Abstract
Background
Delaying plasma separation after phlebotomy (processing delay) can cause perturbations of numerous small molecule analytes. This poses a major challenge to the clinical application of metabolomics analyses. In this study, we further define the analyte changes that occur during processing delays and generate a model for the post hoc detection of this preanalytical error.
Methods
Using an untargeted metabolomics platform we analyzed EDTA-preserved plasma specimens harvested after processing delays lasting from minutes to days. Identified biomarkers were tested on (i) a test-set of samples exposed to either minimal (n=28) or long delays (n=40) and (ii) samples collected in a clinical setting for metabolomics analysis (n= 141).
Results
A total of 149 of 803 plasma analytes changed significantly during processing delays lasting 0–20 h. Biomarkers related to erythrocyte metabolism, e.g., 5-oxoproline, lactate, and an ornithine/arginine ratio, were the strongest predictors of plasma separation delays, providing 100% diagnostic accuracy in the test set. Together these biomarkers could accurately predict processing delays >2 h in a pilot study and we found evidence of sample mishandling in 4 of 141 clinically derived specimens.
Conclusions
Our study highlights the widespread effects of processing delays and proposes that erythrocyte metabolism creates a reproducible signal that can identify mishandled specimens in metabolomics studies.
Keywords: Preanalytical error, Clinical metabolomics, Quality control, Whole blood stability, Phlebotomy
1. Introduction
The majority of clinical testing errors occur during the preanalytical stages of test requisition, sample collection, storage, and transport to the testing facility; this form of error is often underappreciated and can lead to substantial analyte perturbations [1,2]. This issue is particularly relevant to metabolomics analyses [3]. Untargeted mass spectrometry (MS) based metabolomics platforms, in particular, can identify 1000s of small molecule analytes in human plasma, and enable the detection of rare or novel analytes with relevance to diseases such as inborn errors of metabolism [4,5]. It is therefore difficult to anticipate the effects of preanalytical variables on all clinically relevant analytes potentially detectable by untargeted metabolomics studies and thus, it is imperative that preanalytical error be avoided or at least recognized prior to data interpretation. Failure to do so could lead to a missed or incorrect diagnosis.
In a clinical setting, the preanalytical variable of the timing from phlebotomy to plasma separation and storage (processing delay) is difficult to precisely regulate. This poses a major quality control challenge for clinical metabolomics studies, especially when specimen collection occurs across multiple centers working independent of the testing laboratory. Even with the appropriate use of anticoagulants such as EDTA, when whole blood is left at room temperature, the concentrations of a diverse list of clinically important biomarkers change with time, for example: glucose, alanine transaminase, creatine kinase, and cell free DNA [6–8]. Relevant to metabolomics analyses, numerous small molecule analytes have been shown to undergo significant changes in concentration during delays in blood processing, with some analytes showing marked changes when plasma separation is delayed by as a little as 2 h [9–12]. Conversely, many other analytes appear to be stable in whole blood, remaining unchanged over processing delays of at least 24 h [13]. The overall pattern of change may be explained, in part, by continued cellular and enzymatic activity, as well as cell lysis and/or hemoconcentration [14,15].
While it is clear that processing delays should be avoided, our understanding of changes caused by this error remains far from complete and currently there are no proven methods for the detection of plasma processing delays from metabolomics data. A better understanding of this preanalytical variable may improve clinical outcomes and allow individuals to critically review previously generated metabolomics data for sample integrity issues.
In the following study, we hypothesized that preanalytical error could be reliably detected through recurrent signatures of degradation found in metabolomics profiles. To test this hypothesis, we used an untargeted metabolomics platform to catalogue the analyte changes that occur during delays lasting from minutes to days. We mined this dataset to identify the key indicators of processing delays. These markers were then applied to the detection of sample handling errors on (i) a test set of specimens of known sample quality as well as (ii) samples collected in a heterogeneous clinical diagnostic setting for metabolomics analysis. Results of this study demonstrate accurate prediction of samples exposed to processing delays.
2. Materials and methods
2.1. Sample collection
To determine the changes in small molecule analytes that occur during processing delays, we performed 2 time course studies (0–20 h and multi day) using EDTA plasma specimens collected from volunteers within our laboratory. The diagnostic utility of the biomarkers from this initial study was further investigated using additional test specimens (TESTneg and TESTpos). Finally, to explore the real-world application of the discovered biomarkers, we studied these analytes in a sample set (referred to as “clinical”) that was sent to our laboratory for clinical metabolomics analysis with the purpose of screening for inborn errors of metabolism. A further description of the collection protocols for these samples is provided below.
Samples in the 0–20 h time-course assay (n=5) and the optimally handled control sample set (TESTneg; n=28) were drawn from volunteers at the site of this study by venipuncture using 23 G × 3/4″ × 12″ butterfly needles and 6 ml Becton, Dickinson and Company (BD) vacutainer® tubes containing 10.8 mg K2-EDTA. Blood samples remained at room temperature (19–22 °C) for the specified times and then plasma was harvested by centrifugation and placed in a −80 °C freezer for long term storage. For TESTneg samples, no sample experienced a processing delay >15 min. Collection occurred between 9 AM–5 PM and did not exclude participants based on dietary status. An unstructured collection protocol was followed due to volunteer recruitment limitations and also to mimic the clinical sample collection used by our clinical biochemical genetics laboratory. We reasoned that this protocol would capture the variability in dietary status and time-of-collection anticipated in a clinical setting where sampling occurs throughout the day and compliance is an issue. Samples collected for the multi-day assay (n = 4) were collected offsite, using the above methods with the exception that the initial time 0 time point was not harvested and frozen until approximately 1 h after collection, as a result of transportation delays.
Samples from the TESTpos (n = 40) population were collected offsite, preserved in EDTA, and shipped to our laboratory as whole blood at ambient temperature. Samples were in transit for ≤48 h. Upon arrival, samples were stored at 4 °C for an additional 1–3 days prior to harvesting plasma, as described above.
Finally, the clinical sample set (n=141) came from multiple institutions across the US with the requirement that samples be collected in EDTA containing tubes, processed to plasma, and frozen immediately, prior to overnight shipping of frozen samples on dry ice. We presume that most clinical samples were processed within an h of collection; however, compliance with our specimen collection guidelines cannot be guaranteed in this clinical sample set. Retrospective analyses of these previously harvested samples were completed with a waiver of informed consent. All procedures were approved by the Baylor College of Medicine Institutional Review Board.
2.2. Metabolomics analysis
All specimens in this study were subjected to the same untargeted metabolomics analysis described here; this includes not only the samples collected within our laboratory but also those sent to our laboratory from outside sources. General analyte detection rates are described in Table S1. Metabolomics analysis was performed by Metabolon Inc. and was completed essentially as described previously [5,16]. Small molecule analytes (50–1500 Da) were extracted from plasma in an 80% methanol solution that contained 4 standards used to monitor extraction efficiency (tridecanoic acid, 4-Cl-phenylalanine, 2-flurophenylglycine, and d6-cholesterol). Clarified supernatant was split into 5 aliquots and dried to completion under N2. One aliquot was kept as a spare and the remaining 4 aliquots were each resuspended in running buffer and studied in a separate mass spectrometry assay. Aliquot 1 was reconstituted and derivatized using bistrimethyl-silyl-trifluoroacetamide and was analyzed using a Trace DSQ fast-scanning single-quadruple mass spectrometer (Thermo-Finnigan); aliquot 2 was reconstituted in 50 μl of 6.5 mmol/l ammonium bicarbonate, pH 8, and analyzed via liquid chromatography mass spectrometry (LC/MS) in negative ion mode (LCneg); aliquot 3 was reconstituted in 50 μl of 0.1% formic acid in water and analyzed via LC/MS in positive ion mode (LCpos); final, aliquot 4 was reconstituted in 100 μl 85/15 acetonitrile/water in 10 mM ammonium formate, pH 10.8 and analyzed using LCneg. For aliquots 2–4, chromatographic separation was achieved using an ACQUITY UPLC (Waters) equipped with a Waters BEH C18 (aliquot 2 and 3) or HILIC column (aliquot 4) followed by analysis with a QExactive high resolution mass spectrometer (Thermo-Finnigan).
All aliquots were resuspended in buffers that contained instrument internal isotopic standards. These standards were used to monitor performance and serve as retention time/index markers. For negative ion mode analyses, the following standards were used: d7-glucose, d3-methionine, d3-leucine, d8-phenylalanine, d5-tryptophan, Cl-phenylalanine, Br-phenylalanine, d15-octanoic acid, d19-decanoic acid, d27-tetradecanoic acid, and d35-octadecanoic acid. For positive ion mode analyses, the following standards were used: d7-glucose, fluorophenylglycine, d3-methionine, d4-tyrosine, d3-leucine, d8-phenylalanine, d5-tryptophan, d5-hippuric acid, Cl-phenylalanine, Br-phenylalanine, d5-indole acetate, d9-progesterone, and d4-dioctylpthalate. For the polar analyses, the following standards were used: d35-octadecanoic acid, d5-indole acetate, Br-phenylalanine, d5-tryptophan, d4-tyrosine, d3-serine, d3-aspartic acid, d7-ornithine, and d4-lysine. Internal standards were chosen based on their broad chemical structures, biological variety and their elution spectrum on each of the arms of the platform.
Metabolites were identified by matching of chromatographic retention index, accurate mass, and mass spectral fragmentation patterns with a reference library that was created using purified metabolites analyzed under the same analytical procedures as the experimental samples. When an analyte could not be matched to a known reference compound, it was assigned as an “X−” compound, which is a reproducibly detected unnamed molecule.
2.3. Data analysis
Raw integrated intensity values were calculated for each analyte using the area under the chromatographic peak. Raw values for each analyte were then scaled to the median integrated intensity found in an invariant plasma specimen (termed the internal control matrix (ICM) sample) that was independently prepared and analyzed 4–6 times in each MS batch (~36 test samples/batch). The resultant value (termed scaled intensity) allowed for intra-analyte comparisons across batches. Analytes identified in the test specimen but in <2/3 of the ICM specimens were excluded from further analyses.
Analytes that did not have a scaled-intensity value in at least 4 of 5 individuals in time-course #1 (0–20 h) for all time points tested were excluded from subsequent analyses. Similarly for time-course #2 (0–4 days), only analytes found in at least 3 of 4 individuals at all time points tested were studied. Missing values were not imputed. Time-course fold change values were calculated using scaled-intensity values and time 0 as the denominator.
In modeling experiments, analyte values were converted into amore intuitive median-scaled value. This was accomplished by normalizing scaled-intensity values for each analyte to the median-scaled intensity found in our clinical reference population, which, at the time of this study, was comprised of 141 patient specimens that were collected, analyzed, and scaled exactly as described above. Only analytes identified in >50% of all reference population samples were median scaled.
2.4. Statistical analysis
We applied a mixed-effects model in R using the lme function in the nlme package (https://cran.r-project.org/; [17]). In the model, each analyte was treated as a dependent variable and was evaluated for the fixed effect of change over time and random effects were used to model between-subject variability. Analytes with a false discovery rate (FDR) <0.05 were brought forward for further evaluation. Heat maps were created using Java TreeView (vs1.16r2). Linear regression analyses were completed using the default parameters in R. We performed principal components analysis with fully informative analytes using the NIPALS algorithm and calculated components until 99% of variation was explained [18]. Components were evaluated for correlation with time. Subsequently, the unknown samples were run in a blinded analysis, where the NIPALS algorithm was re-applied to the set of controls and the blinded sample. We next prioritized metabolites that showed complete separation over time in our reference sample by calculating the %GAP, which we defined as the minimal gap between the most extreme participant values for TESTneg and TESTpos populations divided by the full range of the values seen across these populations. We examined these metabolites in our time-course data and in a set of 141 patient samples that were sent for clinical testing.
3. Results
3.1. Analyte perturbations during plasma processing delays
To determine the small molecule analyte changes that occur when plasma separation is delayed, we completed a time-course analysis where in whole blood samples from 5 healthy individuals were collected in EDTA-vacutubes and then left at room temperature (19–22 °C) for 0, 0.5, 1, 2, 4, or 20 h prior to harvesting plasma. Untargeted metabolomics analysis was completed on each plasma sample. For each analyte, scaled-intensity values were converted to fold-change values relative to the starting point. Full time-course data were generated for 803 analytes. We did not detect a significant difference in overall analyte identifications or signal intensities between the time points in this assay, thus suggesting that the complete loss of analytes is not a major hallmark of processing delays lasting <1 day (Table S1).
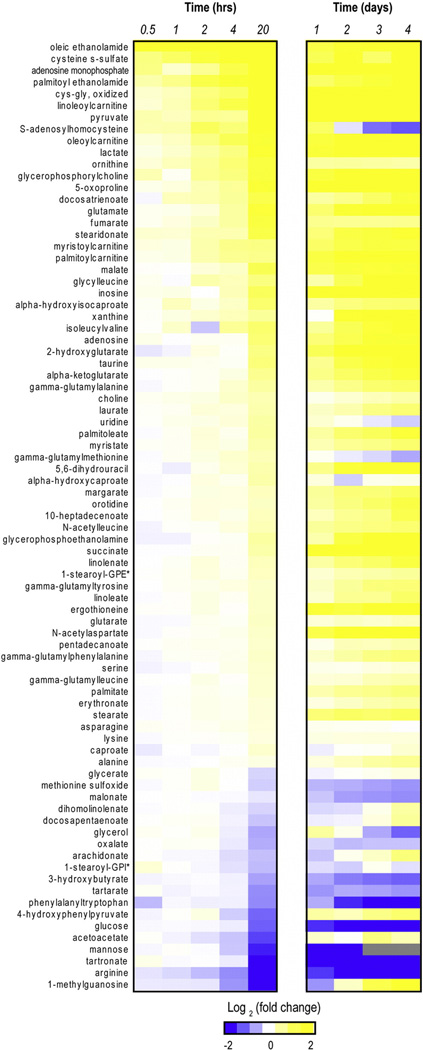
By applying a mixed-effects model, we were able to identify 149 analytes (18.6% of total) that achieved significant consistent perturbation during this time-course (FDR < 0.05; Supplemental dataset). Within this group, the most significantly enriched subpathway perturbations included “glycolysis/gluconeogenesis/and pyruvate metabolism”, “TCA cycle” and “gamma-glutamyl amino acid” classifications, as well as multiple fatty acid subtypes (Table 1). Many unstable analytes followed an apparently linear or logarithmic increase/decrease in analyte concentration from 0 to 20 h (Figs. 1 and S1A–D), and although the patterns of analyte changes were generally similar across individuals, the initial starting concentration and/or rate of change often demonstrated marked inter-individual variation (Fig. S1A–D).
Table 1.
Metabolic pathways perturbed during plasma separation delays.
| Subpathway | # perturbed analytesa |
% of total |
p-Valueb |
|---|---|---|---|
| Polyunsaturated fatty acid (n3 and n6) | 9/14 | 64% | 0.0002 |
| Glycolysis, gluconeogenesis, and pyruvate metabolism |
4/5 | 80% | 0.0047 |
| Long chain fatty acid | 7/14 | 50% | 0.0058 |
| TCA cycle | 4/6 | 67% | 0.0115 |
| Gamma-glutamyl amino acid | 5/9 | 56% | 0.0118 |
| Phospholipid metabolism | 3/4 | 75% | 0.0205 |
| Alanine and aspartate metabolism | 3/4 | 75% | 0.0205 |
| Methionine, cysteine, SAM and taurine metabolism |
6/14 | 43% | 0.023 |
| Ketone bodies | 2/2 | 100% | 0.0342 |
| Endocannabinoid | 2/2 | 100% | 0.0342 |
| Glutathione metabolism | 2/2 | 100% | 0.0342 |
| Purine metabolism, (hypo)xanthine/inosine containing |
3/6 | 50% | 0.0686 |
Total number of subpathway analytes significantly perturbed during the 0 to 20 h time course over the total number of analytes detected in the subpathway.
Calculated using a Fisher's exact test.
Fig. 1.
Analyte perturbations during processing delays. The heat maps show the average fold change values across two independent assays (0–20 h and 0–4 day studies) for a subset of relevant analytes (i.e., named compounds that underwent a significant fold change (FDR < 0.01) during the 0–20 h time course assay). *GPE = glycero-3-phosphoethanolamine; GPI= glycerophosphoinositol.
To confirm our initial findings and to extend the analysis to additional time points, we repeated the above analysis using samples from 4 additional volunteers to study the time points 0, 1, 2, 3, and 4 days delay prior to harvesting plasma. In general, the number and intensity of perturbations were increased over the multi-day time-course compared to the 0–20 h time-course and the pattern of perturbations found in earlier time points persisted over multiple days (Figs. 1 and S2A–C). However, we again noted substantial inter-individual differences in the starting concentrations and depletion/accumulation rates of many analytes. Despite this, a significant correlation was detected between the 20 h and 1 day time points from independent assays (r2 = 0.47; p-value = 2.2 × 10−16), further supporting the reproducibility of these findings (Fig. S2D). Collectively, these results confirm that numerous recurrent perturbations are detectable during plasma separation delays, but these signals may be obscured by inter-individual differences in analyte starting concentrations and/or rates of change.
3.2. Principal components analysis provides accurate detection of processing delays ≥20 h
We were next interested in using the above findings to generate tools for the prospective detection of sample handling error. For this analysis, we used the 0–20 h time-course data in order to perform model-building, and we used a test set comprised of the metabolomics data from optimally handled samples (TESTneg; n=28), as well as from samples exposed to extreme delays prior to plasma separation, lasting 2–5 days (TESTpos; n=40). In our initial analysis of this test set, we performed principal components analysis (PCA) using the NIPALS algorithm on all analytes with no missing values in our dataset (n = 727) [18]. This failed to provide separation between well-handled and poorly-handled specimens; all components had ps > 0.01 when performing regression to time (Fig. S3A). We reasoned that multiple factors such as diet, genetic background, and prandial state could be confounding this analysis. Therefore, we limited the analysis to 68 metabolites that were previously demonstrated to have a fold change of >2 between 0 and 20 h and no missing values. In this limited dataset, the first component, which predicts 38.8% of the variance, was demonstrated to be significantly correlated with time (p < 0.001) (Fig. S3B). No other components were correlated with time. We observed that although data showed an increased trend, it was difficult to separate values from 0 to 2 h. However, the observed increase at 4 h prompted evaluation of principal components regression to accurately predict known, but blinded, samples. We observed that all samples delayed >20 h are predicted accurately by the model. When using cutoffs of 1 and 2 h for the component regression model, we observed inaccurate calling of the TESTneg sample but improvement to 82% when using a 4-h cutoff, as being well-handled (Fig. S3C).
3.3. Analytes related to erythrocyte metabolism provide the strongest biomarkers for processing delays
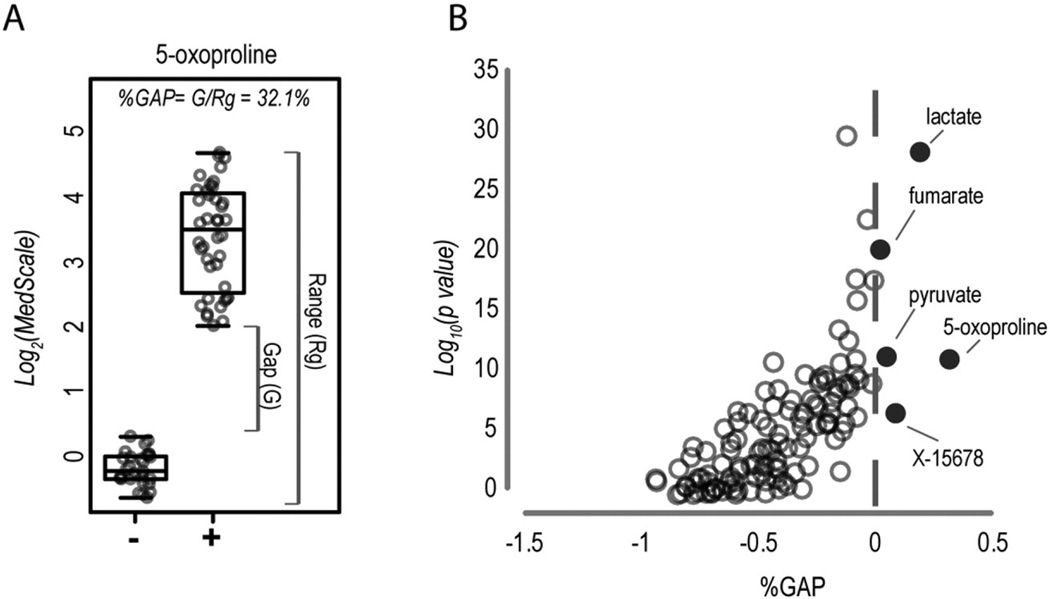
Given the limitations of the PCA-based approach, we explored the diagnostic utility of individual biomarkers identified in the 0–20 h time-course study. To assess the diagnostic utility of a biomarker, it is not fully informative to calculate p-values based on mean differences in test populations (e.g., Student's t-test). We therefore instead calculated a %GAP value for each analyte, which we defined as the minimal gap between the most extreme participant values for TESTneg and TESTpos populations divided by the full range of the values seen across these populations (see example Fig. 2A). In this case, when a biomarker did not provide full separation between TESTneg and TESTpos samples, it received a negative value. We applied the %GAP calculation to our list of 149 analytes that were significantly perturbed during 0–20 h of processing delay. From this analysis, we identified 5 analytes (5-oxoproline, lactate, pyruvate, fumarate, and unknown compound X-15678) that alone provided complete separation in our test set, with 5-oxoproline performing the best (Fig. 2B). A similar prioritization of biomarkers was obtained when using percentile cutoffs as opposed to minimum and maximum values to calculate the %GAP (data not shown).
Fig. 2.
Utility of single biomarkers in the detection of processing delays. (A) The %GAP calculation used to determine test-set diagnostic performance is shown for 5-oxoproline. (−) indicates TESTneg and (+) indicates TESTpos samples. (B) The %GAP was calculated for all biomarkers identified in the 0–20 h time course assay. %GAP values are plotted in terms of p-values (heteroscedastic Student's t-test) to illustrate the utility of the %GAP approach as opposed to standard analyses of population mean differences. Dark circles indicate analytes with a positive %GAP value, that were thus capable of 100% diagnostic accuracy in the test set.
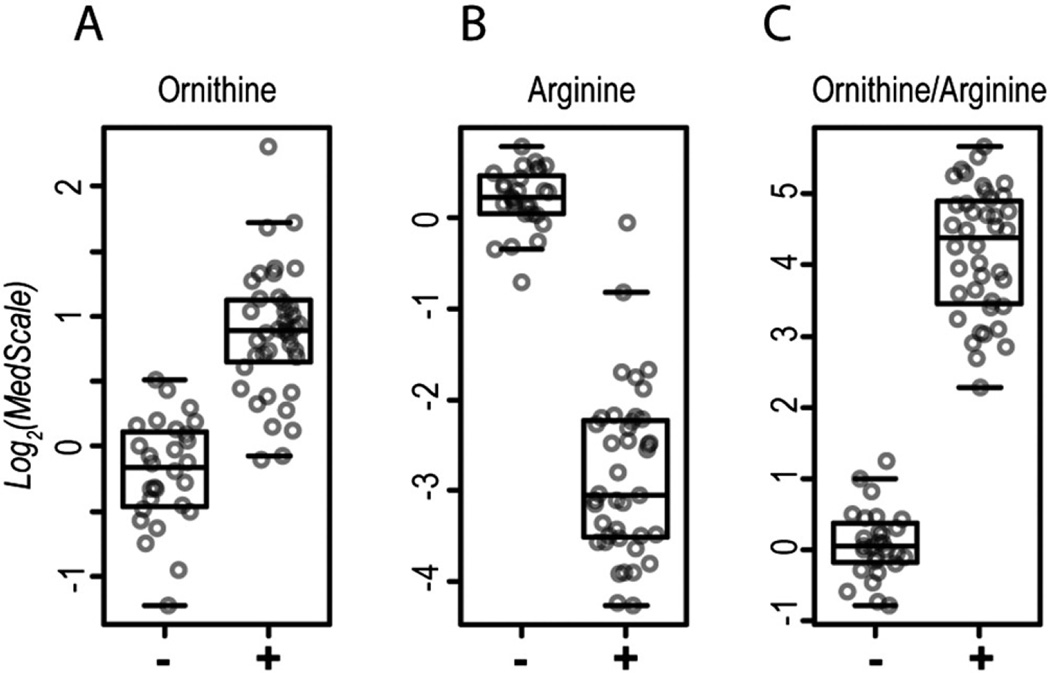
The biomarkers 5-oxoproline, lactate, and pyruvate are all end products of erythrocyte metabolism (see discussion). Therefore, we hypothesized that erythrocyte metabolism was a major driver of analyte perturbations during processing delays and that additional biomarkers of sample degradation may be identified through analyte ratios comprised of start and end-products of erythrocyte metabolic processes. This is exemplified by arginine and ornithine, a substrate and product, respectively, of the erythrocyte-expressed enzyme arginase [19]. Although arginine and ornithine are each highly significant biomarkers for processing delays, neither analyte alone could distinguish between TESTneg and TESTpos samples in all cases (Fig. 3A and B). However, a ratio of these compounds (ornithine/arginine) provided superior results, yielding a complete separation between TESTneg and TESTpos samples (%GAP = 15.9%; Fig. 3C). This finding is consistent with continued erythrocyte arginase enzymatic activity driving plasma depletion of arginine and accumulation of ornithine during processing delays.
Fig. 3.
A ratio of ornithine to arginine is a strong biomarker for processing delays. (A) Ornithine, (B) arginine, and (C) ornithine/arginine values are shown for the test-set. (−) indicates TESTneg and (+) indicates TESTpos samples.
3.4. Pilot application of biomarkers for the detection of processing delays
We were next interested in examining whether the above identified biomarkers could predict preanalytical error in samples exposed to shorter delay times (<20 h) or samples collected under true clinical testing conditions. For this pilot study we used an analyte cutoff of a log2(medscale) increase of 0.5 (1.41 on linear scale) to call mishandled samples. We compared the performance of ornithine/arginine, 5-oxoproline, lactate, pyruvate and fumarate across our test-set (TESTneg and TESTpos) and the one-day time-course data. We also tested these biomarkers on a clinical sample set collected across multiple clinical centers and sent to our laboratory for untargeted metabolomics screening for inborn errors of metabolism (n=141). At the threshold of 1.41× the median, each individual metric had false positive and/or negative predictions, where delays >2 h were considered mishandled and vice versa (Table 2). We were able to improve upon the results using a two-step approach wherein two criteria had to be met in order to call a sample mishandled: (i) ornithine/arginine ratio ≥1.41 × median and (ii) 3 out of 4 individual biomarkers (5-oxoproline, lactate, pyruvate or fumarate) ≥1.41 × median. Using this criterion, we found no false positives for samples experiencing delays ≤1 h and all samples delayed for ≥4 h were identified as poorly handled (50/50) (Table 2). Additionally, with the two-step criterion, 4 of 141 clinical samples were predicted to have experienced sample processing delays.
Table 2.
Prognostic capabilities of individual and combined biomarkers.
| Biomarker | # mishandled samples predicteda (sample processing delay time) |
Clinicalb | ||||
|---|---|---|---|---|---|---|
| <1 h | 1 h | 2 h | 4 h | >20 h | ||
| 5-Oxoproline | 0/38 | 0/5 | 3/5 | 4/5 | 45/45 | 15/141 |
| Lactate | 3/38 | 1/5 | 3/5 | 5/5 | 45/45 | 27/141 |
| Pyruvate | 20/38 | 3/5 | 4/5 | 4/5 | 45/45 | 54/141 |
| Fumarate | 5/38 | 2/5 | 2/5 | 2/5 | 45/45 | 24/141 |
| (I) 3 of 4 metabolitesc | 1/38 | 1/5 | 3/5 | 5/5 | 45/45 | 11/141 |
| (II) ornithine/arginine | 3/38 | 1/5 | 2/5 | 5/5 | 45/45 | 30/141 |
| Two-step prediction (both I and II) | 0/38 | 0/5 | 1/5 | 5/5 | 45/45 | 4/141 |
A threshold of a log2 (medscale) increase of 0.5 (1.41 on a linear scale) was used to predict mishandled samples using the listed biomarkers. Values are presented with the number of samples meeting these criteria over the total number of samples at each time-point.
Samples collected in a multicenter clinical setting. Actual processing time is unknown.
Increases meeting criteria for at least 3 of the 4 biomarkers (5-oxoproline, lactate, pyruvate, fumarate).
4. Discussion
The major conclusion from this study is that during plasma processing delays, recurrent patterns of small molecule analyte perturbations emerge and when used appropriately, these biomarkers can enable the detection of preanalytical error in metabolomics datasets. The strongest recurrent biomarkers identified in our study could be mapped to metabolic pathways of erythrocytes. This is not surprising given that erythrocytes account for 40–45% of the volume of whole blood and greatly out-number any other cell type in this biospecimen. The best performing single biomarker in our study, 5-oxoproline, is the metabolic end product of erythrocyte glutathione metabolism [20]. Consistent with previous studies, we found glucose depletions and lactate and pyruvate elevations to be highly significant biomarkers of processing delays; these compounds are the start and end-products of the truncated glycolytic pathway present in erythrocytes [15,21]. Finally, the predictive success of the ornithine/arginine ratio may be explained by persistent activity of arginase, an enzyme highly expressed in erythrocytes that cleaves arginine, producing ornithine and urea [19]. Collectively, our results indicate that, following phlebotomy, continued erythrocyte activity and/or lysis generates a predictable output that can be used to detect sample processing delays.
Limitations of this study should be considered. A myriad of factors such as diet, prandial state, gender, age, and genetic background can influence plasma levels of small molecule analytes [22–26]. These factors may have unanticipated effects not fully studied here. For example, lactate elevations appeared to be a strong biomarker for separation delays. However, plasma lactate elevations can be precipitated by numerous other factors including intense exercise or mitochondrial disease [27]. As another example, arginine levels can be highly skewed in critically ill individuals or in patients with argininemia-a rare inherited metabolic disease caused by a loss of arginase activity [28,29]. In these cases, an ornithine/arginine ratio may fail to detect preanalytical error. We would therefore caution against the strict use of any single analyte as a marker of preanalytical error and instead propose that any predictions of sample handling error should be based on the observation of multiple perturbed biomarkers and/or ratios of biomarkers.
Our work explored the utility of a principal components regression to detect preanalytical error. PCA and regression have been utilized for quality control procedures across many disciplines [30–32]. However, in the setting of metabolomics data, this would procedurally require establishing reliable time-course controls which may be cumbersome and expensive. If missing data are present, this is typically handled by (i) removal from the dataset, thus decreasing power to detect systematic differences, or (ii) by imputation, which can lead to overall bias for relatively small datasets or those with non-random missing data [33–35].
We found the best predictive outcomes using a model predicated on a small number of strong biomarkers linked to erythrocyte metabolism. Specifically, this model used 2 criteria to identify mishandled specimens (i) moderate elevations of ornithine/arginine and (ii) moderate increases in 3 of the following 4 compounds: 5-oxoproline, lactate, pyruvate and fumarate. Our results clearly demonstrate the utility of these biomarkers for the detection of long processing delays (≥20 h) but additional studies are needed to further define the sensitivity and specificity of these biomarkers for the detection of short term processing delays (<20 h).
When the 2-criteria model was retrospectively applied to analyze metabolomics data from 141 clinically derived samples, we found 4 samples (2.8% of the total) that appeared to have been exposed to long processing delays (>2 h). While the true processing delay time for these samples cannot be known, a ~3% failure rate is in line with our expectations. In our experience, processing times <2 h are difficult for some clinical facilities to achieve. And sample collection requirements are not always strictly followed. For example, we have identified multiple clinical samples lacking EDTA due to the use of an improper specimen collection tube. These preanalytical errors occurred despite strict guidance in the requisition form that accompanied this test. Overall, this highlights some of the preanalytical challenges facing clinical tests and suggests that failure to follow simple collection requirements occurs with some regularity in a clinical setting. If unchecked, this may result in unrecognized clinical testing errors. Based on our results, excessive processing delays could induce a metabolomics profile that might mimic defects of glycolysis, lipid metabolism, or the TCA cycle.
5. Conclusions
Our study demonstrates that widespread changes in small molecule analyte levels begin quickly after phlebotomy. This poses a significant challenge to plasma metabolomics studies, especially those collected in a clinical setting. By measuring the levels of analytes related to erythrocyte metabolism, investigators can probe metabolomics data for clues about sample handling. Based on our results, we would recommend first monitoring 5-oxoproline, lactate, pyruvate and fumarate for elevations, potentially indicating a plasma processing delay. When multiple of these analytes are noted to be increased, we would recommend investigators further test for processing delays by assessing whether the ornithine/arginine ratio is also increased. Using this approach, it is our hope that investigators can critically review published metabolomics datasets and improve quality control in clinical testing.
Supplementary Material
Acknowledgments
This work was funded, in part, by the Medical Genetics Research Fellowship Program (MM, MJ).
Abbreviations
- FDR
false discovery rate
- PCA
principal components analysis
- NIPALS
non-linear iterative partial least squares
Footnotes
Declaration of interest
Mahim Jain, Sarah H. Elsea and Marcus J. Miller are members of the Department of Molecular and Human Genetics at Baylor College of Medicine, and this department, alone or as part of a joint venture with Miraca Holdings, offers a number of clinical tests on a fee-for-service basis, but these in no way conflict with the research reported here. Adam D. Kennedy is an employee of Metabolon, Inc. and, as such, has affiliations with or financial involvement with Metabolon, Inc. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.cca.2017.01.005.
References
- 1.Plebani M, Sciacovelli L, Aita A, Padoan A, Chiozza ML. Quality indicators to detect pre-analytical errors in laboratory testing. Clin. Chim. Acta. 2014;432:44–48. doi: 10.1016/j.cca.2013.07.033. [DOI] [PubMed] [Google Scholar]
- 2.Kellogg MD, Ellervik C, Morrow D, Hsing A, Stein E, Sethi AA. Preanalytical considerations in the design of clinical trials and epidemiological studies. Clin. Chem. 2015;61(6):797–803. doi: 10.1373/clinchem.2014.226118. [DOI] [PubMed] [Google Scholar]
- 3.Yin P, Lehmann R, Xu G. Effects of pre-analytical processes on blood samples used in metabolomics studies. Anal. Bioanal. Chem. 2015;407(17):4879–4892. doi: 10.1007/s00216-015-8565-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patti GJ, Yanes O, Siuzdak G. Innovation: metabolomics: the apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012;13(4):263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller MJ, Kennedy AD, Eckhart AD, Burrage LC, Wulff JE, Miller LA, et al. Untargeted metabolomic analysis for the clinical screening of inborn errors of metabolism. J. Inherit. Metab. Dis. 2015;38(6):1029–1039. doi: 10.1007/s10545-015-9843-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong D, Moturi S, Angkachatchai V, Mueller R, DeSantis G, van den Boom D, et al. Optimizing blood collection, transport and storage conditions for cell free DNA increases access to prenatal testing. Clin. Biochem. 2013;46(12):1099–1104. doi: 10.1016/j.clinbiochem.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 7.Clark S, Youngman LD, Palmer A, Parish S, Peto R, Collins R. Stability of plasma analytes after delayed separation of whole blood: implications for epidemiological studies. Int. J. Epidemiol. 2003;32(1):125–130. doi: 10.1093/ije/dyg023. [DOI] [PubMed] [Google Scholar]
- 8.Bruns DE, Knowler WC. Stabilization of glucose in blood samples: why it matters. Clin. Chem. 2009;55(5):850–852. doi: 10.1373/clinchem.2009.126037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin P, Peter A, Franken H, Zhao X, Neukamm SS, Rosenbaum L, et al. Preanalytical aspects and sample quality assessment in metabolomics studies of human blood. Clin. Chem. 2013;59(5):833–845. doi: 10.1373/clinchem.2012.199257. [DOI] [PubMed] [Google Scholar]
- 10.Townsend MK, Clish CB, Kraft P, Wu C, Souza AL, Deik AA, et al. Reproducibility of metabolomic profiles among men and women in 2 large cohort studies. Clin. Chem. 2013;59(11):1657–1667. doi: 10.1373/clinchem.2012.199133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anton G, Wilson R, Yu ZH, Prehn C, Zukunft S, Adamski J, et al. Pre-analytical sample quality: metabolite ratios as an intrinsic marker for prolonged room temperature exposure of serum samples. PLoS One. 2015;10(3):e0121495. doi: 10.1371/journal.pone.0121495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamlage B, Maldonado SG, Bethan B, Peter E, Schmitz O, Liebenberg V, et al. Quality markers addressing preanalytical variations of blood and plasma processing identified by broad and targeted metabolite profiling. Clin. Chem. 2014;60(2):399–412. doi: 10.1373/clinchem.2013.211979. [DOI] [PubMed] [Google Scholar]
- 13.Breier M, Wahl S, Prehn C, Fugmann M, Ferrari U, Weise M, et al. Targeted metabolomics identifies reliable and stable metabolites in human serum and plasma samples. PLoS One. 2014;9(2):e89728. doi: 10.1371/journal.pone.0089728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyanton BL, Jr., Blick KE. Stability studies of twenty-four analytes in human plasma and serum. Clin. Chem. 2002;48(12):2242–2247. [PubMed] [Google Scholar]
- 15.Hogman CF, Meryman HT. Storage parameters affecting red blood cell survival and function after transfusion. Transfus. Med. Rev. 1999;13(4):275–296. doi: 10.1016/s0887-7963(99)80058-3. [DOI] [PubMed] [Google Scholar]
- 16.Evans AM, Bridgewater BR, Liu Q, Mitchell MW, Robinson RJ, Dai H, et al. High resolution mass spectrometry improves data quantity and quality as compared to unit mass resolution mass spectrometry in high-throughput profiling metabolomics. Metabolomics. 2014;4(2):1–7. [Google Scholar]
- 17.Lindstrom ML, Bates DM. Nonlinear mixed effects models for repeated measures data. Biometrics. 1990;46(3):673–687. [PubMed] [Google Scholar]
- 18.Wold S, Esbensen K, Geladi P. Principal component analysis. Chemom. Intell. Lab. Syst. 1987;2:37–52. [Google Scholar]
- 19.Spector EB, Rice SC, Kern RM, Hendrickson R, Cederbaum SD. Comparison of arginase activity in red blood cells of lower mammals, primates, and man: evolution to high activity in primates. Am. J. Hum. Genet. 1985;37(6):1138–1145. [PMC free article] [PubMed] [Google Scholar]
- 20.Palekar AG, Tate SS, Meister A. Formation of 5-oxoproline from glutathione in erythrocytes by the gamma-glutamyltranspeptidase-cyclotransferase pathway. Proc. Natl. Acad. Sci. U. S. A. 1974;71(2):293–297. doi: 10.1073/pnas.71.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernini P, Bertini I, Luchinat C, Nincheri P, Staderini S, Turano P. Standard operating procedures for pre-analytical handling of blood and urine for metabolomic studies and biobanks. J. Biomol. NMR. 2011;49(3–4):231–243. doi: 10.1007/s10858-011-9489-1. [DOI] [PubMed] [Google Scholar]
- 22.Krug S, Kastenmuller G, Stuckler F, Rist MJ, Skurk T, Sailer M, et al. The dynamic range of the human metabolome revealed by challenges. FASEB J. 2012;26(6):2607–2619. doi: 10.1096/fj.11-198093. [DOI] [PubMed] [Google Scholar]
- 23.Suhre K, Shin SY, Petersen AK, Mohney RP, Meredith D, Wagele B, et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477(7362):54–60. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawton KA, Berger A, Mitchell M, Milgram KE, Evans AM, Guo L, et al. Analysis of the adult human plasma metabolome. Pharmacogenomics. 2008;9(4):383–397. doi: 10.2217/14622416.9.4.383. [DOI] [PubMed] [Google Scholar]
- 25.Shin SY, Fauman EB, Petersen AK, Krumsiek J, Santos R, Huang J, et al. An atlas of genetic influences on human blood metabolites. Nat. Genet. 2014;46(6):543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunn WB, Lin W, Broadhurst D, Begley P, Brown M, Zelena E, et al. Molecular phenotyping of a UK population: defining the human serum metabolome. Metabolomics. 2015;11:9–26. doi: 10.1007/s11306-014-0707-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pavlakis SG, Phillips PC, DiMauro S, De Vivo DC, Rowland LP. Mitochondrial myopathy, encephalopathy, lactic acidosis, and strokelike episodes: a distinctive clinical syndrome. Ann. Neurol. 1984;16(4):481–488. doi: 10.1002/ana.410160409. [DOI] [PubMed] [Google Scholar]
- 28.van Waardenburg DA, de Betue CT, Luiking YC, Engel M, Deutz NE. Plasma arginine and citrulline concentrations in critically ill children: strong relation with inflammation. Am. J. Clin. Nutr. 2007;86(5):1438–1444. doi: 10.1093/ajcn/86.5.1438. [DOI] [PubMed] [Google Scholar]
- 29.Schlune A, Vom Dahl S, Haussinger D, Ensenauer R, Mayatepek E. Hyperargininemia due to arginase I deficiency: the original patients and their natural history, and a review of the literature. Amino Acids. 2015;47(9):1751–1762. doi: 10.1007/s00726-015-2032-z. [DOI] [PubMed] [Google Scholar]
- 30.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 31.Zang H, Wang J, Li L, Zhang H, Jiang W, Wang F. Application of near-infrared spectroscopy combined with multivariate analysis in monitoring of crude heparin purification process. Spectrochim. Acta A. 2013;109:8–13. doi: 10.1016/j.saa.2013.02.018. [DOI] [PubMed] [Google Scholar]
- 32.Anderson KJ, Kalivas JH. Assessment of pareto calibration, stability, and wave-length selection. Appl. Spectrosc. 2003;57(3):309–316. doi: 10.1366/000370203321558227. [DOI] [PubMed] [Google Scholar]
- 33.Barnes SA, Lindborg SR, Seaman JW., Jr. Multiple imputation techniques in small sample clinical trials. Stat. Med. 2006;25(2):233–245. doi: 10.1002/sim.2231. [DOI] [PubMed] [Google Scholar]
- 34.Schafer JL. Multiple imputation: a primer. Stat. Methods Med. Res. 1999;8(1):3–15. doi: 10.1177/096228029900800102. [DOI] [PubMed] [Google Scholar]
- 35.Sterne JA, White IR, Carlin JB, Spratt M, Royston P, Kenward MG, et al. Multiple imputation for missing data in epidemiological and clinical research: potential and pitfalls. BMJ. 2009;338:b2393. doi: 10.1136/bmj.b2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.