Abstract
KIR2DP1 is an inactive member of the human lineage III KIR family, which includes all HLA-C specific receptors. The lethal, and only, defect in KIR2DP1 is a nucleotide deletion in codon 88. Fixed in modern humans, the deletion is also in archaic human genomes. KIR2DP1 is polymorphic, with dimorphism at specificity-determining position 44. By repairing the deletion, we resurrected eleven alleles of KIR2DP1F, the functional antecedent of KIR2DP1. We demonstrate how K44-KIR2DP1F with lysine 44 recognized C1+HLA-C, whereas T44-KIR2DP1F recognized C2+HLA-C. Dimorphisms at twelve other KIR2DP1F residues modulate receptor avidity or signaling. KIR2DP1 and KIR2DL1 are neighbors in the centromeric KIR region, and in tight linkage disequilibrium. Like KIR2DL1, KIR2DP1 contributed to CenA and CenB KIR haplotype differences. Encoded on CenA, C1-specific K44-KIR2DP1F were stronger receptors than the attenuated C2-specific T44-KIR2DP1F encoded on CenB. The last common ancestor of humans and chimpanzees had diverse lineage III KIR that passed on to chimpanzees, but not to humans. Early humans inherited activating KIR2DS4 and an inhibitory lineage III KIR, likely encoding a C1-specific receptor. The latter spawned the modern family of HLA-C receptors. KIR2DP1F has properties consistent with KIR2DP1F having been the founder gene. It first encoded only K44-C1 receptors, but then evolved T44-C2 receptors. The emergence of dedicated KIR2DL2/3 and KIR2DL1 genes encoding C1 and C2 receptors, respectively, could have led to obsolescence of KIR2DP1F. Alternatively, pathogen subversion led to its demise. Preservation of KIR2DP1F functional polymorphism appears a side effect of fixing the deletion in KIR2DP1F by micro gene conversion.
Keywords: Innate immunity, HLA class I, killer-cell immunoglobulin-like receptors, genetic polymorphism, human placentation
Introduction
Selective pressures imposed by variable and rapidly evolving pathogens drive the diversification of genomic regions dedicated to immune defense (1, 2). Epitomizing such genetic diversity are the human killer cell immunoglobulin-like receptor (KIR) genes of the leucocyte receptor complex (3). These genes encode cell-surface receptors that regulate natural-killer (NK) cell and T-cell function through recognition of HLA class I ligands. Amongst the most diverse of human genes, the KIR gene family (the KIR locus) is characterized by extensive structural, haplotypic and allelic polymorphism. The rapid evolution of the KIR gene system is evident from comparison of the human KIR locus with its counterparts in other hominids (4–6).
A key event that distinguished hominid MHC class I genes from those of Old World monkeys, such as rhesus macaque, was emergence of MHC-C as a dominant ligand for KIR (7, 8). This development led to major changes in the KIR locus. The telomeric region of the rhesus macaque KIR locus contains different combinations of 21 lineage II KIR genes (9). These genes encode receptors that recognize Bw4 and structurally related epitopes of Mamu-A and Mamu-B, rhesus macaque homologs of HLA-A and –B. By contrast, in the centromeric region of the rhesus macaque KIR locus there is just one lineage III KIR gene, and its function is not known. The organization of orangutan and chimpanzee KIR loci is the reverse. The centromeric region contains different combinations of nine lineage III KIR genes encoding receptors that each recognize either the C1 or C2 epitope of MHC-C, whereas the telomeric region has one lineage II KIR gene encoding a receptor for Bw4-like epitopes of MHC-A and –B (6). Contributing to these qualitative and quantitative changes in the hominid KIR locus, was an increasing role in reproduction for interactions of MHC-C with lineage III KIR, specifically in formation of the placenta (2, 10).
Although comparison of human with chimpanzee, orangutan and the rhesus macaque shows the evolutionary trajectory of the KIR locus during the last 35 million years (4), such analysis cannot speak to changes that occurred during human-specific evolution in the 6.0–7.2 million years since separation of the human and chimpanzee lines (11). In this context, we have studied KIR2DP1, an inactivated form of a once functional inhibitory human lineage III KIR gene, which we call KIR2DP1-Functional, or KIR2DP1F, for short.
Comparison of human (12, 13), chimpanzee (14), and orangutan KIR haplotype sequences (5) shows KIR2DP1 is a component only of human KIR haplotypes. This property is shared with five of the six expressed lineage III KIR genes: KIR2DL1, KIR2DL2/3, KIR2DS1, KIR2DS2, and KIR2DS3/5. The exception is KIR2DS4, for which there is an ortholog in chimpanzee but not in orangutan or other primate species (15). These observations point to KIR2DP1F and six other lineage III KIR genes being the products of human-specific evolution.
Phylogenetically, KIR2DPIF groups with KIR2DL1 and KIR2DL2/3, the inhibitory receptors that recognize the C1 and C2 epitopes of HLA-C, respectively (6, 16). These epitopes are defined by dimorphism at position 80 in the α1-domain of HLA-C: asparagine 80 giving the C1 epitope and lysine 80 the C2 epitope. Interactions of C2 with KIR2DL1 and C1 with KIR2DL2/3 control the education and response of major subpopulations of human NK cells (17, 18). As originally noted by Vilches et al (19), the key difference that distinguishes KIR2DP1 (previously called KIR15) from KIR2DL1 and KIR2DL2/3 is deletion of nucleotide 325 from codon 88 in exon 4. This deletion changes the reading frame, leading to premature termination in exon 5, 36 codons after the deletion. Consistent with KIR2DP1 having once been a functional gene, the rest of the KIR2DP1 gene has no obvious defect. Like its functional counterparts, KIR2DP1 is highly polymorphic. Since its first description, KIR2DP1 has been sequenced from individuals of various ethnic groups (13). Currently, the sequences of 28 KIR2DP1 alleles are deposited in the Immuno Polymorphism Database (20). That all these alleles have the same nucleotide deletion strongly indicates that the mutation occurred only once, and subsequently spread to fixation. Consequently, our working hypothesis is that KIR2DP1F, the immediate antecedent of KIR2DP1, lacked the deletion and was a functional gene encoding an inhibitory HLA-C receptor. Thus, in the course of human evolution, KIR2DP1 once flourished but was later extinguished. Here we report our investigation to resurrect the KIR2DP1 receptor, define its interactions with HLA class I, and their modulation by the natural polymorphism.
Materials and Methods
Genomic analyses
Lineage III KIR gene sequences were downloaded from the Immuno Polymorphism Database (IPD-KIR Release 2.5.0) (http://www.ebi.ac.uk/ipd/kir/) (21). Sequences were aligned in Geneious 5.4.6 (22) using the MAFFT algorithm (23) with manual correction. Phylogenetic trees were generated using three methods: neighbor-joining (using the Tamura-Nei pairwise substitution), maximum likelihood and parsimony, each with 500 bootstrap replicates. The bootstrap support for each node is indicated when >50. Representative trees are shown in Figure 2 and Supplemental Figure 1. Evolutionary analyses were conducted in MEGA6.0 (24). Complete sequences for 27 KIR haplotypes were downloaded from IPD (13, 21).
Figure 2. Human lineage III KIR have less diversity than chimpanzee lineage III KIR.
(A) Comparison of the chimpanzee (grey, upper panel) and human (blue, lower panel) KIR loci shows that the centromeric part of the chimpanzee KIR locus is both highly diversified and divergent to that of humans. For each locus, alternative gene-content segments are represented on parallel lines, with each gene represented by a colored box. Chimpanzee-specific KIR with no strict human ortholog are indicated with green boxes. Human KIR associated with CenA or CenB are indicated with blue and orange boxes respectively. For both chimpanzee and human lineage III KIR, the amino acid residue found at codon 44 is shown in yellow at the center of the box. Framework genes are shown in grey. Dotted lines indicate orthologs (between species) or alleles (within species).
(B) Phylogenetic analysis of hominoid lineage III KIR genomic sequences (human, blue; chimpanzee, grey; orangutan, green). Shown is a representative neighbor-joining tree. The bootstrap support for each node is indicated: neighbor-joining, maximum-likelihood and parsimony (top to bottom). Black circles represent nodes at which bootstrap support is 100 for each of the three methods. The amino acid residue found at codon 44 is indicated for each gene. The gibbon lineage III KIR2DL1 is used as an out-group in this analysis.
Protein sequence and structure
To generate a consensus sequence for lineage III KIR and an alignment of KIR2DP1 amino acid sequences, all lineage III KIR protein sequences were downloaded from IPD (21). Manual alignment of KIR2DL1–3, KIR2DS1–5 and KIR2DP1 sequences was performed using Seaview (25) v4.4.0 (http://doua.prabi.fr/software/seaview).
Searching Neanderthal and Denisovan genomes for KIR2DP1-specific sequence reads
The genomes of the Neanderthal (26) and Denisovan (27) archaic humans were accessed using the UCSC Genome browser (www.genome.ucsc.edu/Neandertal and www.genome.ucsc.edu/Denisova, respectively) and searched for KIR2DP1 specific sequence reads.
KIR-Fc fusion proteins
KIR-Fc fusion proteins were generated as described (28). Regions encoding the Ig-like domains and stem of KIR2DP1*002, KIR2DL1*003 and KIR2DL3*001 were PCR amplified, fused with the region encoding the Fc portion of human IgG1 and ligated into the pACgp67A transfer vector (BD Biosciences, San Jose, CA). Site-directed mutagenesis was used to generate KIR-Fc corresponding to 12 additional KIR2DP1F allotypes and 13 KIR2DL3-Fc mutants.
DNA constructs were cotransfected into Sf9 insect cells with linearized baculovirus (Baculogold, BD Biosciences), using Cellfectin (Invitrogen, Carlsbad, CA). After two rounds of amplification, Hi5 insect cells were infected with high-titer virus for 72 hours. Sf9 and Hi5 cells were kindly provided by Dr. K. Chris Garcia, Stanford University. Cell supernatant was collected, filtered and stabilized with HEPES-buffer, pH 7.2. After overnight incubation with protein A conjugated to Sepharose beads (Invitrogen), the beads were washed with 500ml of PBS. The KIR-Fc fusion proteins were then eluted from the beads with 0.1 M glycine (pH 2.7), with the eluate being immediately neutralized with 0.2M Tris-base (pH 9.0). Proteins were desalted and concentrated to 500µg/ml.
When commercial sales of the BD Biosciences Baculogold reagent were discontinued, another baculovirus expression system (BestBac 2.0 Cotransfection Kit, Expression Systems) was used to produce KIR-Fc. To compare these different baculovirus expression systems we made KIR2DL1*003-Fc and KIR2DL3*001-Fc using both methods. When the same KIR-Fc was made by the two methods, they had indistinguishable specificity and avidity for HLA class I.
HLA binding assay
KIR-Fc fusion proteins were tested for binding to a panel of microbeads, each coated with one of 97 HLA class I allotypes: 31 HLA-A, 50 HLA-B, and 16 HLA-C (LABScreen Single-Antigen Bead Set; Thermo Fisher Scientific), as described (28, 29). KIR-Fc fusion proteins were diluted in PBS to 100µg/ml and incubated with LABScreen microbeads (60 min at 4°C, shaking). After three washes, secondary staining with anti-human Fc-PE (One Lambda) was performed for 60 min and followed by three washes. Samples were analyzed on a Luminex 100 reader (Luminex, Austin, TX). Beads were also incubated with W6/32, a mouse monoclonal antibody that binds an epitope common to all HLA-A, -B and –C. Specific KIR-Fc binding to a given HLA class I allotype is expressed as a relative fluorescence ratio, calculated using the formula (total binding of KIR-Fc to beads coated with the allotype – KIR-Fc binding to beads not coated with HLA class I) / (W6/32 binding to beads coated with the HLA class I allotype –W6/32 binding to beads not coated with the HLA class I allotype).
Results
KIR2DP1F arose, diversified and became inactivated during the course of human evolution
Consistent with KIR2DP1 pre-dating the divergence of KIR A and KIR B haplotypes, KIR2DP1 is present in the centromeric regions (CenA and CenB) of both haplotype groups (Figure 1A) (6, 13). KIR2DP1 is flanked on the telomeric side by KIR2DL1. On the centromeric side it is flanked by KIR2DL3 on A haplotypes and by KIR2DS3/5 on B haplotypes. Such human-specific evolution is also true for inhibitory KIR2DL1, 2 and 3, which are closely related to KIR2DP1 (Figure 1B and 1C). Distinguishing KIR2DP1 is a deletion at position 325 in exon 4 (codon 88). This deletion causes a change in reading frame and premature termination in exon 5 (Figure 1B) (19).
Figure 1. KIR2DP1: an inactivated human-specific gene of the centromeric KIR region.
(A) A gene map of the centromeric regions (Cen) of KIR A and KIR B haplotypes. On CenA haplotypes, KIR2DP1 is between KIR2DL3 and KIR2DL1, on CenB haplotypes it is between KIR2DS3/5 and KIR2DL1.
(B) Nucleotide deletion at position 325 (codon 88) in the KIR2DP1 coding region changes the reading frame. This produces a stop codon at position 433 (codon 124). When the missing nucleotide is restored, KIR2DP1F encodes a leader sequence (L) of 21 residues and a mature protein of 329 amino acids, comprising five domains: two extracellular ligand-binding domains (D1 and D2), a stem (St), a transmembrane region (Tm) and a cytoplasmic domain (Cyt).
(C) Positions of variation in the KIR2DL1, 2DL2, 2DL3 and 2DP1F sequences. Highlighted yellow are residues unique to KIR2DP1F. (*) indicates a stop codon.
(D) Shows the identity and number of KIR2DP1-specific sequence reads found in the Neanderthal and Denisovan genome data (see also Supplementary Table 1). Listed for each position is the nucleotide (upper) and amino acid (lower, bold) identified in each read. “Del” identifies the deletion variant in KIR2DP1 that results in the inactivating frameshift (FS) mutation. Both genomes contain the inactivating nucleotide deletion at position 325 (codon 88). A subset of the reads has the KIR2DP1-specific threonine 52 and glutamate 71 codons (highlighted in yellow).
We examined the promoter region to identify any changes in transcription factor binding sites that could also have contributed to the inactivation of KIR2DP1. Although there a small number of substitutions that distinguish some KIR2DP1 alleles from other KIR, there are no polymorphisms in any of the functionally conserved parts of the promoter region. These conserved parts include the binding sites for the ETS, CREB, AML1 and SP1 transcription factors.
To define a time frame for the emergence and inactivation of KIR2DP1 we sought for the presence of KIR2DP1-specific sequences in the genomes of Neanderthal (26) and Denisovan (27) archaic humans. Four sequencing reads corresponding to Neanderthal KIR2DP1 and 26 reads corresponding to Denisovan KIR2DP1 (27) were obtained. They both have the same inactivating deletion as modern human KIR2DP1 (Figure 1D and Supplemental Table I). Consistent with this result, the archaic human KIR2DP1 sequences also have cytosine 218 in codon 52 (threonine in KIR2DP1F) and guanine 274 in codon 71 (glutamate in KIR2DP1F), two nucleotide substitutions that distinguish KIR2DP1 from all other human KIR (Figure 1C and 1D). Moreover, the combination of cytosine 218, guanine 274 and the deletion of nucleotide 325 is present in one sequence read generated from a Denisovan DNA fragment (Figure 1D and Supplemental Table I). These results point to KIR2DP1F inactivation predating the separation of modern humans from these archaic humans ~450,000–800,000 years ago (30–32). Thus the window of time for the inactivation of KIR2DP1F is between 6 million and 450,000 years ago. This window also gives a maximum estimate for the lifespan of KIR2DP1F.
Humans inherited less KIR diversity than chimpanzees from their last common ancestor
The last common ancestor of humans and chimpanzees is considered to have been anatomically, physiologically and behaviorally more similar to modern chimpanzees than to modern humans (33–35). Thus the human line underwent greater changes and adaptations, as well as passage through narrower population bottlenecks (36), than the chimpanzee line. Evidence of this difference is seen in the modern human and chimpanzee KIR loci (Figure 2).
The modern chimpanzee KIR locus has four framework genes and ten variable genes, including nine lineage III KIR genes. As KIR haplotypes have between two and eight variable genes, multiple KIR haplotypes were necessary for ancestral chimpanzee populations to sustain KIR diversity. The variable KIR genes form five modules, each of two genes, within the centromeric region (Figure 2A). Within a module there is linkage disequilibrium (LD) between the two genes, but recombination occurs between the modules and has created much of the gene content diversity of hominid KIR haplotypes.
The modern human KIR locus has four framework genes, eleven variable genes and two distinctive forms of KIR haplotype: KIR A and KIR B (Figure 2A). Of the variable genes, five are present on KIR A, the human haplotypes most like chimpanzee KIR haplotypes. Having eight variable genes, KIR B haplotypes are the products of human-specific evolution. Six human KIR genes form three modules, like those in chimpanzee KIR haplotypes. Of note is the module combining genes that encode inactivated inhibitory KIR2DP1 and KIR2DL1, the inhibitory C2 receptor. This module is fixed on KIR A and present on a subset of KIR B haplotypes. That the modern chimpanzee KIR locus has five modules, and the modern human KIR locus has only three modules, is one line of evidence that chimpanzees inherited greater KIR diversity from the last common ancestor than humans.
In a complementary analysis, we constructed phylogenetic trees from the sequences of human, chimpanzee and orangutan lineage III KIR, using a gibbon lineage III KIR as the out-group (Figure 2B). These KIR form five clades. The clade comprising orangutan KIR has the deepest root and divides into two sub-clades corresponding to E44 KIR and K44 KIR. Chimpanzee MHC-C specific KIR distribute between three well-separated clades. One corresponds to C1 receptors with K44 and a second corresponds to C2 receptors with M44. The third clade contains two additional chimpanzee C2-specific KIR, KIR2DL7 with M44 and KIR2DL9 with E44, as well as chimpanzee and human KIR2DS4, which have K44. The clear separation within the tree of the chimpanzee KIR that recognize C1 and the chimpanzee KIR that recognize C2 is further evidence of the considerable KIR diversity that chimpanzees inherited from the last common ancestor. Also apparent is that chimpanzees and humans both inherited KIR2DS4 from the last common ancestor.
In contrast to KIR2DS4, the other human lineage III KIR, which include all HLA-C receptors, show no grouping with individual chimpanzee KIR or groups of chimpanzee KIR. Instead, they form a separate, human-specific, clade with a relatively shallow root. Thus the human C2-specific M44 KIR are no more related to chimpanzee C2-specific M44 KIR than they are to chimpanzee C1-specific K44 KIR. (Likewise, the human C1-specific K44 KIR are no more related to chimpanzee C1-specific K44 KIR than they are to human C2-specific M44 KIR). These results show that the human lineage III KIR has less diversity than chimpanzee lineage III KIR and that much of the ancestral KIR diversity that chimpanzees inherited from the last common ancestor was either not passed on to the human line, or was passed on but subsequently lost. With the exception of KIR2DS4, all the K44, T44 and M44 lineage III KIR now present in the human population appear to be products of an episode of human-specific gene expansion and differentiation that took place soon after the species split (6). These results are consistent with the human line inheriting only two lineage III KIR from the last common ancestor: KIR2DS4 and a ‘founder’ gene that eventually gave rise to all the other human lineage III KIR.
The shallow clade formed by the human lineage III KIR comprises two sub-clades. One includes K44 KIR2DL2, 2DL3, 2DS2 and KIR2DP1F. The other sub-clade includes T44 KIR2DS3/5 and M44 KIR2DS1 and 2DL1. The majority of these KIR (2DL2, 2DS1, 2DS2, 2DS3 and 2DS5) are components of KIR B haplotypes, which were formed after the species split and could not have come from the last common ancestor. This leaves only KIR2DL1, KIR2DL3 and KIR2DP1F as candidates for being most like the ‘founder’ KIR gene.
KIR2DP1F encodes C1-specific receptors that segregate with CenA and C2-specific receptors that segregate with CenB
In a panel of 27 human KIR haplotype sequences (13), 14 haplotypes have CenA and 13 have CenB (Figure 3A). All CenA and seven of the 13 CenB contain KIR2DP1. The other six CenB haplotypes lack the genomic module comprising KIR2DP1 and KIR2DL1 (Figure 3A, lower panel). Each of the ten KIR2DP1 alleles is present on CenA or CenB, but not on both. Six KIR2DP1 alleles (2DP1*002, *003, *004, *005, *007 and *008) are CenA associated and four KIR2DP1 alleles (2DP1*001, *006, *009 and *010) are CenB associated (Figure 3A).
Figure 3. Genomics, polymorphism and evolution of KIR2DP1 variants.
(A) Shows centromeric region gene content for 27 KIR haplotypes that have been sequenced completely at high-resolution (13). Each box represents a KIR gene. For the lineage III, KIR, the allele name is given in the box. Under ‘Codon 44 residue’ is given the residue present in the KIR2DP1F encoded by the haplotype. The B02 haplotypes lack KIR2DP1 as well as KIR2DL1, Centromeric KIR A haplotypes (n=14) are outlined in blue, centromeric KIR, B haplotypes (n=13) are outlined in orange.
(B) Distribution of the KIR2DP1F 44K/44T polymorphism among KIR, A (blue) and KIR, B (orange) haplotypes in the Ga-Adangbe population of Ghana (38).
(C) Statistical analysis showing that the observed segregation of K44- and T44-encoding KIR2DP1F among KIR A and B haplotypes in the Ga-Adangbe deviates significantly from a random distribution (Chi-squared test, p<0.0001). Predicted values were calculated assuming a 50% distribution of K44- and T44- encoding alleles between KIR A (61%) and KIR, B (39%) haplotypes.
(D) Graphical representation of the results of ancestral reconstruction for codon 44 of KIR2DP1F. A phylogenetic tree (shown in Supplemental Figure 2) was constructed using MEGA6.0 (24) (Neighbor joining, partial deletion, Tamura-Nei, 100 bootstrap replicates). Ancestral reconstruction was performed using Parsimony. The dataset contained examples of each known lineage III human and chimpanzee gene as well as the rhesus macaque lineage III gene, Mm1D, as an out-group.
(E) Polymorphisms that distinguish T44-KIR2DP1F and KIR2DS3/5 in threonine codon 44 and its immediate regions upstream and downstream.
(F) Model for the evolution of KIR2DP1F polymorphism. Large circles represent the known KIR2DP1F allotypes, small grey circles represent hypothesized intermediates that have not been observed. Allotypes associated with KIR A and B haplotypes are shown in blue and orange, respectively. Indicated on the lines connecting two allotypes are the amino acid substitutions required to evolve one allotype from the other.
An extraordinary feature of KIR2DP1F is that position 44, the specificity determining position of lineage III KIR (18), exhibits a balanced polymorphism, whereas this residue is monomorphic in all other human, chimpanzee and orangutan lineage III KIR. The CenA-associated KIR2DP1 have K44, the residue that makes KIR2DL3 specific for C1. Conversely, all but one of CenB-associated KIR2DP1 have T44, a residue that conferred C2 specificity when introduced by mutation into KIR2DL3 (37), and which is also present in KIR2DS3/5. The exception is KIR2DP1*009, which is CenB-associated but has K44.
To assess the distribution of KIR2DP1 alleles in an immunogenetically well-defined human population, we examined the Ga-Adangbe of Ghana, who are typed for KIR2DP1 at high resolution (38). In a cohort of 183 individuals, 204 different KIR haplotypes were identified: 90% of 125 Cen A haplotypes, carry K44 KIR2DP1 and 10% carry T44 KIR2DP1 (Figure 3B); 51% of the Cen B haplotypes carry T44 KIR2DP1F, 15% carry K44 KIR2DP1F and 34% lack KIR2DP1. The correlation of Cen A with K44 KIR2DP1 and of Cen B with both T44 KIR2DP1 and loss of KIR2DP1 are statistically significant (Figure 3C). In this large cohort of genetically diverse sub-Saharan Africans the correlations are not as strong as in the small panel of sequenced KIR haplotypes, many of which derive from individuals of European origin (13).
Lysine was the ancestral residue at the specificity determining position
To determine if lysine or threonine was the ancestral residue encoded by codon 44 of KIR2DP1, we constructed a phylogenetic tree of primate lineage III KIR (Supplemental Figure 1). Ancestral reconstruction based on this tree showed that K44 (present in KIR2DL2/3 and Cen A associated KIR2DP1F) is most likely to have been the ancestral residue (Figure 3D). Consequently, methionine (present in KIR2DL1) and threonine (present in KIR2DS3/5 and Cen B associated KIR2DP1F) are derived forms.
KIR2DS3/5 is a CenB gene that encodes an activating receptor with T44. Because T44-KIR2DP1 is next to KIR2DS3/5 in CenB, we considered the possibility that Cen B KIR2DP1F acquired T44 from KIR2DS3/5 by recombination. This proved not to be the case, because the T44 codon is different in KIR2DS3/5 (ACG) and KIR2DP1 (ACA). Giving further support that T44 evolved independently in the two genes, alignment of the immediate upstream and downstream coding regions showed that KIR2DS3/5 and Cen B KIR2DP1F also differ at codons 33, 50, 52 and 56 (Figure 3E). Thus the T44 codons in KIR2DP1F and KIR2DS3/5 were the result of independent mutations in the lysine codon at position 44 (Figure 3F). Both the ACG and ACA threonine codons can be formed by single nucleotide substitutions in a different lysine codon (AAG gave ACG and AAA gave ACA). These events produced an activating receptor, KIR2DS3/5, and an inhibitory receptor, T44-KIR2DP1F with C2 specificity. Selection of these lower avidity C2 receptors is consistent with higher avidity C2-specific KIR with M44, having been lost on the human line. Because KIR2DP1F is the only human lineage III KIR known to have different alleles encoding C1-specific and C2-specific receptors it is a much better candidate for being descended from the founder gene than either KIR2DL1 or KIR2DL3. KIR2DL2, a recombinant of KIR2DL1 and KIR2DL3 is an unlikely candidate, because it is specific to CenB and thus of more recent origin than the other inhibitory lineage III KIR. The position of KIR2DP1F in the phylogenetic tree is also compatible with it being closer to the founder gene (Figure 2B).
Residue 44 determines specificity and influences avidity of KIR2DP1F for HLA-C
To determine the specificity of K44-KIR2DP1F and T44-KIR2DP1F, we made KIR-Fc fusion proteins from K44-KIR2DP1*003F and T44-KIR2DP1*006F, which differ only at position 44 (Supplemental Figure 2). These KIR-Fc were tested for binding to a panel of 97 HLA class I. K44-KIR2DP1*003F-Fc bound to all C1+ HLA-C (Figure 4A) and to HLA-B*46:01 and B*73:01 that have the C1 epitope (29). No binding was observed to HLA-A, other HLA-B and C2+ HLA-C. Conversely, T44-KIR2DP1*006 F-Fc bound to C2+ HLA-C but not to C1+HLA-C, or any HLA-A or -B. These results concur with previous observations that K44 confers specificity for C1 and T44 confers specificity for C2 (37). KIR2DP1*003F-Fc bound the C1 epitope with ~80% the avidity of KIR2DL3*001-Fc, and KIR2DP1*006F-Fc bound C2 with ~65% the avidity of KIR2DL1*003-Fc. Thus KIR2DP1F was a generally weaker receptor than C1-specific KIR2DL3 and C2-specific KIR2DL1. These avidity differences are consistent with KIR2DP1F having D1 and D2 ligand-binding domains that are closer in sequence to those of lower-avidity KIR2DL3 than higher-avidity KIR2DL1 (Figure 1C). Also confirmed, is previous observation that T44 confers lower avidity for HLA-C than either K44 or M44 (37). Thus, the residue at position 44 modulates the specificity and avidity of KIR2DP1F interaction with HLA-C.
Figure 4. Different lineages of KIR2DP1F encode inhibitory receptors for C1 and C2.
(A) Shown is the binding of K44-KIR2DP1*003F-Fc (blue) and T44-KIR2DP1*006F-Fc (orange) to a panel of nine HLA-C1 and seven HLA-C2 allotypes. For comparison, the binding of K44-KIR2DL3*001F-Fc (white) and M44-KIR2DL1*003-Fc (black) are also shown. Avidity is given as percentage of the binding of W6/32, an antibody that binds all HLA class I allotypes with equal avidity. Horizontal bars represent the mean avidities for the nine C1 and seven C2 HLA-C allotypes, respectively.
(B) Shown is the binding of K44-KIR2DP1*003F-Fc (blue), T44-KIR2DP1*006F-Fc (orange), K44-KIR2DL3*001F-Fc (white) and M44-KIR2DL1*003-Fc (black) to nine HLA-C1 allotypes.
(C) Shown is the binding of K44-KIR2DP1*003F-Fc (blue), T44-KIR2DP1*006F-Fc (orange), K44-KIR2DL3*001F-Fc (white) and M44-KIR2DL1*003-Fc (black) to seven HLA-C2 allotypes.
The degree to which KIR2DP1*003F-Fc bound the nine C1+ HLA-C allotypes varied (Figure 4B), as did the binding of KIR2DP1*006F-Fc to the seven C2+HLA-C allotypes (Figure 4C). KIR2DP1*003F-Fc exhibited the same hierarchy of binding to C1+ HLA-C allotypes as the positive control, KIR2DL3*001-Fc. HLA-C*03:02 bound with highest avidity and HLA-C*14:02 with lowest avidity. The relative binding of KIR2DL1*003-Fc and KIR2DP1*006F-Fc to C2+ HLA-C was similar for six of the seven allotypes tested. Exceptional was HLA-C*17:01, an African-specific allotype, which bound KIR2DP1*006F-Fc with avidity <25% that of KIR2DL1*003-Fc (Figure 4C). In summary, K44-KIR2DP1F was a weak C1-specific receptor that bound C1+HLA-C allotypes with variable avidity, and T44-KIR2DP1F was a weak C2-specific receptor that bound C2+HLA-C with variable avidity.
D1 and D2 domain dimorphisms modulate the specificity and avidity of KIR2DP1F for HLA-C
The D1 and D2 domains of lineage III KIR form the HLA-C binding site (39, 40). Sequences of twelve KIR2DP1F allotypes differ at eleven positions in the D1 and D2 domains (Figure 5A). The six K44-KIR2DP1F allotypes differ at four of the eleven positions, whereas the six T44-KIR2DP1F allotypes differ at six other positions (the eleventh is the specificity determining position 44). To determine how these polymorphisms affect recognition of HLA-C by KIR2DP1F, KIR-Fc fusion proteins corresponding to each KIR2DP1F allotype were tested for binding to the panel of HLA class I allotypes. All five K44-KIR2DP1F allotypes exhibited C1 specificity and all eight T44-KIR2DP1F allotypes exhibited C2 specificity (Figure 5B).
Figure 5. D1 and D2 domain polymorphism modulates KIR2DP1F binding to HLA-C.
(A) Shown is an amino acid alignment of polymorphic positions in the D1 and D2 domains of KIR2DP1F allotypes. KIR A haplotype associated allotypes are outlined in blue and KIR B haplotype associated allotypes are outlined in orange.
(B) Shown is the summary of the binding of five K44-KIR2DP1F (KIR2DP1F*003 and KIR2DP1F*009 are identical in the extracellular domain) and six-T44 KIR2DP1F allotypes to seven HLA-C1 and nine HLA-C2 allotypes. The binding of K44-KIR2DP1F-Fc is shown relative to K44-KIR2DL3*001-Fc. The binding of K44-KIR2DP1F-Fc is shown relative to M44-KIR2DL1*003-Fc.
(C) Shows the binding of five K44-KIR2DP1F-Fc, 16 K44-KIR2DL2/3-Fc allotypes and one K44-KIR2DL1-Fc allotype (KIR2DL1*022, shown in gray) to nine HLA-C1 allotypes. The binding of each KIR-Fc is shown relative to K44-KIR2DL3*001F-Fc. Plotted separately are the data for KIR2DL2/3 allotypes encoded by Cen A (blue data points) and the data for KIR2DL2/3 allotypes encoded by CenB (orange data points). K44-KIR2DL2/3-Fc was significantly higher than CenA associated K44 KIR2DP1F (unpaired t-test, p<0.005). The KIR2DL1 binding data are from Hilton et al (45). KIR2DL1*022 is an exceptional KIR2DL1 allotype, found only in southern African populations, that has K44 and is a strong C1-specific receptor (57).
(D) Shows the binding of six T44-KIR2DP1F-Fc and 15 M44-KIR2DL1-Fc allotypes to seven C2+HLA-C allotypes. The binding of each KIR-Fc is shown relative to that of M44-KIR2DL1*003-Fc. Plotted separately are the data for KIR2DL1 allotypes encoded by CenA (blue data points) and the data for KIR2DL1 allotypes encoded by CenB (orange data points). The binding of CenA associated M44-KIR2DL1-Fc is significantly higher than CenB associated T44-KIR2DP1F (unpaired t-test, p<0.05). The KIR2DL1 binding data are from Hilton et al (45).
K44-KIR2DP1F allotypes bound HLA-C1 with 33–89% the avidity of the KIR2DL3*001 control. Highest binding was observed for KIR2DP1F*002 and KIR2DP1F*004: 88% and 89% of control, respectively (Figure 5B). Consequently substitutions at positions 35 and 123, which distinguish KIR2DP1F*002 and KIR2DP1F*004 (Figure 5A), have little effect on KIR2DP1F recognition of HLA-C. KIR2DP1F*013, which has lowest avidity (33% of control), differs from high avidity KIR2DP1F*002 at positions 35 and 49, which are responsible for the 55% difference in binding of these two allotypes (Figure 5B). KIR2DP1F*007 differs from KIR2DP1F*002 at positions 35 and 158, which cause a 19% difference in binding. From these comparisons we see that at least three of the four dimorphisms in the D1 and D2 domains of K44-KIR2DP1F allotypes, alone or in combination with residue 35, influence avidity for C1.
KIR2DP1F*010 bound C2 strongly like KIR2DL1*003. In contrast, KIR2DP1F*00101 bound weakly, one third that of KIR2DL1*003 (Figure 5B). Substitutions at positions 106, 141 and 154 in D2 are responsible for this avidity difference (Figure 5A). The combination of G106, R141 and P154 in KIR2DP1F*010, confers higher avidity, whereas the combination of E106, T141 and T154 in KIR2DP1F*00101 confers lower avidity. Comparison with KIR2DP1F*006, which has E106 as the only low-binding residue, indicates that G106 accounts for 37% of the avidity difference (Figure 5B). Comparison of KIR2DP1F*00101 with KIR2DP1F*00102, which differ only at position 154 (Figure 5A), shows how proline 154 accounts for 22% of the higher binding by KIR2DP1F*00102. By subtraction, R141 accounts for 13% of the binding (Figure 5B). Thus, substitutions at positions 106, 141 and 154 all affect the avidity of T44-KIR2DP1F for C2+ HLA-C.
Comparison of additional allotype pairs that differ by single substitutions identified other functionally important polymorphisms. Comparison of KIR2DP1F*00101 and KIR2DP1F*012 shows that substitution of proline for arginine at position 16 increases binding by ~25%. Similarly, comparison of KIR2DP1F*011 and KIR2DP1F*00102 shows how substitution of glutamine for histidine at position 171 increases binding by ~25%. From these analyses, all six positions of dimorphism in T44-KIR2DP1F are seen to affect the avidity for C2+ HLA-C. In summary, of the 11 dimorphisms that distinguish the ligand-binding domains of KIR2DP1F allotypes, one switched the epitope specificity and all eleven modulated the avidity of epitope recognition. In both the number of positions that modulate HLA-C recognition and the range of the allotypic difference, the effects are greater for T44-KIR2DP1F than K44-KIR2DP1F (Figure 5B).
The mean C1 binding of the K44-KIR2DP1F allotypes is comparable to that of the CenA KIR2DL3 allotypes, but significantly lower than that of the CenB KIR2DL2 allotypes (Figure 5C). The opposite hierarchy is seen for T44-KIR2DP1F allotypes. Mean C2 binding of T44-KIR2DP1F is significantly lower than that of CenA-encoded KIR2DL1, but comparable to that of the CenB-encoded KIR2DL1 (Figure 5D). In summary, the functional effects of the eleven D1 and D2 dimorphisms are consistent with them all having been products of natural selection.
The KIR2DP1F-specific motif N46-T52-E71 differentially affects C1 and C2 recognition
Distinguishing KIR2DP1F from other human lineage III KIR are T52 and E71 in the D1 domain. Also of note, N46 is fixed in KIR2DP1F and KIR2DL1 but absent from KIR2DL2/3 and KIR2DS2, which both have K46 (Figure 6A). To assess how these substitutions affect HLA-C binding, we made KIR2DL3*001-Fc mutants in which residues at positions 46, 52 and 71 were replaced by the corresponding KIR2DP1F residues. All combinations of N46, T52 and E71 were introduced into KIR2DL3*001, giving seven mutants in total: three single mutants, three double mutants and one triple mutant.
Figure 6. KIR2DP1F ligand recognition: C1 and C2 binding are differentially influenced by sequence dimorphisms.
(A) Amino acid sequence alignment of the D1 binding domain of lineage III KIR. KIR2DP1*003F-Fc is set as the consensus sequence. Highlighted in yellow are positions 46, 52 and 71 where KIR2DP1F has a combination of residues not present in other human KIR.
(B and C) Shown is the binding of KIR2DL3*001-Fc and mutants of KIR2DL3*001-Fc to a panel of nine C1+HLA-C1 allotypes (blue bars) and seven C2+HLA-C allotypes (red bars). Mutations were made at positions 44, 46, 52 and 71, where residues present in KIR2DP1*002 were introduced into KIR2DL3*001-Fc in various combinations. In the boxes to the left of the bars, yellow shading denotes residues derived from KIR2DP1F, and no shading denotes residues derived from KIR2DL3*001. The binding of C1-specific KIR-Fc is shown relative to that of KIR2DL3*001-Fc. The binding of C2 specific KIR-Fc is shown relative to KIR2DL1*003-Fc.
Individually, N46 and E71 increased C1 avidity to 145% and 115%, respectively, whereas T52 decreased C1 avidity to 60% of KIR2DL3*001 (Figure 6B). For double mutants, the negative effect of T52 dominated E71 but not N46. The triple mutant, in which K46-I52-Q71 of KIR2DL3*015 was replaced by N46-T52-E71, exhibited 85% the C1 avidity of KIR2DL3*001. Thus the overall effect of the substitutions at position 46, 52 and 171 is relatively small, whereas the individual substitutions have more pronounced effects.
The same set of mutations was introduced into a mutant KIR2DL3*001 with C2 specificity, in which lysine 44 had been replaced by threonine. In the single mutants, N46 increased C2 avidity by more than two fold, whereas T52 and E71 halved C2 avidity. In the double mutants N46 was dominant. In contrast to the results obtained with the K44 C1-specific receptor, replacing K46-I52-Q71 with N46-T52-E71 increased the C2 avidity of the T44 receptor by about one third. The combination of T52 and E71 that is fixed in KIR2DP1F has the effect of reducing the avidity of all KIR2DP1F, irrespective of their C1 or C2 specificity. In contrast, N46 has the effect of increasing the avidity of both C1-specific and, to greater extent, C2-specific KIR2DP1F.
Functional attenuation of CenB associated KIR2DP1F
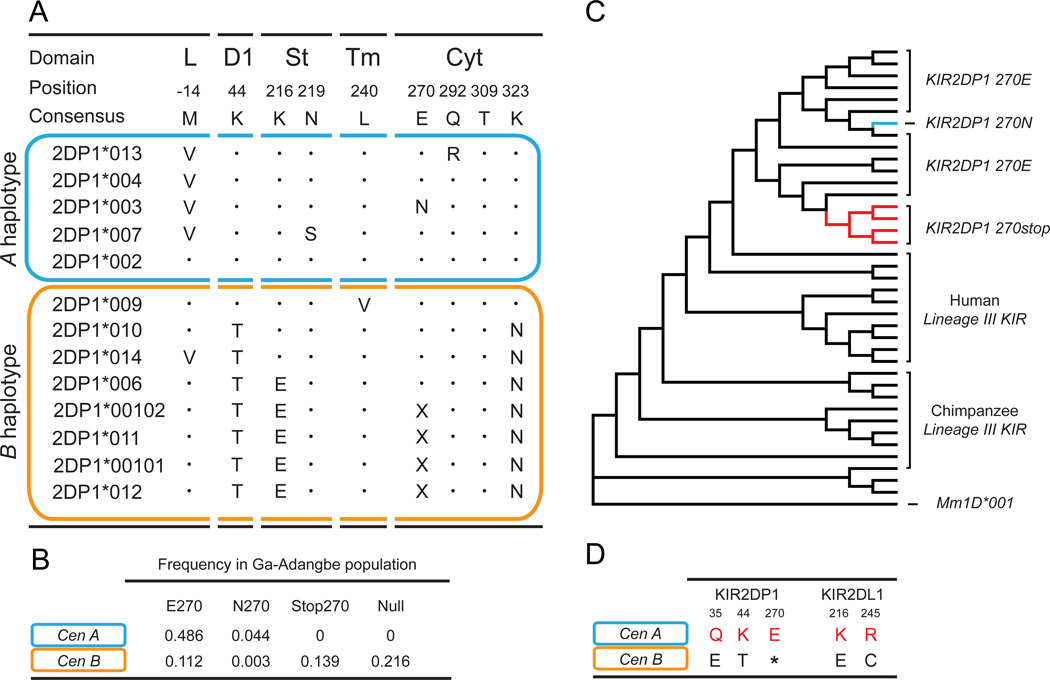
Distinguishing KIR2DP1F from KIR2DL1, 2DL2 and 2DL3 is arginine at position 310, instead of isoleucine (Figure 1C). This fixed substitution in ITIM 2, where the tyrosine of the motif is at position 312, could have had a general attenuating effect on KIR2DP1F signaling (41). Previous studies have shown that ITIM 1, but not ITIM 2, is essential for KIR2DL signaling, but can exert a modulatory role (41–44). We also examined other KIR2DP1F polymorphisms, for their potential effect on receptor function (Figure 7A and supplemental figure 2). Codon 270 in the cytoplasmic tail encodes glutamate, asparagine or a stop codon. We analyzed this trimorphism in the context of the KIR2DP1F allotypes of the Ga-Adangbe population (38). All CenA haplotypes have the KIR2DP1F gene, with 90% of these encoding K44-KIR2DP1F (Figure 3B). The frequency of E270 is more than ten times the frequency of N270 and the frequency of Stop270 is zero (Figure 7B). This distribution contrasts with that of the CenB haplotypes. Almost half of the CenB haplotypes lack the KIR2DP1F gene, as well as KIR2DL1. Of the seven T44-KIR2DP1F allotypes encoded by CenB, four have premature termination at position 270 and consequently lack both ITIMs and all inhibitory signaling function. Among the CenB associated T44-KIR2DP1F allotypes (Figure 7A), stop 270 has the highest frequency, followed by E270 and the rare N270 (Figure 7B). In conclusion we find that all CenA have K44-KIR2DP1F with signaling potential whereas 75% of CenB haplotypes have either lost the KIR2DP1F gene or encode T44-KIR2DP1F with no capacity for inhibitory signaling.
Figure 7. Cen B encoded T44-KIR2DP1F allotypes were subject to loss of signaling function.
(A) Alignment showing polymorphic positions in the leader peptide (L), the stem (St), the transmembrane domain (Tm) and the cytoplasmic tail (Cyt). Also shown is the specificity determining residue at position 44 of the D1 domain. A dot indicates identity with the KIR2DP1*002F consensus sequence and X indicates a stop codon.
(B) Frequencies of E270, N270, Stop270 and absence of KIR2DP1 (Null) associated with CenA (blue) and CenB (orange) in the Ga-Adangbe population (38).
(C) A graphic representation of ancestral reconstruction for codon 270 of primate lineage III KIR. A phylogenetic tree (shown in Figure S2) was constructed using MEGA6.0 (24) (Neighbor joining, partial deletion, Tamura-Nei, 100 bootstrap replicates). Ancestral reconstruction was performed using Parsimony. The dataset contained examples of each known lineage III human and chimpanzee gene as well as the Rhesus macaque lineage III gene, Mm1D, as an out-group.
(D) Table showing predominant residues found at functionally important sites in CenA (blue) and CenB (orange) associated KIR2DP1 and KIR2DL1. Amino acid residues associated with stronger receptor function are shown in red.
To identify the ancestral residue for position 270, we performed ancestral reconstruction analysis of primate lineage III KIR (N=39). The results identify glutamate as the ancestral residue, with N270 and stop270 being derived forms (Figure 7C). The four T44-KIR2DP1F allotypes that terminate at position 270 and lack signaling function (Figure 7A) are capable of recognizing C2+HLA-C (Figure 4D). They are also likely to be surface expressed, as indicated by our previous analysis of KIR2DL1 mutants with truncation at different sites in the transmembrane or cytoplasmic domains (45). Thus the four T44-KIR2DP1F allotypes that terminate at position 270 could have had a function at the cell surface, in mediating adhesion of NK cells to target cells (45).
Extinction of KIR2DP1F by fixation of an inactivating deletion while preserving polymorphism
The evolution of distinctive KIR A and KIR B haplotypes occurred ~6 million years ago (13). This involved formation of distinctive CenA haplotypes with robust receptors and CenB haplotypes with attenuated receptors (Figure 8A). KIR2DP1F contributed to this process prior to its extinction. The signaling capacity of CenB encoded T44-KIR2DP1F allotypes was reduced, while that of CenA encoded K44-KIR2DP1F allotypes was preserved. The deletion that eventually led to the extinction of KIR2DP1F may have initially been a component in the process of Cen B attenuation, like the stop codon at position 270 that eliminated signaling function. At some critical point in time the selection pressure changed from one of balancing selection to a directional, negative selection that favored complete loss of KIR2DP1F function (Figure 8B).
Figure 8. Model for the natural history of KIR2DP1.
(A) This schematic model depicts the divergent evolutionary pathways taken by the chimpanzee and human lineages since their divergence from a common ancestor ~6mya. During the process of speciation and separation from chimpanzee ancestors, the human ancestors passed through population bottlenecks that reduced the diversity of their lineage III, KIR. These ancestral humans appear to have had just two lineage III KIR, KIR2DS4 (blue) and an ancestor of K44-KIR2DP1F, the latter encoding inhibitory C1 receptors. Situated in the centromeric region of the human KIR locus, the lineage III KIR genes underwent gene duplication and differentiation, leading to formation of the KIR A and KIR B haplotypes that define the modern human KIR locus.
(B) Following its emergence, K44-KIR2DP1F (panel I) underwent diversification (panel II), including position 44, which created a second lineage of T44-KIR2DP1F alleles, encoding C2 specific receptors (panel II). Following a period of diversification under balancing selection (panel III), T44-KIR2DP1F became attenuated by loss of signaling function (panel IV). Acquisition of the inactivating mutation occurred in the T44-KIR2DP1F lineage (Panel V) and subsequently spread to other KIR2DP1F alleles by micro gene conversion (panel VI), fixation (panel VII) and permanent inactivation of the KIR2DP1F gene (panel VIII).
That KIR2DP1 retains extensive polymorphism, resembling that of functional KIR2DL1 and KIR2DL2/3, shows how three mechanisms contributed to KIR2DP1F extinction (Figure 8B). The first mechanism was positive selection that increased the frequency of the allele in which the deletion originated. A side effect of this mechanism would have been to reduce the polymorphism of KIR2DP1, which could explain the high frequency of KIR2DP1*00102, a Cen B allele that also has the 270 stop codon. It also suggests that the deletion could have originated in KIR2DP1*00102
The second mechanism is micro gene conversion that acted to move the deletion, and a small amount of surrounding sequence, from the KIR2DP1 allele of origin into other allelic backgrounds, which then became subject to positive selection. This mechanism serves to propagate the deletion, while preserving polymorphism at other positions in KIR2DP1, thus explaining why KIR2DP1 retains considerable polymorphism.
The third mechanism that contributed to the preservation of KIR2DP1 polymorphism is the very strong linkage disequilibrium between KIR2DP1 and KIR2DL1. If the KIR2DP1F allele in which the deletion originated had been fixed by positive selection, its effect would have been to eliminate all KIR2DL1 polymorphism as well as all CenA haplotypes. Because the mechanism of micro gene conversion could have fixed the deletion without approaching these potentially disastrous outcomes, it is possible that microgene conversion had the major role in driving the deletion to fixation. In that case the polymorphism we observe in the KIR2DP1 alleles of modern human populations could well reflect the functional KIR2DP1F polymorphism that was working in early humans several million years ago.
Our analysis also indicates that KIR2DP1 has not acquired random mutations since the time it became non-functional. Such mutations undoubtedly occurred, but they would not have become subject to positive selection and thus able to reach significant frequency in the population. This phenomenon is well illustrated by the database of HLA-A, -B and –C sequences, which contains numerous rare alleles, that differ by one silent substitution from a common allele, and have been detected only in one individual or one family of prospective hematopoietic stem cell donors (46, 47).
Discussion
Following the split of human and chimpanzee ancestors, ~5mya, the chimpanzee line retained much of the lineage III KIR diversity that had evolved in the previous 10–15 million years and was present in the last common ancestor. In contrast, only two lineage III KIR were retained on the human line: one encoding activating KIR2DS4, with unknown function and weak recognition of HLA class I (15), and the other encoding an inhibitory MHC-C receptor (Figure 8A). The latter gene subsequently became subject to a process of gene duplication and differentiation that spawned a family of six human KIR genes and formation of the distinctive CenA and CenB KIR haplotypes. These six genes comprised inhibitory KIR2DL1, 2DL2/3 and KIR2DP1F, and activating KIR2DS1, 2DS2 and 2DS3/5. The implication of this history is that most lineage III KIR, present in the last common ancestor, did not survive the intense selection pressures and population bottlenecks that likely occurred on the human line as speciation was taking place.
That such an erosion of the KIR locus could have been compatible with the survival of early humans can be illustrated by the condition of modern humans. None of the individual KIR is essential for the health and survival of individuals. In today’s population, millions of healthy people either lack C1, and have non-functioning KIR2DL2 and 2DS1 receptors, or lack C2, and have non-functioning KIR2DL2/3 receptors. Likewise, millions of healthy people survive with only KIR A or KIR B haplotypes. A lack of dependence on KIR is possible because the conserved and more ancient interaction between the CD94:NKG2A receptor and its HLA-E ligand can adequately provide for the education of NK cells and control of their major functions in immunity and reproduction (2, 4, 48). The primate KIR and rodent Ly49 systems of NK cell receptors are inherently unstable, because the ligands and receptors are both polymorphic and encoded on different chromosomes (2, 4). This allows for higher diversity of ligand-receptor interactions in populations, but in passing through bottlenecks it can result in receptors losing their ligands and vice versa. The gibbon KIR locus is highly eroded (49), and provides an informative precedent for imagining the state of the KIR locus in early humans following the species split.
That all modern lineage III KIR (apart from KIR2DS4) derive from a single founder gene, implies that early humans either had a C1-specific KIR or a C2-specific KIR gene. Both K44 C1-specific KIR and M44 C2-specific KIR were present in the last common ancestor (Figure 8A). Because the hominid system of MHC-C specific KIR was built around the C1 epitope, and existed and evolved for ~10 million years before emergence of the C2 epitope and its cognate KIR, it seems more likely that an inhibitory C1-specific KIR was selected for survival during the period of the speciation process, than an inhibitory C2-specific KIR. In other words, there is good precedent for species surviving with C1 and without C2, but there is no precedent for a hominid species or population having C2 but not C1. We therefore hypothesize that the modern system of human lineage III KIR evolved from a founder gene encoding K44 C1-specific KIR.
The properties of KIR2DP1F we uncovered in this study are consistent with KIR2DP1F having been the founder gene for the modern family of human lineage III KIR (Figure 8A). We show that KIR2DP1F was first a dedicated inhibitory C1 receptor that encoded K44 allotypes, but later evolved a second set of allotypes with T44 that are inhibitory C2-specific KIR. By this means, the T44-KIR2DP1F allotypes were able to partly replace the inhibitory C2-specific M44-KIR lost during speciation. T44 has only been observed in human KIR, and it first evolved in KIR2DP1F and later, independently emerged in the activating KIR2DS3/5 receptors. The latter are highly attenuated in modern humans but they could have been more effective C2-specific receptors in early humans.
We demonstrate that K44-KIR2DP1F had a higher avidity for C1 than T44-KIR2DP1F had for C2. That hierarchy has been reversed in the modern human system. M44-KIR2DL1 has avidity for C2 that exceeds the avidity of K44-KIR2DL2/3 for C1 (37). The benefit of a stronger inhibitory C2 receptor, could have led to the emergence of KIR2DL1 and thus contributed to the demise of KIR2DP1F. Another potential limitation to KIR2DP1F was that it encoded two functionally different allotypic alternatives. This arrangement maximally provides both inhibitory C1 and C2 both receptors to 50% of the population, but it could also impede the independent evolution and improvement of each receptor. After speciation, with its severe reduction in KIR diversity, provision of both C1 and C2-specific receptors by KIR2DP1F was a major step forward. However, with an increasing number of KIR genes there was greater benefit to be had from inhibitory C1 and C2 receptors encoded by different, dedicated genes. With their emergence, we hypothesize that K44-KIR2DL2/3 began supplanting the functions of K44-KIR2DP1F and that M44-KIR2DL1, with its higher avidity for C2, began to supplant and improve on the functions of T44-KIR2DP1F.
In the modern KIR locus, KIR2DP1 and KIR2DL1 are paired genes that are centrally placed on the centromeric side of the KIR3DP1 and KIR2DL4 framework genes (13). An early feature in human-specific evolution of the locus, was establishment of the KIR A and B haplotype differences in the centromeric region. A striking feature of these differences is that the CenA haplotype encodes strong KIR2DL1 receptors, whereas the CenB haplotypes either encode weak, attenuated receptors or have deleted the KIR2DL1 gene (45). We show that KIR2DP1F had a similar evolutionary trajectory (Figure 8B). The CenA haplotype encodes strong K44-KIR2DP1F receptors that recognize C1, whereas CenB haplotypes either encode weak, attenuated T44-KIR2DP1F that recognize C2 receptors, or have deleted the KIR2DP1F gene. Thus CenB either had, or lacked, both KIR2DL1 and KIR2DP1F. These similar trends show how the genomic module of KIR2DL1 and KIR2DP1F contributed to the genesis of the functionally distinctive CenA and CenB haplotypes, which have been correlated with numerous aspects of human health, disease and therapy (50–54). In comparison to the centromeric region, the telomeric region became populated with variable KIR genes and developed KIR A and B differences at a later stage of human evolution ~1.7mya (13).
The attenuation of KIR2DL1 and KIR2DP1F on CenB involves substitutions that reduce the avidity of ligand binding, reduce or eliminate signal transduction or affect cell-surface expression. In this context, the nucleotide deletion that inactivated KIR2DP1F to give KIR2DP1, could have been introduced as one component, among many, contributing to the attenuation of CenB. But at some point, and with KIR2DP1F functions being superseded by KIR2DL1 and KIR2DL2/3, the balancing selection on KIR2DP1F changed to negative selection. One possible cause of the negative selection was that functions of KIR2DP1F had been taken over by C1-specific KIR2DL2/3 and C2-specific KIR2DL1, rendering it obsolete. A second possible cause was that KIR2DP1F became subverted by a pathogen, or group of pathogens, and became a hindrance to the development of anti-pathogen immunity, rather than an asset.
Because of the strong linkage disequilibrium between KIR2DP1 and KIR2DL1 alleles, the inactivation of KIR2DP1F could not occur by simple fixation of the KIR2DP1F allele in which the deletion arose, because that would have eliminated Cen A and KIR2DL1 polymorphism. Instead, micro gene conversions (55) likely spread the deletion throughout the population of KIR2DP1F alleles, without perturbing KIR2DL1 polymorphism or the balance between CenA and CenB. A side effect of this inactivation mechanism is that KIR2DP1 preserved much functional polymorphism of KIR2DP1F. Consequently, although KIR2DP1 is described as a pseudogene, hence P in its name (19, 56), it is better considered a well-preserved fossil of a versatile HLA-C receptor that educated the NK cells of humans living within the time frame of 6.0 to 0.45 million years ago.
Supplementary Material
Acknowledgments
This study was supported by U.S. National Institutes of Health grants AI22039 and AI17892.
References
- 1.Karlsson EK, Kwiatkowski DP, Sabeti PC. Natural selection and infectious disease in human populations. Nat Rev Genet. 2014;15:379–393. doi: 10.1038/nrg3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parham P, Moffett A. Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nat Rev Immunol. 2013;13:133–144. doi: 10.1038/nri3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrow AD, Trowsdale J. The extended human leukocyte receptor complex: diverse ways of modulating immune responses. Immunological reviews. 2008;224:98–123. doi: 10.1111/j.1600-065X.2008.00653.x. [DOI] [PubMed] [Google Scholar]
- 4.Guethlein LA, Norman PJ, Hilton HH, Parham P. Co-evolution of MHC class I and variable NK cell receptors in placental mammals. Immunol Rev. 2015;267:259–282. doi: 10.1111/imr.12326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guethlein LA, Older Aguilar AM, Abi-Rached L, Parham P. Evolution of killer cell Ig-like receptor (KIR) genes: definition of an orangutan KIR haplotype reveals expansion of lineage III KIR associated with the emergence of MHC-C. J Immunol. 2007;179:491–504. doi: 10.4049/jimmunol.179.1.491. [DOI] [PubMed] [Google Scholar]
- 6.Abi-Rached L, Moesta AK, Rajalingam R, Guethlein LA, Parham P. Human-specific evolution and adaptation led to major qualitative differences in the variable receptors of human and chimpanzee natural killer cells. PLoS Genet. 2010;6:e1001192. doi: 10.1371/journal.pgen.1001192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Older Aguilar AM, Guethlein LA, Adams EJ, Abi-Rached L, Moesta AK, Parham P. Coevolution of killer cell Ig-like receptors with HLA-C to become the major variable regulators of human NK cells. Journal of immunology. 2010;185:4238–4251. doi: 10.4049/jimmunol.1001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guethlein LA, Older Aguilar AM, Abi-Rached L, Parham P. Evolution of killer cell Ig-like receptor (KIR) genes: definition of an orangutan KIR haplotype reveals expansion of lineage III KIR associated with the emergence of MHC-C. Journal of immunology. 2007;179:491–504. doi: 10.4049/jimmunol.179.1.491. [DOI] [PubMed] [Google Scholar]
- 9.Bimber BN, Evans DT. The killer-cell immunoglobulin-like receptors of macaques. Immunological reviews. 2015;267:246–258. doi: 10.1111/imr.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moffett A, Colucci F. Co-evolution of NK receptors and HLA ligands in humans is driven by reproduction. Immunological reviews. 2015;267:283–297. doi: 10.1111/imr.12323. [DOI] [PubMed] [Google Scholar]
- 11.Prado-Martinez J, Sudmant PH, Kidd JM, Li H, Kelley JL, Lorente-Galdos B, Veeramah KR, Woerner AE, O’Connor TD, Santpere G, Cagan A, Theunert C, Casals F, Laayouni H, Munch K, Hobolth A, Halager AE, Malig M, Hernandez-Rodriguez J, Hernando-Herraez I, Prufer K, Pybus M, Johnstone L, Lachmann M, Alkan C, Twigg D, Petit N, Baker C, Hormozdiari F, Fernandez-Callejo M, Dabad M, Wilson ML, Stevison L, Camprubi C, Carvalho T, Ruiz-Herrera A, Vives L, Mele M, Abello T, Kondova I, Bontrop RE, Pusey A, Lankester F, Kiyang JA, Bergl RA, Lonsdorf E, Myers S, Ventura M, Gagneux P, Comas D, Siegismund H, Blanc J, Agueda-Calpena L, Gut M, Fulton L, Tishkoff SA, Mullikin JC, Wilson RK, Gut IG, Gonder MK, Ryder OA, Hahn BH, Navarro A, Akey JM, Bertranpetit J, Reich D, Mailund T, Schierup MH, Hvilsom C, Andres AM, Wall JD, Bustamante CD, Hammer MF, Eichler EE, Marques-Bonet T. Great ape genetic diversity and population history. Nature. 2013;499:471–475. doi: 10.1038/nature12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson MJ, Torkar M, Haude A, Milne S, Jones T, Sheer D, Beck S, Trowsdale J. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci U S A. 2000;97:4778–4783. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pyo CW, Guethlein LA, Vu Q, Wang R, Abi-Rached L, Norman PJ, Marsh SG, Miller JS, Parham P, Geraghty DE. Different patterns of evolution in the centromeric and telomeric regions of group A and B haplotypes of the human killer cell Ig-like receptor locus. PloS one. 2010;5:e15115. doi: 10.1371/journal.pone.0015115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sambrook JG, Bashirova A, Palmer S, Sims S, Trowsdale J, Abi-Rached L, Parham P, Carrington M, Beck S. Single haplotype analysis demonstrates rapid evolution of the killer immunoglobulin-like receptor (KIR) loci in primates. Genome Res. 2005;15:25–35. doi: 10.1101/gr.2381205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graef T, Moesta AK, Norman PJ, Abi-Rached L, Vago L, Older Aguilar AM, Gleimer M, Hammond JA, Guethlein LA, Bushnell DA, Robinson PJ, Parham P. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. J Exp Med. 2009;206:2557–2572. doi: 10.1084/jem.20091010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Traherne JA, Martin M, Ward R, Ohashi M, Pellett F, Gladman D, Middleton D, Carrington M, Trowsdale J. Mechanisms of copy number variation and hybrid gene formation in the KIR immune gene complex. Hum Mol Genet. 2010;19:737–751. doi: 10.1093/hmg/ddp538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winter CC, Long EO. A single amino acid in the p58 killer cell inhibitory receptor controls the ability of natural killer cells to discriminate between the two groups of HLA-C allotypes. J Immunol. 1997;158:4026–4028. [PubMed] [Google Scholar]
- 18.Winter CC, Gumperz JE, Parham P, Long EO, Wagtmann N. Direct Binding and Functional Transfer of NK Cell Inhibitory Receptors Reveal Novel Patterns of HLA-C Allotype Recognition. J Immunol. 1998;161:571–577. [PubMed] [Google Scholar]
- 19.Vilches C, Rajalingam R, Uhrberg M, Gardiner CM, Young NT, Parham P. KIR2DL5, a novel killer-cell receptor with a D0-D2 configuration of Ig-like domains. J Immunol. 2000;164:5797–5804. doi: 10.4049/jimmunol.164.11.5797. [DOI] [PubMed] [Google Scholar]
- 20.Robinson J, Halliwell JA, McWilliam H, Lopez R, Marsh SG. IPD--the Immuno Polymorphism Database. Nucleic Acids Res. 2013;41:D1234–D1240. doi: 10.1093/nar/gks1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SG. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 2015;43:D423–D431. doi: 10.1093/nar/gku1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic acids research. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 26.Green RE, Krause J, Ptak SE, Briggs AW, Ronan MT, Simons JF, Du L, Egholm M, Rothberg JM, Paunovic M, Paabo S. Analysis of one million base pairs of Neanderthal DNA. Nature. 2006;444:330–336. doi: 10.1038/nature05336. [DOI] [PubMed] [Google Scholar]
- 27.Reich D, Green RE, Kircher M, Krause J, Patterson N, Durand EY, Viola B, Briggs AW, Stenzel U, Johnson PL, Maricic T, Good JM, Marques-Bonet T, Alkan C, Fu Q, Mallick S, Li H, Meyer M, Eichler EE, Stoneking M, Richards M, Talamo S, Shunkov MV, Derevianko AP, Hublin JJ, Kelso J, Slatkin M, Paabo S. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–1060. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hilton HG, Moesta AK, Guethlein LA, Blokhuis J, Parham P, Norman PJ. The production of KIR-Fc fusion proteins and their use in a multiplex HLA class I binding assay. Journal of immunological methods. 2015;425:79–87. doi: 10.1016/j.jim.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol. 2008;180:3969–3979. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- 30.Meyer M, Kircher M, Gansauge MT, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prufer K, de Filippo C, Sudmant PH, Alkan C, Fu Q, Do R, Rohland N, Tandon A, Siebauer M, Green RE, Bryc K, Briggs AW, Stenzel U, Dabney J, Shendure J, Kitzman J, Hammer MF, Shunkov MV, Derevianko AP, Patterson N, Andres AM, Eichler EE, Slatkin M, Reich D, Kelso J, Paabo S. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prufer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, Li H, Mallick S, Dannemann M, Fu Q, Kircher M, Kuhlwilm M, Lachmann M, Meyer M, Ongyerth M, Siebauer M, Theunert C, Tandon A, Moorjani P, Pickrell J, Mullikin JC, Vohr SH, Green RE, Hellmann I, Johnson PL, Blanche H, Cann H, Kitzman JO, Shendure J, Eichler EE, Lein ES, Bakken TE, Golovanova LV, Doronichev VB, Shunkov MV, Derevianko AP, Viola B, Slatkin M, Reich D, Kelso J, Paabo S. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mendez FL, Poznik GD, Castellano S, Bustamante CD. The Divergence of Neandertal and Modern Human Y Chromosomes. Am J Hum Genet. 2016;98:728–734. doi: 10.1016/j.ajhg.2016.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young NM, Capellini TD, Roach NT, Alemseged Z. Fossil hominin shoulders support an African ape-like last common ancestor of humans and chimpanzees. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:11829–11834. doi: 10.1073/pnas.1511220112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGrew WC. In search of the last common ancestor: new findings on wild chimpanzees. Philos Trans R Soc Lond B Biol Sci. 2010;365:3267–3276. doi: 10.1098/rstb.2010.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duda P, Zrzavy J. Evolution of life history and behavior in Hominidae: towards phylogenetic reconstruction of the chimpanzee-human last common ancestor. J Hum Evol. 2013;65:424–446. doi: 10.1016/j.jhevol.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 36.Reich DE, Cargill M, Bolk S, Ireland J, Sabeti PC, Richter DJ, Lavery T, Kouyoumjian R, Farhadian SF, Ward R, Lander ES. Linkage disequilibrium in the human genome. Nature. 2001;411:199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- 37.Hilton HG, Vago L, Older Aguilar AM, Moesta AK, Graef T, Abi-Rached L, Norman PJ, Guethlein LA, Fleischhauer K, Parham P. Mutation at positively selected positions in the binding site for HLA-C shows that KIR2DL1 is a more refined but less adaptable NK cell receptor than KIR2DL3. Journal of immunology. 2012;189:1418–1430. doi: 10.4049/jimmunol.1100431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Norman PJ, Hollenbach JA, Nemat-Gorgani N, Guethlein LA, Hilton HG, Pando MJ, Koram KA, Riley EM, Abi-Rached L, Parham P. Co-evolution of human leukocyte antigen (HLA) class I ligands with killer-cell immunoglobulin-like receptors (KIR) in a genetically diverse population of sub-Saharan Africans. PLoS Genet. 2013;9:e1003938. doi: 10.1371/journal.pgen.1003938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boyington JC, Motyka SA, Schuck P, Brooks AG, Sun PD. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature. 2000;405:537–543. doi: 10.1038/35014520. [DOI] [PubMed] [Google Scholar]
- 40.Fan QR, Long EO, Wiley DC. Crystal structure of the human natural killer cell inhibitory receptor KIR2DL1-HLA-Cw4 complex. Nat Immunol. 2001;2:452–460. doi: 10.1038/87766. [DOI] [PubMed] [Google Scholar]
- 41.Burshtyn DN, Lam AS, Weston M, Gupta N, Warmerdam PA, Long EO. Conserved residues amino-terminal of cytoplasmic tyrosines contribute to the SHP-1-mediated inhibitory function of killer cell Ig-like receptors. Journal of immunology. 1999;162:897–902. [PubMed] [Google Scholar]
- 42.Bruhns P, Marchetti P, Fridman WH, Vivier E, Daeron M. Differential roles of N- and C-terminal immunoreceptor tyrosine-based inhibition motifs during inhibition of cell activation by killer cell inhibitory receptors. Journal of immunology. 1999;162:3168–3175. [PubMed] [Google Scholar]
- 43.Fry AM, Lanier LL, Weiss A. Phosphotyrosines in the killer cell inhibitory receptor motif of NKB1 are required for negative signaling and for association with protein tyrosine phosphatase 1C. J Exp Med. 1996;184:295–300. doi: 10.1084/jem.184.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burshtyn DN, Yang W, Yi T, Long EO. A novel phosphotyrosine motif with a critical amino acid at position −2 for the SH2 domain-mediated activation of the tyrosine phosphatase SHP-1. J Biol Chem. 1997;272:13066–13072. doi: 10.1074/jbc.272.20.13066. [DOI] [PubMed] [Google Scholar]
- 45.Hilton HG, Guethlein LA, Goyos A, Nemat-Gorgani N, Bushnell DA, Norman PJ, Parham P. Polymorphic HLA-C Receptors Balance the Functional Characteristics of KIR Haplotypes. J Immunol. 2015;195:3160–3170. doi: 10.4049/jimmunol.1501358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gonzalez-Galarza FF, Takeshita LY, Santos EJ, Kempson F, Maia MH, da Silva AL, Teles e Silva AL, Ghattaoraya GS, Alfirevic A, Jones AR, Middleton D. Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic acids research. 2015;43:D784–D788. doi: 10.1093/nar/gku1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson J, Halliwell JA, McWilliam H, Lopez R, Parham P, Marsh SG. The IMGT/HLA database. Nucleic Acids Res. 2013;41:D1222–D1227. doi: 10.1093/nar/gks949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parham P, Norman PJ, Abi-Rached L, Hilton HG, Guethlein LA. Review: Immunogenetics of human placentation. Placenta. 2012;(33 Suppl):S71–S80. doi: 10.1016/j.placenta.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abi-Rached L, Kuhl H, Roos C, ten Hallers B, Zhu B, Carbone L, de Jong PJ, Mootnick AR, Knaust F, Reinhardt R, Parham P, Walter L. A small, variable, and irregular killer cell Ig-like receptor locus accompanies the absence of MHC-C and MHC-G in gibbons. J Immunol. 2010;184:1379–1391. doi: 10.4049/jimmunol.0903016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cooley S, Weisdorf DJ, Guethlein LA, Klein JP, Wang T, Marsh SG, Spellman S, Haagenson MD, Saeturn K, Ladner M, Trachtenberg E, Parham P, Miller JS. Donor killer cell Ig-like receptor B haplotypes, recipient HLA-C1, and HLA-C mismatch enhance the clinical benefit of unrelated transplantation for acute myelogenous leukemia. Journal of immunology. 2014;192:4592–4600. doi: 10.4049/jimmunol.1302517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hiby SE, Walker JJ, O’Shaughnessy K M, Redman CW, Carrington M, Trowsdale J, Moffett A. Combinations of maternal KIR and fetal HLA-C genes influence the risk of preeclampsia and reproductive success. J Exp Med. 2004;200:957–965. doi: 10.1084/jem.20041214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakimuli A, Chazara O, Hiby SE, Farrell L, Tukwasibwe S, Jayaraman J, Traherne JA, Trowsdale J, Colucci F, Lougee E, Vaughan RW, Elliott AM, Byamugisha J, Kaleebu P, Mirembe F, Nemat-Gorgani N, Parham P, Norman PJ, Moffett A. A KIR B centromeric region present in Africans but not Europeans protects pregnant women from pre-eclampsia. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:845–850. doi: 10.1073/pnas.1413453112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suzuki Y, Hamamoto Y, Ogasawara Y, Ishikawa K, Yoshikawa Y, Sasazuki T, Muto M. Genetic polymorphisms of killer cell immunoglobulin-like receptors are associated with susceptibility to psoriasis vulgaris. J Invest Dermatol. 2004;122:1133–1136. doi: 10.1111/j.0022-202X.2004.22517.x. [DOI] [PubMed] [Google Scholar]
- 54.Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, Cheng J, Goedert JJ, Vlahov D, Hilgartner M, Cox S, Little AM, Alexander GJ, Cramp ME, O’Brien SJ, Rosenberg WM, Thomas DL, Carrington M. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 55.Klitz W, Hedrick P, Louis EJ. New reservoirs of HLA alleles: pools of rare variants enhance immune defense. Trends Genet. 2012;28:480–486. doi: 10.1016/j.tig.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 56.Marsh SG, Parham P, Dupont B, Geraghty DE, Trowsdale J, Middleton D, Vilches C, Carrington M, Witt C, Guethlein LA, Shilling H, Garcia CA, Hsu KC, Wain H. Killer-cell immunoglobulin-like receptor (KIR) nomenclature report, 2002. Immunogenetics. 2003;55:220–226. doi: 10.1007/s00251-003-0571-z. [DOI] [PubMed] [Google Scholar]
- 57.Hilton HG, Norman PJ, Nemat-Gorgani N, Goyos A, Hollenbach JA, Henn BM, Gignoux CR, Guethlein LA, Parham P. Loss and Gain of Natural Killer Cell Receptor Function in an African Hunter-Gatherer Population. PLoS Genet. 2015;11:e1005439. doi: 10.1371/journal.pgen.1005439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.