transient elevation in heart rate accompanying deep inspirations (Figure 5, see also subjects 2, 4, 9, and others in Video 3b). This phenomenon accords with prior reports that short-term heart rate variability correlates with respiration volume and lowered respiration rate (Chang et al., 2013). Lower-frequency modulations of heart rate are present but less frequent; these modulations are often, but not always, obviously linked to the envelope of the respiratory belt waveform (see also subjects 12, 14, 27, 44, and 64 in Video 3b). In short, much variance in heart rate is linked to respiration, and there is relatively little modulation of heart rate that appears to be unrelated (with certainty) to respiratory patterns.
Consistent with reports in the clinical literature (Cannesson et al., 2005), variance in peak amplitude can also be attributed to respiratory cycles, whether as a function of individual respiratory cycles or the envelope of the respiratory waveform (Figure 5, see subjects 17, 58, 65, 79, or 82 in Video 3b for examples of envelope relations). But in many instances low-frequency modulation of peak amplitude is not obviously related to the other traces (see subjects 41 or 45 in Video 3b for examples). If peak amplitude faithfully reflects pulse pressure, unique and perhaps useful information may be gained from variance in this measure, since pulse pressure reflects vascular compliance and stroke volume (among other factors). However, since peak amplitude is also sensitive to motion of the finger within the pulse oximeter (and changes in ambient light), non-cyclic shifts in amplitude may reflect uninteresting effects as well. It is likely that some of the low-frequency peak amplitude changes without obvious correlates in respiratory traces (or fMRI signals) are artifact due to inadequate securing or shielding of the pulse oximeter. Although peak amplitude estimates may not be of sufficient quality in these data to establish reliable relationships to fMRI timeseries, there are sufficient examples of links between peak amplitude, respiration, and fMRI signal changes (e.g., NIH82 in Figure 6) that we nonetheless present peak amplitude data to alert readers to the potential utility of this measure in future studies.
In sum, respiratory belt traces have correlates in heart rate and peak amplitudes derived from pulse oximeter traces, both at the level of individual respiratory cycles spanning 5–10 seconds and at longer intervals of dozens to hundreds of seconds (the respiratory trace envelope). Much of the variance in heart rate, and in peak amplitude (and thus inferred pulse pressure), appears to be a consequence of, or at least correlated with, respiration. Note that when sampled to fMRI-scale intervals (3.5 sec in this case) much of the cyclic variability in cardiac rate and pulse pressure due to individual respiratory cycles is averaged away (see Video 3b).
Respiratory effects are prominent in the global fMRI signal
Figure 6 presents the 3 physiologic traces along with fMRI signals in several NIH subjects. These subjects illustrate effects observed across all NIH subjects. Qualitatively, in subjects with little global signal change (upper left), there is little change in the physiological signals, and in subjects with much global signal change, there is much change in physiological signals. This observation can be quantified by correlating, across subjects, variance in estimated heart rate or variance in the volume of air respired per unit time (RVT), with the standard deviation of the global signal: these correlations are r = 0.55 (p = 5.7e–7) and r = 0.45 (p = 8.1e–5). These relationships are robust: if volumes with motion (FD > 0.5 mm) are excluded from standard deviation calculation in the global signal, the correlations become r = 0.38 (p = 0.001) and r = 0.61 (p = 9.3e–9), and if high-motion subjects (mean FD > 0.2 mm) are also excluded the correlations are r = 0.36 (p = 0.003) and r = 0.60 (p = 1.0e–7).
Deep inspiration precedes large drops in signal across the gray matter, a “typical” global signal modulation. To characterize this common modulation, isolated deep breaths without head motion or other artifact were identified in several subjects and the mean gray matter signal was calculated for 13 timepoints (42 seconds) after the breath. The average derived waveform, which may be considered an “impulse response” to a single large breath, matches well with waveforms from prior reports of such “respiratory response” functions induced by instructed deep breaths (Figures 6 and S7).
In the bottom left and middle panels of Figure 6, changes in the respiratory envelope correlate with large changes in global signals. The bottom middle panel is of particular interest for its 3 periods of (near) apnea. The lack of ventilation during apneic periods contributes to a gradual development of hypercapnia and hypoxia, both of which independently elevate cerebral blood flow. In studies that control blood gas pO2 and pCO2, and that induce conditions similar to those caused by 40–60 s of apnea, middle cerebral artery blood flow increases to approximately 110–160% over its baseline value (Poulin et al., 1996). Much as neural activity enriches local blood oxygenation (Fox and Raichle, 1986) by causing increased blood delivery (yielding increased blood oxygen level dependent (BOLD) signal), hypoxia and hypercapnia cause similar but global increases in blood flow that also enrich blood oxygenation (and increase the BOLD signal). Consistent with these observations, fMRI signal increases markedly during the apneic periods in the bottom middle panel.
To quantify the relative contributions of motion, respiratory patterns, heart rate, and peak amplitude to variability in the global signal, the standard deviation of the global signal was modeled (across subjects) by 7 variables: mean FD, and the mean and standard deviation of RVT (or RV), heart rate, and peak amplitude. If all timepoints are used to calculate global signal standard deviation, ~27% of the variance in global signal variability can be explained by these variables, and the most explanatory variables are motion estimates and the standard deviation and mean of heart rate (Table 3). If, however, high-motion volumes are excluded (which are a small fraction of volumes), the explanatory power of these variables drops and respiratory variables are most responsible for the explained variance. These statistics match the observation that motion, which causes major global signal artifacts, often accompanies deep inspirations that transiently elevate heart rate, and also match the observation that respiration-induced global signal changes last dozens of seconds after such events.
Many common fMRI processing methods do not remove global artifacts
The global fMRI signal contains a variety of undesirable signals, including hardware artifacts, motion artifacts, and variance linked to respiration. These artifacts will degrade modeling of task responses, and will systematically alter the similarity of signals in resting state fMRI analyses. Long-lasting global signal changes, such as those caused by respiratory-related effects, could be misinterpreted as dynamic neural effects if not identified as a nuisance signal.
To accurately assess fMRI data, these global artifacts must be recognized and removed (or somehow controlled for in statistical analyses). Many options exist for denoising fMRI data, and the techniques listed in Table 2 represent many of the common approaches in the field.
We examine nearly all of these methods, alone and in combination, for their ability to remove global fMRI signals. We do not examine local white matter regressors, but instead examine mean white matter signals and principal components of white matter signals, which should better target global signals. We do not examine removal of independent spatiotemporal components because independent component analysis (ICA) is optimized to identify spatially specific signals and is thus unlikely to identify global signals because they have little spatial specificity. This observation is demonstrated in Human Connectome Project data: global variance time-locked to motion and changes in respiration are prominent both before and after FIX-ICA (Figure S8).
Even when applied in combination, none of the examined methods removes global artifacts adequately (apart from techniques that explicitly remove the global signal). Video 4 shows many combinations of methods not shown in Figure 7. Motion regressors remove (some) motion artifact but will be insensitive to delayed effects of respiration or scanner artifact. White matter regressors, when adequately eroded, provide little removal of respiratory artifact though they can remove other artifacts (such as motion or scanner artifacts). Models of respiration-related signal only partially remove respiratory artifact. Note that most datasets cannot leverage physiological models of noise removal since there are no physiological records (i.e., these models are inapplicable to the WU, ABIDE, GSP, and RP datasets).
Readers may examine many combinations of these strategies in all subjects of the NIH cohort in Video 4, such as using principal components from white matter rather than mean white matter signals, or using a somewhat different model of respiratory signals (RV versus RVT). Although there are sometimes modest changes in the variance removed, the fundamental conclusion is unaltered: unwanted global variance time-locked to motion and respiration remains, and is often prominent, no matter the applied combination of these approaches. Video 4 also shows these same denoising combinations (without the physiological models) for the WU, ABIDE, GSP, and RP cohorts, yielding the same conclusion. Video 5 shows the effects of FIX ICA in the “40 Unrelated Subjects” HCP collection, yielding the same conclusion. No combination of examined methods removes global artifacts fully, except those combinations that include explicit removal of brain-wide fMRI signal (e.g., the global signal (GS) regressor at the bottom of Figure 7).
Please note that when we speak of failing to remove global artifacts we are not referring to failing to remove global signals in general but rather about failing to remove specific instances of global signals that are clearly attributable to motion and/or respiration. For example, the black bands in the first 2 columns of Figure 7 are time-locked to respiration and are not adequately removed by any denoising strategy except those that expressly remove all global signals.
Residual global signals
Any investigator who wishes to characterize differences in, or correlates of, global signals in fMRI data should first attempt to denoise the data. But how can one infer whether global signals after denoising represented “signal” or unremoved “noise”?
One strategy would be to examine the temporal and spatial characteristics of plots such as those shown in Figure 7, along with relevant indices of potential artifact, for evidence of artifact and artifact removal.
Another strategy would be to correlate the residual global variance with indices of unwanted variance. In the NIH cohort, global signal variance in raw data correlates with head motion, heart rate variability, and respiratory variability (Figure 8). These correlations remain after any denoising strategy attempted in this paper that does not explicitly remove global signals, no matter whether assessed in all data, only in low-motion subjects, or only in low-motion subjects and low-motion volumes (Figure 8). These relationships are significant when examined separately in “typically developing” (TD) and ASD subjects.
A variation of this procedure is to examine the performance of the model of Table 3 in the context of different denoising approaches (Figure 9). In this model, motion and respiratory variables account for much global signal variability both in raw data and after any examined denoising strategy that doesn’t include explicit removal of global signals. Figure 9 features the statistics of low-motion TD data (since these data likely most reflect other data in the literature), but the effects are similar in ASD subjects and when including all subjects regardless of motion.
Yet another strategy would be to examine the correlations of the residual GSCORR with indices of unwanted variance. For example, one can correlate RVT variability, across subjects, with GSCORR maps created after denoising. Figure 10 presents such a map for the NIH data after the denoising procedure shown in the 4th row of Figure 7, again focusing on low-motion TD subjects. GSCORR is dependent on RVT variability after denoising, and permutation tests indicate that this dependence is both pervasive and highly significant. Such maps are very similar under the other denoising strategies shown in Figure 9.
Discussion
Global fMRI signals play a central role in modern human neuroimaging, whether in the context of clinical findings, cognitive paradigms, or studies of resting state functional connectivity MRI. By studying many scans at the individual level, we were able to identify several common contributors to global fMRI signals, including hardware artifact, head motion, and respiratory-related variance. Most fMRI scans contain prominent global variance, much of which is caused by or correlated with these influences. This variance is often not removed from scans by common denoising techniques. Some implications of these findings are now discussed.
Uses of GSCORR
Maps of GSCORR at the individual level are useful for identifying abnormalities in scans. In this paper, abnormalities in these maps were produced by motion artifact (banding patterns), by scanner artifacts (large, pan-brain elevations in GSCORR), and by malfunctioning scanning equipment (the head coil artifact in NIH scans).
GSCORR maps at the individual and group levels suggest that the global signal is often largely accounted for by pCO2-related signal changes. Cerebral blood flow regulation (e.g., vasodilation in response to hypercapnia) plays an essential role in establishing this spatial pattern. An interesting clinical possibility is that GSCORR maps could reflect regional abnormalities in blood flow regulation (Lv et al., 2013). Recently, Grinband and colleagues have published evidence consistent with this proposition by showing that GSCORR maps can reflect non-enhancing spatial features of brain tumors (Chow et al., 2016).
Interpreting differences in GSCORR across scans, sites, and populations
Because various kinds of global signal have varying spatial properties and qualitatively different GSCORR maps, differences in the amounts and kinds of these signals will cause differences in individual and group-level GSCORR maps. This effect is seen across sites in this paper: the WU scans contained more scanner artifact, uniformly elevating GSCORR maps relative to the other sites (Figure 4). Another example is seen in the NIH scans, where scans with coil artifact contribute to asymmetric GSCORR especially in the frontal lobes (this is one reason why we used median values to create group-level maps, to diminish the effect of such atypical scans). Other expected effects are that group of scans with more motion will be expected to have noisier, less uniform GSCORR maps than a comparable group of scans without motion. Or, a group of scans containing many yawns would be expected to have a more prominent representation of respiratory-related signal (the “typical” GSCORR map) than a comparable set of scans with more regular breathing rates.
Differences in the spatial distribution of the global signal have been reported and examined in various contexts (e.g., ASD vs typical subjects (Gotts et al., 2012), or simulated differences in neural signals (Saad et al., 2012)). Empirical differences in GSCORR could be due to neural signals or the kinds of uninteresting but spatially widespread signals identified in this report. Given the present results, neural interpretations of GSCORR differences should be received with caution until artifactual explanations are convincingly excluded.
The utility of single-subject data in understanding global signals
An important distinction needs to be drawn between variance attributable to a process and variance that can be presently accounted for by models of a process. For example, in the single-subject grayscale plots, changes in respiratory traces are time-locked to a variety of prominent changes in fMRI signals, a relationship that is immediately visually obvious (Figure 6). This observation, joined with the already-existing mechanism for respiration to produce such signal changes, strongly suggests that large portions of the variance in global signals should be attributable to respiration.
However, it is equally visually apparent in the single-subject grayscale plots that respiration-linked variance is not well-removed by existing models of respiratory-related variance (Figure 7). In fact, in existing publications, the variance in timeseries accounted for by respiratory variables is often a only few percent of the total variance (e.g., (Bianciardi et al., 2009; Jo et al., 2010; Shmueli et al., 2007)). These low percentages surely reflect the inability of the respiratory variables and models to capture respiratory variance, rather than respiratory-related variance accounting for a small percentage of variance.
The performance of RVT and other models of respiratory variance is limited both by the quality of respiratory records (which are hard to assess for artifacts post-hoc) and by the validity of the models used to translate these records into anticipated fMRI signals. For example, models of fMRI signal based on “impulse responses” to single deep breaths perform relatively well at removing effects of single breaths, but their performance decreases when more rapid, or deeper breathing patterns are encountered (Birn et al., 2008). As our results illustrate, breathing patterns in the resting state often contain changes in the rate and depth of breathing, and even periods of apnea, complicating the modeling of these signals.
The important point is that the variance explained by a model may not reflect the variance that is intended to be explained by the model. In this paper, single-subject plots routinely provided critical evidence that the respiratory models were not capturing variance that they ought to capture. These limitations accompany not only the use of RVT and RV in timeseries denoising procedures (Figure 7), but also the use of such variables in the models of Figures 8–10 and Table 3. For example, the correlations (or betas) of RVT with global signal variance are already high and significant, but the correlations between an ideal respiratory variable (and/or model) and global signal variance may be even higher.
On removing the influence of global artifact in fMRI scans
Global artifacts – signal changes that are time-locked to respiration and/or motion and that appear throughout the brain – remain in fMRI data following application of many combinations of common denoising techniques. These global artifacts, both before and after such denoising, are often prominent in the timeseries, and will cause at least two major effects. First, global fluctuations will tend to elevate covariance between all signals. Second, global fluctuations may easily be mistaken for neural dynamics. To accurately assess neural effects in fMRI data, such global fluctuations of no interest must be recognized and either be removed from timeseries or statistically accounted for in later analyses. We offer comments on both of these topics.
First, global artifacts arise from multiple sources, and effective removal of such signals will probably require multiple techniques. For example, deep white matter signals are useful for removing scanner artifact, but contain little representation of respiratory-related variance. In contrast, RVT captures some respiratory variance, but not scanner artifact.
Second, it is easy to inadvertently remove the global fMRI signal by inadequately eroding nuisance masks (Jo et al., 2010). Superficial white matter voxels, constituting approximately half of the white matter, have signals that are highly similar to the global signal (Pearson r of ~0.9). Although this effect would seem attributable to partial volume effects in older datasets, it is similar quantitatively in the relatively high-resolution RP and HCP datasets (2.4 and 2.0 mm isotropic voxels at acquisition). Another inadvertent way to remove the global signal is to remove signals that are outside the brain but inside venous sinuses (e.g. note the high GSCORR in the sagittal sinus in Figure 4).
Third, although much attention has recently been given to identifying and removing effects of head motion in fMRI scans, less attention has been devoted to effects of respiration. The prominence of respiratory effects in global signals, and the difficulty of removing these signals, suggests that more work is needed in this area. Relatedly, the pulse pressure traces in the NIH cohort contain notable correlates of respiratory envelopes in many subjects, and these traces may in principle have interesting or useful correlates in fMRI signals. Unfortunately in our data the pulse oximeter traces also contain numerous artifactual changes probably due to shifts of the device on the finger, rendering the peak amplitudes unreliable in many respects. Future studies that use better-secured oximeters may find these signals helpful for explaining fMRI variance.
Fourth, present implementations of ICA, such as FIX-ICA in the Human Connectome Project, tend not to remove respiratory-related global signals (Figure S8), likely because these signals do not meet the criteria for spatial specificity built in to these procedures. Spatial priors based on respiratory-related signal changes may aid the identification of such signals by matrix factorization techniques.
With regard to accounting for shared but unwanted covariance in a statistical manner, some groups have suggested covarying mean GSCORR values from datasets across scans (Saad et al., 2013), or studying only partial covariance between timeseries (e.g., using LASSO techniques). These techniques may help control for artifactual influences, but they also constrain studies in important ways. For example, GSCORR is in many cases likely to covary with effects of interest. Or, if one views signal similarity of “clean” fMRI signals throughout the brain as representing a nested hierarchy of widely distributed processes, then isolating unique variance between signals via partial correlations, the technical challenges aside, is not necessarily a preferred or natural framework for studying brain organization (in other words, some investigators may wish to retain the full nested hierarchy in their representations of the data).
Implications: “neural” global fMRI signals in humans
Work in non-human primates and rodents indicates that “global” neural signals may exist.
In alert macaques, Scholvink and colleagues (Scholvinck et al., 2010) demonstrated an approximately r = 0.25 correlation between band-limited high-gamma (40–80 Hz) power from electrophysiological recordings at multiple sites and fMRI signals 7–8 seconds later at multiple cortical sites. Intriguingly, this study also found that fMRI signals 7–8 seconds preceding neural activity predicted the band-limited power subsequently recorded. Although it is relatively straightforward to postulate mechanisms whereby neural signals elicit global changes in blood flow that manifest as global fMRI signals, it is less clear how or why global fMRI signal changes might systematically precede global neural activity. Two non-mutually-exclusive possibilities for the latter observation are 1) periodic processes, or 2) that pO2 and/or pCO2 (since they govern cerebral blood flow and thus global T2* signal) may systematically affect subsequent neural activity throughout the brain.
Optical imaging in lightly anesthetized mice also suggests the existence of “global” neural signals. Matsui and colleagues (Matsui et al., 2016) noted waves of activity that propagated across the entire mouse cortex, often in a front-to-back pattern, lasting ~5 seconds with a multi-second lag between neural activity and parallel hemodynamic responses. These waves occurred at approximately 0.04 Hz, a frequency within the most-studied pass-band of resting state fMRI signals in humans.
The results of the present study should not be construed as arguments against “neural” global signals in humans or as arguments that the global fMRI signal is entirely artifact. However, from a practical standpoint, the current study does indicate that in humans 1) most of the most prominent global fMRI signal changes are artifacts that can be attributed to head motion or respiration and 2) methods to selectively identify and successfully remove those artifacts are lacking at this time.
Implications: respiratory effects and sensorimotor patterns in fMRI data
“Functional” organization accompanies some artifacts: respiration-related variance affects all gray matter but is especially prominent in occipital cortex, peri-Rolandic cortex, and in the superior posterior insula, all “sensory” or “motor” brain regions. Results related to such spatial patterns may fit respiratory-related explanations just as well as “functional” explanations.
Sensorimotor-patterned correlates of cognitive, behavioral, and clinical phenomena should be interpreted in light of this observation. For example, many fMRI studies of descent into sleep detect spatial patterns of increased variance that are notably similar to the GSCORR and pCO2 patterns shown in Figure 4 – changes most prominent in visual and somatomotor regions (see, e.g., Figure 3 of (Horovitz et al., 2008), Figure 2 of (Tagliazucchi et al., 2013), or Figure 5 of (Tagliazucchi and Laufs, 2014)). Such findings may reflect slowed, deepened breathing as subjects fall asleep. Similar sensorimotor patterns have discriminated schizophrenic patients from control subjects (Kaufmann et al., 2015).
Another potential manifestation of respiratory effects is the following. With respect to the global fMRI signal itself, Wong and colleagues have reported that caffeination and increased vigilance in subjects are correlated with decreased variance in the global fMRI signal, using denoising similar to the physiological modeling procedure highlighted in Figures 7–10 (Wong et al., 2012, 2013). Given that these denoising procedures only partially remove respiratory variance, which contributes strongly to global signal variance, and that caffeinated and/or vigilant subjects are a priori less likely to fall asleep in the scanner, it seems possible that respiratory effects contribute to global signal properties that are contingent on caffeination and vigilance.
Implications: clinical reports of altered global signals in fMRI data
Several studies have reported changed variance or changed spatial distributions of the global signal in clinical conditions. For example, Hahamy and colleagues (Hahamy et al., 2014) removed motion, mean CSF, and mean WM signals from fMRI data (similar to the 3rd row of Figure 7). This study found large differences in voxel degree between schizophrenic patients and typical subjects, and that these differences were abolished when the global signal was regressed from the data. Following similar denoising procedures, Yang and colleagues (Yang et al., 2014) found increased global variance in schizophrenic patients relative to controls. And using denoising similar to the 4th row of Figure 7, Gotts and colleagues reported different distributions in GSCORR between autistic and control subjects (Figure S2 of (Gotts et al., 2012)).
Based on the present findings, it does not appear that any of these studies can exclude unremoved artifact as a source of differences in global fMRI signal properties, whether in terms of global signal variance or GSCORR distribution. Given the present difficulties of separating global fMRI artifacts from global “neural” signals, it seems that non-fMRI data (e.g., intracranial recordings) may be critical for substantiating global neural phenomena in humans.
Implications: global signal regression
As mentioned in the Introduction, the topic of global signal regression (GSR) is one of the most controversial issues in neuroimaging. Readers should note that this paper does not adopt any position on this topic, which is extraneous to the present results and conclusions. Nevertheless the present results should inform these debates and we offer several comments below.
Much of the field is reluctant to explicitly remove global fMRI signals from datasets, due in part to claims about clinical or cognitive correlates of the global signal (Gotts et al., 2012; Yang et al., 2014), and due in part to arguments made against removing a mean signal that represents mixtures of signals (Gotts et al., 2013; Murphy et al., 2009; Saad et al., 2012). As discussed above, we find the evidence for global “neural” signals in clinical conditions to be inconclusive, since it cannot be determined whether those studies were reporting neural global effects or the kinds of artifacts shown in this report. The conceptual arguments against global signal regression take two forms: first, that negative correlations are “induced” by GSR (Murphy et al., 2009; Weissenbacher et al., 2009), and second, that differences in covariance structure (i.e., the composition of the global signal, GSCORR) will confound comparisons of covariance if mean signals are regressed (Gotts et al., 2013; Saad et al., 2012). Both of these arguments are valid but depend critically on the dimensionality of the signals entering the global fMRI signal, and on the presence or absence of truly global signals. This article contributes to these debates by showing 1) that truly global signals exist, 2) that truly global signals are prominent features of most scans, and 3) that truly global signals frequently arise from respiration and less frequently from head motion and/or hardware malfunction. The calculated global signal is thus not just a mixture of focal signals but is instead a mixture of focal signals plus a truly global signal with multiple sources, and the truly global component is not negligible under most real-world circumstances. Further, the artifactual aspects of the truly global component are not removed by most denoising strategies.
The theoretical concerns about GSR are valid, but must be weighted alongside concerns about unremoved artifact in datasets. For example, it has been found that lagged high gamma power in intracranial electrophysiological recordings in humans better approximates fMRI signal patterns after removing the global fMRI signal (Keller et al., 2013). In the study by Keller and colleagues, the fMRI analysis approaches being contrasted were regression of 6 motion parameters plus mean white matter and mean ventricle signals from eroded brain masks, without or with an additional global signal regressor. This strategy is approximately that shown in the 3rd row of Figure 7 compared to the bottom row of Figure 7. While the additional consequences of explicitly removing the global signal remain unclear, it seems reasonable to think that the enhanced correspondence of electrophysiological signals and fMRI signals upon removal of the global signal was mainly due to the removal of widespread and uninteresting variance of the kinds described in this report.
Conclusion
Functional connectivity MRI studies can be deceptively simple: there is no experimental paradigm and covariance between timeseries is easily computed. But many unwanted factors can influence observed covariance and it takes considerable effort to identify and control for these confounds. It should be evident that changes in global signal variance and localization are expected simply due to artifact or other factors of no interest. Many standard denoising strategies do not guarantee the removal of significant artifact from fMRI datasets at the time of analysis. Denoising strategies that isolate and remove artifactual global variance while preserving potential “neural” global variance are needed.
Supplementary Material
Highlights.
Demonstrates brain-wide (global) fMRI signals, individually, in 1000+ scans from 8 sites
Global signals often reflect artifact caused by head motion, respiration, or hardware problems
Most existing fMRI denoising methods do not adequately remove global artifacts
Global artifacts mimic dynamic neural activity and modulate signal correlations
Studies reporting global fMRI signal effects must carefully account for artifact
Acknowledgments
We thank Kevin Tran, David Godlove, and the Biowulf staff at the NIH for their computing support. We thank Steve Petersen and Brad Schlaggar for contributing the WU data for analysis. We thank Alan Anticevic and our Reviewers for suggestions that improved the manuscript. This work was supported by the Intramural Research Program, National Institute of Mental Health/NIH (ZIAMH002920; NCT01031407).
Footnotes
The authors have no conflicts of interest to declare with regard to this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguirre GK, Zarahn E, D'Esposito M. Empirical analyses of BOLD fMRI statistics. II. Spatially smoothed data collected under null-hypothesis and experimental conditions. Neuroimage. 1997;5:199–212. doi: 10.1006/nimg.1997.0264. [DOI] [PubMed] [Google Scholar]
- Aguirre GK, Zarahn E, D'Esposito M. The inferential impact of global signal covariates in functional neuroimaging analyses. Neuroimage. 1998;8:302–306. doi: 10.1006/nimg.1998.0367. [DOI] [PubMed] [Google Scholar]
- Bianciardi M, Fukunaga M, van Gelderen P, Horovitz SG, de Zwart JA, Shmueli K, Duyn JH. Sources of functional magnetic resonance imaging signal fluctuations in the human brain at rest: a 7 T study. Magn Reson Imaging. 2009;27:1019–1029. doi: 10.1016/j.mri.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birn RM, Diamond JB, Smith MA, Bandettini PA. Separating respiratory-variation-related fluctuations from neuronal-activity-related fluctuations in fMRI. Neuroimage. 2006;31:1536–1548. doi: 10.1016/j.neuroimage.2006.02.048. [DOI] [PubMed] [Google Scholar]
- Birn RM, Smith MA, Jones TB, Bandettini PA. The respiration response function: the temporal dynamics of fMRI signal fluctuations related to changes in respiration. Neuroimage. 2008;40:644–654. doi: 10.1016/j.neuroimage.2007.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannesson M, Attof Y, Rosamel P, Desebbe O, Joseph P, Metton O, Bastien O, Lehot JJ. Respiratory variations in pulse oximetry plethysmographic waveform amplitude to predict fluid responsiveness in the operating room. Anesthesiology. 2007;106:1105–1111. doi: 10.1097/01.anes.0000267593.72744.20. [DOI] [PubMed] [Google Scholar]
- Cannesson M, Besnard C, Durand PG, Bohe J, Jacques D. Relation between respiratory variations in pulse oximetry plethysmographic waveform amplitude and arterial pulse pressure in ventilated patients. Crit Care. 2005;9:R562–568. doi: 10.1186/cc3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell F, Bellec P, Shmuel A. Global and system-specific resting-state fMRI fluctuations are uncorrelated: principal component analysis reveals anti-correlated networks. Brain Connect. 2011;1:496–510. doi: 10.1089/brain.2011.0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Cunningham JP, Glover GH. Influence of heart rate on the BOLD signal: the cardiac response function. Neuroimage. 2009;44:857–869. doi: 10.1016/j.neuroimage.2008.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Glover GH. Relationship between respiration, end-tidal CO2, and BOLD signals in resting-state fMRI. Neuroimage. 2009;47:1381–1393. doi: 10.1016/j.neuroimage.2009.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Metzger CD, Glover GH, Duyn JH, Heinze HJ, Walter M. Association between heart rate variability and fluctuations in resting-state functional connectivity. Neuroimage. 2013;68:93–104. doi: 10.1016/j.neuroimage.2012.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow DS, Horenstein CI, Canoll P, Lignelli A, Hillman EM, Filippi CG, Grinband J. Glioblastoma Induces Vascular Dysregulation in Nonenhancing Peritumoral Regions in Humans. AJR Am J Roentgenol. 2016;206:1073–1081. doi: 10.2214/AJR.15.14529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci U S A. 2005;102:9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Zhang D, Snyder AZ, Raichle ME. The global signal and observed anticorrelated resting state brain networks. J Neurophysiol. 2009;101:3270–3283. doi: 10.1152/jn.90777.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox PT, Raichle ME. Stimulus rate dependence of regional cerebral blood flow in human striate cortex, demonstrated by positron emission tomography. J Neurophysiol. 1984;51:1109–1120. doi: 10.1152/jn.1984.51.5.1109. [DOI] [PubMed] [Google Scholar]
- Fox PT, Raichle ME. Focal physiological uncoupling of cerebral blood flow and oxidative metabolism during somatosensory stimulation in human subjects. Proc Natl Acad Sci U S A. 1986;83:1140–1144. doi: 10.1073/pnas.83.4.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, Xu J, Jbabdi S, Webster M, Polimeni JR, Van Essen DC, Jenkinson M Consortium WUMH. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage. 2013;80:105–124. doi: 10.1016/j.neuroimage.2013.04.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover GH, Li TQ, Ress D. Image-based method for retrospective correction of physiological motion effects in fMRI: RETROICOR. Magn Reson Med. 2000;44:162–167. doi: 10.1002/1522-2594(200007)44:1<162::aid-mrm23>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Gordon EM, Laumann TO, Adeyemo B, Huckins JF, Kelley WM, Petersen SE. Generation and Evaluation of a Cortical Area Parcellation from Resting-State Correlations. Cereb Cortex. 2016;26:288–303. doi: 10.1093/cercor/bhu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotts SJ, Saad ZS, Jo HJ, Wallace GL, Cox RW, Martin A. The perils of global signal regression for group comparisons: a case study of Autism Spectrum Disorders. Front Hum Neurosci. 2013;7:356. doi: 10.3389/fnhum.2013.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotts SJ, Simmons WK, Milbury LA, Wallace GL, Cox RW, Martin A. Fractionation of social brain circuits in autism spectrum disorders. Brain. 2012;135:2711–2725. doi: 10.1093/brain/aws160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Krasnow B, Reiss AL, Menon V. Functional connectivity in the resting brain: a network analysis of the default mode hypothesis. Proc Natl Acad Sci U S A. 2003;100:253–258. doi: 10.1073/pnas.0135058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahamy A, Calhoun V, Pearlson G, Harel M, Stern N, Attar F, Malach R, Salomon R. Save the global: global signal connectivity as a tool for studying clinical populations with functional magnetic resonance imaging. Brain Connect. 2014;4:395–403. doi: 10.1089/brain.2014.0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes AJ, Hollinshead MO, O'Keefe TM, Petrov VI, Fariello GR, Wald LL, Fischl B, Rosen BR, Mair RW, Roffman JL, Smoller JW, Buckner RL. Brain Genomics Superstruct Project initial data release with structural, functional, and behavioral measures. Sci Data. 2015;2:150031. doi: 10.1038/sdata.2015.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horovitz SG, Fukunaga M, de Zwart JA, van Gelderen P, Fulton SC, Balkin TJ, Duyn JH. Low frequency BOLD fluctuations during resting wakefulness and light sleep: a simultaneous EEG–fMRI study. Hum Brain Mapp. 2008;29:671–682. doi: 10.1002/hbm.20428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz B, Duara R, Rapoport SI. Intercorrelations of glucose metabolic rates between brain regions: application to healthy males in a state of reduced sensory input. J Cereb Blood Flow Metab. 1984;4:484–499. doi: 10.1038/jcbfm.1984.73. [DOI] [PubMed] [Google Scholar]
- Jo HJ, Saad ZS, Simmons WK, Milbury LA, Cox RW. Mapping sources of correlation in resting state FMRI, with artifact detection and removal. Neuroimage. 2010;52:571–582. doi: 10.1016/j.neuroimage.2010.04.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastrup A, Li TQ, Takahashi A, Glover GH, Moseley ME. Functional magnetic resonance imaging of regional cerebral blood oxygenation changes during breath holding. Stroke. 1998;29:2641–2645. doi: 10.1161/01.str.29.12.2641. [DOI] [PubMed] [Google Scholar]
- Kaufmann T, Skatun KC, Alnaes D, Doan NT, Duff EP, Tonnesen S, Roussos E, Ueland T, Aminoff SR, Lagerberg TV, Agartz I, Melle IS, Smith SM, Andreassen OA, Westlye LT. Disintegration of Sensorimotor Brain Networks in Schizophrenia. Schizophr Bull. 2015;41:1326–1335. doi: 10.1093/schbul/sbv060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller CJ, Bickel S, Honey CJ, Groppe DM, Entz L, Craddock RC, Lado FA, Kelly C, Milham M, Mehta AD. Neurophysiological investigation of spontaneous correlated and anticorrelated fluctuations of the BOLD signal. J Neurosci. 2013;33:6333–6342. doi: 10.1523/JNEUROSCI.4837-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumann TO, Gordon EM, Adeyemo B, Snyder AZ, Joo SJ, Chen MY, Gilmore AW, McDermott KB, Nelson SM, Dosenbach NU, Schlaggar BL, Mumford JA, Poldrack RA, Petersen SE. Functional System and Areal Organization of a Highly Sampled Individual Human Brain. Neuron. 2015;87:657–670. doi: 10.1016/j.neuron.2015.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Y, Margulies DS, Cameron Craddock R, Long X, Winter B, Gierhake D, Endres M, Villringer K, Fiebach J, Villringer A. Identifying the perfusion deficit in acute stroke with resting-state functional magnetic resonance imaging. Ann Neurol. 2013;73:136–140. doi: 10.1002/ana.23763. [DOI] [PubMed] [Google Scholar]
- Matsui T, Murakami T, Ohki K. Transient neuronal coactivations embedded in globally propagating waves underlie resting-state functional connectivity. Proc Natl Acad Sci U S A. 2016 doi: 10.1073/pnas.1521299113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K, Birn RM, Bandettini PA. Resting-state fMRI confounds and cleanup. Neuroimage. 2013;80:349–359. doi: 10.1016/j.neuroimage.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K, Birn RM, Handwerker DA, Jones TB, Bandettini PA. The impact of global signal regression on resting state correlations: are anti-correlated networks introduced? Neuroimage. 2009;44:893–905. doi: 10.1016/j.neuroimage.2008.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldrack RA, Laumann TO, Koyejo O, Gregory B, Hover A, Chen MY, Gorgolewski KJ, Luci J, Joo SJ, Boyd RL, Hunicke-Smith S, Simpson ZB, Caven T, Sochat V, Shine JM, Gordon E, Snyder AZ, Adeyemo B, Petersen SE, Glahn DC, Reese Mckay D, Curran JE, Goring HH, Carless MA, Blangero J, Dougherty R, Leemans A, Handwerker DA, Frick L, Marcotte EM, Mumford JA. Long-term neural and physiological phenotyping of a single human. Nat Commun. 2015;6:8885. doi: 10.1038/ncomms9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin MJ, Liang PJ, Robbins PA. Dynamics of the cerebral blood flow response to step changes in end-tidal PCO2 and PO2 in humans. J Appl Physiol (1985) 1996;81:1084–1095. doi: 10.1152/jappl.1996.81.3.1084. [DOI] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 2012;59:2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, Vogel AC, Laumann TO, Miezin FM, Schlaggar BL, Petersen SE. Functional network organization of the human brain. Neuron. 2011;72:665–678. doi: 10.1016/j.neuron.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Mitra A, Laumann TO, Snyder AZ, Schlaggar BL, Petersen SE. Methods to detect, characterize, and remove motion artifact in resting state fMRI. Neuroimage. 2014a;84:320–341. doi: 10.1016/j.neuroimage.2013.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Schlaggar BL, Lessov-Schlaggar CN, Petersen SE. Evidence for hubs in human functional brain networks. Neuron. 2013;79:798–813. doi: 10.1016/j.neuron.2013.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Schlaggar BL, Petersen SE. Studying brain organization via spontaneous fMRI signal. Neuron. 2014b;84:681–696. doi: 10.1016/j.neuron.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JDPM, Kundu P, Bandettini P, Martin A. Head motion is underestimated in fMRI scans following common preprocessing steps submittedA. [Google Scholar]
- Saad ZS, Gotts SJ, Murphy K, Chen G, Jo HJ, Martin A, Cox RW. Trouble at rest: how correlation patterns and group differences become distorted after global signal regression. Brain Connect. 2012;2:25–32. doi: 10.1089/brain.2012.0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad ZS, Reynolds RC, Jo HJ, Gotts SJ, Chen G, Martin A, Cox RW. Correcting brain-wide correlation differences in resting-state FMRI. Brain Connect. 2013;3:339–352. doi: 10.1089/brain.2013.0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimi-Khorshidi G, Douaud G, Beckmann CF, Glasser MF, Griffanti L, Smith SM. Automatic denoising of functional MRI data: combining independent component analysis and hierarchical fusion of classifiers. Neuroimage. 2014;90:449–468. doi: 10.1016/j.neuroimage.2013.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite TD, Elliott MA, Gerraty RT, Ruparel K, Loughead J, Calkins ME, Eickhoff SB, Hakonarson H, Gur RC, Gur RE, Wolf DH. An improved framework for confound regression and filtering for control of motion artifact in the preprocessing of resting-state functional connectivity data. Neuroimage. 2013;64:240–256. doi: 10.1016/j.neuroimage.2012.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholvinck ML, Maier A, Ye FQ, Duyn JH, Leopold DA. Neural basis of global resting-state fMRI activity. Proc Natl Acad Sci U S A. 2010;107:10238–10243. doi: 10.1073/pnas.0913110107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmueli K, van Gelderen P, de Zwart JA, Horovitz SG, Fukunaga M, Jansma JM, Duyn JH. Low-frequency fluctuations in the cardiac rate as a source of variance in the resting-state fMRI BOLD signal. Neuroimage. 2007;38:306–320. doi: 10.1016/j.neuroimage.2007.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman AE, Hu X, Jerosch-Herold M. Functional MRI of brain during breath holding at 4 T. Magn Reson Imaging. 1995;13:893–897. doi: 10.1016/0730-725x(95)00037-h. [DOI] [PubMed] [Google Scholar]
- Tagliazucchi E, Laufs H. Decoding wakefulness levels from typical fMRI resting-state data reveals reliable drifts between wakefulness and sleep. Neuron. 2014;82:695–708. doi: 10.1016/j.neuron.2014.03.020. [DOI] [PubMed] [Google Scholar]
- Tagliazucchi E, von Wegner F, Morzelewski A, Brodbeck V, Jahnke K, Laufs H. Breakdown of long-range temporal dependence in default mode and attention networks during deep sleep. Proc Natl Acad Sci U S A. 2013;110:15419–15424. doi: 10.1073/pnas.1312848110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenbacher A, Kasess C, Gerstl F, Lanzenberger R, Moser E, Windischberger C. Correlations and anticorrelations in resting-state functional connectivity MRI: a quantitative comparison of preprocessing strategies. Neuroimage. 2009;47:1408–1416. doi: 10.1016/j.neuroimage.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Wise RG, Ide K, Poulin MJ, Tracey I. Resting fluctuations in arterial carbon dioxide induce significant low frequency variations in BOLD signal. Neuroimage. 2004;21:1652–1664. doi: 10.1016/j.neuroimage.2003.11.025. [DOI] [PubMed] [Google Scholar]
- Wong CW, DeYoung PN, Liu TT. Differences in the resting-state fMRI global signal amplitude between the eyes open and eyes closed states are related to changes in EEG vigilance. Neuroimage. 2016;124:24–31. doi: 10.1016/j.neuroimage.2015.08.053. [DOI] [PubMed] [Google Scholar]
- Wong CW, Olafsson V, Tal O, Liu TT. Anti-correlated networks, global signal regression, and the effects of caffeine in resting-state functional MRI. Neuroimage. 2012;63:356–364. doi: 10.1016/j.neuroimage.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CW, Olafsson V, Tal O, Liu TT. The amplitude of the resting-state fMRI global signal is related to EEG vigilance measures. Neuroimage. 2013;83:983–990. doi: 10.1016/j.neuroimage.2013.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang GJ, Murray JD, Repovs G, Cole MW, Savic A, Glasser MF, Pittenger C, Krystal JH, Wang XJ, Pearlson GD, Glahn DC, Anticevic A. Altered global brain signal in schizophrenia. Proc Natl Acad Sci U S A. 2014;111:7438–7443. doi: 10.1073/pnas.1405289111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarahn E, Aguirre GK, D'Esposito M. Empirical analyses of BOLD fMRI statistics. I. Spatially unsmoothed data collected under null-hypothesis conditions. Neuroimage. 1997;5:179–197. doi: 10.1006/nimg.1997.0263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.