Abstract
IL-10 is an immunoregulatory cytokine that has broad effects across the immune system. In T helper cell subsets, Th2 cells produce considerable amounts of IL-10. The transcription factors that regulate IL-10 production in Th2 cells are still incompletely described. In this report, we demonstrate that the ETS family transcription factor Etv5 regulates IL-10 production in Th2 cells. T cell-specific Etv5-deficient and littermate control mice demonstrated that IL-10 production and gene expression were significantly decreased in the absence of Etv5. In an Aspergillus fumigatus extract-induced inflammation model, IL-10 producing CD4+ T cells in BAL and lung were significantly decreased in mice that lacked Etv5 in T cells, compared to control mice. We showed that Etv5 directly binds to the Il10 locus CNS3 site and that it activates gene expression in a luciferase reporter assay and following retroviral transduction. Etv5-deficiency did not affect the expression of other transcription factors known to be important for expression of IL-10, including Jun family members, GATA3, E4BP4, and IRF4. Yet, in the absence of Etv5, binding of these transcription factors to the Il10 locus was dramatically reduced. Ectopic Etv5 expression in Th2 cells that lack Etv5 restored IL-10 production and the binding of IL-10-inducing transcription factors including E4BP4, IRF4, and GATA3. Taken together, we conclude that Etv5 plays a crucial role in regulating IL-10 production in Th2 cells by facilitating the binding of IL-10-inducing transcription factors at the Il10 locus.
Introduction
CD4+T cells play an essential role in adaptive immune system (1). Naïve CD4+T cells differentiate into T helper cells with help of antigen presenting cells (APCs) to regulate host defense and inflammatory responses (2). APCs provide three signals inducing lineage specific transcription factors that are required for proper differentiation of naïve CD4+T cells; the first is a T cell receptor (TCR) signal initiated by antigen peptide-MHC complexes on APCs, the second, a co-stimulatory signal through CD28, and the third signal is provided by cytokines that activate the signal transducer and activator of transcription (STAT) signaling pathway (2). CD4+T helper cell subsets express transcription factors and have specific effector functions mediated by specific cytokines. T helper 2 (Th2) cells express GATA3 and produce IL-4, IL-5 and IL-13 that stimulate other immune cells and tissue structural cells (3, 4). Th2 cells also express IL-10 (5).
IL-10, an anti-inflammatory cytokine, suppresses inflammatory responses at mucosal surfaces (6). IL-10 was initially identified as a Th2 cell specific cytokine, but subsequent reports showed that most T helper subsets can produce IL-10 (7). IL-10 prevents inflammatory and autoimmune pathologies, but increased IL-10 production prevents clearance of pathogens resulting in chronic infection. Therefore, IL-10 production needs to be strictly regulated. In Th2 cells, IL-10 expression is induced by IL-4/STAT6 signaling, and GATA3 is required for active chromatin in the Il10 locus (5). Several other transcription factors promote Il10 expression in Th2 cells including E4BP4 (NF-IL3), IRF4, and Jun family members (8-10).
Etv5, or Ets variant gene 5, belongs to the ETS transcription factor family. Like other ETS proteins, Etv5 recognizes and binds GGAA/T, the Ets specific consensus sequence, through the Ets domain (11). Many studies showed that Etv5 is involved in cell development (12-14), but the roles of Etv5 in immune system are still not fully understood. Initial reports suggested that Etv5 expression is increased by IL-12/STAT4 signaling, not by IFNγ-STAT1 signaling during Th1 cell development (15). However, we reported that in experiments with Etv5 conditional mutant mice found no defect in Th1 cytokine production, but a distinct decrease in STAT3-dependent IL-17 production (16). We further reported that Etv5 promotes IL-9 production in Th9 cells (17). Increased Etv5 expression was observed in IL-10 secreting Th1 cells, although no function was tested (18). In this report, we showed that Etv5 increases IL-10 production in vitro and in vivo by binding the Il10 locus and recruiting IL-10 inducing transcription factors.
Methods
Mice
Etv5fl/fl CD4-Cre+ mice (13, 16) were previously described, and Cre-negative littermates were used as control mice. Mice were maintained under specific pathogen-free conditions. All experiments were performed with the approval of the Indiana University Institutional Animal Care and use Committee.
In vitro T cell differentiation
Naïve CD4+CD62L+ T cells were positively selected from the enriched CD4+ T cells from spleen and lymph nodes using MACS beads and columns (Miltenyi Biotec). Naive CD4+CD62L+ T cells were activated with plate-bound anti-CD3 (2 ug/ml 145-2C11 ; Bio X Cell) and soluble anti-CD28 (2 ug/ml ; BD Pharmingen) to generate Th0 or with additional cytokines (all from PeproTech) and antibodies (Bio X Cell) to generate Th2 (20 ng/ml IL-4; and 10 ug/ml anti-IFNγ, XMG), and Th1 (20 ng/ml IL-12; 10U hIL-2; and 10 ug/ml anti-IL-4, 11B11) culture conditions. Cells were expanded after 3 days with same concentration of the original cytokines in fresh medium and in the absence of anti-CD3. Cells were harvested on day 5 for analysis.
Aspergillus fumigatus extract-induced allergic airway inflammation
Mice were challenged intranasally with Aspergillus fumigatus extract (Greer Laboratories) every other day for 21 days. 100 μg of powdered A. fumigatus extract was diluted with 50 μl PBS and administered intranasally. Mice were sacrificed 1 day after the final intranasal challenge. Bronchoalvelor lavage (BAL) cells were collected with 1ml PBS. BAL cells and the single cell suspension from lungs were stimulated with PMA and Ionomycin for 6 hours to assess cytokine analysis using intracellular staining. Cytokine production of BAL cells was measured by ELISA.
Analysis of gene expression, ELISA and flow cytometry
Quantitative Reverse Transcriptase (qRT)-PCR and ELISA were performed as previously described (19). Gene expression was normalized to housekeeping gene expression (β2-microglobulin). The relative gene expression was calculated by the change-in-threshold (−ΔΔCT) method. For cytokine staining, CD4+ T cells were stimulated with PMA and ionomycin for 2 hours followed by monensin for a total 6 hours, fixed, permeabilized with 0.2% saponin, and stained for IFNγ (PerCP Cy5.5), IL-4 (Alexa Fluor 647), IL-10 (FITC or PE) (BD Pharmingen). Flow cytometric analysis was performed on cells gated for lymphocyte size and granularity and on CD4 expression.
Chromatin Immunoprecipitation (ChIP)
ChIP assay was performed as described (20). In brief, activated Th cells were cross-linked for 10 min with 1% formaldehyde and lysed by sonication. After pre-clearing with salmon sperm DNA, bovine serum albumin, and Protein Agarose bead slurry (50%), cell extracts were incubated with rabbit polyclonal Etv5 (H-100), p300 (N-15), E4BP4 (V-19 or C-18), IRF4 (M-17) (all from Santa Cruz), GATA3 (D13C9) (Cell signaling), H3K14ac, H3K4me3, or normal rabbit IgG (all from Millipore) overnight at 4°C. The immunocomplexes were precipitated with Protein Agarose beads at 4°C for 1 hour, washed, eluted and cross-links reversed at 65°C overnight. DNA was purified, resuspended in H2O and analyzed by qRT-PCR. The percentage input was calculated by subtracting the amount of immunoprecipitated DNA from the IgG control from the amount of immunoprecipated DNA from the specific antibody and normalized against the amount of input DNA. ChIP primers were used as described (21)
Luciferase reporter assay
293 T cells were grown in DMEM 1640 with 10%FBS and co-transfected with 1ug of the Il10 promoter, CNS3 or mutated CNS3 luciferase reporter vectors and control or Etv5 expressing vector through lipofectamin 2000 (Life technologies). After 24 hours, cell extracts were obtained through passive lysis buffer and luciferase activities were measured using dual luciferase reporter assay system (Promega) according to the manufacturer's instructions. The Etv5 binding site in the CNS3 region was mutated (from “a” to “t”; −6282,−6283,−6289,−6290) using QuickChange II Site-Directed Mutagenesis Kit (Agilent Technologies).
Retroviral transduction
Retroviral transduction was performed as described (16). In brief, activated CD4+ T cells were infected on day1 with retrovirus containing control or expressing interest gene by using centrifugation at 2300 rpm at 32 °C for 90 minutes in the presence of 8 ug/ml polybrene (Sigma-Aldrich). After spin infection, viral supernatant was replaced with the new Th1 or Th2 conditioned media. Cells were expanded on day 3 and analyzed on day 5. Transduction efficiency was generally >80%. Transduced cells were gated on EGFP+ and hCD4+ populations for analysis of cytokine staining.
Statistical Analysis
A two-tailed Student's t-test was used to generate p-values for all pairwise comparisons. p ≤ 0.05 was considered statistically significant.
Results
Etv5 promotes lL-10 production in Th1 and Th2 cells
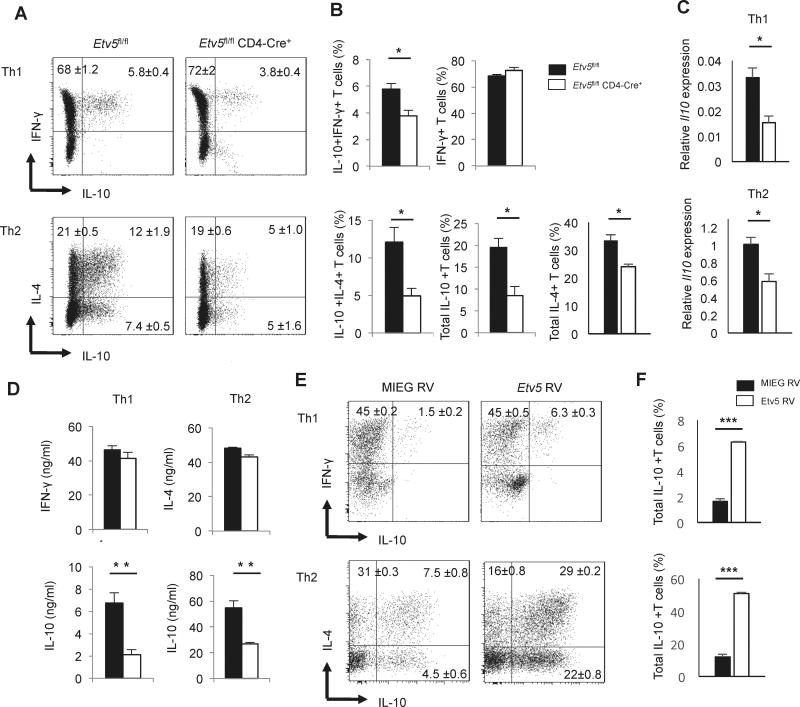
Our previous studies have demonstrated that Etv5 promotes the production of IL-17A, IL-17F, and IL-21 in Th17 cells, while it had a modest repressive effect on Th2 cytokines (16). Interestingly, a recent study showed that Etv5 is highly expressed in IL-10-producing Th1 cells (18). To determine if Etv5 had a role in regulating IL-10 production, we isolated naïve CD4+ T cells from Etv5fl/fl CD4-Cre mice and Cre-negative littermate control and cultured them under Th1 and Th2 cell polarizing conditions before measuring IFN-γ, IL-4 and IL-10 production using intracellular cytokine staining. Although total IFN-γ-producing Th1 cells or IL-4-producing Th2 cell populations were either unaffected or modestly affected by Etv5 deficiency, we observed a significant decrease of IL-10-producing Th1 and Th2 cells in the absence of Etv5 (Figure 1A-B). Consistent with these results, Il10 gene expression and secreted IL-10 concentrations were significantly decreased in Etv5 deficient Th1 and Th2 cells (Figure 1C-1D). To further test the function of Etv5, we introduced Etv5 into Th1 and Th2 cells using retroviral transduction. Ectopic expression of Etv5 strongly enhanced IL-10 production in both Th1 and Th2 cells (Figure 1E). However, the total number of Th1 or Th2 cells were not affected (data not shown). Taken together, these data suggested that Etv5 positively regulates IL-10 production in both Th1 and Th2 cells without affecting lineage specific cytokine production. The slight decrease or similarity in IL-4 production between wild type and Etv5-deficient Th2 cells (Fig. 1B and D) was surprising because our previous report demonstrated increased IL-4 in the absence of Etv5 (16). We found that the difference in result is due to changes in the Th2 cell culture condition (Supplementary Figure 1). Thus, increased co-stimulation and IL-4 signaling alter the effects of Etv5-deficiency on IL-4, but not on IL-10.
Figure 1.
Etv5 promotes IL-10 but does not affect IL-4 and IFN-γ production. A-D, Naïve CD4+ T cells from control and Etv5fl/fl CD4 Cre+ mice were cultured under Th1 or Th2 cell conditions for 5 days. A, On day 5, Th1 and Th2 cells were stimulated with PMA/Ionomycin for 6 hours to measure cytokine production using intracellular staining. B, Average percentage of IL-10-producing Th1 and Th2 cells. C-D, Th1 and Th2 cells were restimulated with anti-CD3 for 6 hours or overnight to measure Il10 gene expression by using qRT-PCR (C) or to assess cytokine production by means of ELISA (D). E, Naïve CD4+ T cells from wild type mice were differentiated under Th1 and Th2 cell conditions. Twenty four hours after initiation of culture, cells were transduced with control or Etv5 expressing retrovirus. On day 5, cells were stimulated with PMA/Ionomycin for 6 hours to measure cytokine production using intracellular staining. F, Average percentage of IL-10 producing Th1 and Th2 cells after retroviral transduction. Data are mean ± SEM of 4 independent experiments (n=4/experiment). *p < 0.05, **p <0.005,***p <0.005
Etv5 regulates IL-10 production in vivo
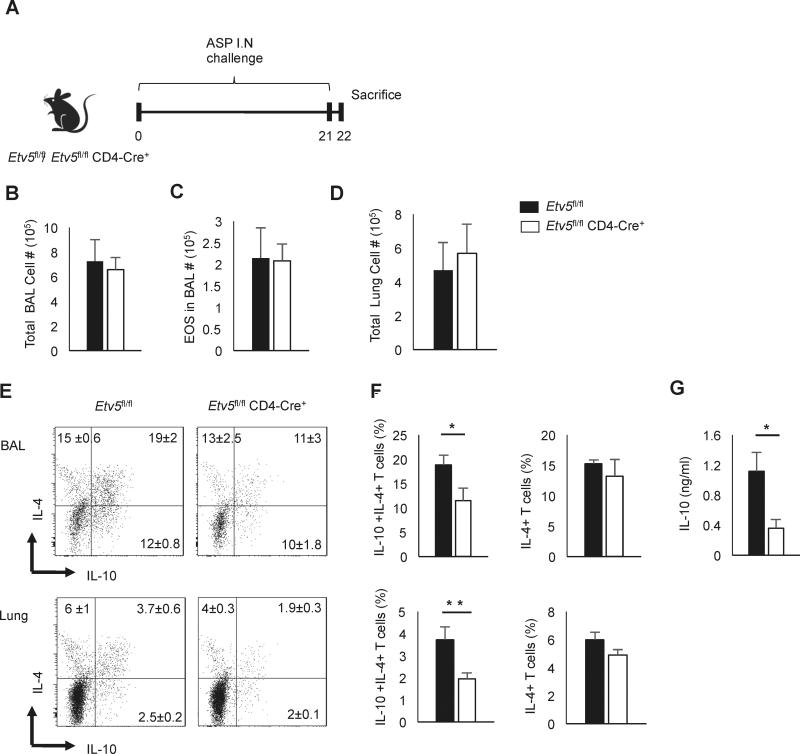
A. fumigatus challenged Il10−/− mice showed exaggerated airway inflammation with significant increased production of Th2 cytokines (22). To define the effect of Etv5 on Th2 cells in vivo, we sensitized mice with Aspergillus fumigatus extract (A. fumigatus) every other day for 21 days to induce type 2 inflammation (Fig. 2A). One day following the final challenge, we collected and counted bronchoalveolar lavage (BAL) and lung cells. There was no difference in numbers of total cells in the BAL, eosinophils in the BAL, or total cells in the lung between control mice and mice that had Etv5-deficient T cells (Figure 2B-2D). However, using intracellular cytokine staining, the IL-10-producing CD4 T cell population was significantly decreased in mice that had Etv5-deficient T cells in both BAL and lung, compared to control mice (Figure 2E-2F). This result was consistent with decreased IL-10 present in the BAL fluid in mice with Etv5-deficient T cells (Fig. 2G). However, Etv5-deficiency did not affect production of IL-4 in vivo (Fig. 2E-F). These data demonstrated that Etv5 plays a crucial role in regulating IL-10 production in vivo.
Figure 2.
Etv5 deficient Th2 cells produce less IL-10 in Aspergillus fumigatus extract-induced airway inflammation. A, Control and Etv5fl/fl CD4 Cre+ mice were intranasally challenged with A.fumigatus extract every other day for 21days. B-D, Total cell count (B), eosinophils (C) in the BAL and total cell count in the lung (D) of A.fumigatus extract-challenged control and Etv5fl/fl CD4 Cre+ mice. E-F, Lung and BAL cells were stimulated with PMA/Ionomycin for 6 hours to measure cytokine production using intracellular staining. Representative dot plots (E) and average percentage of IL-10 producing Th2 cells in BAL and lung (F) are indicated. Cells for flow cytometric analysis were gated on lymphocyte size and granularity, and expression of CD4 and TCRβ. G, IL-10 production of BAL cells was measured using ELISA. Data are mean ± SEM of 2 independent experiments (n=6/experiment). *p < 0.05, **p <0.005
Etv5 directly binds Il10 locus and promotes IL-10 production
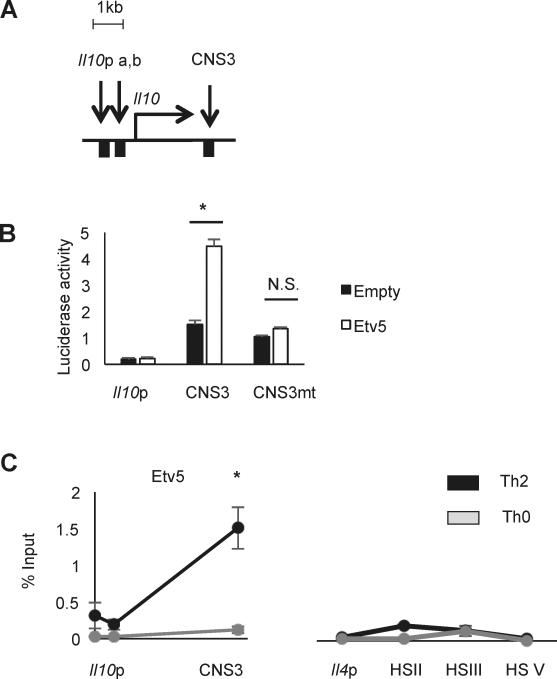
The Il10 gene locus consists of a promoter and several CNS regions (7). Previous studies reported that several IL-10 inducing transcription factors bind to Il10 locus and promote Il10 expression (5, 8-10). Specifically, CNS3 is a primary target region of IL-10-inducing transcription factors and critical for IL-10 regulation (10)(Fig. 3A). To test the ability of Etv5 to directly activate gene expression from Il10 regulatory elements, we co-transfected 293 T cells with Etv5 expressing vectors and reporter vectors containing the Il10 promoter or CNS3 region. Etv5 significantly increased CNS3 reporter activity but did not activate the Il10 promoter reporter (Fig. 3B). To define the Etv5 binding element, we mutated and ETS binding site in CNS3 region from “GGAAGTGGGAA” to “GGTTGTGGGTT”. The mutation of the Etv5 binding site in the CNS3 region eliminated the ability of Etv5 to transactivate the promoter (Figure 3B). We then tested whether Etv5 directly binds to the Il10 locus at the promoter and CNS3 region using ChIP assay. Etv5 strongly bound to CNS3 regions in Th2 cells but not in Th0 cells that have minimal expression of IL-10 (Figure 3C and data not shown). As a control, Etv5 does not bind to several regulatory elements in the Il4 locus, consistent with a lack of altered IL-4 production in Th2 cells lacking Etv5 in these cultures (Figure 3C). These data suggest that Etv5 directly binds to Il10 CNS3 region and promotes gene expression from binding this element.
Figure 3.
Etv5 binds the Il10 locus in Th2 cells. A, Schematic of Il10 locus. B, luciferase activity of 293T cells transfected with Etv5 expressing vector or control vector with Il10 locus reporter vectors. C, ChIP analysis of Etv5 binding on Il10 and Il4 locus in Th2 or Th0 cells. Data are mean ± SEM of 2 independent experiments (n=4/experiment). *p < 0.05, N.S., Not significant
Etv5 recruits IL-10-inducing transcription factors onto the Il10 locus
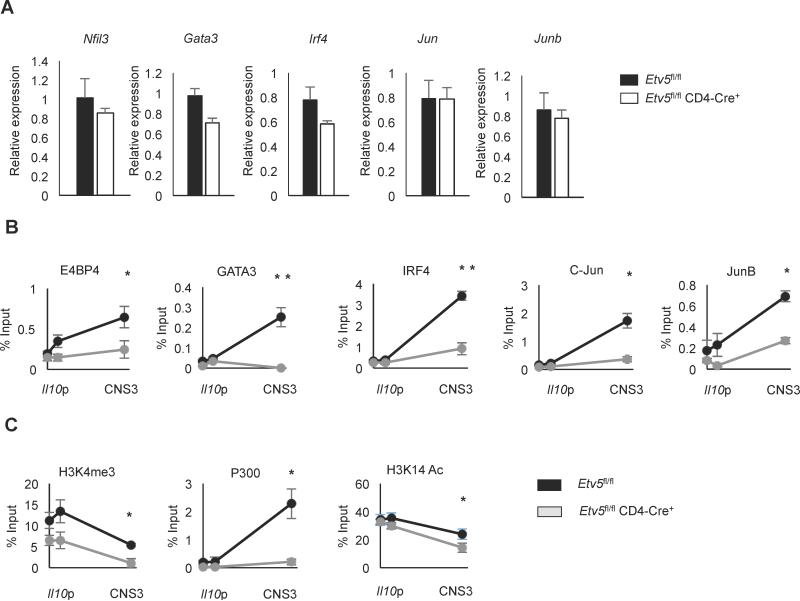
Although the reporter assay would suggest that Etv5 can directly activate the Il10 gene, it is also possible that Etv5 regulates the expression of other transcription factors that are required for IL-10 expression. To determine whether Etv5 regulates expression of those transcription factors, we measured gene expression of the transcription factors in control or Etv5-deficient Th2 cells. Expression of Nfil3, a global IL-10 regulator that encodes E4BP4, was not affected by Etv5 deficiency (Figure 4A). Similarly, gene expression of other transcription factors was not affected by Etv5 deficiency (Figure. 4A).
Figure 4.
Etv5 enhances the binding of IL-10-inducing transcription factors. A, Expression of IL-10-inducing transcription factors in control and Etv5fl/fl CD4 Cre+ Th2 cells were measured using qRT-PCR. Th2 cells were restimulated with anti-CD3 for 6 hours followed by cDNA synthesis. B-C, ChIP analysis of IL-10 inducing transcription factors (B), or histone modifications and p300 (C) at the Il10 locus in WT or Etv5-deficient Th2 cells. Data are mean ± SEM of 2 independent experiments (n=4/experiment). *p < 0.05, **p <0.005
Despite normal expression of other IL-10-inducing transcription factors, it was still possible that they were not able to bind Il10 in the absence of Etv5. To test whether factors that bound the Il10 locus were still binding the Il10 locus in the absence of Etv5, we used ChIP assays to assess binding in control and Etv5-deficient Th2 cells. In the CNS3 region, recruitment of all transcription factors was significantly decreased in Etv5 deficient Th2 cells (Figure 4B). Etv5-deficient Th2 cells also showed decreased Histone 3 lysine 4 tri-methylation, an active promoter marker (Figure 4C). P300, a histone acetyltransferase, strongly bound to the CNS3 region, but binding was greatly decreased in Etv5-deficient Th2 cells (Figure 4C). Histone 3 lysine 14 acetylation, linked to p300 activity, was also significantly decreased in Etv5 deficient Th2 cells (Figure 4C). Thus, Etv5 is required for the maximal binding of IL-10-inducing transcription factors to the CNS3 region in Th2 cells.
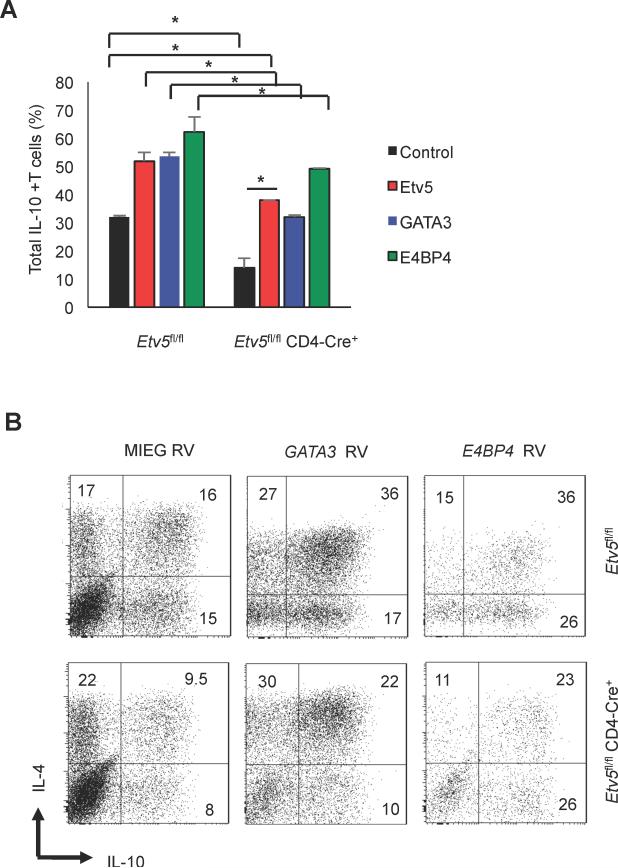
Etv5 functionally cooperates with GATA3 and E4BP4
The diminished binding of multiple transcription factors to the Il10 locus suggests that transduction of those factors into Etv5-deficient T cells would also have diminished effects. To test this directly, we ectopically introduced Etv5, E4BP4, or GATA3 into control or Etv5-deficient Th2 cells using retroviral transduction and measured IL-10 production using intracellular staining. In control Th2 cells, ectopic expression of each factor significantly promoted IL-10 production (Figure 5A-B). In Etv5-deficient Th2 cells, ectopic Etv5 expression rescued IL-10 production to control levels (Figure 5A-B). Ectopic expression of GATA3 and E4BP4 increased IL-10-producing cells, but effects were still decreased in absence of Etv5, compared to control cells. These data further support an important role for Etv5 in the appropriate function of GATA3 and E4BP4 in Il10 regulation.
Figure 5.
Etv5 deficiency decreases the ectopic expression effects of IL-10 inducing transcription factors. A-C, WT and Etv5-deficient naïve CD4+ T cells were cultured under Th2 cell condition for 5 days. After one day of culture, cells were transduced with control or transcription factor-expressing retrovirus. On day 5, cells were stimulated with PMA/Ionomycin for 6 hours to assess IL-10 production using intracellular staining analysis. (A) Average percentage of IL-10 producing cells of GFP- or hCD4-positive (transduced) and (B) representative flow cytometry dot plots of data in (A). Data are mean ± SEM of 2 independent experiments (n=4/experiment). *p < 0.05
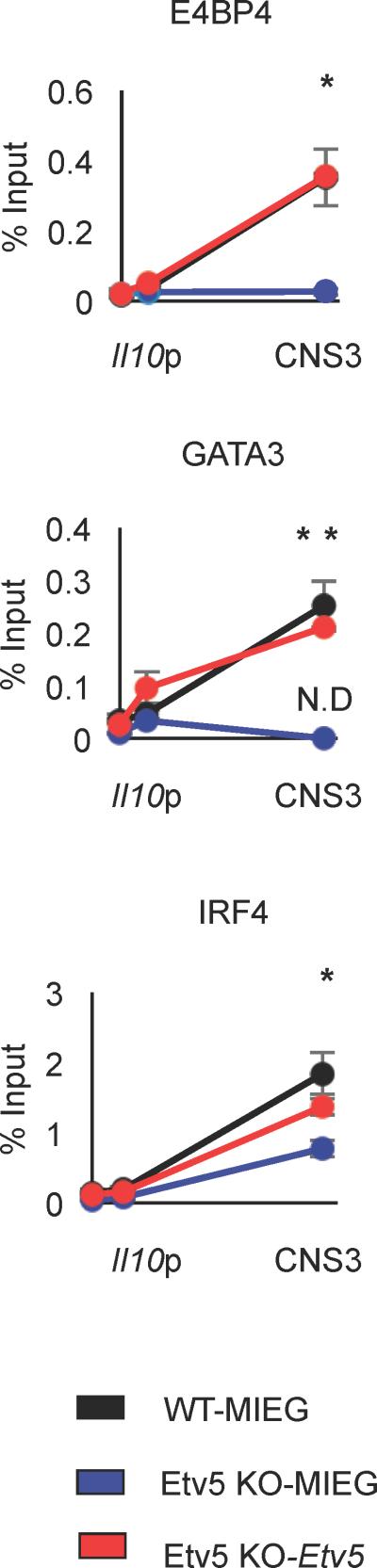
Ectopic Etv5 expression restores its function in Etv5 deficient Th2 cells
Our data suggests that IL-10-inducing transcription factors require Etv5 for optimal binding and IL-10 induction (Figure 4 and 5). To test whether re-introduction of Etv5 rescues binding of other transcription factors to the Il10 locus, we introduced Etv5 into Etv5 deficient Th2 cells using retroviral transduction followed by ChIP assay. Etv5 deficient Th2 cells infected by empty virus showed decreased binding of IRF4, GATA3 and E4BP4 at the CNS3 region, compared to control, Th2 cells infected by the same virus (Figure 6), consistent with data in Figure 4C using non-transduced cells. Importantly, ectopic Etv5 expression in Etv5-deficient Th2 cells restored binding of all three transcription factors at the CNS3 region indicating that Etv5 plays a pivotal role in regulating the binding of transcription factors to the Il10 locus.
Figure 6.
Ectopically introduced Etv5 in Etv5-deficient Th2 cells increases transcription factor binding to the Il10 locus. After one day of culture, Etv5-deficient Th2 cells were transduced with control or Etv5 expressing retrovirus. On day 5 of culture, ChIP assay was performed to examine transcription factor binding at the Il10 locus. Data are mean ± SEM of 2 independent experiments (n=4/experiment). *p < 0.05, **p <0.005
Discussion
IL-10 is a critical regulatory cytokine. In the absence of IL-10, mice develop spontaneous autoimmune inflammation (23). Thus, transcriptional control of the Il10 gene is essential in maintaining disease-free immune homeostasis (7). In this report we identify Etv5 as a regulator of IL-10 in Th1 and Th2 cells, in vivo and in vitro. Etv5 binds directly to the Il10 locus and facilitates the binding of a number of other transcription factors that regulate IL-10 production.
The regulation of IL-10 production may be distinct among subsets of Th cells. Previous reports identified several transcription factors that are involved in Il10 regulation in Th2 cells. GATA3 remodels the Il10 locus independently of IL-4 production in Th2 cells (5). JunB and c-Jun proteins bind to the CNS3 region in the Il10 locus and induce IL-10 production in Th2 cells (10). In Th2 cells, IRF4 directly binds Il10 locus and promotes IL-10 production (8). E4BP4 is reported as a universal IL-10 regulator in all Th cell subsets, and E4BP4 deficiency cause impairment of IL-10 production, but does not affect IL-13 production in Th2 cells(9). In this report, we add Etv5 to the list of factors that promote IL-10 production, and show that it works in concert with other factors by facilitating their binding to the Il10 locus. It is not clear if Etv5 functions by directly interacting with other transcription factors bound to the locus, or if it remodels chromatin allowing other transcription factors to bind more efficiently. Nevertheless, Etv5 is required for the other factors to optimally induce IL-10 production.
Etv5 adds to a subset of ETS family transcription factors that regulate Il10. Ets-1 represses IL-10 expression in Th1 cell by recruiting histone deacetyltransferase (HDAC) to the Il10 promoter and intronic regions (21). Polymorphisms in the human Il10 gene increase binding of the ETS factor Elk1 (24). PU.1 also negatively regulates IL-10 in Th2 and Th9 cells (8, 25). At least some of the mechanism of PU.1 function in Th2 cells is by interfering with IRF4 binding to Il10 locus regulatory elements (8). As each of these factors seems to function through different mechanisms, and having different effects on IL-10, it seems unlikely that there would competition among them.
Etv5 does not work in a subset specific manner, regulating IL-10 production in both Th1 and Th2 cells. Interestingly, IL-27 could induce IL-10 in both wild type and Etv5-deficient T cells, suggesting that it functions in an Etv5-independent manner (data not shown). Etv5 did not dramatically affect the lineage identifying cytokines for these Th subsets. This is in contrast to Etv5 function in Th17 where it is required for optimal expression of the lineage associated cytokines (16). We detected little IL-10 production from Th17, Th9 and Treg cells in our culture conditions (data not shown), making it impossible to determine if Etv5 is contributing to Il10 expression in these subsets. Additionally, even though we could detect IL-10 producing Th1 cells in our Th1 cell conditions, the number of these cells was quite low and not enough to study in more detail. This function might best be addressed in vivo in disease models where there are more IL-10-producing Th1, Th9 and Th17 cells.
It is still not entirely clear how Etv5 acts at the Il10 locus. It is more likely that it functions by modifying chromatin, and opening the locus to allow other factors to bind, rather than physically interacting with each of the factors examined. However, this remains to be formally tested. Relevant to this point, transduction of Etv5-deficient Th2 cells with GATA3 or E4BP4 was able to increase IL-10 production. There could be several reasons for this, but it is likely that Etv5 is acting to increase efficiency of binding of the other factors, and that they do have partial function in the absence of Etv5. This is supported by the fact that IL-10 production and binding of some transcription factors is only partially diminished in the absence of Etv5. When cells are transduced and transcription factors are ectopically expressed, it shifts the equilibrium further towards binding by increasing the concentration of transcription factor within the cell. Importantly, even though GATA3 and E4BP4 can increase IL-10 in the absence of Etv5, production is not increased to the percentage of cells observed when wild type cells are transduced. Thus, while increasing transcription factor concentration can increase IL-10 production, the effects of Etv5-deficiency are still observed.
Mice that have Etv5-deficient T cells clearly do not behave as IL-10-deficient mice. The Etv5fl/fl CD4 Cre+ mice do not develop spontaneous autoimmune inflammation. Moreover, A. fumigatus challenged Il10−/− mice showed exaggerated airway inflammation and excessive Th2 cytokines in BAL (22). In our studies, A. fumigatus extract-challenged Etv5fl/fl CD4 Cre+ mice demonstrated no difference in the overall inflammation, but had a reduced number of IL-10-producing T cells in the BAL and lung. This is consistent with the partial role for Etv5 in IL-10 production that we observe in vitro and in vivo. The approximate 50% decrease in IL-10 production is not sufficient to result in the more dramatic effects of IL-10-deficiency in vivo. Additionally, in non-T cells, where Etv5 is not deleted, IL-10 production is likely normal, further attenuating the effects of the T cell-specific deficiency of Etv5 in vivo.
Our studies identify Etv5 as a promoter of IL-10 regulation in Th subsets. It binds the Il10 CNS3 element, and facilitates the binding of several other IL-10-inducing factors that together control IL-10 production. This latest component of the Il10 enhancer complex provides new insight into regulation of immunoregulatory cytokine production in Th subsets and could provide another potential target for modulating IL-10 production in vivo.
Supplementary Material
Acknowledgments
This work was supported by Public Health Service grants AI045515 and AI057459 to MHK. MMH was supported by PHS T32 AI060519. Support provided by the HB Wells Center was in part from the Riley Children's Foundation.
Abbreviations
- CNS
conserved non-coding sequence
- ETV
ETS variant
- HS
hypersensitivity site
- IRF
interferon regulatory factor
References
- 1.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 2.Chaudhry A, Rudensky AY. Control of inflammation by integration of environmental cues by regulatory T cells. J Clin Invest. 2013;123:939–944. doi: 10.1172/JCI57175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 4.Zhu J, Min B, Hu-Li J, Watson CJ, Grinberg A, Wang Q, Killeen N, Urban JF, Jr., Guo L, Paul WE. Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nat Immunol. 2004;5:1157–1165. doi: 10.1038/ni1128. [DOI] [PubMed] [Google Scholar]
- 5.Shoemaker J, Saraiva M, O'Garra A. GATA-3 directly remodels the IL-10 locus independently of IL-4 in CD4+ T cells. J Immunol. 2006;176:3470–3479. doi: 10.4049/jimmunol.176.6.3470. [DOI] [PubMed] [Google Scholar]
- 6.Groux H, Cottrez F. The complex role of interleukin-10 in autoimmunity. J Autoimmun. 2003;20:281–285. doi: 10.1016/s0896-8411(03)00044-1. [DOI] [PubMed] [Google Scholar]
- 7.Saraiva M, O'Garra A. The regulation of IL-10 production by immune cells. Nat Rev Immunol. 2010;10:170–181. doi: 10.1038/nri2711. [DOI] [PubMed] [Google Scholar]
- 8.Ahyi AN, Chang HC, Dent AL, Nutt SL, Kaplan MH. IFN regulatory factor 4 regulates the expression of a subset of Th2 cytokines. J Immunol. 2009;183:1598–1606. doi: 10.4049/jimmunol.0803302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Motomura Y, Kitamura H, Hijikata A, Matsunaga Y, Matsumoto K, Inoue H, Atarashi K, Hori S, Watarai H, Zhu J, Taniguchi M, Kubo M. The transcription factor E4BP4 regulates the production of IL-10 and IL-13 in CD4+ T cells. Nat Immunol. 2011;12:450–459. doi: 10.1038/ni.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang ZY, Sato H, Kusam S, Sehra S, Toney LM, Dent AL. Regulation of IL-10 gene expression in Th2 cells by Jun proteins. J Immunol. 2005;174:2098–2105. doi: 10.4049/jimmunol.174.4.2098. [DOI] [PubMed] [Google Scholar]
- 11.Oh S, Shin S, Janknecht R. ETV1, 4 and 5: an oncogenic subfamily of ETS transcription factors. Biochim Biophys Acta. 2012;1826:1–12. doi: 10.1016/j.bbcan.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z, Verheyden JM, Hassell JA, Sun X. FGF-regulated Etv genes are essential for repressing Shh expression in mouse limb buds. Dev Cell. 2009;16:607–613. doi: 10.1016/j.devcel.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z, Sui P, Dong A, Hassell J, Cserjesi P, Chen YT, Behringer RR, Sun X. Preaxial polydactyly: interactions among ETV, TWIST1 and HAND2 control anterior-posterior patterning of the limb. Development. 2010;137:3417–3426. doi: 10.1242/dev.051789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu X, Goodyear SM, Tobias JW, Avarbock MR, Brinster RL. Spermatogonial stem cell self-renewal requires ETV5-mediated downstream activation of Brachyury in mice. Biol Reprod. 2011;85:1114–1123. doi: 10.1095/biolreprod.111.091793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ouyang W, Jacobson NG, Bhattacharya D, Gorham JD, Fenoglio D, Sha WC, Murphy TL, Murphy KM. The Ets transcription factor ERM is Th1-specific and induced by IL-12 through a Stat4-dependent pathway. Proc Natl Acad Sci U S A. 1999;96:3888–3893. doi: 10.1073/pnas.96.7.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pham D, Sehra S, Sun X, Kaplan MH. The transcription factor Etv5 controls TH17 cell development and allergic airway inflammation. J Allergy Clin Immunol. 2014;134:204–214. doi: 10.1016/j.jaci.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koh B, Hufford MM, Pham D, Olson MR, Wu T, Jabeen R, Sun X, Kaplan MH. The ETS Family Transcription Factors Etv5 and PU.1 Function in Parallel To Promote Th9 Cell Development. J Immunol. 2016;197:2465–2472. doi: 10.4049/jimmunol.1502383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neumann C, Heinrich F, Neumann K, Junghans V, Mashreghi MF, Ahlers J, Janke M, Rudolph C, Mockel-Tenbrinck N, Kuhl AA, Heimesaat MM, Esser C, Im SH, Radbruch A, Rutz S, Scheffold A. Role of Blimp-1 in programing Th effector cells into IL-10 producers. J Exp Med. 2014;211:1807–1819. doi: 10.1084/jem.20131548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stritesky GL, Muthukrishnan R, Sehra S, Goswami R, Pham D, Travers J, Nguyen ET, Levy DE, Kaplan MH. The transcription factor STAT3 is required for T helper 2 cell development. Immunity. 2011;34:39–49. doi: 10.1016/j.immuni.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu Q, Thieu VT, Kaplan MH. Stat4 limits DNA methyltransferase recruitment and DNA methylation of the IL-18Ralpha gene during Th1 differentiation. EMBO J. 2007;26:2052–2060. doi: 10.1038/sj.emboj.7601653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee CG, Kwon HK, Sahoo A, Hwang W, So JS, Hwang JS, Chae CS, Kim GC, Kim JE, So HS, Hwang ES, Grenningloh R, Ho IC, Im SH. Interaction of Ets-1 with HDAC1 represses IL-10 expression in Th1 cells. J Immunol. 2012;188:2244–2253. doi: 10.4049/jimmunol.1101614. [DOI] [PubMed] [Google Scholar]
- 22.Grunig G, Corry DB, Leach MW, Seymour BW, Kurup VP, Rennick DM. Interleukin-10 is a natural suppressor of cytokine production and inflammation in a murine model of allergic bronchopulmonary aspergillosis. J Exp Med. 1997;185:1089–1099. doi: 10.1084/jem.185.6.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 24.Sakurai D, Zhao J, Deng Y, Kelly JA, Brown EE, Harley JB, Bae SC, Alarcomicronn-Riquelme ME, Biolupus, G. networks. Edberg JC, Kimberly RP, Ramsey-Goldman R, Petri MA, Reveille JD, Vila LM, Alarcon GS, Kaufman KM, Vyse TJ, Jacob CO, Gaffney PM, Sivils KM, James JA, Kamen DL, Gilkeson GS, Niewold TB, Merrill JT, Scofield RH, Criswell LA, Stevens AM, Boackle SA, Kim JH, Choi J, Pons-Estel BA, G. Argentine Collaborative. Freedman BI, Anaya JM, Martin J, Yu CY, Chang DM, Song YW, Langefeld CD, Chen W, Grossman JM, Cantor RM, Hahn BH, Tsao BP. Preferential binding to Elk-1 by SLE-associated IL10 risk allele upregulates IL10 expression. PLoS Genet. 2013;9:e1003870. doi: 10.1371/journal.pgen.1003870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang HC, Zhang S, Thieu VT, Slee RB, Bruns HA, Laribee RN, Klemsz MJ, Kaplan MH. PU.1 expression delineates heterogeneity in primary Th2 cells. Immunity. 2005;22:693–703. doi: 10.1016/j.immuni.2005.03.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.