Abstract
HIV-infected patients of all ages frequently underperform in responsiveness to seasonal influenza vaccination despite virologic control of HIV. Molecular mechanisms governing this impairment as well as predictive biomarkers for responsiveness remain unknown. This study was performed in pre-vaccination samples (T0) of HIV-infected children who received the 2012–2013 seasonal influenza vaccine. Response status was determined based on established criteria of hemagglutination inhibition (HAI) titer; participants with HAI ≥ 1:40 plus ≥ 4-fold increase over T0 at three weeks post-vaccination (T1) were designated as responders. All children had a history of prior influenza vaccinations. At T0, frequencies of CD4 T cell subsets, including peripheral T follicular helper (pTfh) cells which provide help to B cells for developing into Ab secreting cells were similar between responders and non-responders. However, in response to in vitro stimulation with H1N1 antigen, differential gene expression related to pTfh function was observed by Fluidigm high density RT-PCR between responders and non-responders. In responders, H1N1 stimulation at pre-vaccination also resulted in CXCR5 induction (mRNA and protein) in CD4 T cells and IL21 gene induction in pTfh cells that strongly associated with H1N1-specific B cell responses post-vaccination. In contrast, CD4 T cells of non-responders exhibited increased expression of IL2 and STAT5 genes which are known to antagonize pTfh function. These results suggest that the quality of pTfh at the time of immunization are important for influenza vaccine responses and provide a rationale for targeted, ex vivo antigen-driven molecular profiling of purified immune cells to detect predictive biomarkers of vaccine response.
Introduction
Understanding how the immune systems of immunocompromised individuals respond to current vaccines is important in order to develop tailored vaccines. Influenza viruses cause illnesses with higher morbidity and mortality in patients with acquired immunodeficiencies and can lead to deadly pandemics due to antigenic drift. Seasonal vaccination is recommended in young children, the elderly, and in immunocompromised individuals to prevent influenza infection and its complications. Protection from influenza is mediated through neutralizing antibodies against viral surface proteins hemagglutinin (HA) and neuraminidase (NA). The dilution of serum capable of blocking the hemagglutination reaction between HA and red blood cells defines the titer of influenza antibodies. A titer of ≥1:40 is considered protective in healthy adults, and together with at least a four-fold increase in titer post vaccination represents a positive response to vaccination. Antibody responses to the influenza vaccine are generated following the germinal center (GC) reaction that occurs between B cells and T follicular helper (Tfh) cells, a CD4 T cell subset that is critical for affinity maturation and somatic hypermutation in antigen (Ag)-primed B cells (1). Binding of the surface molecule CXC chemokine receptor type 5 (CXCR5) on Tfh to its ligand CXCL13 is required for the homing of Tfh to lymphoid follicles. The cytokine interleukin (IL)-21 is an important secretory product of Tfh and plays a dominant role in the GC reaction (2, 3). In the peripheral circulation, a subset of circulating memory CD4 T cells that express CXCR5 are referred to as peripheral Tfh (pTfh) cells and manifest functional properties of the GC Tfh cells, including the capacity for IL-21 secretion which currently represents the strongest correlate of Tfh function in the peripheral blood (4). Previous studies have demonstrated a relationship between pTfh and B cell function in vaccine responders at 3–4 weeks post-influenza vaccination, including expansion of pTfh, concurrently with influenza H1N1 antigen-induced production of IL-21in pTfh in vitro and ‘help’ by purified pTfh to autologous B cells in co-culture experiments for H1N1-specific IgG production (5–7).
In patients with HIV infection, seasonal influenza vaccination has emerged as a useful model for probing immune competency by evaluation of serologic response to the vaccine (5, 6, 8–10). In patients on combination antiretroviral therapy (cART), we have observed pTfh functional deficiencies in response to H1N1 flu antigen that worsen with aging (8). However, repeated immunizations with antigens such as H1N1, which has been retained in the vaccine since its introduction in 2009 to address the H1N1 pandemic, has altered the serologic landscape, and baseline titers can be high (11–13). Although informative biomarkers of influenza vaccine response and efficacy such as early transcriptional changes in blood (14–16) and serum antibodies to NA post-vaccination are under investigation (17), the immunocompromised populations, such as HIV-infected, will require independent testing and validation in order to design appropriate vaccination regimens (18–20).
In the study described herein, we employed a targeted multiplex RT-PCR approach to evaluate pre-vaccination gene expression patterns by relevant cell types (i.e. CD4 T and pTfh cells) in HIV-infected children and adolescents. Our study was focused on pre-vaccination samples that were stimulated with influenza vaccine antigen (H1N1) in vitro to explore predictive measures of vaccine response. We hypothesized that the pattern of expression of a selected set of genes associated with immune function in CD4 and pTfh would provide unique insight into the quality of pre-existing memory pTfh cells in the clinical setting and especially in the context of vaccine responses, regardless of pre-existing titers of HAI due to previous vaccinations. Our results suggest that ex-vivo antigen stimulation is a useful approach to dissect molecular mechanisms of H1N1 memory responses and for identifying novel correlates of response.
Materials and Methods
Study Participants and Immunization
Participants were recruited from Children’s Hospital Bambino Gesù (Rome, Italy). ART-treated HIV-1 vertically infected individuals and HIV-negative age-matched controls were enrolled between September and November 2012. Written informed consent was obtained from all subjects or parents/legal guardians before enrollment and the ethical committees of the Bambino Gesù Children’s Hospital approved the study. Participants were immunized with one single dose of Inactivated Influenza Vaccine Trivalent Types A and B (Split Virion) VAXIGRIP® (Sanofi Pasteur). The strains for the 2012–2013 season include: A/California/7/2009 (H1N1) pdm09-like strain (abbreviated as H1N1), A/Victoria/361/2011 (H3N2)-like strain (abbreviated as H3N2) and B/Wisconsin/1/2010-like strain (abbreviated as B). Peripheral blood mononuclear cells (PBMC), serum and plasma were collected pre- (T0) and post-vaccination (21 days; T1). An accurate clinical survey has been performed over the course of the vaccination trial in order to exclude natural infection.
Hemagglutination inhibition (HAI) assay
Antibody titers to H1N1, H3N2, and B influenza strains were evaluated separately by HAI assay. The HAI assay was performed as previously described (21). HAI titers are expressed as the reciprocal of the highest serum dilution at which hemagglutination was prevented.
ELISpot Assay
PBMC from T0 and T1 were thawed and polyclonally activated in vitro for 5 days with CpG (2.5mg/ml) as described (13). Briefly, ELISpot 96-well filtration plates (Millipore) were coated with purified H1N1, H3N2 and B influenza inactivated virus particles and subsequently loaded with 2 × 105 cells/well. Processed membranes were scanned with an Eli.Scan and counted with the ELISpot Analysis Software V5.1 (all from A.EL.VIS).
Enzyme-linked immunosorbent assay (ELISA)
Plasma IL-21 titers were measured using the human IL-21 platinum ELISA kit (eBioscience, San Diego, CA, USA), following the manufacturer’s instructions.
FACS Sorting and RNA Extraction
A single vial of PBMC (T0) was thawed, counted with Countess Automated Cell counter (Life Technologies), and reconstituted in complete RPMI medium at a concentration of 5 x 106 PBMC/mL. Cells were cultured in the presence or absence of H1N1 (A/California/09) peptides (5ug/mL) overnight (16 hours). Following the culture period, cells were labeled with monoclonal antibodies against CD3 (AmCyan), CD4 (PerCP Cy5.5), CD45RO (ECD), CCR7 (Alexafluor 700), and CXCR5 (Alexafluor 647) and a live/dead marker (ViViD, Molecular Probes). All antibodies were previously titrated. 4-way sorting mode was used on FACS Aria II (BD Biosciences) to sort cell populations of interest. 500 cells per subset were sorted directly into 1.1ml tubes containing CellsDirect one-step PCR buffer and pooled TaqMan gene expression assays (2X CellsDirect Reaction mix 5ul, Superscript III + Taq polymerase 0.5ul, 0.2X TaqMan primer pool 2.5ul, Resuspension Buffer 1ul). After sorting, tubes containing cells were immediately centrifuged (3,000 RPM for 3 minutes) and kept on ice. Samples were subsequently transferred to PCR tubes and reverse transcription and target-specific pre-amplification was performed on a C1000 Thermal Cycler (Bio Rad) with the following scheme (50°C for 20min, 95°C for 2min, 95°C for 15sec, 60°C for 4min, last 2 steps repeated for 18 cycles). Resulting cDNA was diluted 1:1 with TE Buffer (10mM Tris, 1mM EDTA, pH 8.0) and stored at −20°C until further analysis.
Fluidigm BioMark RT-PCR
Previously amplified samples were loaded onto 96.96 dynamic array IFC (Fluidigm) according to the manufacturer’s instructions. Briefly, the assay pre-mix was prepared with 3ul 20X TaqMan Gene Expression Assay (Applied Biosystems) and 3ul 2X Assay Loading Reagent (Fluidigm). The sample pre-mix was prepared with 3ul TaqMan Universal PCR Master Mix (2X, Applied Biosystems), 0.3ul 20X GE Sample Loading Reagent (Fluidigm), and 2.7ul of diluted cDNA (67.5 cell equivalent). Total volume of 5ul was loaded into appropriate sample and assay wells. The IFCs were primed in the Fluidigm Controller HX unit prior to loading onto BioMark HD instrument for standard RT-PCR. The panel of TaqMan gene expression assays have been qualified on Human PBMC and T lymphocytes as previously described (22). Cycle threshold (Ct) values derived from BioMark experiments were normalized to 500 cells in cases where the sorted population did not reach 500 according to the following calculations: (1) determine y using 67.5/500 = y/x, where x is the number of cells sorted and y is the cDNA cell equivalent loaded onto the IFC. (2) Normalized Ct= Reported Ct − log2 (67.5/y). Expression threshold (Et) values or log2 expression were used for all analysis and were determined using Et = 40 − Ct.
Bioinformatics and Statistical analysis
Et values were analyzed using SingulaR analysis toolset 3.0 (Fluidigm) package in R (software R 3.0.2 GUI 1.62). Expression plot of samples, expression plot overviews and identification of outliers was performed through this software as well as principle component analysis and analysis of variance (ANOVA) which was used to identify differentially expressed genes between participant groups and between cell subsets of the same group. Fold change was determined using the mean Et for each group. Automated paired analysis between unstimulated and stimulated samples from the same individual was carried out using student’s T test. Correlation matrix analysis and multivariate linear regression models were performed using Mast and Stats packages also in R. Kolmogorov-Smirnov normality test was performed on single gene data sets and on total 96 gene sets in order to select statistical analysis to perform for correlation matrix analysis. Pearson correlation coefficient was performed if data was normally distributed and Spearman correlation if non-parametric test was needed. Linear regression plots were generated using Graphpad (Prism 6.0).
Results
HIV-infected participants exhibit heterogeneous serological response to seasonal influenza vaccine
We measured serologic response to influenza vaccination in 22 HIV perinatally infected youth (Table 1) by hemagglutination inhibition (HAI) assay with each influenza strain in the vaccine in serum samples collected before (T0) and 3wks after vaccination (T1). Higher baseline titers against H1N1 were observed in HIV-infected compared to HIV-uninfected, age-matched controls, indicative of repeated vaccination during the preceding years (Figure 1A). Baseline titers against the other strains, H3N2 and B, were not different between the HIV-infected and –uninfected groups. Increases in antibody titer following vaccination were more robust to H1N1 and H3N2 (Figure 1B), consistent with poor immunogenicity of the Influenza B strain (23). Serum titers to H1N1 increased at least 4-fold in 90% (9/10) of HIV uninfected and only 50% (11/22) HIV-infected individuals. Those participants who demonstrated good antibody responses as measured by HAI titer tended to have greater responses to the other vaccine antigens as well (Supplemental Figure 1A). Vaccinees were characterized as responders (R) if they exhibited a serum H1N1 titer of at least 1:40 and a 4-fold increase above baseline at T1, and non-responders (NR) if they did not meet the selection criteria. Response to H1N1 was confirmed by memory B cell ELISpot assay performed at T1 which also showed greater response in R compared to NR (Supplemental Figure 1B).
Table 1.
Characteristics of Study Participants
| HIV negative | HIV+ Responder | HIV+ Non-Responder | |
|---|---|---|---|
| Age years mean (SEM) | 14.3 (3.4) | 13.7 (2.4) | 15.2 (2.2) |
| Participants, n (female) | 10 (5) | 11 (6) | 11 (8) |
| Lymphocytes/mm3 (SEM) | 3063 (428) | 2961 (363) | 2494 (279) |
| CD4+T cells (%) mean (SEM) | 29.8 (6.3) | 32.5 (6.1) | 38.0 (4.9) |
| HIV RNA <50c/ml, n | NA | 10 | 11 |
| Prior Influenza vaccinations between 2009–2012 (1/2/3) | NA | 5/4/2 | 3/6/3 |
Figure 1. Serological Response to Influenza Vaccine Strains.
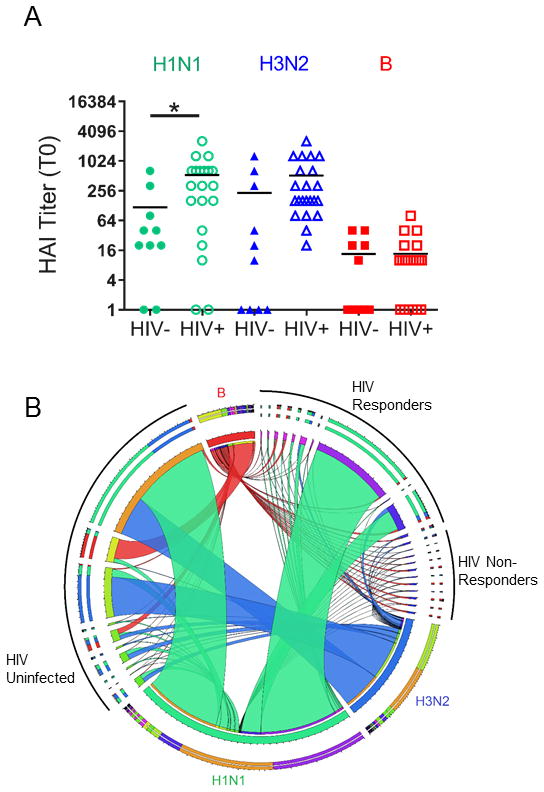
A) Baseline serum titers from HIV-infected (n=22) and –uninfected (n=10) individuals for each of the influenza strains contained in the 2012–2013 vaccine as determined by HAI Assay. Each symbol represents one person and horizontal lines show mean of the data. Statistical significance was determined by student’s T test, *denotes p-value<0.05. B) Circos plot (49) showing fold change in serum titer as determined by HAI assay for each participant and influenza strain. Ribbons originate from a node representing each participant and connect to individual vaccine strains (Green, H1N1; Blue, H3N2; Red, B). Ribbon thickness represents the fold increase from T0 to T1. Study participants were clustered into 3 groups: HIV Uninfected, HIV responders and HIV non-responders indicated by black arcs outside the circus plot.
CD4 T cells from HIV+ vaccine responders upregulate CXCR5 in response to H1N1 stimulation
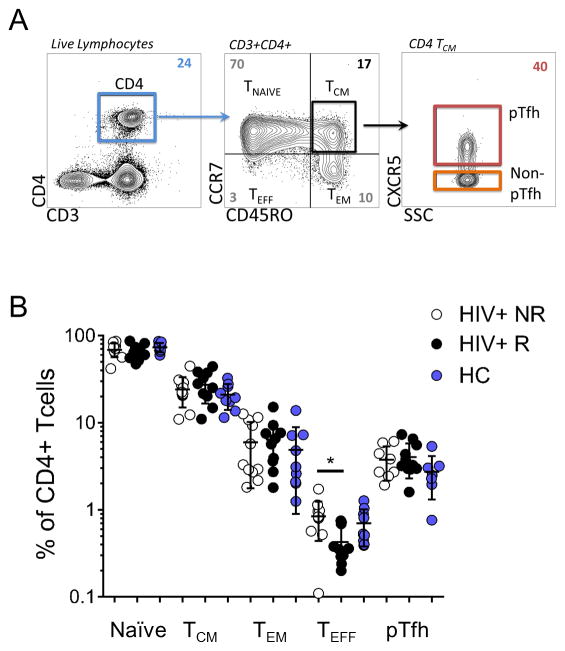
Phenotypic analysis of CD4 T cells was performed by flow cytometry following stimulation with or without H1N1 antigen to determine subset frequencies in the participant groups (Figure 2A). Comparable frequencies were observed for CD4 T cells (Table 1) and maturation subsets with the exception of effector/terminally differentiated subset (Teff; CD45ROneg CCR7neg), which was significantly higher in NR compared to R (p=0.034, Figure 2B) though it represented less than 2% of the CD4 population. Within the T Central Memory compartment (TCM, CD45RO+CCR7+), pTfh were characterized as CXCR5+ and exhibited similar frequencies in HIV+R, HIV+NR and HC.
Figure 2. Characterization of Pre-vaccination CD4 T cell subset frequencies in Study Participants.
A) Representative dot plots of flow cytometry gating scheme for identifying T cell subsets and pTfh cells. Colored boxes designate populations of interest in sorting experiments. B) Summary data for frequencies of CD4 T cell subsets from study participant groups (HC, Healthy Control; HIV+ R, HIV Responders; HIV+NR, HIV Non-Responders). Frequencies shown are out of Live CD3+CD4+ and statistical significance was determined by student’s T test, *denotes p-value <0.05.
Next we examined the ability of pTfh cells from HIV+ study participants to respond to in vitro antigen stimulation. Paired analysis of CXCR5 expression on TCM showed a significant increase following in vitro stimulation with H1N1 antigen overnight (16hr) only in HIV+ R (Figure 3). These data suggest that memory CD4 from HIV+ NR might harbor functional defects that could impair vaccine responsiveness.
Figure 3. Central memory cells from vaccine responders induce CXCR5 expression in response to H1N1.
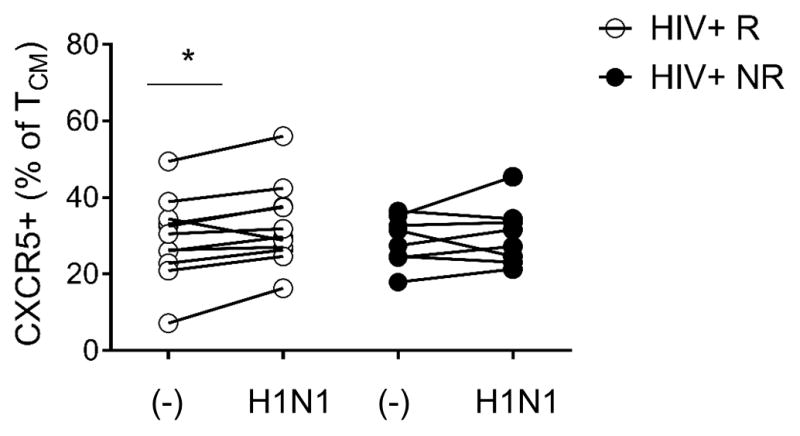
Frequency of CD4 TCM (CD45RO+CCR7+) expressing CXCR5 with (H1N1) and without (−) overnight in vitro H1N1 antigen (5μg/ml) stimulation in each study group. Lines connect measurements obtained from the same individual, statistical significance was determined by paired t test, *denotes p-value<0.05.
Targeted transcriptional analysis in T cell subsets distinguish HIV+ vaccine responders and non-responders
To evaluate potential T cell-derived molecular biomarkers for predicting seasonal influenza vaccine responses and to dissect transcriptional mechanisms driving the upregulation of CXCR5 expression in H1N1-stimulated CD4 T cells, we performed targeted gene expression analysis in purified CD4 T cells, pTfh, and non-pTfh cells (TCM CXCR5neg) (Supplemental Figure 2A). The Fluidigm BioMark platform was used to analyze expression of a custom panel of genes involved in immune activation and inflammation, Tfh function, TCR signaling, cytokine and chemokine signaling, and anti-viral and interferon (IFN)-inducible factors (Supplemental Table 1). Using this gene panel, cell subsets (PBMC, CD4 T, pTfh, and non-pTfh) distinctly segregated from one another by principle component analysis, though clustering was not evident between subsets from HIV+ and HIV negative individuals (Supplemental Figure 2B).
For analysis of gene expression levels in pre-vaccination samples, first we directly compared R and NR groups using analysis of variance (ANOVA) and identified genes exhibiting differential expression between the 2 groups for each cell type. In the absence of any in vitro stimulation, more differentially expressed genes (DEGs) were identified in CD4 T cells (R vs. NR) compared to purified subsets (pTfh and non-pTfh) and few were overlapping between total CD4 and subsets (Figure 4A). This observation demonstrates that changes in gene expression from purified CD4 T cell subsets are not detectable in analysis of total CD4 T cells. This concept was also revealed when evaluating DEGs in PBMC compared to CD4 T cells and/or subsets; only 2 genes (BCL6 and IL6ST) overlapped between PBMC and CD4, and they showed enrichment in different groups (Supplemental Figure 3A). Differential gene expression in PBMC between R and NR was dominated by genes with higher expression in NR, while in CD4 T cell analysis higher number of DEGs were expressed in R, highlighting divergent functions for different cell types (Figure 4A). In pTfh and non-pTfh, there were few DEGs that reached significance (p<0.05), and the top 5 DEGs according to p-value were used for the next analysis to explore the relationship between baseline gene expression in CD4 TCM subsets and vaccine response.
Figure 4. Differential gene expression analysis in Pre-Vaccination CD4, pTfh and Non-pTfh cells between HIV+ Flu vaccine responders and Non-responders.
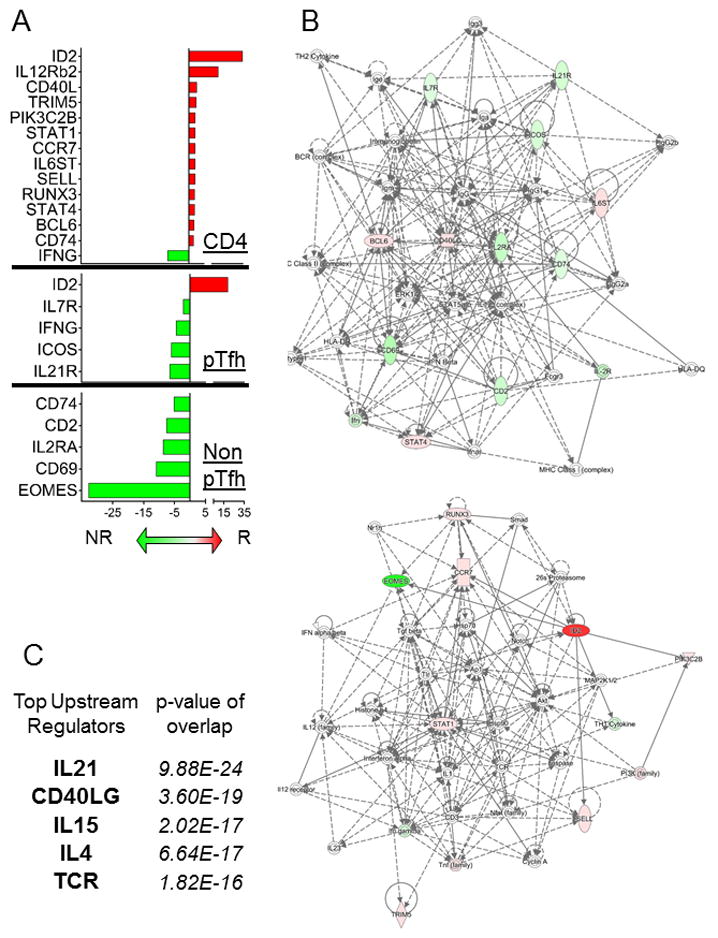
A) Bar graph showing the average fold change in gene expression between R and NR for CD4 T cells (top), pTfh (middle), and non-pTfh (bottom) using fold change cutoff >1.5. Red bars correspond to gene expression that was higher in R compared to NR and Green bars indicate higher gene expression in NR compared to R. B) Gene networks were generated using Ingenuity Pathway Analysis (IPA) software and combined DEGs from the 3 cell types in (A) as input. Colors in the network are defined as follows: red indicates up-regulation in R vs NR, green indicates up-regulation in NR vs R, and white indicates the molecule was added from the Ingenuity Knowledge Base. Solid lines indicate direct interaction and dashed lines indicate indirect interaction. C) The top 5 upstream regulators were determined using IPA software and combined DEGs from the 3 cell types in (A) as input. The overlap p-values show statistically significant overlap by Fisher’s exact test between dataset genes and the genes that are known to be regulated by a particular molecule.
Network analysis was performed on enriched genes identified in CD4 T cells and subsets from Figure 4A using Ingenuity Pathway Analysis (IPA), shown in Figure 4B. IPA generated two networks for the enriched genes; one contained mostly genes enriched in NR and the other had mostly genes enriched in R. IL-12 complex, STAT5, and ERK1/2 were major nodes in the network displaying NR enriched genes. Multiple markers of immune activation such as ICOS, IL2RA, CD69, and IFNG were enriched in NR in the absence of stimulation. The genes enriched in R contained multiple transcription factors such as RUNX3, STAT1, STAT4, BCL6, and ID2. Inhibitor of DNA binding (ID) 2 and ID3 have been shown to be involved in T lineage developmental progression (24). ID2 was strongly enriched in CD4 (34-fold) and pTfh (19-fold) from R vaccine participants, while ID3 was not differentially expressed in these samples. The top 5 upstream regulators of enriched genes were IL21, CD40LG, IL15, IL4, and TCR (Figure 4C).
IL21 gene expression in pTfh following H1N1 stimulation defines vaccine responders
In the next set of analyses, we sought to evaluate the activation potential of immune cells from pre-vaccination samples. We compared gene expression from CD4 T, pTfh, and Non-pTfh cells from R and NR following in vitro stimulation with H1N1 to identify DEGs that may be directly related to vaccine response (Figure 5A). As expected, the IL21 gene was expressed in pTfh cells and was not detectable in non-pTfh or CD4 T cells. Importantly IL21 expression was higher in pTfh from R following in vitro stimulation together with upregulation of other critical Tfh functional markers CD40LG, ICOS, and IL6ST. Non-pTfh cells exclusively exhibited DEGs that were expressed higher in NR, suggesting immune inhibitory roles for IFNG, DUSP4, RUNX3, and IL21R in this cell type. In CD4 T cells, expression of the chemokine receptor CXCR4 was strongly associated with NR status. Increased expression of HIV co-receptors on CD4 T cells is a concern in HIV-infected individuals because they could enhance de novo infection of target cells. Similar analysis in PBMC revealed 3 DEGs between R and NR following H1N1 stimulation: IL21R, IRF4, and IL2 (Supplementary Figure 3B). Of these genes only IL21R was observed in DEG analysis following in vitro stimulation in non-pTfh cells. Network Analysis of DEGs from H1N1-stimulated CD4 T, pTfh and non-pTfh cells from the two vaccine response groups revealed 2 gene networks; the first related to TCR signaling with CD4, ZAP70 and NFAT complex as major nodes, and the second related to IL21 signaling (Figure 5B). Increased CXCR5 gene expression in CD4 T cells following H1N1 stimulation supports the flow cytometry data showing an increase in CXCR5 expression in central memory T cells (Figure 3). ID2 expression was maintained in CD4 and pTfh cells at high levels in R compared to NR, suggesting that its expression is not related to activation status but rather differentiation or maturation state of the cell. Both transcription regulators ID2 and ID3 were included in the top 5 upstream regulators for the DEGs identified, along with IL21, CD40LG, and TCR (Figure 5C).
Figure 5. Differential gene expression analysis in H1N1-Stimulated Pre-Vaccination CD4, pTfh and Non-pTfh cells between HIV+ Flu vaccine responders and Non-responders.
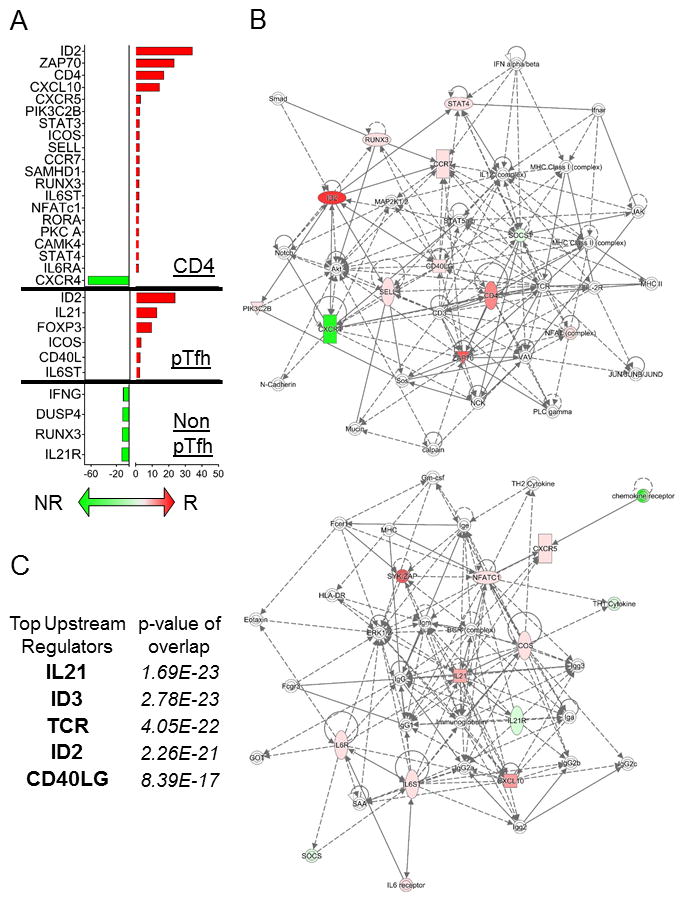
A) Bar graph showing the average fold change in gene expression between R and NR for CD4 T cells (top), pTfh (middle), and non-pTfh (bottom) using ANOVA p-value <0.05 and fold change >1.5 as cutoffs. Red bars correspond to gene expression that was higher in R compared to NR and Green bars indicate higher gene expression in NR compared to R. B) Gene networks were generated using Ingenuity Pathway Analysis (IPA) software and combined DEGs from the 3 cell types in (A) as input. Colors in the network are defined as follows: red indicates up-regulation in R vs NR, green indicates up-regulation in NR vs R, and white indicates the molecule was added from the Ingenuity Knowledge Base. Solid lines indicate direct interaction and dashed lines indicate indirect interaction. C) The top 5 upstream regulators were determined using IPA software and combined DEGs from the 3 cell types in (A) as input. The overlap p-values show statistically significant overlap by Fisher’s exact test between dataset genes and the genes that are known to be regulated by a particular molecule.
Responders and non-responders exhibit distinct H1N1-induced gene expression patterns
To further dissect gene expression changes following H1N1 stimulation in CD4 T cells and pTfh, we performed paired analysis using data from each participant to examine how gene expression changed on an individual level following H1N1 stimulation. This analysis revealed 12 common genes whose expression was significantly induced in CD4 T cells from both R and NR in response to H1N1 stimulation. These genes consisted of IFN-inducible and antiviral genes (OAS1, IFIT2, STAT1, MX1, FAS, IRF4, BST2) (Figure 6A), immune activation genes (CD74, ICOS, IL2RA) and transcription factors (FOXP3, BCL6). Nineteen genes were differentially induced (termed DIGs for differentially induced genes) in CD4 T cells and included 16 that were upregulated exclusively in R and 3 in NR. DIGs in R included chemokine signaling molecules (CCR2, CXCL10, CCR7) along with classical activation markers (CD69, CD38). DIGs observed only in NR included IL2, STAT5A, and IL21R.
Figure 6. Gene induction in response to H1N1-stimulation in Pre-vaccination CD4 and pTfh cells in HIV+ responders and Non-responders to Influenza Vaccine.

Dual column heatmaps show average fold change in gene expression from H1N1-stimulated cells compared to unstimulated cells in A) CD4 T cells and B) pTfh cells. Values were derived by calculating the delta for each study participant such that (Et_H1N1) − (Et_Unstim) =Delta, if the group of Deltas was found to be significant (p-value<0.05) for the group (R or NR) then the average fold change was calculated and displayed in the heatmap as a color based on the scale provided to the right of each heatmap.
Similar to analysis in CD4 T cells, DIG expression patterns in pTfh cells showed induction of IFN-inducible genes (IFIT2, IRF4, MX1, OAS1, STAT1) that were common in R and NR (Figure 6B). However, genes that exhibited significant changes only in R were mostly different than what was observed in the analysis of CD4 T cells, such as ABCB1, DUSP4, BST2, and IL21. Induction of SAMHD1 was observed in both CD4 and pTfh cells from R. IL21 is the signature cytokine for Tfh function required for high affinity antibody production from B cells, thus IL21 induction following antigen stimulation in pTfh from R supports the notion that pTfh can exhibit function similar to LN Tfh. ABCB1 and DUSP4 were the only genes that showed reduced expression following H1N1 stimulation suggesting regulatory roles for these genes. ABCB1 (MDR1) encodes P-glycoprotein, a membrane transporter enriched in pathogenic Th17 cells (25). DUSP4 is a phosphatase and negative regulator of the mitogen-activated protein kinase (MAPK) superfamily and has been shown to contribute to dampened CD4 T memory responses in the elderly (26). The genes induced in NR in response to H1N1 stimulation were FAS, CCR5, LAG3, and IL2. CCR5 is a chemokine receptor upregulated in response to inflammation that plays a role in homing to sites of inflammation, and is also a co-receptor for HIV entry. LAG3 is an immune checkpoint receptor which dampens T cell activation through competing with CD4 for MHC binding (27, 28). Finally, IL2 is a crucial cytokine for immune cell development and function which is produced primarily by activated CD4+ T cells. IL2 signaling, however, has been shown to reduce antigen-specific humoral immunity by directly suppressing Tfh cell formation (29). IL2 induction was observed in PBMC, CD4, and pTfh cells from NR pointing to a global increase in IL-2 being associated with poor vaccine responses. Overall, these data show that in vitro H1N1-stimulated induction of gene expression in pTfh at baseline can produce a distinct molecular signature in vaccine responders.
Baseline molecular signatures correlate with post-vaccination antibody responses
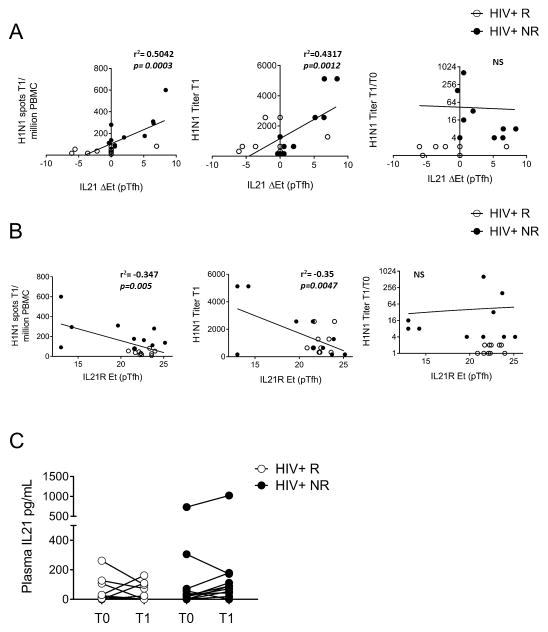
In order to identify the most important variables in our analysis for antibody responses to the influenza vaccine in HIV-infected children and adolescents, we performed pairwise spearman correlations between the aforementioned observations (54 DEGs and 45 DIGs) and immunological correlates of response to the vaccine [i.e. H1N1 serum antibody titer at T1, fold change antibody titer increase (T1/T0), and memory B cell H1N1 Elispot data at T1]. This analysis allowed us to reduce the number of observations from 99 to 33 DEGs and DIGs. The significantly correlated observations were then subjected to cluster analysis to identify highly correlative gene expression patterns in our data set (Figure 7). A cluster of DEGs from H1N1-stimulated CD4 T cells revealed correlations between RORA, RUNX3, PIK3C2B, STAT3, STAT4, and ICOS following antigen stimulation in the HIV+ vaccinees. All of these genes were expressed higher in R compared to NR suggesting a functional relationship between gene expression in CD4 T cells and vaccine response. A second cluster contained IFN-inducible genes expressed in pTfh which were significantly induced in both groups with H1N1 stimulation suggesting co-regulation of transcription of these genes, as would be anticipated. Interestingly, IL21 gene induction (DIG) and IL21R gene expression (DEG) correlated with post-vaccination vaccine response parameters (H1N1 titer and Elispot).
Figure 7. Cluster analysis of DEGs and DIGs with serological and immunological correlates of TIV response.
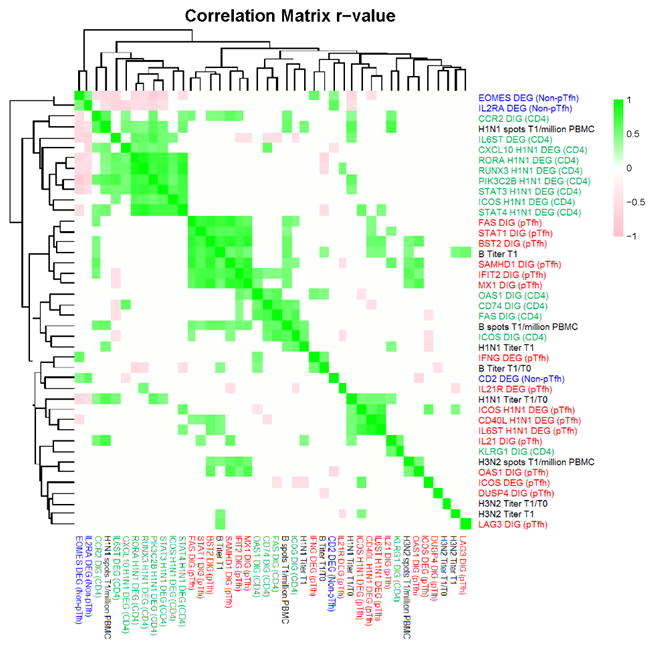
Heatmap showing pairwise Spearman correlations for a group of parameters including 33 DEGs and DIGs and 9 serological and immunological correlates were calculated against each other using R statistical software. Spearman correlations that resulted in p-value >0.05 were assigned an r-value of 0 (white), while significant correlations were clustered and visualized as a heatmap depicting r-values on a scale of −1 (pink) to 1 (green). The parameters tested are color-coded as follows: blue font designates parameters from Non-pTfh cells, green from CD4 T cells, red from pTfh cells, and black font designates Serum Titer and Elispot results for each vaccine antigen.
We performed univariate linear regression analysis to further evaluate the relationship between IL21 signaling molecules at baseline and memory B cell responses after vaccination (Figure 8). We observed a strong relationship between H1N1-induced IL21 expression and H1N1 Elispot T1 (r2=0.504, p=0.0003) and a similar association with H1N1 Titer T1 (r2=0.432, p=0.0012) as shown in Figure 8A. Conversely, pre-vaccination IL21R expression in pTfh cells (in absence of stimulation) showed a significant negative linear relationship with the same parameters (Figure 8B). Linear relationships were not observed for IL21 DIG or IL21R DEG parameters with the H1N1 Titer fold change (T1/T0) (Figure 8A and B). Given the strong association of IL21 and IL21R gene expression with immunological correlates of vaccine response, we measured plasma levels of IL21 in the study participants before and after vaccination. No differences were observed between R and NR at either timepoint (T0 or T1), nor did we observe a significant change in plasma IL21 in response to the vaccine in any group (Figure 8C) suggesting that gene expression of IL21 is more informative as a biomarker for response to Flu vaccination than plasma levels of IL21 cytokine.
Figure 8. Gene expression from IL21 signaling pathway in Pre-vaccination pTfh correlates with post-vaccination serological and immunological responses to Influenza vaccination.
X-Y plots show linear regression of A) IL21R expression (without stimulation) and B) Delta IL21 expression in pTfh (as determined by Delta= [Et_H1N1] − [Et_Unstim]) with H1N1 Spots T1/million PBMC (left), H1N1 Titer T1 (middle), and H1N1 Titer T1/T0 (right). R- and p-values are shown on the graphs, open circles designate non-responders while closed circles designate responders. C) IL-21 plasma levels before (T0) and after influenza vaccination (T1) in HIV+ responders and non-responders.
Because we were unable to identify a single observation (DEG or DIG) with a strong relationship to the fold change in H1N1 titer, we performed multivariate regression analysis to explore whether the combined effect of any of these transcriptional parameters could be governing vaccine responses. Using the results from the cluster analysis in Figure 7, we reduced the number of observations for multivariate analysis from 33 to 18 by retaining a single observation from each cluster of correlated observations. The best-fit model following a bidirectional stepwise regression contained the pTfh IL21 DIG parameter with a significant positive effect on the fold change in serum titer response to H1N1 (Table 2). Other significant parameters in the model were baseline CXCL10 expression in CD4 T cells after H1N1-stimulation (DEG) and the pTfh STAT1 DIG with negative effects, while CD4 IL6ST DEG exhibited a positive relationship. Taken together, our findings support a model in which Tfh-related gene expression by memory CD4 T cells in the peripheral blood is an indicator of vaccine response, while Th1-related gene expression (IFN-gamma induced genes CXCL10 and STAT1) are indicators of poor responses.
Table 2.
Pre-Vaccination Biomarkers associated with H1N1 Titer Fold Change (T1/T0).
| Biomarker | Estimate | Std. Error | Pr(>|t|) |
|---|---|---|---|
| MX1 DIG (pTfh) | 44.3948 | 23.6437 | 0.081419 |
| IL6ST DEG (CD4) | 100.6927 | 26.95345 | 0.002215 |
| STAT1 DIG (pTfh) | −73.1667 | 28.01986 | 0.020523 |
| IL21 DIG (pTfh) | 29.34022 | 11.29875 | 0.02111 |
| ICOS H1N1 DEG (CD4) | −39.9732 | 21.93624 | 0.089845 |
| CXCL10 H1N1 DEG (CD4) | −6.67798 | 2.538514 | 0.019761 |
| KLRG1 DIG (CD4) | −13.8312 | 8.429431 | 0.123101 |
Multiple R-squared: 0.6191, Adjusted R-squared: 0.4287, p-value: 0.02878
Discussion
Tfh cells residing in lymphoid follicles play a critical role in B cell maturation and the formation of robust antibody responses (1), but the relationship between Tfh and circulating Tfh-like cells (pTfh) remains unclear. The data presented herein regarding pTfh cells are, however, in line with two recent studies showing the value of circulating IL-21-producing CD4 T cells as biomarkers for vaccine induced responses (4, 30). Our study is novel in that it demonstrates the importance of IL-21 gene expression in response to antigen stimulation in memory CD4 T cells even before vaccination rather than days to weeks after vaccination. We used the Fluidigm BioMark platform to interrogate gene expression of a curated panel of genes from several cell populations, including total CD4 T and pTfh (CXCR5+CD4 Tcm) isolated from HIV-infected virologically suppressed children and adolescents on ART prior to seasonal Flu vaccination. This approach, while biased compared to whole transcriptome analyses (e.g. RNA Sequencing or Microarray), allowed us to obtain a large amount of transcriptional information on highly relevant cell types (e.g. pTfh CD4 T cells) from relatively low cell numbers (1–2 million PBMC) obtained from pediatric samples (31). Moreover, as this patient group was vaccinated on a yearly basis and had high pre-existing Ab titers, we employed an overnight stimulation with H1N1 antigen and compared gene expression in cells with or without stimulation, in order to gain insight into the functional capabilities of CD4 T cells before vaccination. Ex vivo stimulation with H1N1 led to highly correlative IL21 gene expression in pTfh cells from responders which was not detectable in total PBMC or CD4 T cells. On the other hand, non-responders exhibited increased IL2 gene expression from pTfh, total PBMC and CD4 T cells. These data coupled with the observation that pTfh frequencies are equivalent between responders and non-responders further suggest that the quality of pTfh is important for antibody responses to Flu vaccination.
IL-21 is a pleiotropic cytokine with effects on lymphoid and myeloid cells. Dysregulation of IL-21 signaling can lead to detrimental immunosuppression (cancer) or immune stimulation (autoimmunity) depending on the biological context (32). IL-21 has previously been studied as an immunotherapeutic modality for treatment of several cancers and SIV (model of HIV) based on its immunomodulatory effects on cytotoxic T cells (33–35). CD4 T cells express IL-21R at low levels constitutively that increase upon TCR activation. Interestingly, in our study, higher expression of IL21R in the absence of antigen stimulation on pTfh, non-pTfh and PBMC was always associated with vaccine nonresponsiveness pointing to a necessity for tightly regulated IL-21 signaling for productive vaccine responses.
Our study revealed important insights into biology of pTfh and immune responses. ID2 was highly expressed in CD4 T cells (and pTfh) in vaccine responders both with and without in vitro stimulation. ID2 has been shown in murine models to play roles in T cell differentiation and immune responses (24, 36, 37) and overexpression of ID2 in T cell lymphomas and other cancers (38, 39) suggests a role in regulation of cell survival and proliferation. Further studies are required to explore the role of ID2 in antibody-driven responses in humans. In addition to the classic Th1 cytokine, IL2, IFNG was also associated with poor responses in our cohort. IFNG expression was enriched in non-responders in total CD4 T cells (6-fold) and pTfh (4-fold); excessive IFN-γ production by T cells has been associated with autoimmunity and is attributed to impaired quality of the T cell response, accumulation of Tfh cells and overactive GC responses (40). The increased transcription of both IL2 and STAT5A in CD4 T cells from vaccine non-responders supports the hypothesis that IL-2 production by CD4 T cells after antigen exposure may antagonize pTfh function. In fact, it is known that the IL2 signaling pathway inhibits Tfh differentiation through STAT5 mediated expression of PRDM1 (Blimp-1) (41, 42).
Tfh cells are of intense interest in the HIV field as they are shown to harbor latent and active HIV reservoirs during chronic HIV infection, even in patients under virological control achieved through either natural immunity or anti-retroviral therapy (43–46). pTfh have increased susceptibility to HIV infection in vitro (47). We did not observe differences in pTfh frequency or gene expression between HIV-infected and uninfected participants, however it was evident that gene induction in pTfh was variable within HIV-infected participants following in vitro stimulation leading to differential response to influenza vaccination. The question remains whether HIV infection of pTfh cells could be playing a role in this context and needs further investigation.
Overall, we noted that differential gene expression at baseline (in the absence of stimulation) was skewed towards enrichment of deleterious immune activation/inflammation genes in non-responders rather than “good” gene expression identified in responders. Participants classified as vaccine responders were characterized by gene expression indicating low baseline immune activation, while PBMC and CD4 Tcm subsets (pTfh and non-pTfh) from vaccine non-responders displayed molecular signatures consistent with a state of chronic immune activation (CD69, IL2RA, IFNG, ICOS), despite being under stable cART and virological control. The results for ICOS gene expression in CD4 and pTfh concurred with a previous report showing higher baseline ICOS levels in elderly HIV-uninfected individuals with poor responses to influenza vaccination who demonstrated stunted ICOS induction 7 days post-vaccination (7).
Statistical data integration revealed significant and biologically meaningful correlations between gene expression from H1N1-stimulated pTfh before vaccination and H1N1-specific antibody production post-vaccination. The definitions imposed on influenza vaccinees for responder status, while accepted in the field, can be problematic when individuals have a protective titer at baseline. Higher baseline titers result in lower fold increases following vaccination (48). Thus, by integrating gene expression results with the post-vaccination antibody production rather than fold change we were able to identify features that may be predictive for influenza vaccine responses in HIV-infected individuals. The finding of IL21 expression as a highly correlative marker of antibody responses post-vaccination, supported by recent findings by Schultz, et al regarding HIV-specific IL-21-producing CD4 T cells in the RV144 HIV vaccine trial (4), strongly implicate IL-21 as a biomarker for antibody-driven vaccine responses. Additionally, identification of molecular targets prior to vaccination as demonstrated in this study, opens up the possibility for therapeutic intervention to improve immune function. This approach has great potential to advise studies on immune responses to other vaccine antigens, particularly therapeutic vaccines in which vaccine efficacy involves stimulating a memory response (e.g. HIV, Cancer).
Supplementary Material
Acknowledgments
Financial Support: This publication was made possible by support for the Miami Center for AIDS Research (CFAR) at the University of Miami, Miller School of Medicine funded by a grant (P30AI073961) and to S.P. (R01AI108472) from the National Institutes of Health (NIH). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We would like to acknowledge all patients and guardians who consented to participate in the study. We thank Rajendra Pahwa for helpful comments on this project; Melanie Weiss, Jennifer Faudella and Quita Nimrod for their administrative assistance; and Mario Roederer and Pratip Chattopadhyay for useful discussions and suggestions during the preliminary phase of the study.
Footnotes
Author Contributions: L.R.D., N.C., Su.P., P.P., and S.P conceived the study and designed the experiments; L.R.D, N.C., M.C.S., and A.C. performed the experimental procedures; L.R.D drafted the first version of the paper; all authors participated in writing, review and editing of the paper; L.R.D, N.C., L.P, S.R., and L.G performed statistical analysis; supervision and resources were provided by P.R., P.P., and S.P.
References
- 1.Schaerli P, Willimann K, Lang AB, Lipp M, Loetscher P, Moser B. CXC chemokine receptor 5 expression defines follicular homing T cells with B cell helper function. The Journal of experimental medicine. 2000;192:1553–1562. doi: 10.1084/jem.192.11.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bryant VL, Ma CS, Avery DT, Li Y, Good KL, Corcoran LM, de Waal Malefyt R, Tangye SG. Cytokine-mediated regulation of human B cell differentiation into Ig-secreting cells: predominant role of IL-21 produced by CXCR5+ T follicular helper cells. Journal of immunology. 2007;179:8180–8190. doi: 10.4049/jimmunol.179.12.8180. [DOI] [PubMed] [Google Scholar]
- 3.Shulman Z, Gitlin AD, Weinstein JS, Lainez B, Esplugues E, Flavell RA, Craft JE, Nussenzweig MC. Dynamic signaling by T follicular helper cells during germinal center B cell selection. Science. 2014;345:1058–1062. doi: 10.1126/science.1257861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schultz BT, Teigler JE, Pissani F, Oster AF, Kranias G, Alter G, Marovich M, Eller MA, Dittmer U, Robb ML, Kim JH, Michael NL, Bolton D, Streeck H. Circulating HIV-Specific Interleukin-21(+)CD4(+) T Cells Represent Peripheral Tfh Cells with Antigen-Dependent Helper Functions. Immunity. 2016;44:167–178. doi: 10.1016/j.immuni.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 5.Pallikkuth S, Parmigiani A, Silva SY, George VK, Fischl M, Pahwa R, Pahwa S. Impaired peripheral blood T-follicular helper cell function in HIV-infected nonresponders to the 2009 H1N1/09 vaccine. Blood. 2012;120:985–993. doi: 10.1182/blood-2011-12-396648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pallikkuth S, Pilakka Kanthikeel S, Silva SY, Fischl M, Pahwa R, Pahwa S. Upregulation of IL-21 receptor on B cells and IL-21 secretion distinguishes novel 2009 H1N1 vaccine responders from nonresponders among HIV-infected persons on combination antiretroviral therapy. Journal of immunology. 2011;186:6173–6181. doi: 10.4049/jimmunol.1100264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herati RS, Reuter MA, Dolfi DV, Mansfield KD, Aung H, Badwan OZ, Kurupati RK, Kannan S, Ertl H, Schmader KE, Betts MR, Canaday DH, Wherry EJ. Circulating CXCR5+PD-1+ response predicts influenza vaccine antibody responses in young adults but not elderly adults. Journal of immunology. 2014;193:3528–3537. doi: 10.4049/jimmunol.1302503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.George VK, Pallikkuth S, Parmigiani A, Alcaide M, Fischl M, Arheart KL, Pahwa S. HIV infection Worsens Age-Associated Defects in Antibody Responses to Influenza Vaccine. The Journal of infectious diseases. 2015;211:1959–1968. doi: 10.1093/infdis/jiu840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pallikkuth S, Kanthikeel SP, Silva SY, Fischl M, Pahwa R, Pahwa S. Innate immune defects correlate with failure of antibody responses to H1N1/09 vaccine in HIV-infected patients. The Journal of allergy and clinical immunology. 2011;128:1279–1285. doi: 10.1016/j.jaci.2011.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parmigiani A, Alcaide ML, Freguja R, Pallikkuth S, Frasca D, Fischl MA, Pahwa S. Impaired antibody response to influenza vaccine in HIV-infected and uninfected aging women is associated with immune activation and inflammation. PloS one. 2013;8:e79816. doi: 10.1371/journal.pone.0079816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sasaki S, He XS, Holmes TH, Dekker CL, Kemble GW, Arvin AM, Greenberg HB. Influence of prior influenza vaccination on antibody and B-cell responses. PloS one. 2008;3:e2975. doi: 10.1371/journal.pone.0002975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frasca D, Diaz A, Romero M, Blomberg BB. The generation of memory B cells is maintained, but the antibody response is not, in the elderly after repeated influenza immunizations. Vaccine. 2016;34:2834–2840. doi: 10.1016/j.vaccine.2016.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rinaldi S, Zangari P, Cotugno N, Manno EC, Brolatti N, Castrucci MR, Donatelli I, Rossi P, Palma P, Cagigi A. Antibody but not memory B-cell responses are tuned-down in vertically HIV-1 infected children and young individuals being vaccinated yearly against influenza. Vaccine. 2014;32:657–663. doi: 10.1016/j.vaccine.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 14.Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, Haining WN, Means AR, Kasturi SP, Khan N, Li GM, McCausland M, Kanchan V, Kokko KE, Li S, Elbein R, Mehta AK, Aderem A, Subbarao K, Ahmed R, Pulendran B. Systems biology of vaccination for seasonal influenza in humans. Nature immunology. 2011;12:786–795. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan Y, Tamayo P, Nakaya H, Pulendran B, Mesirov JP, Haining WN. Gene signatures related to B-cell proliferation predict influenza vaccine-induced antibody response. European journal of immunology. 2014;44:285–295. doi: 10.1002/eji.201343657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakaya HI, Hagan T, Duraisingham SS, Lee EK, Kwissa M, Rouphael N, Frasca D, Gersten M, Mehta AK, Gaujoux R, Li GM, Gupta S, Ahmed R, Mulligan MJ, Shen-Orr S, Blomberg BB, Subramaniam S, Pulendran B. Systems Analysis of Immunity to Influenza Vaccination across Multiple Years and in Diverse Populations Reveals Shared Molecular Signatures. Immunity. 2015;43:1186–1198. doi: 10.1016/j.immuni.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Memoli MJ, Shaw PA, Han A, Czajkowski L, Reed S, Athota R, Bristol T, Fargis S, Risos K, Powers JH, Davey RT, Jr, Taubenberger JK. Evaluation of Antihemagglutinin and Antineuraminidase Antibodies as Correlates of Protection in an Influenza A/H1N1 Virus Healthy Human Challenge Model. mBio. 2016:7. doi: 10.1128/mBio.00417-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cotugno N, Douagi I, Rossi P, Palma P. Suboptimal immune reconstitution in vertically HIV infected children: a view on how HIV replication and timing of HAART initiation can impact on T and B-cell compartment. Clinical & developmental immunology. 2012;2012:805151. doi: 10.1155/2012/805151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cagigi A, Cotugno N, Giaquinto C, Nicolosi L, Bernardi S, Rossi P, Douagi I, Palma P. Immune reconstitution and vaccination outcome in HIV-1 infected children: present knowledge and future directions. Human vaccines & immunotherapeutics. 2012;8:1784–1794. doi: 10.4161/hv.21827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cagigi A, Rinaldi S, Di Martino A, Manno EC, Zangari P, Aquilani A, Cotugno N, Nicolosi L, Villani A, Bernardi S, Donatelli I, Pahwa S, Rossi P, Palma P. Premature immune senescence during HIV-1 vertical infection relates with response to influenza vaccination. The Journal of allergy and clinical immunology. 2014;133:592–594. doi: 10.1016/j.jaci.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Cagigi A, Pensieroso S, Ruffin N, Sammicheli S, Thorstensson R, Pan-Hammarstrom Q, Hejdeman B, Nilsson A, Chiodi F. Relation of activation-induced deaminase (AID) expression with antibody response to A(H1N1)pdm09 vaccination in HIV-1 infected patients. Vaccine. 2013;31:2231–2237. doi: 10.1016/j.vaccine.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 22.Dominguez MH, Chattopadhyay PK, Ma S, Lamoreaux L, McDavid A, Finak G, Gottardo R, Koup RA, Roederer M. Highly multiplexed quantitation of gene expression on single cells. Journal of immunological methods. 2013;391:133–145. doi: 10.1016/j.jim.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DiazGranados CA, Denis M, Plotkin S. Seasonal influenza vaccine efficacy and its determinants in children and non-elderly adults: a systematic review with meta-analyses of controlled trials. Vaccine. 2012;31:49–57. doi: 10.1016/j.vaccine.2012.10.084. [DOI] [PubMed] [Google Scholar]
- 24.Miyazaki M, Miyazaki K, Chen S, Itoi M, Miller M, Lu LF, Varki N, Chang AN, Broide DH, Murre C. Id2 and Id3 maintain the regulatory T cell pool to suppress inflammatory disease. Nature immunology. 2014;15:767–776. doi: 10.1038/ni.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramesh R, Kozhaya L, McKevitt K, Djuretic IM, Carlson TJ, Quintero MA, McCauley JL, Abreu MT, Unutmaz D, Sundrud MS. Pro-inflammatory human Th17 cells selectively express P-glycoprotein and are refractory to glucocorticoids. The Journal of experimental medicine. 2014;211:89–104. doi: 10.1084/jem.20130301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu M, Li G, Lee WW, Yuan M, Cui D, Weyand CM, Goronzy JJ. Signal inhibition by the dual-specific phosphatase 4 impairs T cell-dependent B-cell responses with age. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E879–888. doi: 10.1073/pnas.1109797109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Workman CJ, Cauley LS, Kim IJ, Blackman MA, Woodland DL, Vignali DA. Lymphocyte activation gene-3 (CD223) regulates the size of the expanding T cell population following antigen activation in vivo. Journal of immunology. 2004;172:5450–5455. doi: 10.4049/jimmunol.172.9.5450. [DOI] [PubMed] [Google Scholar]
- 28.Huard B, Prigent P, Pages F, Bruniquel D, Triebel F. T cell major histocompatibility complex class II molecules down-regulate CD4+ T cell clone responses following LAG-3 binding. European journal of immunology. 1996;26:1180–1186. doi: 10.1002/eji.1830260533. [DOI] [PubMed] [Google Scholar]
- 29.Ballesteros-Tato A, Leon B, Graf BA, Moquin A, Adams PS, Lund FE, Randall TD. Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity. 2012;36:847–856. doi: 10.1016/j.immuni.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spensieri F, Siena E, Borgogni E, Zedda L, Cantisani R, Chiappini N, Schiavetti F, Rosa D, Castellino F, Montomoli E, Bodinham CL, Lewis DJ, Medini D, Bertholet S, Del Giudice G. Early Rise of Blood T Follicular Helper Cell Subsets and Baseline Immunity as Predictors of Persisting Late Functional Antibody Responses to Vaccination in Humans. PloS one. 2016;11:e0157066. doi: 10.1371/journal.pone.0157066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cotugno N, De Armas L, Pallikkuth S, Rossi P, Palma P, Pahwa S. Paediatric HIV infection in the ‘omics era: defining transcriptional signatures of viral control and vaccine responses. Journal of virus eradication. 2015;1:153–158. [PMC free article] [PubMed] [Google Scholar]
- 32.Leonard WJ, Wan CK. IL-21 Signaling in Immunity. F1000 Research. 2016:5. doi: 10.12688/f1000research.7634.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White L, Krishnan S, Strbo N, Liu H, Kolber MA, Lichtenheld MG, Pahwa RN, Pahwa S. Differential effects of IL-21 and IL-15 on perforin expression, lysosomal degranulation, and proliferation in CD8 T cells of patients with human immunodeficiency virus-1 (HIV) Blood. 2007;109:3873–3880. doi: 10.1182/blood-2006-09-045278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strbo N, de Armas L, Liu H, Kolber MA, Lichtenheld M, Pahwa S. IL-21 augments natural killer effector functions in chronically HIV-infected individuals. Aids. 2008;22:1551–1560. doi: 10.1097/QAD.0b013e3283089367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pallikkuth S, Micci L, Ende ZS, Iriele RI, Cervasi B, Lawson B, McGary CS, Rogers KA, Else JG, Silvestri G, Easley K, Estes JD, Villinger F, Pahwa S, Paiardini M. Maintenance of intestinal Th17 cells and reduced microbial translocation in SIV-infected rhesus macaques treated with interleukin (IL)-21. PLoS pathogens. 2013;9:e1003471. doi: 10.1371/journal.ppat.1003471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi YS, Gullicksrud JA, Xing S, Zeng Z, Shan Q, Li F, Love PE, Peng W, Xue HH, Crotty S. LEF-1 and TCF-1 orchestrate T(FH) differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nature immunology. 2015;16:980–990. doi: 10.1038/ni.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin YY, Jones-Mason ME, Inoue M, Lasorella A, Iavarone A, Li QJ, Shinohara ML, Zhuang Y. Transcriptional regulator Id2 is required for the CD4 T cell immune response in the development of experimental autoimmune encephalomyelitis. Journal of immunology. 2012;189:1400–1405. doi: 10.4049/jimmunol.1200491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lasorella A, Boldrini R, Dominici C, Donfrancesco A, Yokota Y, Inserra A, Iavarone A. Id2 is critical for cellular proliferation and is the oncogenic effector of N-myc in human neuroblastoma. Cancer research. 2002;62:301–306. [PubMed] [Google Scholar]
- 39.Yu S, Zhou X, Steinke FC, Liu C, Chen SC, Zagorodna O, Jing X, Yokota Y, Meyerholz DK, Mullighan CG, Knudson CM, Zhao DM, Xue HH. The TCF-1 and LEF-1 transcription factors have cooperative and opposing roles in T cell development and malignancy. Immunity. 2012;37:813–826. doi: 10.1016/j.immuni.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee SK, Silva DG, Martin JL, Pratama A, Hu X, Chang PP, Walters G, Vinuesa CG. Interferon-gamma excess leads to pathogenic accumulation of follicular helper T cells and germinal centers. Immunity. 2012;37:880–892. doi: 10.1016/j.immuni.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 41.Nurieva RI, Podd A, Chen Y, Alekseev AM, Yu M, Qi X, Huang H, Wen R, Wang J, Li HS, Watowich SS, Qi H, Dong C, Wang D. STAT5 protein negatively regulates T follicular helper (Tfh) cell generation and function. The Journal of biological chemistry. 2012;287:11234–11239. doi: 10.1074/jbc.M111.324046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnston RJ, Choi YS, Diamond JA, Yang JA, Crotty S. STAT5 is a potent negative regulator of TFH cell differentiation. The Journal of experimental medicine. 2012;209:243–250. doi: 10.1084/jem.20111174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Connick E, Folkvord JM, Lind KT, Rakasz EG, Miles B, Wilson NA, Santiago ML, Schmitt K, Stephens EB, Kim HO, Wagstaff R, Li S, Abdelaal HM, Kemp N, Watkins DI, MaWhinney S, Skinner PJ. Compartmentalization of simian immunodeficiency virus replication within secondary lymphoid tissues of rhesus macaques is linked to disease stage and inversely related to localization of virus-specific CTL. Journal of immunology. 2014;193:5613–5625. doi: 10.4049/jimmunol.1401161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Connick E, Mattila T, Folkvord JM, Schlichtemeier R, Meditz AL, Ray MG, McCarter MD, Mawhinney S, Hage A, White C, Skinner PJ. CTL fail to accumulate at sites of HIV-1 replication in lymphoid tissue. Journal of immunology. 2007;178:6975–6983. doi: 10.4049/jimmunol.178.11.6975. [DOI] [PubMed] [Google Scholar]
- 45.Perreau M, Savoye AL, De Crignis E, Corpataux JM, Cubas R, Haddad EK, De Leval L, Graziosi C, Pantaleo G. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. The Journal of experimental medicine. 2013;210:143–156. doi: 10.1084/jem.20121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Banga R, Procopio FA, Noto A, Pollakis G, Cavassini M, Ohmiti K, Corpataux JM, de Leval L, Pantaleo G, Perreau M. PD-1+ and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals. Nature medicine. 2016 doi: 10.1038/nm.4113. [DOI] [PubMed] [Google Scholar]
- 47.Pallikkuth S, Sharkey M, Babic DZ, Gupta S, Stone GW, Fischl MA, Stevenson M, Pahwa S. Peripheral T Follicular Helper Cells Are the Major HIV Reservoir within Central Memory CD4 T Cells in Peripheral Blood from Chronically HIV-Infected Individuals on Combination Antiretroviral Therapy. Journal of virology. 2015;90:2718–2728. doi: 10.1128/JVI.02883-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cagigi A, Cotugno N, Rinaldi S, Santilli V, Rossi P, Palma P. Downfall of the current antibody correlates of influenza vaccine response in yearly vaccinated subjects: Toward qualitative rather than quantitative assays. Pediatric allergy and immunology : official publication of the European Society of Pediatric Allergy and Immunology. 2016;27:22–27. doi: 10.1111/pai.12483. [DOI] [PubMed] [Google Scholar]
- 49.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome research. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.