Abstract
Recent work has made important advances in describing the large-scale systems-level organization of human cortex by analyzing functional magnetic resonance imaging (fMRI) data averaged across groups of subjects. However, new findings have emerged suggesting that individuals’ cortical systems are topologically complex, containing small but reliable features that cannot be observed in group-averaged datasets, due in part to variability in the position of such features along the cortical sheet. This previous work has reported only specific examples of these individual-specific system features; to date, such features have not been comprehensively described. Here we used fMRI to identify cortical system features in individual subjects within three large cross-subject datasets and one highly sampled within-subject dataset. We observed system features that have not been previously characterized, but 1) were reliably detected across many scanning sessions within a single individual, and 2) could be matched across many individuals. In total, we identified forty-three system features that did not match group-average systems, but that replicated across three independent datasets. We described the size and spatial distribution of each non-group feature. We further observed that some individuals were missing specific system features, suggesting individual differences in the system membership of cortical regions. Finally, we found that individual-specific system features could be used to increase subject-to-subject similarity. Together, this work identifies individual-specific features of human brain systems, thus providing a catalog of previously unobserved brain system features and laying the foundation for detailed examinations of brain connectivity in individuals.
Keywords: individual variability, brain systems, functional connectivity, fMRI
1. INTRODUCTION
The human cortex is organized into large-scale, spatially-distributed systems. These systems can be described in vivo using a functional magnetic resonance imaging (fMRI)-based technique known as resting state functional connectivity (RSFC), which relies on the observation that in the absence of any task, spatially distant regions of cortex exhibit highly correlated patterns of BOLD activity (Biswal et al., 1995). This is posited to at least partly reflect the statistical history of interactions between regions (Dosenbach et al., 2007). RSFC-based approaches have consistently identified around 10–17 brain systems that replicate across multiple datasets and analysis strategies (Power et al., 2011; Yeo et al., 2011). The spatial characterization of these systems has enabled the identification of plausible links between brain organization and cognitive function by associating specific systems identified during the resting state with sets of regions activated during cognitive processes (Bertolero et al., 2015; Dosenbach et al., 2007; Laird et al., 2011; Smith et al., 2009).
Previous descriptions of cortical systems using RSFC have usually been derived from data averaged across many individuals. While RSFC correlation patterns calculated in single individuals are broadly similar across people (Shehzad et al., 2009; Van Dijk et al., 2010), inter-individual variability can nonetheless be observed in these patterns (Laumann et al., 2015; Mueller et al., 2013; D. Wang et al., 2015). This variability has been presumed to reflect individual differences in the strength of communication between brain areas. Thus, many investigators test whether measured differences in RSFC relate to group membership or to individual differences in cognitive function. However, an implicit assumption in this approach is that a given section of cortex always represents the same brain area—or at least the same brain system—across individuals (Dubois and Adolphs, 2016; Satterthwaite and Davatzikos, 2015; Wang and Liu, 2014).
Recent work has suggested that this may not be the case. Detailed descriptions of RSFC-derived brain systems in single individuals suggest that inter-subject variability in these systems is not solely driven by varying strengths of network connections. Rather, the topological features of individual-level systems can vary in shape from one individual to the next (Gordon et al., 2015; Langs et al., 2015; Laumann et al., 2015; D. Wang et al., 2015), such that, for example, a portion of cortex that is in the Default system in one subject may be more strongly connected to the Fronto-Parietal system in another.
Critically, systems of individuals also tend to have focal topological features that cannot be observed in group-averages (Harrison et al., 2015; Laumann et al., 2015; D. Wang et al., 2015). As a result, the brain systems present at a given cortical location can vary dramatically and categorically between an individual and a group-average (Laumann et al., 2015), as well as between one individual and the next (Gordon et al., 2015). These variable brain system features can be consistently observed across many— but not all—individuals (Gordon et al., 2015).
Why do these system features appear in individual data but not in group averages? Laumann et al. (2015) proposed that two factors may account for this observation. First, some features of brain systems may be sufficiently spatially variable (relative to the morphological features of cortex used for cross-subject alignment) that they do not overlap well across individuals. This would cause such features to be “smeared out” when averaged across individuals at each cortical location, creating the appearance of reduced topological complexity in group-average systems. Second, some features of brain systems may be “missing” from some individuals due to specific cortical areas being connected to different brain systems in different individuals. If only a minority of individuals have a strong connection between a brain system and a given cortical area, that connection will not be evident in group-average data.
If this conceptualization of individual variability in brain systems is correct, then it is likely that the system features observed in individuals may represent detailed aspects of the systems-level organization of the brain that, due to reliance on group-average data, have not previously been described. Identifying and characterizing such variable, individual-specific features of brain organization across many individuals would thus advance our understanding of the brain’s functional organization.
In the present study we attempted a comprehensive description of the features in individual-specific brain systems that do not emerge in group-average systems. This was accomplished by using a template matching technique (Gordon et al., 2015) to identify features of brain systems in individuals. We first established the within-subject reliability of this technique by examining the consistency of observed brain features across five hours of resting-state fMRI data collected from a single subject. We then identified brain system features that were common across many subjects by applying the technique to many individuals and matching the discrete features of identified brain systems across individuals. We performed several analyses of these commonly-present features. First, we characterized the size, spatial distribution, and frequency of occurrence of each system feature. Second, we determined whether we could observe system features that appeared consistently across subjects, but that were not present in group-average systems. Third, we examined whether these system features reliably emerged across three independent datasets, and whether they reliably emerged when a different set of brain system templates was used for systems definition. Finally, we tested whether matching system features across subjects increased the similarity of functional connectivity patterns.
2. METHODS
2.1 Single Subject dataset
2.1.1
Data was collected from a single healthy, right-handed, young adult male subject, age 34 (author ND). Informed consent was obtained from the subject. The study was approved by the Washington University School of Medicine Human Studies Committee and Institutional Review Board. These data were previously described in the Supplemental Material of Laumann et al(2015).
2.1.2 Data Acquisition
Imaging was performed on a MAGNETOM Trio Tim 3.0T Scanner (Erlangen, Germany) with a Siemens 12 channel Head Matrix Coil. Four T1-weighted images (sagittal, 224 slices, 0.8mm isotropic resolution, TE=3.74ms, TR=2400ms, TI=1000ms, flip angle=8°) and four high-resolution T2-weighted images (sagittal, 224 slices, 0.8 mm isotropic resolution, TE=479ms, TR=3200ms) were obtained. Thirty contiguous minutes of resting state data were collected in ten separate sessions, each on a different day (total time = 300 minutes). The subject visually fixated on a white crosshair presented against a black background. Functional imaging was performed using a gradient-echo EPI sequence (TR=2.2s, TE=27ms, flip angle=90°, voxel size = 4×4×4 mm, 36 slices). In each session, a gradient echo field map sequence was acquired with the same prescription as the functional images.
2.1.3 Distortion correction
Mean field map creation: A mean field map was generated based on the field maps collected in each session, and was then applied to all sessions for distortion correction. See the Supplemental Materials and Laumann et al. (2015) for details on this procedure.
2.1.4 Preprocessing
Functional data were preprocessed to reduce artifact and to maximize cross-session registration. All sessions underwent intensity normalization to a whole brain mode value of 1000 and within run correction for head movement. Atlas transformation was computed by registering the mean intensity image from a single BOLD session to atlas space via the average high-resolution T2-weighted image (n = 4) and average high-resolution T1-weighted image (n = 4). All subsequent BOLD sessions were linearly registered to this first session. Atlas transformation, distortion correction, and resampling to an isotropic 3-mm atlas space (Talairach and Tournoux, 1988) were combined into a single interpolation using FSL’s applywarp tool (Smith et al., 2004).
2.2 Washington University (Wash U) Dataset
2.2.1 Subjects
Data was collected from 120 healthy young adult subjects during relaxed eyes-open fixation (60 females, mean age = 25 years, age range = 19–32 years). All subjects were native speakers of English and right-handed. Subjects were recruited from the Washington University community and were screened with a self-report questionnaire to ensure that they had no current or previous history of neurological or psychiatric diagnosis. Informed consent was obtained from all subjects. The study was approved by the Washington University School of Medicine Human Studies Committee and Institutional Review Board.
2.2.2 Data Acquisition
Structural and functional MRI data were obtained with a Siemens MAGNETOM Trio Tim 3.0T Scanner (Erlangen, Germany) and a Siemens 12 channel Head Matrix Coil. A T1-weighted sagittal magnetization-prepared rapid acquisition gradient echo (MP-RAGE) structural image was obtained (TE=3.08ms, TR(partition)=2.4s, TI=1000ms, flip angle=8°, 176 slices with 1×1×1mm voxels) (Mugler and Brookeman, 1990). An auto align pulse sequence protocol provided in the Siemens software was used to align the acquisition slices of the functional scans parallel to the anterior commissure-posterior commissure (AC-PC) plane and centered on the brain. This plane is parallel to the slices in the Talairach atlas (Talairach and Tournoux, 1988).
During functional MRI data acquisition, subjects were instructed to relax while fixating on a black crosshair that was presented against a white background. Functional imaging was performed using a blood oxygenation level-dependent (BOLD) contrast sensitive gradient echo echo-planar sequence (TE=27ms, flip angle=90°, in-plane resolution=4×4 mm). Whole brain EPI volumes (MR frames) of 32 contiguous, 4 mm-thick axial slices were obtained every 2.5 seconds. A T2-weighted turbo spin echo structural image (TE=84ms, TR=6.8s, 32 slices with 1×1×4 mm voxels) in the same anatomical planes as the BOLD images was also obtained to improve alignment to an atlas. An average of 336 volumes (14.0 mins) were collected from each subject (range: 184–729 frames).
2.2.3 Preprocessing
Functional images were first processed to reduce artifacts (Miezin et al., 2000). These steps included: (i) correction of odd vs. even slice intensity differences attributable to interleaved acquisition without gaps, (ii) correction for head movement within and across runs and (iii) across-run intensity normalization to a whole brain mode value of 1000. Atlas transformation of the functional data was computed for each individual using the MP-RAGE scan. Each run was then re-sampled to an isotropic 3-mm atlas space (Talairach and Tournoux, 1988), combining movement correction and atlas transformation in a single cubic spline interpolation (Lancaster et al., 1995; Snyder, 1996).
2.3 Dartmouth Dataset
2.3.1 Subjects
Data were collected from 104 healthy young adult subjects during relaxed eyes-open fixation (67 females, mean age = 21 years, age range = 18–32 years). Subjects were recruited from the Dartmouth College community and were screened with a self-report questionnaire to ensure that they had no neurological problems, were not using psychoactive medications and had normal or corrected to normal vision. Participants were given course credit or monetary compensation in exchange for their participation and were provided informed consent in accordance with the guidelines set by the Committee for the Protection of Human Subjects at Dartmouth College. These subjects were selected as the subjects with minimal in-scan head motion from a larger cohort of 746 subjects.
2.3.2 Data Acquisition
Structural and functional MRI data were obtained with a Philips Achieva 3.0 Tesla scanner and a thirty-two channel phased array coil. A T1-weighted sagittal magnetization-prepared rapid acquisition gradient echo (MP-RAGE) structural image was obtained (TE=4.6ms, TR=9.9ms, flip angle=8°, 160 slices with 1×1×1mm voxels.
During functional MRI data acquisition, subjects were instructed to relax while fixating on a white crosshair that was presented against a black background. Functional imaging was performed using a blood oxygenation level-dependent (BOLD) contrast sensitive gradient echo echo-planar sequence (TE=35ms, flip angle=90°, in-plane resolution=3×3 mm, sense factor = 2). Whole brain EPI volumes (MR frames) of 36 3.5 mm-thick axial slices were obtained every 2.5 seconds with .5mm skip between slices. Two 5:00 minute runs (240 volumes total) were collected from each subject.
2.3.3 Preprocessing
Preprocessing procedures were identical to the Wash U dataset.
2.4 Human Connectome Project (HCP) Dataset
2.4.1 Subjects
80 subjects (40 females; mean age=29 years, age range=22–36) were retrieved from the publicly available Human Connectome Project (HCP) dataset (Van Essen et al., 2013, 2012b). These subjects were selected as the subjects with minimal in-scan head motion estimates from the S500 data release.
2.4.2 Data Acquisition
See Van Essen et al. (2012b) and Smith et al. (2013) for details of the data acquisition procedures. Briefly, structural and functional MRI data were acquired on a custom Siemens Skyra 3T scanner. T1-weighted MP-RAGE images were acquired at .7×.7×.7mm resolution. Four 15-minute resting-state BOLD runs were acquired at 2×2×2mm resolution, with images collected every .7 seconds; the two runs (one each of left-right/right-left phase-encoding) with minimal in-scan motion were selected for each subject.
2.4.3 Preprocessing
Functional images were processed using the HCP minimal preprocessing pipeline (Glasser et al., 2013), which includes head motion correction, intensity normalization, bias field correction, and transformation to an isotropic 2-mm MNI atlas space.
Further analysis of the three datasets was identical except where specified. Note that the different volumetric atlas spaces used in the different datasets do not impede inter-dataset comparisons because of the subsequent registration to a common surface atlas (see “Surface processing and CIFTI creation” below).
2.5 Functional connectivity processing
Additional preprocessing steps to reduce spurious variance unlikely to reflect neuronal activity were executed as recommended in (Power et al., 2014). Temporal masks were created to flag motion-contaminated frames. Motion contaminated volumes were identified by frame-by-frame displacement (FD, described in Power et al. (2012)), calculated as the sum of absolute values of the differentials of the 3 translational motion parameters and 3 rotational motion parameters.
In the HCP dataset, this FD metric did not appear to have a “noise floor” enabling clear definition of an FD threshold for volume censoring across subjects. We found that temporally filtering the movement parameters allowed identification of this threshold. Thus, volumes with FD > 0.2 mm (Single Subject and Wash U datasets)/FD > .25mm (Dartmouth dataset)/filtered FD > .04mm (HCP dataset), as well as uncensored segments of data lasting fewer than 5 contiguous volumes, were flagged for removal. In the Single Subject dataset, this mask censored 11.4% of the data, with 7250 volumes retained. In the Wash U dataset, these masks censored 16% ± 14% (range: 0.7% – 66%) of the data across subjects; on average, subjects retained 279 ± 107 volumes (range: 151 – 719). In the Dartmouth dataset, these masks censored 9% ± 4% (range: 4% – 29%) of the data across subjects; on average, subjects retained 220 ± 10 volumes (range: 170 – 230). In the HCP dataset, these masks censored 3% ± 3% (range: 0% – 16%) of the data across subjects; on average, subjects retained 2318 ± 76 volumes (range: 2018 – 2399). We note that the mean percentage of data lost was higher in the Single Subject and Wash U datasets because subjects in the other two datasets were preselected to have minimal movement.
After computing the temporal masks for high motion frame censoring, the data were processed with the following steps: (i) demeaning and detrending, (ii), multiple regression including: whole brain, ventricular and white matter signals, and motion regressors derived by Volterra expansion (Friston et al., 1996), with censored data ignored during beta estimation, (iii) interpolation across censored frames using least squares spectral estimation of the values at censored frames (Power et al., 2014) so that continuous data can be passed through (iv) a band-pass filter (0.009 Hz < f < 0.08 Hz) without contaminating frames near high motion frames (Carp, 2013; Power et al., 2012). Censored frames were then excised from the data for all subsequent analyses.
2.6 Surface processing and CIFTI creation
Surface generation and sampling of functional data to anatomical surfaces followed the procedures described in (Gordon et al., 2016, 2015; Laumann et al., 2015). First, following volumetric registration, anatomical surfaces were generated from each subject’s MP-RAGE image using FreeSurfer’s default recon-all processing pipeline. This pipeline included brain extraction, segmentation, generation of white matter and pial surfaces, inflation of the surfaces to a sphere, and surface shape-based spherical registration of the subject’s ‘native’ surface to the fsaverage surface (Dale et al., 1999; Dale and Sereno, 1993; Fischl et al., 1999; Ségonne et al., 2005, 2004). The fsaverage-registered left and right hemisphere surfaces were then brought into register with each other (Van Essen et al., 2012a) and resampled to a resolution of 164,000 vertices using Caret tools (Van Essen et al., 2001). Finally, each subject’s surface was down-sampled to a 32,492 vertex surface (fs_LR 32k), a computationally tractable surface space that allows for quantitative analysis across subjects. A script for this procedure is available on the Van Essen Lab website (Freesurfer_to_fs_LR Pipeline, http://brainvis.wustl.edu/wiki/index.php/Caret:Operations/Freesurfer_to_fs_LR).
Surface processing of the BOLD data proceeded through the following steps. First, the BOLD volumes were sampled to each subject’s individual ‘native’ midthickness surface (generated as the average of the white and pial surfaces) using the ribbon-constrained sampling procedure available in Connectome Workbench 1.0, which samples data from voxels within the gray matter ribbon (i.e. between the white and pial surfaces) (Glasser and Van Essen, 2011). Voxels with a timeseries coefficient of variation 0.5 standard deviations higher than the mean coefficient of variation of nearby voxels (within a 5 mm sigma Gaussian neighborhood) were excluded from the volume to surface sampling (Glasser et al., 2013). After being sampled to the ‘native’ surface, timecourses were deformed and resampled to the 32k fs_LR surface described above. Finally, the time courses were smoothed along the 32k fs_LR surface using a Gaussian smoothing kernel (σ = 2.55mm).
These surfaces are then combined with volumetric subcortical and cerebellar data into the CIFTI format using Connectome Workbench (Glasser et al., 2013), creating full brain timecourses that exclude non-gray matter tissue. Subcortical (including accumbens, amygdala, caudate, hippocampus, pallidum, putamen, and thalamus) and cerebellar voxels were selected based on a mask generated separately in each dataset by finding the modal assignment of voxels by Freesurfer segmentation across subjects within each dataset. Volumetric data was smoothed within this mask with a Gaussian kernel (σ = 2.55mm).
2.7 Template matching for systems identification in individual subjects
Many different techniques have been proposed to identify brain systems at the individual level (Filippini et al., 2009; Harrison et al., 2015; Langs et al., 2015; Laumann et al., 2015; D. Wang et al., 2015). In the present study, we elected to use a template-matching technique we have described in previous work (Gordon et al., 2015). This approach was chosen for two reasons. First, our technique describes brain systems by comparing individual RSFC patterns to a set of brain system priors. Using priors ensures that individual subject versions of brain systems are “labeled” and thus easily comparable to one another (something that is not guaranteed using data-driven approaches). Second, our technique makes comparisons between individual RSFC patterns and the priors based on the strongest positive FC connections only. This approach follows graph-theory-based analyses of both functional and diffusion MRI that conceptualize the brain as a network of nodes communicating via relatively sparse connections; this requires removal of all negative and weak positive connections in functional data (Rubinov and Sporns, 2010).
2.7.1 Calculating templates for each brain system
The template-matching procedure was conducted using the following steps. First, we built a connectivity template for each system in a previously described group-average systems map (Figure 3, top; described in detail in (Gordon et al., 2015; Laumann et al., 2015)); these templates represented the average RSFC pattern of each system across many individuals. For each subject in the Wash U dataset, we calculated the average timecourse across cortical vertices within each system. Then, we correlated that timecourse against all other gray matter timecourses in that subject to obtain a subject-specific system connectivity map. We then applied the Fisher transform and averaged these maps across subjects. Finally, we thresholded and binarized each averaged system map at the top 5% of connectivity strengths (as calculated across systems), which was Z(r) >= .383. These binarized maps serve as templates for individual subject matching. The template maps used are identical to the templates used in our previous work (Gordon et al., 2015), with the exception that two additional templates, in anterior and posterior medial temporal lobe, were also included. See Supplemental Figure 1 for template maps. We note that the particular binarization threshold used here (5%) was chosen as to be approximately in the middle of the threshold range explored in previous work (Power et al., 2011). However, because template and individualized maps are thresholded to the same extent before matching (see below), the particular value chosen for this threshold has minimal effect on the systems identified (see Supplemental Figure 2 for a demonstration of this in the Single Subject dataset). Note also that this single set of templates, which was derived from the Wash U dataset, was used to investigate all four datasets.
Figure 3.
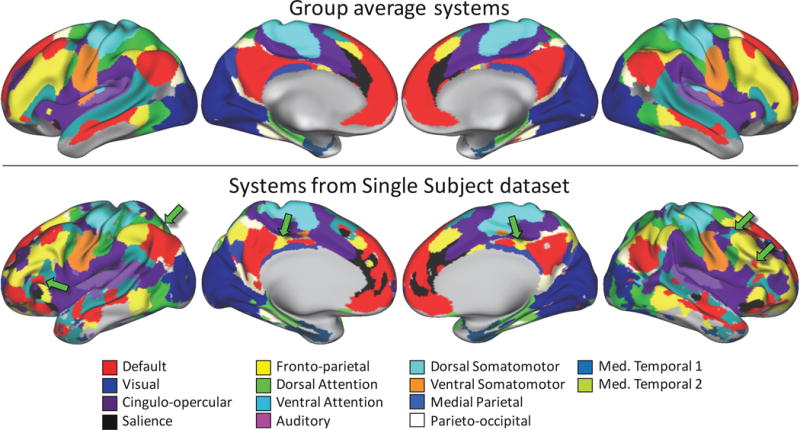
Topological features of large-scale brain systems can be observed in resting-state fMRI data collected from individuals, but are not present in group-average data. Top: Brain systems derived from group-average data (described in Gordon et al., 2015; Laumann et al., 2015). Bottom: Brain systems derived from an individual with a large quantity of resting-state data (300 minutes) are generally similar to group-average systems, but also have numerous topological features not found in the group-average systems (see green arrows for examples).
2.7.2 Conducting the template-matching procedure in each subject to identify individual-specific brain systems
In each subject, we identified that subject’s versions of group-average systems by matching each vertex’s subject-specific connectivity pattern to the set of templates described above. See Figure 1A for a graphical illustration of this procedure. We first correlated the vertex’s timecourse with all other gray matter timecourses in that subject’s data, Fisher transformed the resulting connectivity map, and then thresholded and binarized the resulting map at the top 5% of connectivity values (as calculated across vertices). This resulted in a binarized map of regions with high connectivity to the chosen cortical vertex. We then calculated the Dice coefficient of overlap between the binarized vertex connectivity map and each binarized template map, excluding from the calculation data from all vertices within 30mm geodesic distance from the selected vertex. The template with the highest overlap was judged to be the best match, and that template’s system identity was assigned to that vertex in that subject. We note that the distance-based vertex exclusion step is critical in voxel- or vertex-wise network detection analyses because correlation maps are known to be artifactually dominated by very strong local correlations (Power et al., 2011). Artifactually elevated local correlations are problematic for any network detection procedure, but are especially problematic for those which employ priors. As the vertices near a given seed vertex always have the strongest correlations in the brain, these local connections will strongly bias comparisons with the prior.
Figure 1.
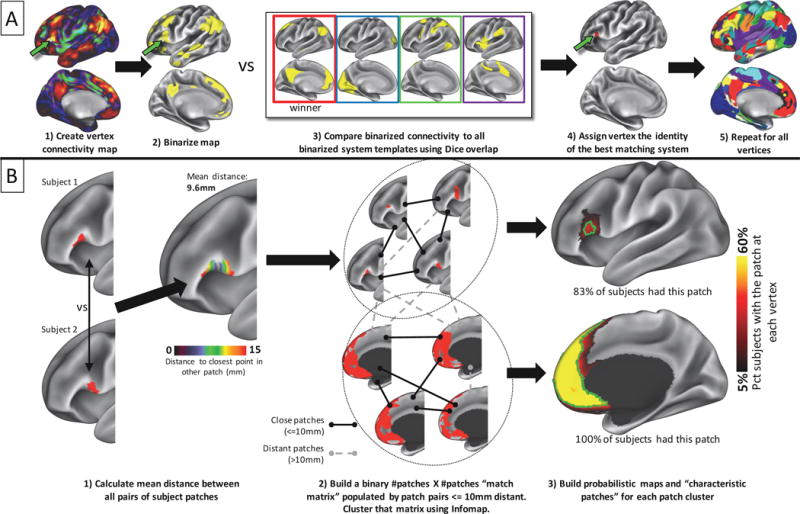
Visual depiction of methods employed for identifying brain systems and matching system patches across subjects. A) a functional connectivity seedmap is generated for each cortical point in each subject. This map is binarized to the top 5% of values across seedmaps. Each seedmap is compared to a series of templates using Dice coefficient of overlap. The template identity with the highest overlap is assigned to that point. This procedure is repeated across the cortex to generate a map of brain systems in the individual. B) Discrete contiguous patches of brain systems are extracted from an individual’s systems map. Left: For each brain system, the physical distance is assessed between all patches in one subject and all patches in all other subjects. Distance is calculated as the geodesic distance between each point within one subject’s patch and the nearest point in the other subject’s patch; these distances are then averaged across all points within both patches to get an overall distance between the patches. Middle: Subject patches are “matched” to each other if their pairwise distances are below 10mm (black links), but not matched if the distances are above 10mm (gray dotted links). A community detection algorithm is applied to the resulting graph of pairwise matches to identify clusters of similar system patches (dotted circles). Right: probabilistic maps are created for each cluster of patches. The green outline indicates the “characteristic patch”, defined as an object with the median surface area of all individual patches that follows the topography of the probabilistic map. While the majority of individuals had both pictured patches, overlap across subjects was very different in the two patches: in the top patch, no cortical vertex was within the patch in more than 40% of individuals.
Finally, given the resolution of our datasets (ranging from 4×4×4mm voxels to 2×2×2mm voxels, smoothed on the cortical surface with σ = 2.55mm), surface objects below 30 mm2 are likely smaller than the approximate effective resolution of our data. Thus, contiguous surface patches smaller than 30 mm2 were removed from subsequent analysis.
Within the Single Subject dataset, systems derived from this technique were observed to both have strong correspondence with a widely used graph-theory based approach for community detection (infomap; (Rosvall and Bergstrom, 2008) (Supplemental Figure 3) and to have within-subject reliabilities comparable to previously published approaches for systems identification (D. Wang et al., 2015) (Supplemental Figures 4 and 5); see Supplemental Materials for details.
2.8 Matching system patches across individuals and sessions
Discrete pieces of subject-level system maps (“system patches”) are expected to vary across subjects, both in their position on the cortical surface and in their size (Frost and Goebel, 2012; Gordon et al., 2015; Harrison et al., 2015; Laumann et al., 2015). However, despite this variance, it is possible to compare system patches between individuals based on their spatial position and overall shape. Here, we matched system patches within and across individuals by modifying the approach for subject-group comparisons described in Gordon et al. (2015). This procedure was conducted separately for the Single Subject dataset (in which system patches from individual scanning sessions were matched to each other) and for each of the three cross-subject datasets (in which patches from individual subjects were matched to each other).
A graphical summary of this procedure can be seen in Figure 1B. We first identified the discrete, discontiguous patches of each brain system in each individual. For each pair of subjects, we compared each patch of a given system identity in one subject with all patches of the same system identity in the other subject. ‘Matches’ were determined based on the geodesic distance between two patches in different subjects, calculated as the average distance between each vertex and the nearest vertex in the other subject’s patch. Thus, vertices that overlapped between the two patches would have distances of zero, while vertices that did not overlap would have nonzero distances, with the exact distance depending on the proximity to the closest vertex in the other subject’s patch. If many vertices overlapped or were proximal in the two patches, the average distance would be small. This distance-based approach thus indexes the spatial similarity between two local, binarized maps by testing for proximity rather than simple overlap. Because we observed that a single patch in one individual could be split into multiple patches in another individual, multiple-to-one matches were allowed if adding additional patches reduced the average distance of the match. Potential patch-to-patch matches were considered successful if the average minimum distance of the match was less than 10mm.
This procedure matched system patches to each other in a pairwise subject-to-subject fashion. To combine these matches across all subjects, we represented all patch-to-patch matches in a large binary [# subject patches] X [# subject patches] matrix, in which a “1” represented a successful match between patches and a “0” represented no match. We then clustered this matrix using the Infomap community detection algorithm (Rosvall and Bergstrom, 2008), where each community represented patches that tended to be matched to each other across subjects. This approach allows system patches to be sorted into clusters, even if not every subject patch in the cluster was successfully matched to every other subject patch. Thus, commonalities across patches are identified in a way that is robust to spatial variability across subjects and to low data quality of any given subject.
While system patches can be expected to vary substantially across individuals, they should vary only minimally across scanning sessions within a single subject. To test this, we also applied the patch matching procedure described above to the various sessions of the Single Subject dataset.
The threshold of 10mm to define a patch-to-patch match conforms with work by Fischl et al. (2008), who estimated the subject-to-subject spatial displacement of most architectonic cortical areas to be within 10mm. While this previous work was conducted on very different types of data, making exact application of this threshold somewhat dubious, we observed that varying this threshold across a substantial range had effectively no impact on the identification of common clusters of systems patches across sessions in the Single Subject dataset (see Supplemental Figure 6), or the Wash U dataset (see Supplemental Figure 7).
2.9 Characterizing system patches that were matched across individuals
For each patch identified across the subjects in each dataset, we characterized four features of the patch. First, we calculated the percent of subjects that had that patch. Second, we characterized the spatial distribution of the individual versions of the patch by summing each binarized patch vertexwise across subjects and dividing by the number of subjects that had that patch. This created a probabilistic map of the patch. In the Single Subject dataset, these characterizations represent the detectability and spatial reliability of each patch. In the cross-subject datasets, they represent the similarity of patches across subjects.
Third, we calculated the median surface area of the patch. Fourth, we created a ‘characteristic’ patch that was the same size as the median patch and followed the contours of the probabilistic spatial distribution map; this object can be thought of as the central tendency of that patch. Figure 1B shows examples of system patches that were matched (left of figure) and grouped into clusters (middle of figure), as well as resulting probabilistic maps (right of figure, red underlays) and characteristic patches (right of figure, green outline overlays), from the Wash U dataset.
2.10 Identifying system patches present and absent from group-average systems
In the Wash U dataset, we visually identified all system patches that did and did not represent patches present in the group-average systems, based on the spatial pattern of their probabilistic maps. In order to avoid making claims about the presence of brain systems in regions with poor BOLD signal, we conservatively excluded any patches that were primarily in regions known to have low signal-to-noise (SNR) ratios (defined as regions with mean BOLD signal < 750, consisting primarily of orbitofrontal cortex, ventral/anterior temporal lobe, and occipital pole; see (Ojemann et al., 1997; Wig et al., 2014)).
We observed that many system patches (e.g., the bottom probabilistic map in Figure 1B) appeared to represent patches present in the group-average systems map (Figure 3, top). By contrast, many other patches did not appear to correspond to anything in the group systems (e.g., green arrows in Figure 3, bottom; the top probabilistic map in Figure 1B). It is possible that some of these patches may represent features of large-scale systems that are “washed out” of group-average data by spatial variability across subjects. However, many of these patches may also represent the chance overlap of features present in noisy, relatively low-data system estimates. For example, 606 clusters of system patches were calculated in the Wash U dataset that did not appear similar to anything in the group-average systems, but 324 of the clusters had patches in fewer than 10% of subjects. Clusters with small numbers of subjects very likely represent artifactual patches driven by noise To be confident about which patches represent noise and which represent the convergence of real system features across subjects, we developed an empirical null model of patch clustering.
2.10.1 Surface rotation-based null model of patch clustering
To determine which non-group patches were matched across more subjects than could be expected by chance, we developed a null model of non-group patch locations. This procedure follows the logic that system patches that emerge purely because of noise will be randomly located on the cortex. Randomly located patches should not match across subjects as well as true system patches that are in moderately variable, but generally consistent, locations across subjects. Thus, in this procedure, we generate randomly placed versions of non-group patches, cluster them based on the spatial distance criterion, and determine how many subjects are grouped into each cluster. We can then compare the degree of matching across subjects for real patches to the null expectation of matching across subjects for random patches.
A graphical illustration of this procedure can be seen in Figure 2. To create these randomly rotated patches, we first removed the subject-specific versions of each “group” patch from each subject’s systems map (see Figure 2A, left for an example). We then rotated each hemisphere of each subject’s non-group patches randomly around each of the x, y, and z axes on the spherical expansion of the 32k fs_LR cortical surface. This procedure randomly relocated each parcel (see Figure 2B, left for an example). In any random rotation, some patches will inevitably be rotated into the ventral medial wall of the cortical surface, where the original system maps have no data. Any patch rotated into this medial wall was randomly re-rotated until it was entirely within data-containing regions of cortex and did not overlap any other rotated patches.
Figure 2.
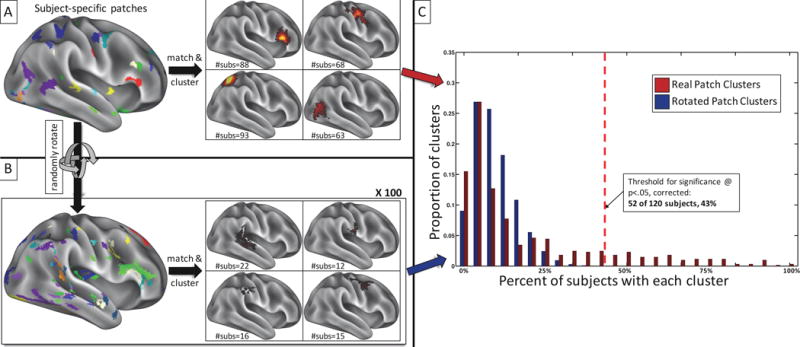
Randomly rotating patch clusters around the cortical surface provides an empirical description of the null expectation for clustering the system patches that are not observed in group-average system maps. A) All system patches that do not match the group-average systems are identified in each subject (see example on the left) and matched across subjects via the distance-based patch-matching procedure. Matched patches are then clustered across subjects (see examples on the right), and the number of subjects who have a patch in each cluster is noted. B) A null expectation of these patch clusters is created by taking the non-group system patches in A) and randomly rotating them around the cortex in each subject, producing patches of the same size and shape but random locations (see example on the left). These randomly rotated patches are then matched across subjects and clustered as in A), and the number of subjects who have a patch in each cluster is noted. This procedure is iterated 100 times. C) The distributions of subject counts for clusters of real (red) and rotated (blue) patches. Many clusters of real patches are entirely outside the distribution of random patch clusters. The red dotted line indicates the threshold set to determine real patch clusters significantly (p<.05, Bonferroni-corrected for multiple comparisons) outside the distribution of random patch clusters.
These randomly-placed patches were matched and clustered across subjects, as above (Sections 2.7 and 2.8), and the number of subjects in each cluster of random patches was recorded. This procedure was iterated 100 times. In total, over 140,000 clusters that had system patches from at least two subjects were generated.
The distribution of subject counts in these randomly rotated clusters represents a null expectation for convergence of features across subjects. We tested each cluster of patches derived from real (i.e. non-rotated) data against this null expectation. Real clusters were determined to match across subjects better than random if they had patches from more subjects than the top Xth percentile of the distribution of random subject counts, where X represents a threshold for “significance”. We conservatively set the percentile threshold for significance at 5% divided by the number of real clusters tested (corresponding to p<.05, Bonferroni corrected). Thus, for the 606 clusters of patches found in the Wash U dataset, clusters were not accepted as significantly different from random noise unless they were found in more subjects than 99.992% of randomly rotated clusters. In the Wash U dataset, this threshold corresponded to patches being found in at least 52 of 120 subjects (43% of subjects); see Figure 2C. Similar thresholds for significance were observed in both the Dartmouth dataset (48 of 104 subjects, 46%) and the HCP dataset (37 of 80 subjects, 46%); see Supplemental Figure 8.
2.11 Reliability of cross-subject system patches across datasets and system templates
If the system patches described here represent real objects in the brain, they should replicate across multiple independent cross-subject datasets. We examined whether the patches described above from the Wash U dataset could also be observed in both the Dartmouth and HCP datasets by visually comparing the probabilistic maps across datasets that were found to be significantly different from random noise.
Further, if the system patches described here represent real objects in the brain, they should be observed regardless of the template system maps used. A widely used set of system maps was released by (Yeo et al., 2011); the 17-cluster version of these “Yeo” systems have many features in common with the Infomap-derived systems used above, but also exhibit some notable differences (see Figure 9, top). Similarly, the Human Connectome Project has released parcellations derived by conducting Independent Components Analysis (ICA) on the data from the 900 subjects release and assigning each data point to the strongest ICA component at that point in a winner-take-all fashion (Smith et al., 2013). The 50-component ICA map has 29 components that each cover at least 500 vertices on the cortex; these 29 “ICA” systems also have many similar features to the Infomap-derived system features used above (Supplemental Figure 10, top), but divide the brain more finely. The remaining, excluded components constitute five small components in occipital pole and sixteen components in subcortical structures. We generated templates using these 29 retained components as system maps and repeated the template-matching and patch clustering procedures for the Wash U dataset using both the Yeo- and the ICA-derived templates. We then examined whether system patches observed using the Infomap-derived templates could also be observed using the Yeo- and ICA-derived templates. We then quantitatively compared similar patches (determined via maximal spatial correlation) by correlating their median sizes, by correlating the percent of subjects in which they were found, and by calculating the Dice overlap of their characteristic patches.
Figure 9.
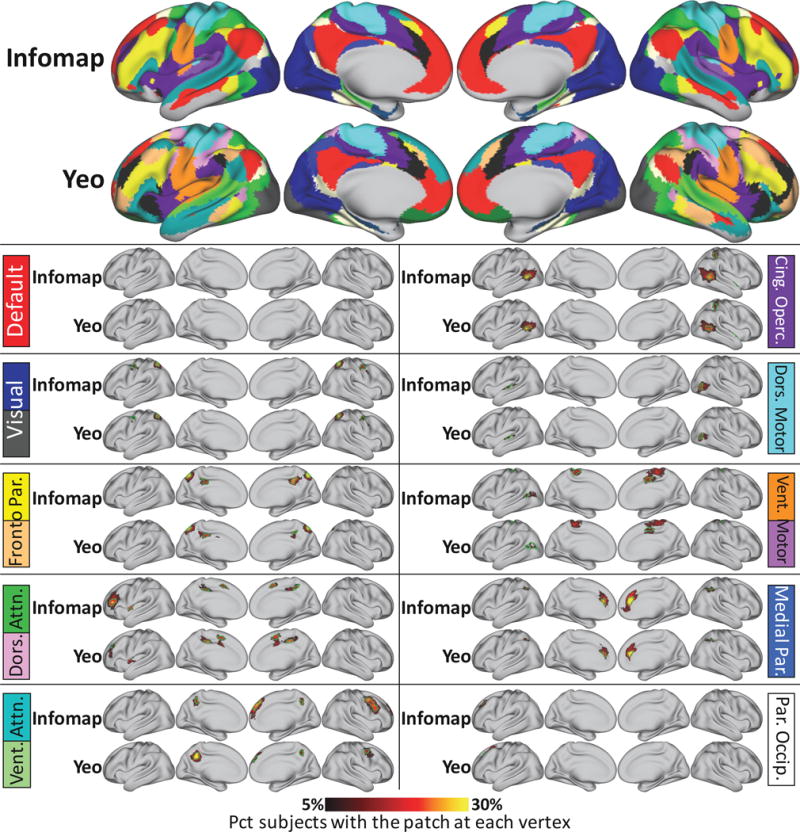
System features are similar when different group-average maps are used as template systems. Probabilistic maps and characteristic patches of non-group system patches identified using the Infomap-derived group-average systems (top row) and using the 17 “Yeo” group-average systems previously reported by (Yeo et al., 2011) (second row). Yeo systems have been recolored to match the Infomap systems for easy comparison. 33 of 43 non-group patches that replicated across datasets were also identified when using the Yeo systems as templates. All probabilistic maps within a system are summed here for display purposes. The green outlines indicate “characteristic patches”, defined as objects that have the median surface area of each patch across subjects in each dataset and follow the topography of the patch’s probabilistic map. The color block next to each map indicates the brain system of the displayed patches; multiple colors are displayed in cases where one system in one map was split into two systems in the other map. Note that for the medial temporal and orbitofrontal systems, no non-group patches were detected outside of low-SNR regions (inferior temporal and orbitofrontal cortex).
Because the Yeo and Infomap templates were relatively similar and had similar dimensionality, we were able to make comprehensively direct comparisons between them. In some cases, two systems in one set of templates were grouped together and treated as one system, whereas the other set of templates had only one equivalent system (e.g. two Dorsal Attention systems in Yeo but only one in Infomap; a separation between Ventral Somatomotor and Auditory systems in Infomap but no such separation in Yeo). By contrast, because the ICA systems were of a substantially higher dimensionality than the Infomap systems, comprehensive comparisons between templates were more difficult, as it was not always evident which systems corresponded with which. Instead, we focused on identifying specific patches derived from systems that were easily comparable.
2.12 Using matched system patches to improve comparisons across subjects
If the matching procedure described here is effective in describing the spatial variability of system features at across individuals, then calculating functional connectivity across matched patches should remove some or all of the cross-subject variability in functional connectivity that can be attributed to spatial variance. Therefore, patch-seeded functional connectivity patterns should be more similar between two subjects if matched patches are used as seeds than if the same patch is used as a seed in every individual.
To test this hypothesis, we generated matched system patches using data from all three cohorts of subjects. For each subject, we calculated the whole-brain functional connectivity seeded from the matched patches in each subject, as well as seeded from the same “characteristic patch” in each subject. Functional connectivity maps were computed by averaging the functional timecourses of all points within each patch and then correlating that timecourse against all other timecourses in the brain. We note that the characteristic patches are an ideal comparison for the subject-specific patches, as they represent objects that are the same median size (across subjects) as the subject-specific patches.
To quantify the overall effectiveness of the patch-matching procedure in improving subject-to-subject similarity, the similarity between functional connectivity matrices based on subject-specific patches was compared to the similarity between functional connectivity matrices based on ‘characteristic’ patches. The functional connectivity matrices were generated by computing the Fisher-z transformed, Pearson r correlation between the average functional timecourses within each patch. Both group and non-group characteristic patches were used in this analysis (seen in Figure 7A). For each pair of subjects, the similarity of the subjects’ connectivity matrices was computed as the correlation between the two functional connectivity matrices, generating a matrix of subject-to-subject similarities. For the subject-specific-specific similarity matrix, patches that were not identified in both subjects were not considered in this correlation. For the ‘characteristic’ patches similarity matrix, all patches were included in each correlation, as they were identical for every subject.
Figure 7.
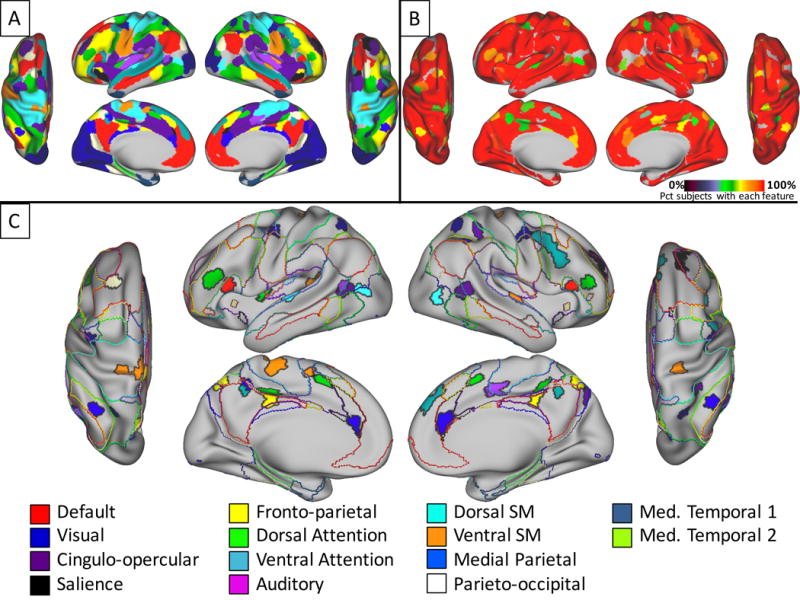
All system features that replicated across three datasets. A) Characteristic patches of all replicating system features are colored according to their system membership. In cases where two patches overlapped, the smaller patch is shown on top of the larger one. B) Detection rates among subjects in the Wash U dataset for all of the system features shown in A). C) Features that were not present in group-average systems. Filled regions indicate replicable characteristic patches colored according to their system membership. Colored outlines indicate borders of group-average brain systems (see Figure 3, top). See Table 2 for MNI coordinates of these non-group patches.
2.13 Testing hub-like behavior of system patches
It is possible that these non-group system patches, which are inconsistently detected across subjects, represent regions which have strong functional relationships with multiple brain systems, allowing for integration of information across discrete systems. Such regions can be conceptualized as “hubs” of the networked brain, and they previously have been posited to be critical for maintaining the integrity of the brain network and for performance of complex cognitive tasks (Bertolero et al., 2015; Gratton et al., 2012; Warren et al., 2014). We tested whether non-group patches preferentially tend to be brain hubs in individual subjects by comparing the participation coefficients (PCs) of individual-specific non-group patches to the PCs of individual-specific versions of group average patches. PC, a measure of the distribution of connectivity across sub-systems within a large network, has previously been proposed as a measure of hub-ness (Guimerà et al., 2007; Guimerà and Nunes Amaral, 2005).
In each subject in the Wash U dataset, we calculated the patch-to-patch correlation matrix (as above) and thresholded this matrix at the top 5% of connections. We then used the patch system identities to sort patches into “communities”, and calculated the PC of each patch following (Guimerà and Nunes Amaral, 2005). For each patch, we then averaged the PC values of matched patches across subjects (omitting subjects who did not have a particular patch), and we used a two-sample t-test to determine whether patches absent from the group average systems had higher PC values than those present in the group average systems.
3. RESULTS
3.1 System patches were reliably detected within a single subject
We identified discrete, contiguous features (hereafter known as “system patches”) in the brain system map derived from all concatenated data in the Single Subject dataset. Notably, we observed many system patches in this map that did not match any feature of the group-average systems identification (Figure 3, green arrows), a finding that agrees with several recent reports (Harrison et al., 2015; Laumann et al., 2015; D. Wang et al., 2015).
We then determined how reliably system patches could be identified in single sessions. We identified system patches in the maps of each 30-minute session from the Single Subject dataset and then matched and clustered spatially proximal patches across sessions to identify the patches commonly present across sessions, as well as to characterize their frequency of detection and their spatial distribution (see Figure 1B). Finally, we matched these probabilistically detected patches to patches from all concatenated data.
We observed that the majority of system patches present in the maps derived from all the Single Subject data could be reliably detected in single 30-minute sessions (Figure 4A, middle), including perfect detection of all system patches greater than 350mm2. Smaller patches were not always perfectly detected in all sessions, but were very commonly detected more than 80% of the time, including small patches that did not exist in group-average systems (see Figure 4B, middle row for examples). Only very small patches near the size threshold of 30mm2 (see Methods) were detected less than half the time. The spatial distributions of patches from single sessions were also very tight, echoing findings in previous work (D. Wang et al., 2015).
Figure 4.
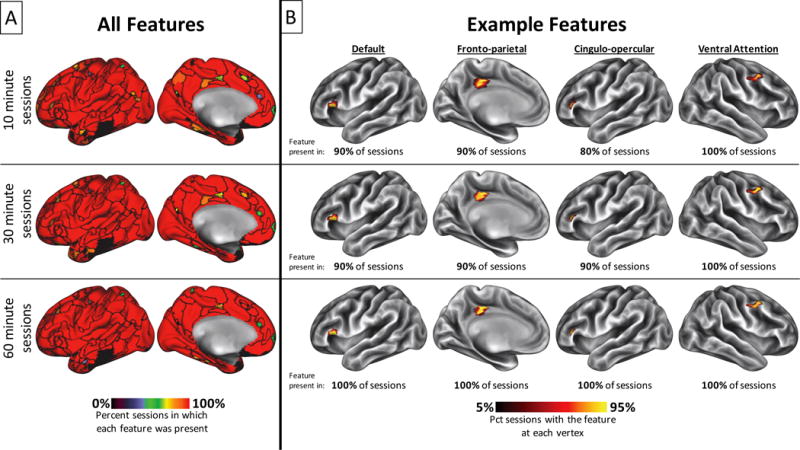
Within a single subject, most brain system features can be detected with high probability even in relatively short sessions. A) Percent of sessions in which each brain system feature was detected. Features were defined in data concatenated across all sessions in the Single Subject dataset and are delineated by black outlines. Detection rates are shown for the original ten 30-minute sessions (middle), as well as for thirty 10-minute subdivisions of those sessions (top) and for five 60-minute sessions built by combining two temporally adjacent 30-minute sessions (bottom). B) Spatial distributions and percent detectability across sessions for four example features. See labels above top row for system membership of these features. These features were not present in group-average systems (see Figure 3), but were detected in 80% or more of 10 minute sessions, 90% or more of 30 minute sessions, and 100% of 60 minute sessions; and further, they had tight spatial distributions across all session lengths.
We further examined how the reliability of system patches from single sessions changed as a function of session length. Notably, we observed that dividing the original sessions into 10-minute segments only slightly decreased the reliability of detecting a patch (Figure 4A, top), and decreases were only observed for medium-sized and small patches. Similarly, combining the sessions into 60-minute segments only slightly increased the reliability of detecting a patch (Figure 4A, bottom). Further, the spatial distributions of detected patches, including small patches that were not present in group-average systems, were largely invariant to session length. Figure 4B shows examples of this phenomenon in 10-minute sessions (top row), 30-minute sessions (middle row), and 60-minute sessions (bottom row). We conclude that the patch-matching methods we employ here can reliably detect most individual-specific features of brain systems even from a relatively short amount of data (though we note that the contours of these features on the cortex are not reliable when low amounts of data are used; see Supplemental Figure 4).
3.2 System patches matched across subjects
We used the template-matching algorithm to estimate brain systems in each individual in each of three different datasets (the “Wash U” dataset, the “Dartmouth” dataset, and the “HCP” dataset). We then matched spatially proximal system patches across subjects.
In the Wash U dataset, we observed that some of the system patches that matched across subjects represented patches in the group-average systems, while others represented patches absent from the group systems. Eighty-two patches were found that closely conformed to patches in the group-average systems (Table 1; see example in Figure 1B, bottom probabilistic map). Seventy-four of these patches were present in at least 90% of individual subjects. The median sizes of these patches varied from 70mm2 to 4410mm2.
Table 1.
System patches matched across subjects that were similar to group-average system patches
| System | % subjects with patch | Median patch size (mm2) | Hemisphere | MNI coordinates of vertex with highest subject overlap | Location |
|---|---|---|---|---|---|
| Default | 100% | 4410mm2 | left | −7 26 61 | Medial Frontal |
| Default | 100% | 3766mm2 | right | 4 37 14 | Medial Frontal |
| Default | 100% | 1562mm2 | left | −59 −47 −6 | Middle Temporal |
| Default | 100% | 1431mm2 | right | 65 −41 2 | Middle Temporal |
| Default | 100% | 1443mm2 | right | 6 −43 33 | Medial Parietal |
| Default | 100% | 1723mm2 | left | −4 −44 32 | Medial Parietal |
| Default | 100% | 349mm2 | left | −44 40 −16 | Lateral Orbital |
| Default | 97% | 327mm2 | right | 47 43 −12 | Lateral Orbital |
| Default | 100% | 1116mm2 | left | −50 −53 46 | Angular |
| Default | 99% | 884mm2 | right | 57 −48 31 | Angular |
| Visual | 100% | 7398mm2 | right | 26 −87 21 | Medial Occipital |
| Visual | 100% | 7157mm2 | left | −18 −91 21 | Medial Occipital |
| FrontoPar | 100% | 2720mm2 | left | −39 50 6 | Lateral Frontal |
| FrontoPar | 100% | 3058mm2 | right | 39 50 8 | Lateral Frontal |
| FrontoPar | 100% | 494mm2 | left | −59 −47 −6 | Middle Temporal |
| FrontoPar | 100% | 468mm2 | right | 63 −36 −10 | Middle Temporal |
| FrontoPar | 100% | 879mm2 | left | −48 −53 49 | Inferior Parietal |
| FrontoPar | 100% | 816mm2 | right | 53 −42 44 | Inferior Parietal |
| FrontoPar | 99% | 425mm2 | right | 7 10 58 | Dorsal Medial Frotnal |
| FrontoPar | 99% | 323mm2 | left | −7 8 52 | Dorsal Medial Frotnal |
| FrontoPar | 92% | 154mm2 | left | −36 11 0 | Anterior Insula |
| FrontoPar | 88% | 178mm2 | right | 38 15 0 | Anterior Insula |
| FrontoPar | 66% | 70mm2 | left | −2 −8 34 | Middle Cingulate |
| FrontoPar | 59% | 78mm2 | right | 3 −11 33 | Middle Cingulate |
| DorsalAttn | 100% | 2857mm2 | right | 34 −42 55 | Superior Parietal |
| DorsalAttn | 100% | 2519mm2 | left | −35 −44 57 | Superior Parietal |
| DorsalAttn | 100% | 771mm2 | left | −45 −80 4 | Middle Occipital |
| DorsalAttn | 100% | 736mm2 | right | 44 −68 0 | Middle Occipital |
| DorsalAttn | 100% | 613mm2 | left | −33 −2 49 | Dorsal Precentral |
| DorsalAttn | 100% | 423mm2 | right | 49 9 14 | Inferior Precentral |
| DorsalAttn | 100% | 610mm2 | right | 34 0 51 | Dorsal Precentral |
| DorsalAttn | 100% | 430mm2 | left | −47 0 43 | Inferior Precentral |
| VentAttn | 100% | 2054mm2 | left | −43 −53 16 | Superior Temporal |
| VentAttn | 100% | 1826mm2 | right | 51 −45 14 | Superior Temporal |
| VentAttn | 100% | 730mm2 | left | −7 26 61 | Superior Frontal |
| VentAttn | 100% | 916mm2 | left | −50 33 7 | Inferior Frontal |
| VentAttn | 99% | 581mm2 | right | 49 37 8 | Inferior Frontal |
| VentAttn | 99% | 224mm2 | left | −52 −5 49 | Middle Precentral |
| VentAttn | 91% | 193mm2 | right | 8 19 64 | Superior Frontal |
| Salience | 98% | 283mm2 | left | −29 23 −11 | Ventral Anterior Insula |
| Salience | 98% | 219mm2 | right | 37 24 −13 | Ventral Anterior Insula |
| Salience | 83% | 159mm2 | left | −7 24 35 | Dorsal Anterior Cingulate |
| Salience | 84% | 132mm2 | right | 7 31 34 | Dorsal Anterior Cingulate |
| Salience | 74% | 82mm2 | right | 12 5 68 | Superior Frontal |
| Salience | 68% | 62mm2 | left | −10 3 69 | Superior Frontal |
| CingOperc | 100% | 1526mm2 | left | −5 −26 43 | Dorsomedial Prefrontal/Paracentral |
| CingOperc | 100% | 1897mm2 | right | 6 −26 43 | Dorsomedial Prefrontal/Paracentral |
| CingOperc | 100% | 1800mm2 | left | −36 −21 2 | Middle Insula |
| CingOperc | 100% | 1936mm2 | right | 38 −18 0 | Middle Insula |
| CingOperc | 100% | 575mm2 | right | 25 49 34 | Anterior Middle Frontal |
| CingOperc | 100% | 822mm2 | right | 58 −39 41 | Supramarginal |
| CingOperc | 100% | 473mm2 | left | −29 50 22 | Anterior Middle Frontal |
| CingOperc | 100% | 592mm2 | left | −58 −45 40 | Supramarginal |
| CingOperc | 96% | 312mm2 | right | 52 −3 50 | Middle Precentral |
| CingOperc | 92% | 241mm2 | left | −46 −2 50 | Middle Precentral |
| MortorHand | 100% | 5064mm2 | right | 20 −41 69 | Dorsal Pre/Postcentral |
| MortorHand | 100% | 4901mm2 | left | −19 −41 96 | Dorsal Pre/Postcentral |
| MortorHand | 96% | 271mm2 | right | 33 −23 17 | Dorsal Posterior Insula |
| MortorHand | 90% | 202mm2 | left | −33 −25 19 | Dorsal Posterior Insula |
| MotorMouth | 100% | 1670mm2 | left | −52 −7 46 | Ventral Pre/Postcentral |
| MotorMouth | 100% | 1393mm2 | right | 52 −6 46 | Ventral Pre/Postcentral |
| MotorMouth | 88% | 148mm2 | right | 40 −4 16 | Dorsal Middle Insula |
| Auditory | 100% | 2715mm2 | right | 33 −23 17 | Posterior Insula |
| Auditory | 100% | 2768mm2 | left | −33 −25 19 | Posterior Insula |
| MTL1 | 100% | 1288mm2 | right | 34 10 −28 | Anterior Temporal |
| MTL1 | 100% | 1302mm2 | left | −34 7 −26 | Anterior Temporal |
| MTL2 | 100% | 576mm2 | right | 27 −36 −6 | Medial Temporal |
| MTL2 | 100% | 535mm2 | left | −27 −38 −5 | Medial Temporal |
| MedPar | 98% | 311mm2 | right | 3 −40 31 | Posterior Cingulate |
| MedPar | 99% | 312mm2 | left | −9 −69 29 | Parieto-Occipital Sulcus |
| MedPar | 98% | 319mm2 | right | 12 −65 30 | Parieto-Occipital Sulcus |
| MedPar | 99% | 275mm2 | left | −2 −40 29 | Posterior Cingulate |
| ParOccip | 98% | 239mm2 | left | −6 −47 46 | Dorsal Precuneus |
| ParOccip | 99% | 385mm2 | right | 29 −46 −6 | Lingual |
| ParOccip | 99% | 441mm2 | right | 8 −53 8 | Retrosplenial |
| ParOccip | 98% | 191mm2 | right | 7 −47 45 | Dorsal Precuneus |
| ParOccip | 98% | 479mm2 | left | −16 −39 −11 | Lingual |
| ParOcc | 100% | 260mm2 | left | −31 −85 30 | Dorsal Middle |
By contrast, eighty-one patches were found that did not match anything in the group-average systems, but were present in significantly more subjects than could be expected due to random chance (i.e., at least 52 subjects). Probabilistic maps of these non-group patches can be seen in Figure 5. Every brain system was found to have multiple non-group patches (though for the medial temporal 1 and 2 systems, all non-group patches were in low-SNR areas and so are omitted from the reported results). Across all systems, the median sizes of these patches varied from 44mm2 to 332mm2. The percent of subjects who had each patch varied from 43% (i.e. just above the threshold for inclusion) to 100%; see Figure 2C for the distribution of these values across patches.
Figure 5.
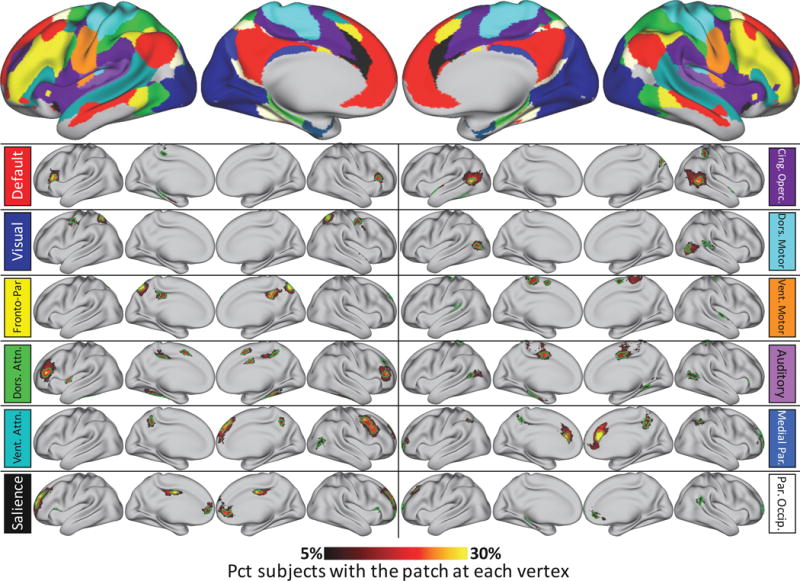
Probabilistic maps and characteristic patches of all system patches identified in at least 52 individuals (corresponding to p<.05, Bonferroni corrected) in the Wash U dataset, but not similar to anything in the group-average systems (top). All probabilistic maps within a system are summed here for display purposes. The green outlines indicate “characteristic patches”, defined as objects with the median surface area of all individual subject versions of each patch that follow the topography of the patch’s probabilistic map. The color block next to each map indicates the brain system of the displayed patches. Note that for the two medial temporal systems, no non-group patches were detected outside of low-SNR regions (inferior temporal and orbitofrontal cortex). Note the change in color map thresholds relative to Figures 1 and 4; this is necessary in order to demonstrate the dynamic range of the spatial distributions.
In general, non-group patches were spatially variable. Figure 5 illustrates this variability. Note that the color map threshold was dramatically reduced in Figure 5 compared to Figure 4, in order that the dynamic range of spatial variability could be better shown. A critical point here is that the color maps represent the degree of physical overlap across subjects, not the existence of the patch across subjects; the spatial variability of these patches mean that their physical overlap is always substantially lower than the total number of subjects who have a patch. For example, more than eighty percent of subjects had a Dorsal Attention patch in the left and right inferior frontal sulcus (regions that do not contain any Dorsal Attention representation in most group-average network descriptions (Power et al., 2011; Yeo et al., 2011). However, these commonly-present patches did not overlap across more than forty-five percent of subjects in any single vertex, indicating variability in the spatial positions of the patches.
3.3 System patches replicate across datasets
We tested whether the system patches we observed in the Wash U dataset could also be observed in the Dartmouth and HCP datasets. We found that every patch matching the group-average system map (Table 1) replicated robustly across datasets. Further, forty-three of the eighty-one non-group patches described above, which have not been described in previous work, replicated across both the Dartmouth and HCP datasets (Table 2). Examples of these replicating patches can be seen in Figure 6. Visually, similar patches appeared to have nearly identical spatial distributions in different datasets, indicating a robust central tendency of the patches’ spatial variability that consistently emerges in large groups.
Table 2.
System patches matched across subjects that replicated across datasets but were not similar to any group-average system patches
| System | % subjects with patch | Median patch size (mm2) | Hemisphere | MNI coordinates of vertex with highest subject overlap | Patch location |
|---|---|---|---|---|---|
| Default | 85% | 131mm2 | left | −53 27 16 | Inferior Frontal |
| Default | 73% | 97mm2 | right | 55 29 13 | Inferior Frontal |
| Visual | 78% | 107mm2 | right | 27 −56 55 | Superior Parietal |
| Visual | 78% | 74mm2 | left | −26 −58 54 | Superior Parietal |
| Visual | 57% | 128mm2 | right | 44 −10 49 | Middle Precentral |
| Visual | 55% | 113mm2 | left | −43 −10 55 | Middle Precentral |
| FrontoPar | 82% | 108mm2 | right | 9 −72 53 | Posterior Cingulate |
| FrontoPar | 78% | 116mm2 | left | −7 −72 52 | Posterior Cingulate |
| FrontoPar | 70% | 123mm2 | right | 6 −37 44 | Posterior Precuneus |
| FrontoPar | 58% | 84mm2 | left | −3 −31 35 | Posterior Precuneus |
| Dorsal Attn | 82% | 155mm2 | left | −38 34 15 | Middle Frontal |
| Dorsal Attn | 80% | 103mm2 | right | 37 37 13 | Middle Frontal |
| Dorsal Attn | 62% | 75mm2 | right | 12 −43 48 | Dorsal Posterior Cingulate |
| Dorsal Attn | 46% | 84mm2 | left | −36 −7 14 | Middle Indula |
| Dorsal Attn | 48% | 111mm2 | left | −13 −33 39 | Dorsal Posterior Cingulate |
| Dorsal Attn | 45% | 58mm2 | left | −7 7 52 | Dorsomedial Prefrontal |
| Dorsal Attn | 43% | 56mm2 | right | 8 6 53 | Dorsomedial Prefrontal |
| VentAttn | 93% | 288mm2 | right | 10 51 42 | Medial Superior Frontal |
| VentAttn | 93% | 185mm2 | right | 47 5 48 | Dorsolateral Prefrontal |
| VentAttn | 54% | 60mm2 | left | −8 −50 45 | Dorsal Precuneus |
| VentAttn | 49% | 44mm2 | right | 8 −55 44 | Dorsal Precuneus |
| Salience | 98% | 218mm2 | right | 29 49 18 | Anterior Prefrontal |
| CingOperc | 93% | 159mm2 | left | −54 −61 7 | Posterior Middle Temporal |
| CingOperc | 92% | 196mm2 | right | 54 −57 7 | Posterior Middle Temporal |
| CingOperc | 53% | 60mm2 | right | 20 −74 39 | Dorsal Parieto-Occipital |
| CingOperc | 53% | 75mm2 | right | 54 11 −7 | Superior Temporal |
| CingOperc | 57% | 88mm2 | right | 35 −40 48 | Intraparietal |
| Mortor Hand | 63% | 88mm2 | right | 49 −75 4 | Middle Occipital |
| Mortor Hand | 51% | 89mm2 | left | −43 −71 8 | Middle Occipital |
| Mortor Hand | 53% | 108mm2 | left | −58 −17 6 | Superior Temporal |
| Motor Mouth | 62% | 189mm2 | left | −23 −31 58 | Dorsal Central |
| Motor Mouth | 68% | 158mm2 | right | 23 −30 60 | Dorsal Central |
| Motor Mouth | 43% | 52mm2 | left | −4 −3 59 | Medial Superior Frontal |
| Motor Mouth | 48% | 77mm2 | right | 66 −12 1 | Superior Temporal |
| Motor Mouth | 43% | 82mm2 | left | −54 −41 19 | Posterior Insula |
| Auditory | 73% | 124mm2 | right | 8 −5 42 | Middle Cingulate |
| Auditory | 63% | 71mm2 | left | −51 −60 14 | Posterior Middle Temporal |
| MedPar | 88% | 232mm2 | right | 4 35 19 | Anterior Cingulate |
| MedPar | 74% | 138mm2 | left | −3 33 19 | Anterior Cingulate |
| MedPar | 72% | 82mm2 | left | −38 −55 42 | Inferior Parietal |
| MedPar | 73% | 91mm2 | right | 46 −48 46 | Inferior Parietal |
| MedPar | 57% | 45mm2 | left | −6 −46 50 | Dorsal Precuneus |
| ParOccip | 83% | 91mm2 | left | −27 29 39 | Superior Frontal |
Figure 6.
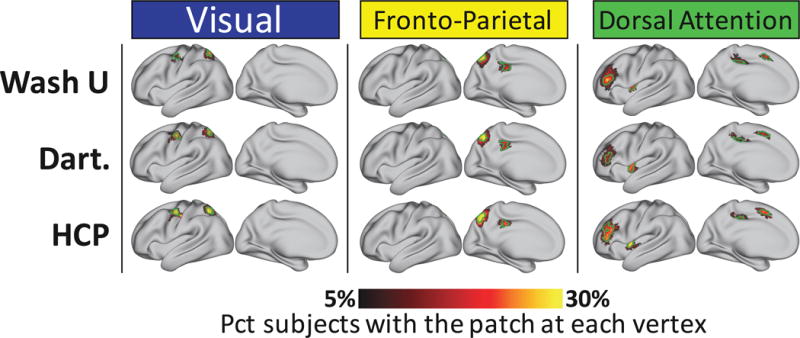
Non-group system features replicate across datasets. Probabilistic maps and characteristic patches of example system patches identified in all three of the Wash U, Dartmouth, and HCP datasets, but not similar to anything in the group-average systems (Figure 3, top). 43 out of 84 novel patches identified in the Wash U dataset were also found in both Dartmouth and HCP datasets. All probabilistic maps within a system are summed here for display purposes. The green outlines indicate “characteristic patches”, defined as objects with the median surface area of all individual subject versions of each patch in each dataset that follow the topography of the patch’s probabilistic map. The color block above each map indicates the brain system of the displayed patches. See Supplemental Figure 9 for replicating patches in all systems.
Across datasets, we found that these visually similar patches tended to have both similar median sizes and a similar proportion of individuals with the patch. For non-group patches, the dataset-to-dataset correlations of median patch size were all highly significant (Wash U vs Dartmouth: r = .77; Wash U vs HCP: = .47; Dartmouth vs HCP: R = .57; all ps<.005), as were the rank correlations of the percent of subjects who had the patch (Wash U vs Dartmouth: ρ = .73; Wash U vs HCP: ρ = .48; Dartmouth vs HCP: ρ = .51; all ps<.005).
Figure 7A illustrates the characteristic position of all patches that replicated across datasets, including those both present and absent in the group-average systems. Note the extremely high similarity between many of these characteristic patches and the group-average systems (Figure 3). Figure 7B illustrates the detection rate in the Wash U dataset of each of the replicated patches. Finally, the central tendencies of all replicated, non-group patches are overlaid on the group-average systems in Figure 7C. This figure represents a first-pass estimation of all features of large-scale brain systems that replicate robustly across datasets, but are too infrequently present and/or spatially variable relative to their size to emerge in group-average data.
3.4 “Missing” system patches reflect both methodological limitations and true functional variability
Some subjects were found to be “missing” patches that were identified across many other subjects (Figure 2C). These missing patches may represent a significant form of functional variability: cortical regions that are functionally connected to different systems in different individuals, as suggested in two very high-data subjects in Laumann et al. (2015). However, we also observed that many of the patches that were most commonly missing in individuals in the cross-subject datasets, as well as in the individual sessions from the Single Subject dataset, were also the smallest patches. This observation raises the possibility that missing patches may represent a methodological limitation of the technique. Specifically, it could be the case that all system patches exist in all individuals, but that small patches in particular may be more difficult to detect, primarily because of the explicit size threshold we imposed on system patches.
We reasoned that if a patch exists in an individual but is unnoticed because it is below the set detection threshold, then there should still be some evidence of the patch in the Dice maps describing the similarity of connectivity to the templates. To examine this possibility, we examined the Dice similarity maps in 30-minute sessions from the Single Subject dataset, as well as in select individuals with relatively large amounts of data (>25 minutes). This examination was restricted to high-data subjects so that we could be relatively confident of the reliability of our RSFC estimates (Anderson et al., 2011; Laumann et al., 2015).
We found that some individuals demonstrated very small sections of cortex that might be a patch, but were not identified as such using our template matching procedure, likely because the identified section of cortex was very small and/or only weakly similar to the system template. Similar observations were made for specific sessions in the Single Subject dataset. However, we also found individuals for whom no indication of similarity to the relevant system could be found. In some cases, the ‘missing’ patches were relatively large, with median sizes many times the detection threshold of 30mm2, indicating that near-threshold size is not the major determinant of patch existence. This phenomenon is illustrated in Figure 8 for two example patches, which were chosen because they were detected in ~50% of individuals and were of moderate size (median sizes: 84 and 89 mm2). These findings indicate that, while methodological limitations may prevent detection of patches in some specific scanning sessions, at least some individuals are truly missing system patches. By contrast, no 30-minute session in the Single Subject dataset appeared to have patches that were present in some sessions but completely missing in others (i.e. there were “missing” patches, but they appeared to represent detection failure rather than the complete absence of the patch). This suggests the presence of variable brain system membership of cortical regions across individuals, but not across scanning sessions within an individual.
Figure 8.
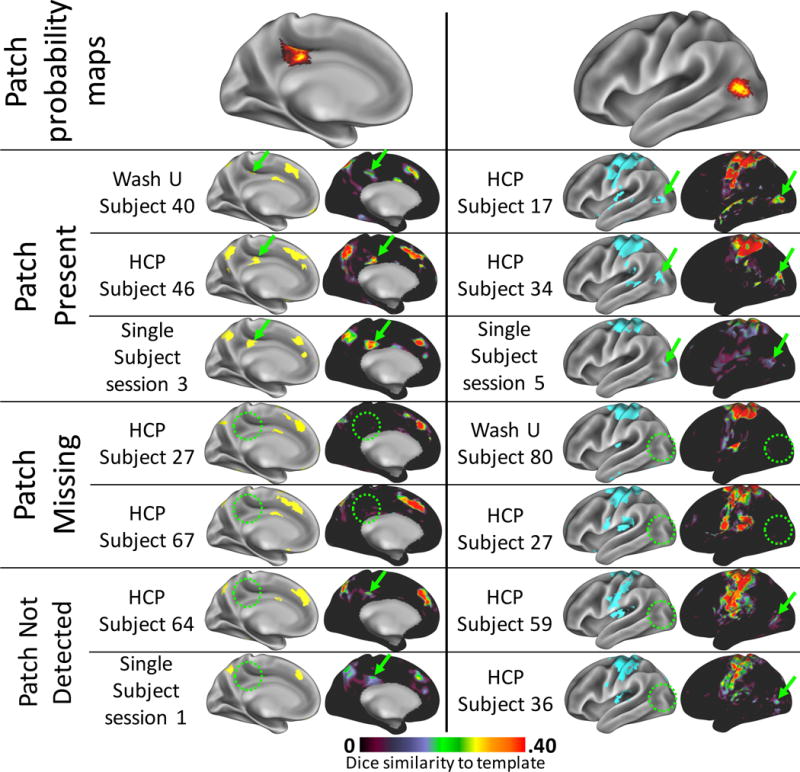
“Missing” patches appeared to be truly absent in some subjects, but failed to be identified in others. Top row: two example patches, one from the Fronto-parietal system (left) and one from the Dorsal Motor system (right), which were detected in ~50% of subjects. Rows 2–4: example high-data subjects and Single Subject dataset sessions in whom the patch was detected (green arrows). On the left are all patches in the brain system derived from the individual’s/session’s data. On the right are the Dice similarity maps to the template brain system, indicating how well each cortical vertex’s connectivity pattern matches the template system’s connectivity pattern. Regions of high Dice similarity can be clearly matched to the identified brain system patches. Rows 5–6: example high-data subjects in whom the patch was not detected (green circles). The Dice maps confirm that no vertices in that area were at all similar to the template systems, suggesting a truly missing system patch. No sessions from the Single Subject dataset could be found that were truly missing patches present in other sessions. Rows 7–8: Example high-data subjects and Single Subject sessions in whom the patch was not detected but may exist below the threshold for detection. Small, relatively weak regions of Dice similarity (green arrows) were present that could represent the patch, but were not identified as such (green circles).
3.5 System patches replicated across different system templates
We examined non-group system patches derived using two alternate sets of priors: a publicly-available group-average set of 17 systems derived via a clustering technique (Yeo et al., 2011) (“Yeo” systems; Figure 9, top), and a publicly available group-average set of 50 “systems” derived by applying a winner-take-all procedure to 50-dimensional ICA results; we restricted these to 29 template systems after removing very small components and those located entirely in non-cortical regions.
We found that of the forty-three replicating non-group patches that were found using the Infomap system templates (described above), thirty-three similar patches were also found using the Yeo system templates. These patches can be seen in Figure 9. This finding suggests that similar patches that are not present in group-average systems can be identified regardless of the template systems used. We quantitatively compared the overlap of the similar patches by calculating the Dice coefficient between the characteristic patches for each pair of similar patches. On average, the Dice overlap was .63 ± .23 (std deviation), indicating good overlap between similar patches. Similarly, we found that twenty-six replicating non-group patches found using the Infomap system templates were also found using the ICA system templates, with an average Dice overlap between matched patches of .63 ± .15 (std deviation). See Supplemental Figure 10 for examples of these matched ICA patches.
Further, we also observed that two other non-group patches found using the Infomap system templates represented regions that were actually included in the Yeo system maps (i.e. they were not non-group patches according to the Yeo templates). Similarly, of the patches defined using the Yeo systems, fifteen non-group patches corresponded with regions included in the Infomap systems. This finding, which is illustrated in Supplemental Figure 11, suggests that each set of systems describes real aspects of brain organization that are not well described by the other.
3.6 Using matched system patches as seeds increases the similarity of functional connectivity measures across subjects
To investigate whether using matched subject-specific system patches improves comparisons across subjects, we examined how whole-brain connectivity maps differed across subjects if they were seeded from matched subject-specific system patches rather than from the same region of interest in every subject.
Across all three datasets, when using matched system patches as seeds, the average pairwise similarity between subjects’ patch-to-patch correlation matrices was r = .62 ± .06. By contrast, when using the “characteristic patches” as seeds (which were identical for every subject), the average similarity between subjects’ patch-to-patch correlation matrices was r = .50 ± .06 (Figure 10A). Further, correlation matrices derived from matched system patches were more similar to each other than matrices derived from characteristic patches for 98.6% of pairwise subject-to-subject comparisons (Figure 10B). These findings suggest that matching system patches across subjects increases the specificity of regions used as seeds in functional connectivity analysis by accounting for the spatial variance of network features across subjects.
Figure 10.
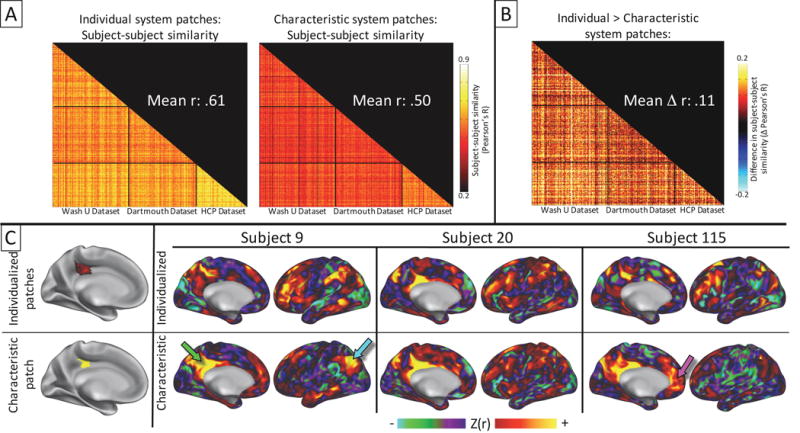
Across all three datasets, single-subject seed-based RSFC maps are more similar to each other if individualized system features are used as seeds than if common seeds are used, indicating that the identification and cross-subject matching of individualized system patches reduces variability in functional connectivity patterns. A) Pairwise subject-to-subject similarity of seed-to-seed correlation matrices calculated using matched system patches (left) and “characteristic” patches representing the central tendency of each patch location across subjects (right). Each row or column represents one subject, while correlation values represent the similarity between two subjects’ seed-to-seed correlation matrices. Black lines indicate divisions between the three different datasets. B) Improvement in pairwise subject-to-subject similarities using matched system patches vs. characteristic patches, calculated as the left similarity matrix from A) minus the right similarity matrix. 98.6% of these differences were positive, indicating that individuals were almost always more similar to each other when individual system patches were used as seeds than when characteristic patches were used. C) Examples of increased similarity of connectivity patterns when individualized seeds are used. For three subjects from the Wash U dataset, whole-brain seed-connectivity maps were calculated by seeding from matched, individual-specific Fronto-parietal patches in middle cingulate cortex, as well as from the characteristic patch. All maps seeded from the individualized seeds (top of each subject) demonstrated strong connectivity in canonical Fronto-parietal regions. However, some maps seeded from the characteristic patch (bottom of each subject) had less robust Fronto-parietal connections, but strong connectivity in non-Fronto-parietal regions such as posterior cingulate cortex (green arrow), angular gyrus (light blue arrow), or ventromedial prefrontal cortex (magenta arrow).
This phenomenon is illustrated for a specific patch in Figure 10C. For each subject, whole-brain connectivity maps seeded from the subject-specific, matched patches were compared to maps seeded from the corresponding characteristic patch. We observed that maps seeded from subject-specific Fronto-parietal system patches near middle cingulate cortex always had strong connectivity in classical Fronto-parietal regions, such as lateral frontal, inferior parietal, and occipito-temporal cortex. By contrast, maps seeded from the same patch across subjects sometimes differed substantially, demonstrating weaker connectivity in Fronto-parietal regions, but stronger connectivity in classical Default system regions such as posterior cingulate (green arrow), angular gyrus (light blue arrow), or ventromedial prefrontal cortex (see magenta arrow).
3.7 Non-group patches are not more hub-like than group patches
In each subject, we calculated participation coefficients (PCs) for all patches observed in the group average systems, as well as for all patches absent from the group systems, and we averaged PC values across subjects. We found that the PC values for non-group patches did not differ from those of group patches (non-group: mean PC = .26 ± .12 (std dev); group: mean PC = .26 ± .09 (std dev); t(177) = .22; p = .82), indicating that that these non-group patches are not more likely to be hubs than group patches.
4. DISCUSSION
4.1 Features of brain systems are spatially variable across individuals
In this work we characterized features of large-scale systems specific to individual human brains. We found that individuals’ brain systems had features that did not appear in group-averaged systems maps, but that could be reliably detected within an individual and, importantly, could be matched across many individuals much more frequently than would be expected by a spatially random distribution of features (Figure 2C). The fact that these individual-specific system features systematically appear across subjects in specific locations rather than randomly across the cortex suggests that they represent genuine features of the brain’s functional organization.
Several previous studies have made important progress in describing inter-individual variance in RSFC patterns (Mennes et al., 2010; Mueller et al., 2013), including demonstrating that such variance can uniquely identify individuals from each other (Finn et al., 2015). Further, recent individual-level investigations have observed that the brain systems of individuals vary categorically, rather than continuously, from each other by the presence, absence, or spatial variability of specific system features that cannot be observed in group-average data (Gordon et al., 2015; Harrison et al., 2015; Laumann et al., 2015; Satterthwaite and Davatzikos, 2015; D. Wang et al., 2015). The present work expands on these findings by demonstrating both the reliability of such individual-specific features within a single subject and the convergence of such features across subjects. In identifying and characterizing such individual-specific features across the entire cortex (Figure 7C, Table 2), these findings are an important reference for future work making claims about the system identity of a region of cortex.
Within a single individual, the spatial locations of these individual-specific features were reliable across scanning sessions (Figure 4). However, across individuals, many of these features also had large spatial distributions relative to their size, indicating that they were in different locations on the aligned cortical surface from individual to individual (Figure 5). High spatial variability of a system feature has been hypothesized to impair detection of that feature in group-average data, as vertex-wise averaging will blur that feature with different system features in other subjects (Laumann et al., 2015).
Spatial variability of system features is likely influenced by two independent factors. First, these features may be spatially distributed because of imperfect inter-individual registration techniques that may register data from different anatomical regions to the same spatial point. The surface-based alignment approach employed here (Glasser et al., 2013; Van Essen et al., 2012a) is the current state-of-the-art anatomical registration, but is still imperfect, particularly in anatomically variable regions such as prefrontal cortex. However, we have shown in previous work that the quality of this registration does not substantially explain variance in brain system estimates (Gordon et al., 2015).
Second, system features may be spatially distributed because they reflect the variable positions of cytoarchitectonic cortical areas, which are the discrete units of cortex that interact to form large-scale systems (Sejnowski and Churchland, 1989). Like the system features described here, cortical areas are spatially variable across individuals relative to gyral anatomy (Caspers et al., 2008; Fischl et al., 2008; Malikovic et al., 2007; Van Essen et al., 2012a). Further, published probabilistic maps of specific highly variable cortical areas (e.g., area 44 (Fischl et al., 2008); area hOc5 (Van Essen et al., 2012a)) appear to correspond with some of the probabilistic maps of system features described here (respectively, Ventral Attention system, right hemisphere inferior frontal gyrus; and Dorsal Motor system, bilateral lateral occipital cortex; see Figure 5). The possibility that some of these system features may represent subject-specific cortical areas remains to be explicitly tested in the future.
4.2 Features of brain systems were not always observed in every subject
Beyond being spatially variable, brain systems were also topologically variable across individuals, such that some system features were absent in a given individual. Indeed, some system features clustered together across subjects significantly better than random, but still could be detected in fewer than half of subjects (Figure 2C, Table 2). While detection of system features may have failed in some scanning sessions due to methodological choices and/or intra-individual variability in connectivity strengths (see Figure 8, bottom for examples), system features appeared to be truly absent in many other subjects (Figure 8, middle).
One possible explanation for a system feature being present in one individual but absent in another is that the regions of cortex underlying that feature (potentially cortical areas, as discussed above) may be functionally related to different systems in the two individuals. Such variant function has previously been observed in individuals with developmental experiences that differ dramatically from the general population. For example, extrastriate visual cortex responds during auditory tasks in congenitally blind individuals (Amedi et al., 2003; Gougoux et al., 2009; Röder et al., 2002), and demonstrates elevated connectivity with widespread regions in frontal cortex (X. Wang et al., 2015; Heine et al., 2015). The present data suggest less extreme versions of the same effect may be present within a healthy population, such that an individual with different genetics or life experiences could demonstrate variant functional connectivity patterns.
Together, these findings strongly suggest that small features of functional brain systems detected during the resting state not only vary across subjects in their size, shape, and spatial position, but also in their function (i.e., which brain system they are coupled to). Notably, this work converges with recent findings by Tavor et al. (2016) showing that individual variability in the size, shape, spatial position, and local presence of functional task responses can be predicted by applying machine-learning techniques to RSFC data. We hypothesize that the features of the RSFC data that predict task responses in that work are the same variable system features we describe here.
4.3 Individual-level brain system features replicated across multiple datasets and template systems
Most identified system features—including those not observed in group-average systems— replicated across three independent datasets collected with different sequences on different scanners and processed with different pipelines. This replication was observed not only in the cortical location of the features, but also in the shape of the features’ spatial distribution (Figure 6, Supplemental Figure 9), in the median sizes of the features, and in the percent of subjects in which the features were observed.
A similar result was observed when comparing system features identified using three different sets of system priors. The first prior was generated by applying the Infomap procedure to a group-average of the Wash U dataset; the second prior is a publicly available set of 17 brain systems (Yeo et al., 2011); and the third prior is a publicly available set of ICA components calculated from Human Connectome Project data (Smith et al., 2013). Many similar system features absent from the templates were detected regardless of which template was used (Figure 9; Supplemental Figure 10). In some cases, any set of system templates could be used to identify individual-specific system features present in one but not all template sets (Supplemental Figure 11). This suggests that each brain system map may describe real features of individual-specific brain systems that are not well described by the others, though this hypothesis should ideally be tested in individual subjects using a more data-driven approach (see “Limitations” below).
Together, these results indicate that the individual-specific system features described here represent meaningful functional objects that are similarly probabilistically present and spatially distributed in any sufficiently large group of healthy adults, and that they can be detected using multiple priors.
4.4 Matching system features can help control for inter-individual variance in brain systems
Individual variability in brain systems is an important consideration for neuroimaging studies (Gordon et al., 2015; Laumann et al., 2015; Satterthwaite and Davatzikos, 2015; Wang and Liu, 2014). If neurobiological objects of interest are spatially variable across individuals, then in any given subject it is unclear whether a weaker observed response in a brain region indicates altered function, or whether it indicates a poorly aligned system feature. We showed that allowing seed regions of interest to vary across individuals according to features of their brain systems maps results in more similar seed-based connectivity patterns across individuals and datasets (Figure 10). This suggests that utilizing information about subject-specific features of brain systems can help reduce the portion of inter-subject variability in functional connectivity that is driven by variable spatial positions of functional units.
Several recent publications have focused on developing techniques for similarly reducing or eliminating spatial variability by registering individual brains to each other based on functional responses, including RSFC patterns—in other words, conducting a functional registration. Techniques developed by Sabuncu et al. (2010) and Conroy et al. (2013) warp pre-anatomically-aligned fMRI data along the cortical sheet by maximizing the similarity of functional responses, while approaches developed by Frost and Goebel (2013) and Robinson et al. (2014) integrate the functional alignment with the anatomical alignment procedure to accomplish a single-step cross-subject functional/anatomic registration. Other approaches have discarded the idea of registering subjects into an anatomical space, instead using fMRI responses to warp individuals into an abstract, high-dimensional space that can be projected back into the anatomical space of any individual subject (Guntupalli et al., 2016; Haxby et al., 2011; Langs et al., 2015). These methods have all demonstrated successful improvements in inter-subject alignment. However, by rigorously aligning functional features across individuals, these approaches do not account for the possibility that some brain system features might not be identifiable in all individuals (whether due to technical limitations, intra-individual variability, or true neurobiological differences).
By contrast, the inter-subject feature-matching procedure described here is not in and of itself a functional registration, as it does not put all subjects into a true common space—all matching is directly subject-to-subject. However, this procedure could form the basis for a functional registration that both removes inter-individual spatial variance and accounts for the possibility of similar functional regions demonstrating variant function across individuals, thus allowing better direct comparison of functionally similar regions.
4.5 Post-hoc correspondence of unusual system features with known cortical functions
Several of the features described here appear to represent the presence of brain systems in unusual cortical locations. For example, we demonstrated that more than half of our subjects have patches of Visual system outside of occipital cortex, in both bilateral superior parietal and bilateral precentral gyrus. We further observed patches of Fronto-parietal system that consistently emerged in bilateral posterior cingulate cortex, an area usually thought of as the core of the Default system.
While these regions seem at first to be dubious locations for such systems to emerge, we argue that that the observed unexpected system identities are plausibly explained by previous work at the single-subject level. For example, Meier et al. (2008) demonstrated that a region of precentral gyrus of almost the exact same size, shape, and position as our precentral Visual system patch was engaged by the motor control of squinting behaviors in single subjects. Sereno et al. (2001) demonstrated the existence of a retinotopic map preferentially engaged by attention-demanding stimuli that is in the same area of superior parietal cortex as our patch of Visual system; this finding has been confirmed by Hagler et al. (2007) and Swisher et al. (2007). And finally, Tavor et al. (2016) recently demonstrated that, along with the rest of the Fronto-parietal system, a region in posterior cingulate cortex of nearly the exact size, shape, and position as our Fronto-parietal patch activated more to a math task than a language task, but only in specific subjects. See Figure 11 for illustrations of these corresponding regions. The fact that some of the system features we identify here correspond closely with localized cortical regions of known function supports the idea that these features may represent real, discrete, functionally meaningful regions on the cortex. However, future work comparing across multiple highly sampled single subject data sets is needed to confirm that the system features described here overlap with functional responses elicited in the same subjects.
Figure 11.
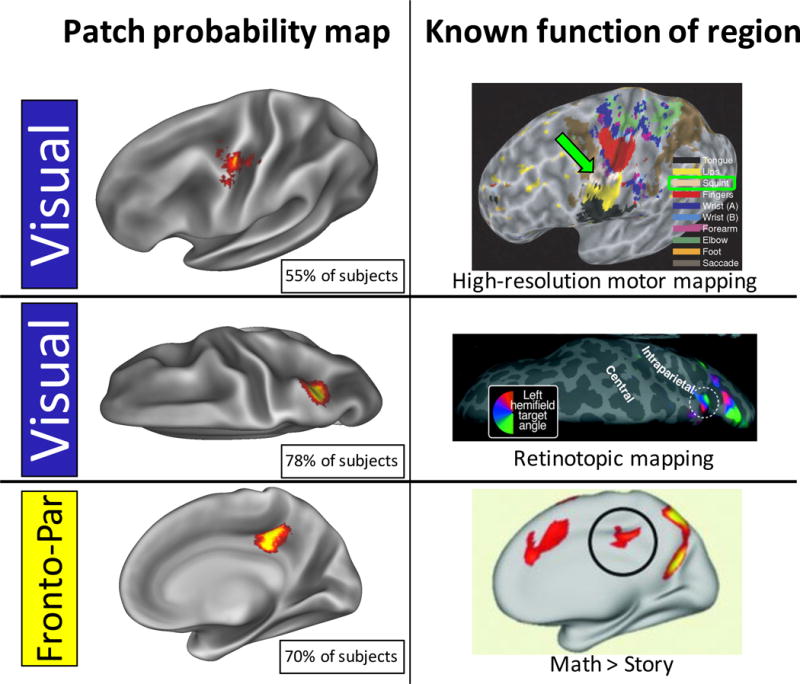
Probability maps of several system features correspond to previous findings demonstrating subject-specific localization of various functions. Top row: a Visual system patch in precentral gyrus (left) appears very similar to a single-subject map of squinting (right, beige colored; green arrow) published in Meier et al., 2008. Middle row: a Visual system patch in posterior intraparietal sulcus (left) appears in a very similar location to a subject-specific, attention-dependent retinotopic map (right) described by Sereno et al. (2001). Bottom row: A Fronto-parietal system patch in posterior cingulate cortex (left) has a very similar location and shape to a region in a subject-specific map of increased activation during a math task compared to a language task (right), as described by Tavor et al. (2016). Figures adapted with permission from Meier et al., 2008, Sereno et al., 2001, and Tavor et al., 2016.
It is noteworthy that these previous findings all come from examinations of single subjects. Indeed, in each of these previous findings, illustration of the same effect in multiple subjects has demonstrated either a poor overlap of the functional response across subjects (Hagler et al., 2007; Meier et al., 2008; Sereno et al., 2001; Swisher et al., 2007) or an absence of detectable response in some subjects in that region of cortex (Tavor et al., 2016). This is very similar to our finding that these small system patches both overlap poorly across subjects and are not present in all subjects, and it reinforces the idea that some features of brain systems cannot be easily detected except via detailed examination of single subjects.
4.6 Limitations
The approaches described here are primarily intended to identify regularities in brain systems across individuals. As such, we do not make strong claims about the accuracy of systems identified in any single subject in our cross-subject datasets. We selected a template-matching approach to estimate cortical systems in these individual subjects because 1) it assigns systems based on strong positive connectivities only, following graph-theory based conceptualizations of networks; and 2) it enforces identical brain systems across individuals, which is required for the cross-subject comparisons employed here. However, despite their popularity, it is possible that these specific graph-theory approaches may not be the best way to describe brain systems. Further, the use of a strong prior in the present technique may be obscuring some of the more subtle aspects of individual-specific brain systems. In particular, it is possible that this approach may slightly bias the systems detected in individuals to look more like the group-derived systems; and further, since the templates were derived from only the Wash U dataset, it may subtly bias individuals in other datasets to look more like the Wash U subjects. Accordingly, we do not claim that this technique is the best possible description of individual brain systems. However, we note that such potential biases would not account for the observation of system features not present in any prior. Further, given the convergence we observed between template matching-derived systems and systems derived from a widely employed data-driven approach (see Supplemental Figure 3), we argue that the template matching approach is at least an adequate technique for brain systems identification, and that resulting system maps are well-suited for matching across subjects. We hope that future work will clarify which of many proposed methods (e.g., (Beckmann and Smith, 2004; Filippini et al., 2009; Gordon et al., 2015; Hacker et al., 2013; Harrison et al., 2015; Laumann et al., 2015; D. Wang et al., 2015) are optimal for brain system identification in single individuals.
The accuracy of system estimation in a specific subject may also be affected by data limitations. Reliable estimation of single-subject RSFC patterns requires upwards of 25 minutes of data (Anderson et al., 2011; Laumann et al., 2015; Mueller et al., 2015), which only a minority of our subjects have. Accordingly, our analyses of within-subject reliability suggested that system maps derived from 10 minutes of data (an amount similar to many of our lower-data subjects from the Wash U and Dartmouth datasets) have reduced vertex-wise reliability (Supplemental Figure 4). While we did find that most system features present in 300 minutes of data were reliably detected even in 10-minute sessions (i.e. system feature detection has relatively high sensitivity), we cannot rule out the possibility of “extra” system features appearing in noisy, low-data RSFC estimates (i.e., low specificity). Further, the sensitivity of this detection procedure was enhanced by comparing it to a robust set of system estimates derived from 300 minutes of data; comparing 10-minute sessions to 10-minute sessions would have resulted in lower reliability of system feature detection. For the purposes of matching system features across subjects, we anticipate that relatively low amounts of data could be used, but that lower data quantity will produce noisier, less accurate network maps with worse cross-subject agreement in each patch. We note, for example, how in Figure 6, cross-subject overlaps appear markedly better in the higher-data HCP dataset than in the lower-data Wash U and Dartmouth datasets.
To reduce the influence of such noise on identified cross-subject features, we required a minimum percentage of subjects to have any given system feature, as empirically determined by comparison to a null model developed by rotating real system patches randomly around the cortical surface. This thresholding appeared effective, as features identified in the low-data Wash U. and Dartmouth datasets were extremely similar to those identified in the higher-data HCP dataset. However, this cutoff makes it impossible to identify possible unique or very rare features that might be present in only a few subjects, which would not be easily distinguishable from noise. Future work may characterize rare features of brain systems by increasing the reliability of system estimates using extensive single-subject sampling, as in our Single Subject dataset (Laumann et al., 2015).
4.7 Conclusions
Here we describe individual-specific features of large-scale brain systems that emerged consistently within and across individuals, datasets, and choice of priors for systems definition. The exact significance of these system features is not yet clear, but one exciting possibility is that they may represent cortical areas, or combinations of adjacent cortical areas, found in individual brains. If this is true—a possibility that should be examined in future work—it opens the door for detailed investigations of functionally aligned cortical areas in individual humans, including investigating the possibility that some cortical areas may have variant functional relationships with large-scale brain systems across individuals.
Code to perform the template matching procedure for single-subject system identification, cross-subject patch-matching and clustering, and the rotation-based null model for significance testing are available at http://www.nil.wustl.edu/labs/petersen/Resources.html.
Supplementary Material
Highlights.
Features of brain systems identified in individuals are absent from group averages
These features were both reliable within a single subject and present across subjects
These features were observed across three independent datasets
Some subjects were “missing” system features, suggesting variable system connections
Matching system features between individuals increased inter-individual similarity
Acknowledgments
This work was supported by the National Institutes of Health (grant numbers NS061144 and NS046424 to S.E.P.; MH100872 to T.O.L; NS088590 to N.U.F.D.; and MH091657 to David Van Essen); the McDonnell Foundation (Collaborative Action Award to S.E.P.); the Simons Foundation (Award 95177 to S.E.P.); the Mallinckrodt Institute of Radiology (Pilot Grant to N.U.F.D.); the Child Neurology Foundation (Scientific Research Award to N.U.F.D.); and the National Science Foundation Graduate Research Fellowship Program (grant number DGE-1143954 to A.W.G.). We would further like to acknowledge Bill Kelley and Jeremy Huckins for providing the Dartmouth Dataset; Matt Glasser for contributions to the procedures to generate cortical surfaces and map data to the surface; and Russell Poldrack for providing examples of high-quality single-subject data that motivated this investigation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amedi A, Raz N, Pianka P, Malach R, Zohary E. Early “visual” cortex activation correlates with superior verbal memory performance in the blind. Nat Neurosci. 2003;6:758–766. doi: 10.1038/nn1072. [DOI] [PubMed] [Google Scholar]
- Anderson JS, Ferguson MA, Lopez-Larson M, Yurgelun-Todd D. Reproducibility of single-subject functional connectivity measurements. Am J Neuroradiol. 2011;32:548–555. doi: 10.3174/ajnr.A2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann CF, Smith SM. Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans Med Imaging. 2004;23:137–52. doi: 10.1109/TMI.2003.822821. [DOI] [PubMed] [Google Scholar]
- Bertolero MA, Yeo BTT, D’Esposito M. The modular and integrative functional architecture of the human brain. Proc Natl Acad Sci. 2015;112:E6798–E6807. doi: 10.1073/pnas.1510619112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal BB, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34:537–41. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Carp J. Optimizing the order of operations for movement scrubbing: Comment on Power et al. NeuroImage. 2013;76:436–438. doi: 10.1016/j.neuroimage.2011.12.061. [DOI] [PubMed] [Google Scholar]
- Caspers S, Eickhoff SB, Geyer S, Scheperjans F, Mohlberg H, Zilles K, Amunts K. The human inferior parietal lobule in stereotaxic space. Brain Struct Funct. 2008;212:481–495. doi: 10.1007/s00429-008-0195-z. [DOI] [PubMed] [Google Scholar]
- Conroy BR, Singer BD, Guntupalli JS, Ramadge PJ, Haxby JV. Inter-subject alignment of human cortical anatomy using functional connectivity. NeuroImage. 2013;81:400–411. doi: 10.1016/j.neuroimage.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI. Cortical Surface-Based Analysis: I. Segmentation and Surface Reconstruction. NeuroImage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Dale AM, Sereno MI. Improved Localizadon of Cortical Activity by Combining EEG and MEG with MRI Cortical Surface Reconstruction: A Linear Approach. J Cogn Neurosci. 1993;5:162–176. doi: 10.1162/jocn.1993.5.2.162. [DOI] [PubMed] [Google Scholar]
- Dosenbach NUF, Fair DA, Miezin FM, Cohen AL, Wenger KK, Dosenbach RAT, Fox MD, Snyder AZ, Vincent JL, Raichle ME, Schlaggar BL, Petersen SE. Distinct brain networks for adaptive and stable task control in humans. Proc Natl Acad Sci. 2007;104:11073–11078. doi: 10.1073/pnas.0704320104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois J, Adolphs R. Building a Science of Individual Differences from fMRI. Trends Cogn Sci. 2016 doi: 10.1016/j.tics.2016.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippini N, MacIntosh BJ, Hough MG, Goodwin GM, Frisoni GB, Smith SM, Matthews PM, Beckmann CF, Mackay CE. Distinct patterns of brain activity in young carriers of the APOE-ε4 allele. Proc Natl Acad Sci U S A. 2009;106:7209–14. doi: 10.1073/pnas.0811879106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn ES, Shen X, Scheinost D, Rosenberg MD, Huang J, Chun MM, Papademetris X, Constable RT. Functional connectome fingerprinting: identifying individuals using patterns of brain connectivity. Nat Neurosci advance online publication. 2015 doi: 10.1038/nn.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Rajendran N, Busa E, Augustinack J, Hinds O, Yeo BTT, Mohlberg H, Amunts K, Zilles K. Cortical Folding Patterns and Predicting Cytoarchitecture. Cereb Cortex. 2008;18:1973–1980. doi: 10.1093/cercor/bhm225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale AM. Cortical Surface-Based Analysis: II: Inflation, Flattening, and a Surface-Based Coordinate System. NeuroImage. 1999;9:195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Williams S, Howard R, Frackowiak RSJ, Turner R. Movement-Related effects in fMRI time-series. Magn Reson Med. 1996;35:346–355. doi: 10.1002/mrm.1910350312. [DOI] [PubMed] [Google Scholar]
- Frost MA, Goebel R. Functionally informed cortex based alignment: an integrated approach for whole-cortex macro-anatomical and ROI-based functional alignment. NeuroImage. 2013;83:1002–1010. doi: 10.1016/j.neuroimage.2013.07.056. [DOI] [PubMed] [Google Scholar]
- Frost MA, Goebel R. Measuring structural–functional correspondence: Spatial variability of specialised brain regions after macro-anatomical alignment. NeuroImage. 2012;59:1369–1381. doi: 10.1016/j.neuroimage.2011.08.035. [DOI] [PubMed] [Google Scholar]
- Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, Xu J, Jbabdi S, Webster M, Polimeni JR, Van Essen DC, Jenkinson M, WU-Minn HCP Consortium The minimal preprocessing pipelines for the Human Connectome Project. NeuroImage. 2013;80:105–124. doi: 10.1016/j.neuroimage.2013.04.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Van Essen DC. Mapping human cortical areas in vivo based on myelin content as revealed by T1- and T2-weighted MRI. J Neurosci. 2011;31:11597–11616. doi: 10.1523/JNEUROSCI.2180-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon EM, Laumann TO, Adeyemo B, Huckins JF, Kelley WM, Petersen SE. Generation and Evaluation of a Cortical Area Parcellation from Resting-State Correlations. Cereb Cortex. 2016;26:288–303. doi: 10.1093/cercor/bhu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon EM, Laumann TO, Adeyemo B, Petersen SE. Individual Variability of the System-Level Organization of the Human Brain. Cereb Cortex Advance access. 2015 doi: 10.1093/cercor/bhv239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gougoux F, Belin P, Voss P, Lepore F, Lassonde M, Zatorre RJ. Voice perception in blind persons: a functional magnetic resonance imaging study. Neuropsychologia. 2009;47:2967–2974. doi: 10.1016/j.neuropsychologia.2009.06.027. [DOI] [PubMed] [Google Scholar]
- Gratton C, Nomura EM, Pérez F, D’Esposito M. Focal Brain Lesions to Critical Locations Cause Widespread Disruption of the Modular Organization of the Brain. J Cogn Neurosci. 2012;24:1275–1285. doi: 10.1162/jocn_a_00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimerà R, Nunes Amaral LA. Functional cartography of complex metabolic networks. Nature. 2005;433:895–900. doi: 10.1038/nature03288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimerà R, Sales-Pardo M, Amaral LAN. Classes of complex networks defined by role-to-role connectivity profiles. Nat Phys. 2007;3:63–69. doi: 10.1038/nphys489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guntupalli JS, Hanke M, Halchenko YO, Connolly AC, Ramadge PJ, Haxby JV. A Model of Representational Spaces in Human Cortex. Cereb Cortex. 2016;26:2919–2934. doi: 10.1093/cercor/bhw068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker CD, Laumann TO, Szrama NP, Baldassarre A, Snyder AZ, Leuthardt EC, Corbetta M. Resting state network estimation in individual subjects. Neuroimage. 2013;82:616–633. doi: 10.1016/j.neuroimage.2013.05.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagler DJ, Riecke L, Sereno MI. Parietal and superior frontal visuospatial maps activated by pointing and saccades. NeuroImage. 2007;35:1562–1577. doi: 10.1016/j.neuroimage.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SJ, Woolrich MW, Robinson EC, Glasser MF, Beckmann CF, Jenkinson M, Smith SM. Large-scale Probabilistic Functional Modes from resting state fMRI. NeuroImage. 2015;109:217–231. doi: 10.1016/j.neuroimage.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haxby JV, Guntupalli JS, Connolly AC, Halchenko YO, Conroy BR, Gobbini MI, Hanke M, Ramadge PJ. A common, high-dimensional model of the representational space in human ventral temporal cortex. Neuron. 2011;72:404–416. doi: 10.1016/j.neuron.2011.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine L, Bahri MA, Cavaliere C, Soddu A, Laureys S, Ptito M, Kupers Ron. Prevalence of increases in functional connectivity in visual, somatosensory and language areas in congenital blindness. Front Neuroanat. 2015;9:86. doi: 10.3389/fnana.2015.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird AR, Fox PM, Eickhoff SB, Turner JA, Ray KL, McKay DR, Glahn DC, Beckmann CF, Smith SM, Fox PT. Behavioral Interpretations of Intrinsic Connectivity Networks. J Cogn Neurosci. 2011;23:4022–4037. doi: 10.1162/jocn_a_00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster JL, Glass TG, Lankipalli BR, Downs H, Mayberg H, Fox PT. A modality-independent approach to spatial normalization of tomographic images of the human brain. Hum Brain Mapp. 1995;3:209–223. doi: 10.1002/hbm.460030305. [DOI] [Google Scholar]
- Langs G, Wang D, Golland P, Mueller S, Pan R, Sabuncu MR, Sun W, Li K, Liu H. Identifying Shared Brain Networks in Individuals by Decoupling Functional and Anatomical Variability. Cereb Cortex Advance Access. 2015 doi: 10.1093/cercor/bhv189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumann TO, Gordon EM, Adeyemo B, Snyder AZ, Joo SJ, Chen MY, Gilmore AW, McDermott KB, Nelson SM, Dosenbach NUF, Schlaggar BL, Mumford JA, Poldrack RA, Petersen SE. Functional System and Areal Organization of a Highly Sampled Individual Human Brain. Neuron. 2015;87:657–670. doi: 10.1016/j.neuron.2015.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malikovic A, Amunts K, Schleicher A, Mohlberg H, Eickhoff SB, Wilms M, Palomero-Gallagher N, Armstrong E, Zilles K. Cytoarchitectonic Analysis of the Human Extrastriate Cortex in the Region of V5/MT+: A Probabilistic, Stereotaxic Map of Area hOc5. Cereb Cortex. 2007;17:562–574. doi: 10.1093/cercor/bhj181. [DOI] [PubMed] [Google Scholar]
- Meier JD, Aflalo TN, Kastner S, Graziano MSA. Complex Organization of Human Primary Motor Cortex: A High-Resolution fMRI Study. J Neurophysiol. 2008;100:1800–1812. doi: 10.1152/jn.90531.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennes M, Kelly C, Zuo XN, Di Martino A, Biswal BB, Castellanos FX, Milham MP. Inter-individual differences in resting-state functional connectivity predict task-induced BOLD activity. NeuroImage. 2010;50:1690–1701. doi: 10.1016/j.neuroimage.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miezin FM, Maccotta L, Ollinger JM, Petersen SE, Buckner RL. Characterizing the Hemodynamic Response: Effects of Presentation Rate, Sampling Procedure, and the Possibility of Ordering Brain Activity Based on Relative Timing. NeuroImage. 2000;11:735–759. doi: 10.1006/nimg.2000.0568. [DOI] [PubMed] [Google Scholar]
- Mueller S, Wang D, Fox MD, Pan R, Lu J, Li K, Sun W, Buckner RL, Liu H. Reliability correction for functional connectivity: Theory and implementation. Hum Brain Mapp. 2015;36:4664–4680. doi: 10.1002/hbm.22947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S, Wang D, Fox MD, Yeo BTT, Sepulcre J, Sabuncu MR, Shafee R, Lu J, Liu H. Individual Variability in Functional Connectivity Architecture of the Human Brain. Neuron. 2013;77:586–595. doi: 10.1016/j.neuron.2012.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugler JP, 3rd, Brookeman JR. Three-dimensional magnetization-prepared rapid gradient-echo imaging (3D MP RAGE) Magn Reson Med. 1990;15:152–157. doi: 10.1002/mrm.1910150117. [DOI] [PubMed] [Google Scholar]
- Ojemann JG, Akbudak E, Snyder AZ, McKinstry RC, Raichle ME, Conturo TE. Anatomic localization and quantitative analysis of gradient refocused echo-planar fMRI susceptibility artifacts. NeuroImage. 1997;6:156–167. doi: 10.1006/nimg.1997.0289. [DOI] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. NeuroImage. 2012;59:2142–54. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, Vogel AC, Laumann TO, Miezin FM, Schlaggar BL, Petersen SE. Functional Network Organization of the Human Brain. Neuron. 2011;72:665–678. doi: 10.1016/j.neuron.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Mitra A, Laumann TO, Snyder AZ, Schlaggar BL, Petersen SE. Methods to detect, characterize, and remove motion artifact in resting state fMRI. NeuroImage. 2014;84:320–341. doi: 10.1016/j.neuroimage.2013.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson EC, Jbabdi S, Glasser MF, Andersson J, Burgess GC, Harms MP, Smith SM, Van Essen DC, Jenkinson M. MSM: a new flexible framework for Multimodal Surface Matching. NeuroImage. 2014;100:414–426. doi: 10.1016/j.neuroimage.2014.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röder B, Stock O, Bien S, Neville H, Rösler F. Speech processing activates visual cortex in congenitally blind humans. Eur J Neurosci. 2002;16:930–936. doi: 10.1046/j.1460-9568.2002.02147.x. [DOI] [PubMed] [Google Scholar]
- Rosvall M, Bergstrom CT. Maps of random walks on complex networks reveal community structure. Proc Natl Acad Sci. 2008;105:1118–1123. doi: 10.1073/pnas.0706851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M, Sporns O. Complex network measures of brain connectivity: Uses and interpretations. NeuroImage. 2010;52:1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Sabuncu MR, Singer BD, Conroy B, Bryan RE, Ramadge PJ, Haxby JV. Function-based Intersubject Alignment of Human Cortical Anatomy. Cereb Cortex. 2010;20:130–140. doi: 10.1093/cercor/bhp085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite TD, Davatzikos C. Towards an Individualized Delineation of Functional Neuroanatomy. Neuron. 2015;87:471–473. doi: 10.1016/j.neuron.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ségonne F, Dale AM, Busa E, Glessner M, Salat D, Hahn HK, Fischl B. A hybrid approach to the skull stripping problem in MRI. NeuroImage. 2004;22:1060–1075. doi: 10.1016/j.neuroimage.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Ségonne F, Grimson E, Fischl B. A Genetic Algorithm for the Topology Correction of Cortical Surfaces. In: Christensen GE, Sonka M, editors. Information Processing in Medical Imaging Presented at the 19th International Conference; IPMI, Springer, Glenwood Springs, CO, USA. 2005. p. 393. [DOI] [PubMed] [Google Scholar]
- Sejnowski TJ, Churchland PS. Brain and cognition. In: Posner MI, editor. Foundations of Cognitive Science. MIT Press; Cambridge, MA: 1989. p. 888. [Google Scholar]
- Sereno MI, Pitzalis S, Martinez A. Mapping of Contralateral Space in Retinotopic Coordinates by a Parietal Cortical Area in Humans. Science. 2001;294:1350–1354. doi: 10.1126/science.1063695. [DOI] [PubMed] [Google Scholar]
- Shehzad Z, Kelly AMC, Reiss PT, Gee DG, Gotimer K, Uddin LQ, Lee SH, Margulies DS, Roy AK, Biswal BB, Petkova E, Castellanos FX, Milham MP. The Resting Brain: Unconstrained yet Reliable. Cereb Cortex. 2009;19:2209–2229. doi: 10.1093/cercor/bhn256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Beckmann CF, Andersson J, Auerbach EJ, Bijsterbosch J, Douaud G, Duff E, Feinberg DA, Griffanti L, Harms MP, Kelly M, Laumann T, Miller KL, Moeller S, Petersen S, Power J, Salimi-Khorshidi G, Snyder AZ, Vu AT, Woolrich MW, Xu J, Yacoub E, Uğurbil K, Van Essen DC, Glasser MF. Resting-state fMRI in the Human Connectome Project. NeuroImage, Mapping the Connectome. 2013;80:144–168. doi: 10.1016/j.neuroimage.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, Filippini N, Watkins KE, Toro R, Laird AR, Beckmann CF. Correspondence of the brain’s functional architecture during activation and rest. Proc Natl Acad Sci. 2009;106:13040–13045. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TEJ, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, Matthews PM. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage. 2004;23:S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Snyder AZ. Difference image versus ratio image error function forms in PET-PET realignment. In: Myer R, Cunningham VJ, Bailey DL, Jones T, editors. Quantification of Brain Function Using PET. Academic Press; San Diego, CA: 1996. pp. 131–137. [Google Scholar]
- Swisher JD, Halko MA, Merabet LB, McMains SA, Somers DC. Visual Topography of Human Intraparietal Sulcus. J Neurosci. 2007;27:5326–5337. doi: 10.1523/JNEUROSCI.0991-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talairach J, Tournoux P. Co-planar stereotaxic atlas of the human brain. Thieme Medical Publishers, Inc; New York: 1988. [Google Scholar]
- Tavor I, Jones OP, Mars RB, Smith SM, Behrens TE, Jbabdi S. Task-free MRI predicts individual differences in brain activity during task performance. Science. 2016;352:216–220. doi: 10.1126/science.aad8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk KRA, Hedden T, Venkataraman A, Evans KC, Lazar SW, Buckner RL. Intrinsic Functional Connectivity As a Tool For Human Connectomics: Theory, Properties, and Optimization. J Neurophysiol. 2010;103:297–321. doi: 10.1152/jn.00783.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Drury HA, Dickson J, Harwell J, Hanlon D, Anderson CH. An Integrated Software Suite for Surface-based Analyses of Cerebral Cortex. J Am Med Inform Assoc. 2001;8:443–459. doi: 10.1136/jamia.2001.0080443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Glasser MF, Dierker DL, Harwell J, Coalson T. Parcellations and Hemispheric Asymmetries of Human Cerebral Cortex Analyzed on Surface-Based Atlases. Cereb Cortex. 2012a;22:2241–2262. doi: 10.1093/cercor/bhr291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Smith SM, Barch DM, Behrens TEJ, Yacoub E, Ugurbil K, WU-Minn HCP Consortium The WU-Minn Human Connectome Project: an overview. NeuroImage. 2013;80:62–79. doi: 10.1016/j.neuroimage.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Ugurbil K, Auerbach E, Barch D, Behrens TEJ, Bucholz R, Chang A, Chen L, Corbetta M, Curtiss SW, Della Penna S, Feinberg D, Glasser MF, Harel N, Heath AC, Larson-Prior L, Marcus D, Michalareas G, Moeller S, Oostenveld R, Petersen SE, Prior F, Schlaggar BL, Smith SM, Snyder AZ, Xu J, Yacoub E. The Human Connectome Project: A data acquisition perspective. NeuroImage, Connectivity Connectivity. 2012b;62:2222–2231. doi: 10.1016/j.neuroimage.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Buckner RL, Fox MD, Holt DJ, Holmes AJ, Stoecklein S, Langs G, Pan R, Qian T, Li K, Baker JT, Stufflebeam SM, Wang K, Wang X, Hong B, Liu H. Parcellating cortical functional networks in individuals. Nat Neurosci. 2015;18:1853–1860. doi: 10.1038/nn.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Liu H. Functional Connectivity Architecture of the Human Brain Not All the Same. The Neuroscientist. 2014;20:432–438. doi: 10.1177/1073858414543290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Peelen MV, Han Z, He C, Caramazza A, Bi Y. How Visual Is the Visual Cortex? Comparing Connectional and Functional Fingerprints between Congenitally Blind and Sighted Individuals. J Neurosci. 2015;35:12545–12559. doi: 10.1523/JNEUROSCI.3914-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren DE, Power JD, Bruss J, Denburg NL, Waldron EJ, Sun H, Petersen SE, Tranel D. Network measures predict neuropsychological outcome after brain injury. Proc Natl Acad Sci. 2014;111:14247–14252. doi: 10.1073/pnas.1322173111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wig GS, Laumann TO, Petersen SE. An approach for parcellating human cortical areas using resting-state correlations. NeuroImage. 2014;93:276–291. doi: 10.1016/j.neuroimage.2013.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo BTT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zöllei L, Polimeni JR, Fischl B, Liu H, Buckner RL. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106:1125–1165. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


