Abstract
Post-translational modifications (PTMs) of proteins enable fast modulation of protein function in response to metabolic and environmental changes. Phosphorylation is known to play a major role in regulating distribution of light energy between the Photosystems (PS) I and II (state transitions) and in PSII repair cycle. In addition, thioredoxin-mediated redox regulation of Calvin cycle enzymes has been shown to determine the efficiency of carbon assimilation. Besides these well characterized modifications, recent methodological progress has enabled identification of numerous other types of PTMs in various plant compartments, including chloroplasts. To date, at least N-terminal and Lys acetylation, Lys methylation, Tyr nitration and S-nitrosylation, glutathionylation, sumoylation and glycosylation of chloroplast proteins have been described. These modifications impact DNA replication, control transcriptional efficiency, regulate translational machinery and affect metabolic activities within the chloroplast. Moreover, light reactions of photosynthesis as well as carbon assimilation are regulated at multiple levels by a number of PTMs. It is likely that future studies will reveal new metabolic pathways to be regulated by PTMs as well as detailed molecular mechanisms of PTM-mediated regulation.
Keywords: chloroplast, phosphorylation, photosynthesis, post-translational modification, redox regulation
Introduction
Chloroplasts are sites of versatile metabolism. In addition to photosynthetic reactions, chloroplasts host a number of other processes, such as nitrogen and sulfur assimilation, amino acid and fatty acid biosynthesis as well as accumulation of pigments, photoreceptors, and hormones. Chloroplasts are surrounded by the envelope membrane, and the majority of nuclear-encoded chloroplast proteins are imported through the envelope into the plastid via the Toc/Tic machinery. The subchloroplastic destination of a specific protein is determined by the information buried within the primary amino acid sequence, either in the form of cleavable transit peptide or as an internal targeting signal. Due to their endosymbiotic origin, biosynthesis and function of chloroplasts is not only dependent on nuclear control, but also on the expression of approximately 120 plastome encoded genes, mostly involved in photosynthesis and plastid gene expression (Sugiura, 1992; Green, 2011). Obviously, coordination of gene expression between these compartments as well as integration of plastid metabolism with the rest of the cell are required to induce appropriate physiological responses to various environmental stimuli, thereby enabling successful growth and reproduction of the plants. This coordination takes place at many different levels, including the control of nuclear and plastid transcription, RNA processing and translation, protein translocation and assembly of protein complexes as well as functional adjustments of specific enzymes and/or pathways.
Recent interest and methodological progress on PTMs of non-histone proteins has revealed that also a great number of chloroplast proteins are post-translationally modified, which denotes for covalent processing of a mature protein. The most well-studied chloroplast protein modifications: (de)phosphorylation, conveyed by kinases and phosphatases, and oxidation-reduction (including disulfide-thiol exchange of Cys residues, regulated via thioredoxins) have been extensively reviewed (e.g., Buchanan and Balmer, 2005; Tikkanen and Aro, 2012; Michelet et al., 2013; Rochaix, 2013) and thus are not described in detail in the present article. Other PTM types, such as acetylation, methylation, glycosylation, nitration and nitrosylation, sumoylation, and glutathionylation have been identified in chloroplast proteins much later. As only limited information is available for these PTMs, it is currently not possible to conclude whether a given PTM is found in the chloroplasts of all plant and algal species, or whether it is specific for a certain group of organisms. In addition to the PTMs modifying a given amino acid, recent studies have shown that a number of chloroplast proteins are prone to N-terminal trimming resulting in different N-termini or N-terminal acetylation (Lehtimäki et al., 2015; Rowland et al., 2015). It is intriguing that both nuclear- and chloroplast-encoded proteins may be subjected to these modifications (Lehtimäki et al., 2015). In most cases the site of the PTM (cytosol or plastid) and/or the responsible enzymes have remained obscure. PTMs alter the physicochemical properties and thus the function of proteins in different ways depending on the modification and the molecular environment. The molecular structures of the different chloroplast PTM are presented in Figure 1. Here, we will draw together the current understanding of the PTMs regulating distinct metabolic processes in chloroplasts, and review the known physiological effects of these modifications.
FIGURE 1.
Molecular structures of chloroplast Post-translational modifications (PTMs). (A) N-terminal acetylation (Ac denotes acetyl group and N-term. the N-terminal amino acid of a protein). (B) Lys acetylation. (C) Lys mono-, di- and trimethylation (Me denotes methyl group). (D) Ser and Thr phosphorylation (P denotes phospho group). (E) Tyr nitration. (F) S-nitrosylation. (G) Disulfide formation (-SS- denotes disulfide bridge). (H) Glutathionylation of Cys (GSH denotes reduced and G oxidized glutathione). (I) Lys sumoylation. (J) N-glycosylation of Asn (-N-X denotes N-linked glycosyl group). Dash line indicates where structures have been cut off.
Chloroplast Machinery for DNA Replication and Gene Expression
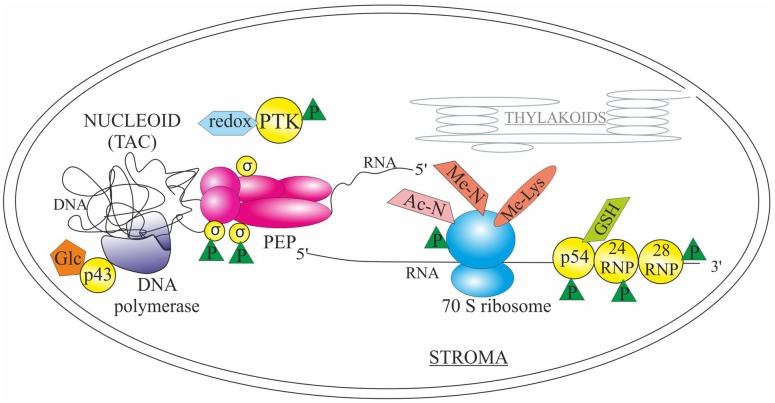
Organellar genomes are organized as nucleoids (also called as transcriptionally active chromosomes or TACs), DNA-protein complexes, which have been identified as the sites for both DNA replication and transcription (Melonek et al., 2016). Recently, proteomic analyses have suggested that also mRNA processing, splicing, editing, and ribosome assembly occur in association with the nucleoid, which supports the idea of co-transcriptional translation of plastid-encoded genes (Majeran et al., 2012). Although only few examples are thoroughly studied, PTMs of various types have been shown to regulate chloroplast genome replication and gene expression at multiple levels (Figure 2).
FIGURE 2.
Post-translational modifications in the regulation of plastid machinery for DNA replication and gene expression. Glycosylation (Glc) of chloroplast protein p43 induces the activity of DNA polymerase. The sigma factors (σ) associated with the plastid encoded RNA polymerase PEP are regulated by (de)phosphorylation (P) conveyed by the plastid transcription kinase (PTK), which is autophosphorylated and redox-regulated. Glutathione-mediated redox regulation (GSH) and phosphorylation of endoribonuclease p54 affects the processing activity of trnK and rps16 transcripts. Phosphorylation of RNA-binding proteins 24RNP and 28RNP affect the binding capacity to the 3′ end of chloroplast transcripts. RPL11 subunit of the 70S ribosome is trimethylated (Me-Lys), and ribosomes are also subjected to acetylation and monomethylation of N-terminal amino acid (Ac-N and Me-N, respectively), as well as phosphorylation. See text for details.
Even if a glycosylation machinery has been identified only in endoplasmic reticulum, some chloroplast proteins have been found to be glycosylated suggesting an existence of vesicular Toc/Tic independent chloroplast protein import route (Villarejo et al., 2005). One of the glycosylated proteins is the pea chloroplast protein p43, which associates with and activates the chloroplast DNA polymerase (Chen et al., 1996). Specifically, the N-terminal domain of p43 is highly O-arabinosylated (Gaikwad et al., 1999). Glycosylation of the protein is required for the induction of polymerization activity, although DNA binding is retained even if the protein is deglycosylated (Gaikwad et al., 1999, 2000). In addition to DNA replication, transcriptional activity of chloroplast genes is (partly) regulated by PTMs. Two different types of RNA polymerases, the plastid-encoded polymerase PEP, and the nuclear-encoded polymerase NEP, are responsible for the transcription of plastid-encoded genes (Shiina et al., 2005). The core subunits of PEP polymerase are associated with nuclear-encoded sigma factors, which are regulated by (de)phosphorylation (Link, 2003; Shimizu et al., 2010). Ser phosphorylation of the sigma factors (SIG6 being the most well studied one) is at least partly conveyed by the plastid transcription kinase (PTK), which is a chloroplast Ser/Thr protein kinase (Baginsky et al., 1997; Baena-González et al., 2001; Ogrzewalla et al., 2002; Salinas et al., 2006; Schweer et al., 2010). The kinase itself is regulated via autophosphorylation and glutathione-dependent redox regulation (Baginsky et al., 1997, 1999; Ogrzewalla et al., 2002). Effect of sigma factor phosphorylation on transcription depends on the sigma factor and the transcribed gene in question: for instance phosphorylation of the Thr170 in SIG1 inhibits transcription of the psaA gene (Shimizu et al., 2010), while phosphorylation of Ser94/95 and/or Ser174 in SIG6 enhances transcription of the atpB and trnK genes with no apparent effect on the transcription of the psbA gene (Schweer et al., 2010).
Processing of the chloroplast transcripts is also affected by phosphorylation and redox regulation of RNA binding proteins. For instance phosphorylation of endoribonuclease p54, which is responsible for the 3′ processing of the plastid trnK and rps16, affects the RNA processing activity but not the cleavage specificity (Nickelsen and Link, 1993; Liere and Link, 1994). Additionally, the processing activity of p54 was modulated by glutathione (Liere and Link, 1994). Phosphorylation of 24 kDa (24RNP) and 28 kDa (28RNP) RNA-binding proteins, associated with a complex regulating the maturation of the 3′ end of chloroplast transcripts (Hayes et al., 1996), has been shown to affect the affinity of the proteins to RNA. Specifically, phosphorylation of 24RNP increased its binding capacity to petD and psbA 3′ UTR (Loza-Tavera et al., 2006), whereas phosphorylation of 28RNP resulted in decreased affinity to RNA (Lisitsky and Schuster, 1995). Recently, it was shown that phosphorylation status of the 24RNP and 28RNP (and apparently other unidentified RNA binding proteins) mediates the interplay between the petD mRNA stability and processing (Vargas-Suarez et al., 2013).
The translational machinery of the chloroplast is composed of prokaryotic-type 70S ribosomes organized in small (Yamaguchi et al., 2000, 2003) and large (Yamaguchi and Subramanian, 2000) subunits. Chloroplast ribosomes contain rRNA and proteins, which are encoded both by the nuclear and chloroplast genomes (Carroll, 2013). Several ribosomal proteins in chloroplasts are targets of extensive PTMs, including formyl group or formyl methionine removal, N- and C-terminal processing, acetylation and monomethylation of N-terminal amino acids, trimethylation of Lys (Kamp et al., 1987; Schmidt et al., 1992; Yamaguchi and Subramanian, 2000; Yamaguchi et al., 2000; Alban et al., 2014) as well as phosphorylation (Guitton et al., 1984; Posno et al., 1984; Wagner et al., 2006). Recently, the enzyme responsible for the trimethylation of the internal Lys in Arabidopsis plastid ribosomal protein L11 (RPL11) has been identified as PrmA-like (Protein Arg methyltransferase-like) protein (Alban et al., 2014; Mazzoleni et al., 2015). Although depletion of Arabidopsis PRMA-like gene did not result in any phenotypic effects, mapping of the trimethylated Lys on the surface of the RPL11 protein allows hypothesizing that methylation might influence the stalk region, which is responsible for the recruitment of initiation, elongation and release factors (Mazzoleni et al., 2015).
A special case in the chloroplast gene expression processes is the regulation of psbA gene expression, which has been under intense study for decades. The psbA gene encodes the light-sensitive PSII core subunit D1, which is constantly degraded and resynthesized in a light-responsive PSII repair cycle (Aro et al., 1993; Mulo et al., 2008). It has been shown that in chloroplasts of green algae and higher plants psbA gene expression is mainly controlled at post-transcriptional levels (Mulo et al., 2012). In Chlamydomonas reinhardtii, ADP-dependent phosphorylation of the cPDI (chloroplast protein disulfide isomerase or RB60) protein in darkness leads to release of the protein from the 5′ UTR of psbA mRNA and cessation of translation (Danon and Mayfield, 1994). Additionally, binding of RB47 to the psbA mRNA is controlled via redox regulation of disulfide groups in RB60 (Danon and Mayfield, 1994; Alergand et al., 2006). It has also been hypothesized that phosphorylation of the spinach 28RNP (in addition to participating in 3′ UTR processing, see above) and ribosomal protein(s) might provide a light-dependent translation control mechanism for the chloroplast, especially during the repair cycle of PSII (Lisitsky and Schuster, 1995; Trebitsh et al., 2000; Yamaguchi and Subramanian, 2003).
Light Reactions of Photosynthesis
Light reactions of photosynthesis, i.e., capture of light energy by the light harvesting complex (LHC) for the production of reducing power (NADPH) occur at the thylakoid membrane via the thylakoid-embedded pigment-protein complexes, namely PSII, Cyt b6f, and PSI. Concomitantly, protons are pumped into the thylakoid lumen, and ADP is photophosphorylated to ATP upon release of the generated proton gradient via the ATP synthase (Figure 3). NADPH and ATP, in turn, are used for numerous reactions, carbon assimilation being the major process. PSII functions as an oxygen-plastoquinone oxidoreductase, which is prone to light-induced photoinhibition (Aro et al., 1993; Tyystjärvi, 2013). The PSII core proteins D1 and D2 as well as the inner antenna protein CP43 and a minor PSII subunit PsbH are targets for light-dependent Thr phosphorylation (Figure 3) catalyzed mainly by the STN8 kinase (Bellafiore et al., 2005; Bonardi et al., 2005; Fristedt and Vener, 2011), while the PSII CORE PHOSPHATASE is responsible for the reverse reaction (i.e., dephosphorylation; Samol et al., 2012). PSII protein phosphorylation is involved in the folding of the thylakoid membrane, which affects the lateral migration of damaged D1 protein from grana stacks to stroma lamellae for degradation and resynthesis (Tikkanen et al., 2008; Fristedt et al., 2009). Another well-studied phosphorylation process is involved in the balancing electron transfer between PSII and PSI according to ambient environmental cues (i.e., light quality and quantity). Phosphorylation of the light harvesting proteins Lhcb1, Lhcb2 and Lhcb4 is catalyzed by the STN7 kinase (Depege et al., 2003; Bellafiore et al., 2005), and instead of PSII, the phosphorylated LHC trimers deliver excitation energy to PSI (so called state transitions) to adjust the absorption cross sections of the two PSs (Rochaix, 2014). Dephosphorylation of LHC by the PPH1/TAP38 (chloroplast protein phosphatase/thylakoid associated phosphatase of 38 kDa) protein phosphatase, in turn, results in redistribution of excitation energy toward PSII (Pribil et al., 2010; Shapiguzov et al., 2010). The STN7 kinase is activated by the binding of plastoquinol to the Qo site of Cyt b6f complex (Vener et al., 1997; Lemeille et al., 2009) and inhibited by stromal reductants (Rintamäki et al., 2000).
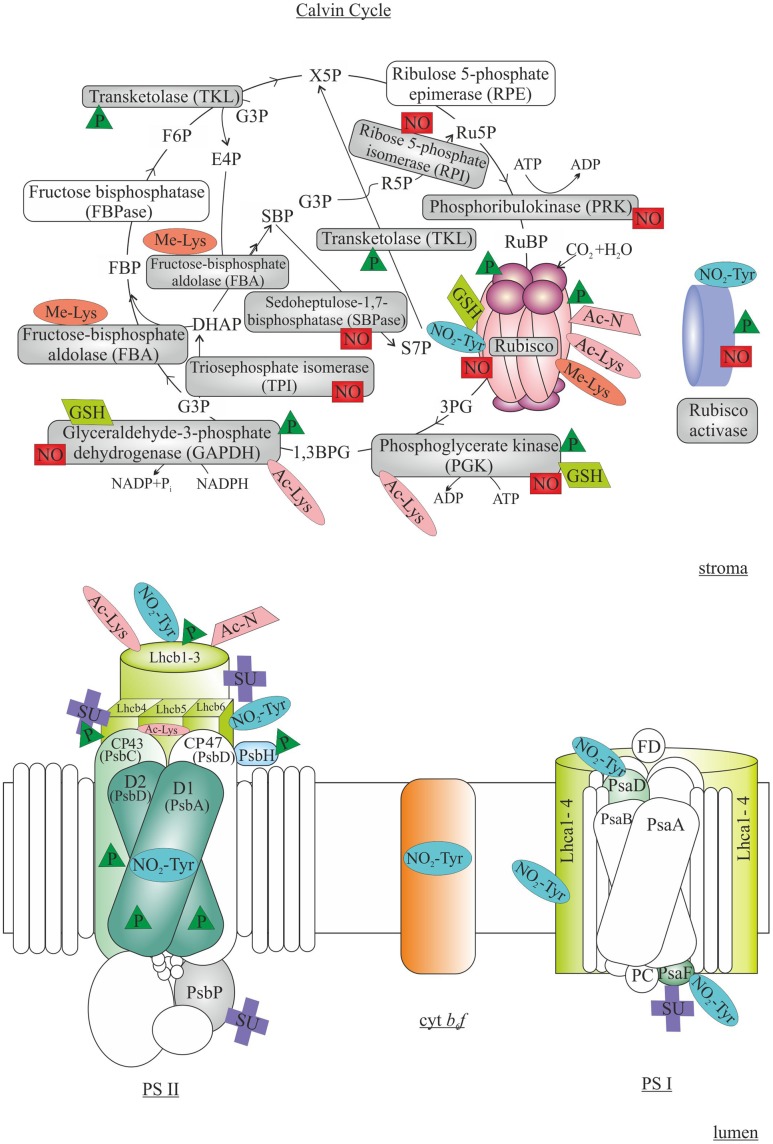
FIGURE 3.
Post-translational modifications in regulation of photosynthetic light reactions (Lower) and Calvin cycle (Upper). In light reactions, phosphorylation (P) of Photosystem (PS) II subunits D1, D2, CP43 and PsbH are involved in PSII repair cycle, while phosphorylation of light harvesting proteins (Lhcb) is required for state transitions. Tyr nitration (NO2-Tyr), N-terminal and Lys acetylation (Ac-N and Ac-Lys, respectively) and sumoylation (SU) of various PSII, Cytochrome b6f (Cyt b6f) and PSI subunits have been detected. In Calvin cycle, the function of Rubisco is controlled by a multitude of PTMs, including phosphorylation, Tyr-nitration, acetylation, Lys methylation (Me-Lys), nitrosylation (NO) and glutathionylation (GSH). Additionally, several other enzymes functioning in the Calvin cycle and activation of Rubisco are targets of various PTMs. The subchloroplastic sites of the PTMs are not indicated in the figure. 3PG, 3-phosphoglycerate; 1,3BPG, 1,3-bisphosphoglycerate; G3P, glyceraldehyde 3-phosphate; DHAP, dihydroxyacetone phosphate; FBP, fructose 1,6-bisphosphate; F6P, fructose 6-phosphate; X5P, xylulose 5-phosphate; R5P, ribose 5-phosphate; Ru5P, ribulose 5-phosphate; RuBP, ribulose 1,5-bisphosphate; S7P, sedoheptulose 7-phosphate; SBP, sedoheptulose 1,7-bisphosphate; E4P, erythrose 4-phosphate. The modified proteins are indicated with colors, while the non-modified proteins are shown as transparent. See text for details.
In addition to phosphorylation, LHC proteins are prone to various other PTMs (Figure 3), such as N-terminal acetylation (Michel et al., 1991; Wu et al., 2011; Rowland et al., 2015), Lys-acetylation (Finkemeier et al., 2011; Wu et al., 2011), Tyr-nitration (Galetskiy et al., 2011b) and sumoylation (Elrouby and Coupland, 2010; López-Torrejón et al., 2013). As acetylation neutralizes the positive charge either on the protein N-terminus or on Lys residue, it has numerous implications in biologic processes including determination of enzyme activity, protein stability and mediation of protein–protein interactions (Hwang et al., 2010; Bienvenut et al., 2011; Scott et al., 2011; Hoshiyasu et al., 2013). Accordingly, acetylation of the Lhcb1 and Lhcb2 proteins appear to be involved in the regulation of LHC attachment to the PSII complexes: the peripheral LHC antenna loosely bound to PSII showed higher level of Lys acetylation than the PSII-LHCII supercomplexes (Wu et al., 2011). In contrast to phosphorylation, acetylation status did not respond to changes in illumination (Wu et al., 2011). It is also worth noting that only the N-terminally trimmed form of Lhcb5 starting with Leu38 (other forms starting with Phe39 or Ser40) were reported to be Lys acetylated, indicating a cross-talk between N-terminal processing and acetylation of chloroplast proteins (Wu et al., 2011). Neither the chloroplast acetylation machinery (Dinh et al., 2015) nor the enzymes responsible for N-terminal processing (Rowland et al., 2015) have been thoroughly characterized yet. Also Tyr nitration of proteins representing PSII (including D1), Cyt b6f, PSI as well as LHC has been detected (Galetskiy et al., 2011a,b). Protein Tyr nitration is a marker of nitrosative stress, and it can irreversibly modify the conformation of proteins thus affecting the catalytic activity and susceptibility to proteolysis (Corpas et al., 2007). Indeed, changes in light conditions resulted in variation in nitration levels in different PSII-LHCII complexes, suggesting that nitration might be involved in photodamage, disassembly of complexes and subsequent degradation of proteins (Galetskiy et al., 2011a). It has also been found that LHC may be post-translationally modified by sumoylation (Elrouby and Coupland, 2010; López-Torrejón et al., 2013), which refers to covalent binding of the small ubiquitin-like modifier (SUMO) protein (Miura et al., 2007). Sumoylation has been implicated in the regulation of protein localization, interactions and catalytic activity (Vierstra, 2012). Obviously, the exact effects of these PTMs on the function of LHC require further studies.
Carbon Assimilation and Starch Metabolism
The photosynthetic carbon reduction cycle, i.e., the Calvin cycle, is a multistep pathway in which redox equivalents and chemical energy (NADPH and ATP) originating from the light reactions is utilized for the reduction of atmospheric carbon dioxide into organic compounds. Calvin cycle involves 11 stromal enzymes, which catalyze 13 distinct reactions. In the first step, inorganic CO2 is fixed by ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco), producing 3-phosphoglycerate (3PG), which is first phosphorylated and then reduced into glyceraldehyde-3-phosphate (G3P). G3P then exits from the Calvin cycle and is further used for the synthesis of more complex sugars, including starch that is the most abundant storage polyglucan in nature (Tetlow and Emes, 2014). Several Calvin cycle enzymes have been reported to be activated in light upon reduction of specific disulfide bonds by thioredoxin (Pedersen et al., 1966; Jensen and Bassham, 1968; Buchanan and Wolosiuk, 1976; Wolosiuk and Buchanan, 1976; Buchanan, 1980). In addition to redox regulation, all steps of CO2 fixation and starch metabolism are carefully controlled by multiple (PTM-dependent) mechanisms which balance the rate of starch synthesis with the availability of energy and carbon in different plant tissues and under various environmental conditions (Figure 3).
Rubisco
In terrestrial plants and green alga, Rubisco exists as a holocomplex composed of eight nuclear-encoded small subunits (RBCS) and eight plastid-encoded large subunits (RBCL). Among the other enzymes involved in Calvin cycle Rubisco has been reported as a target of reversible phosphorylation in many plant species (Figure 3) (Reiland et al., 2009, 2011; Facette et al., 2013; Wang et al., 2014; Roitinger et al., 2015), RBCL being phosphorylated in response to light (Budde and Randall, 1990; Wang et al., 2014). The RBCL and RBCS subunits of Rubisco have been shown to contain multiple phosphorylation sites (Cao et al., 2011; Wang et al., 2014). Phosphorylation of the highly conserved RBCL residues Ser208, Thr246, Tyr239 and Thr330, located in the close proximity to RuBP binding site, might affect the catalytic activity of the enzyme (Lohrig et al., 2009; Hodges et al., 2013). Indeed, dephosphorylation of RBCL has been shown to result in decreased activity of the enzyme (Chen et al., 2011), perhaps via affecting the interaction between Rubisco and RA (Guitton and Mache, 1987; Aggarwal et al., 1993; Hodges et al., 2013). Moreover, it has been suggested that dephosphorylation of RBCL and/or RBCS may lead to dissociation of Rubisco holocomplex (Guitton and Mache, 1987; Aggarwal et al., 1993; Hodges et al., 2013).
Rubisco has also been found as a target of both N-terminal acetylation and Lys acetylation (Figure 3). In spinach, RBCL is post-translationally processed by removal of Met1 and Ser2 followed by the acetylation of the penultimate amino acid (Mulligan et al., 1988). Although N-acetylation of proteins in general is known to modify their activity and stability, the detailed significance and mechanism of RBCL N-termini modification remains unknown (Mulligan et al., 1988; Houtz et al., 1992; Zybailov et al., 2008). Lys acetylation of the Rubisco subunits has been identified only recently, and it has been reported as a dynamic modification in response to the changes in the energy status in plants under different light conditions (Gao et al., 2016). The Rubisco holocomplex contains multiple Lys acetylation sites (e.g., nine in Arabidopsis and thirteen in wheat; Finkemeier et al., 2011), which are localized either in the catalytic center of Rubisco (Cleland et al., 1998; Finkemeier et al., 2011), at the interface between the two RBCL subunits (Knight et al., 1990; Finkemeier et al., 2011) or at the site crucial for the formation of tertiary structure of Rubisco (Knight et al., 1990). Therefore, Lys-acetylation has been suspected to affect Rubisco activity and interactions between the subunits and with other molecules, and indeed recent studies have shown negative regulation of Rubisco activity by Lys acetylation (Finkemeier et al., 2011; Gao et al., 2016). Thus, acetylation of Rubisco might provide a mechanism to coordinate the function of light reactions and carbon assimilation with the carbon status of the cell.
In addition to acetylation, Lys residues of RBCL may be methylated (Figure 3). In many organisms (e.g., pea and tobacco), RBCL is considered as the main stromal methylprotein (Alban et al., 2014). Trimethylation of RBCL at Lys14 has been found in several plant species (Alban et al., 2014; Ma et al., 2016) as a modification catalyzed by the large subunit Rubisco methyltransferase (LSMT), a highly conserved SET-domain protein lysine methyltransferase found in all plant species (Dirk et al., 2006). Despite numerous studies, the role of Lys14 trimethylation of RBCL (as well as the role of methylation for other chloroplastic methylproteins) has not been identified (Clarke, 2013; Ma et al., 2016). Interestingly, in Arabidopsis, spinach, and wheat plants RBCL is not methylated at Lys14 indicating species-specific differences in regulatory mechanisms (Houtz et al., 1992; Mininno et al., 2012; Ma et al., 2016). The methylation of chloroplast proteins seems to be biologically important, as a mutant impaired in PTAC14 (plastid-located SET-domain methyltransferase) exhibits defects in chloroplast differentiation and shows an albino phenotype (Steiner et al., 2011). On the other hand, the LSMT knockdown plants do not show any decrease in CO2 assimilation and growth (Mininno et al., 2012).
Intriguingly, some Calvin cycle enzymes, including Rubisco, have been reported to be modified by peroxynitrite (Figure 3) (Cecconi et al., 2009; Lozano-Juste et al., 2011; Barroso et al., 2013). It has been suggested that Tyr-nitration of RBCL (and RA) might act as a modulator of plant defense-related responses including hypersensitive responses (Cecconi et al., 2009). On the other hand, Tyr-nitration of abundant proteins such as those involved in carbon metabolism might function as a non-specific scavenging system for reactive nitrogen forms under stress conditions. Reversibility of Tyr-nitration is still discussed, thus additionally raising new questions about a potential function as a specific signaling event (Souza et al., 2008; Baudouin, 2011).
The reversible S-nitrosylation of Rubisco Cys residues has been reported both in vitro and in vivo for several plant species in response to nitric oxide (NO) -releasing compounds or to abiotic stresses (Figure 3) (Abat et al., 2008; Abat and Deswal, 2009; Fares et al., 2011; Vanzo et al., 2016). As the redox-active thiols in Cys residues can be modified by the covalent binding of NO resulting in the formation of S-nitrosothiol (Lindermayr et al., 2005), it is plausible that S-nitrosylation of Cys residues adjacent to the Rubisco active site in Arabidopsis might regulate the activity of the enzyme and degradation of the protein (Takahashi and Yamasaki, 2002; Marcus et al., 2003; Romero-Puertas et al., 2008). Indeed, recent enzymatic activity assays have revealed that Rubisco inactivation in response to S-nitrosylation is probably the main cause of reduction in carbon fixation upon various stress conditions (Clark et al., 2000; Abat et al., 2008; Abat and Deswal, 2009).
Another modification of Cys residues is protein S-glutathionylation, a well-described mechanism of signal transduction and protein regulation in mammals (Chrestensen et al., 2000). S-glutathionylation is a reversible post-translational formation of a mixed disulfide between the Cys residue of protein and glutathione. Previously, three Cys residues in RBCL and one in RBCS have been identified as targets of S-glutathionylation in plants (Rouhier et al., 2005), green alga (Zaffagnini et al., 2012a) and cyanobacteria (Sakr et al., 2013; Chardonnet et al., 2015). Protein S-glutathionylation probably protects specific Cys residues against irreversible oxidation under stress conditions (Ito et al., 2003; Zaffagnini et al., 2012b), but this PTM can also result in modulation of protein activity (Klatt and Lamas, 2000; Fratelli et al., 2004) and localization (Chardonnet et al., 2015). Nevertheless, the functional significance of Rubisco S-glutathionylation is not known yet.
Activation and Function of the Calvin Cycle
Although PTMs of Rubisco have been extensively studied, also numerous other enzymes involved in carbon assimilation have been shown to possess multiple PTMs (Figure 3). As RA is responsible for removing inhibitors from Rubisco active center and thus contributes to initiation of carbon fixation, the stimuli affecting the RA activity is reflected in the yield of the entire carbon assimilation cycle. In green alga C. reinhardtii, RA is phosphorylated at Ser53 by the thylakoid-localized Stn7 ortholog Stt7 kinase (see above; Lemeille et al., 2010). RA is mainly localized in the stroma, but a smaller portion of the enzyme has been found in association with the thylakoid membrane (Jin et al., 2006). It has been suggested that phosphorylation of RA increases the attachment of RA to the membrane, protecting Stt7 against proteolysis (Lemeille et al., 2009, 2010). The relocation could also be a mechanism reducing the activity of Rubisco under specific environmental conditions (Lemeille et al., 2010). In Arabidopsis plants, RA is phosphorylated at two sites, Thr78 and Ser172 (Boex-Fontvieille et al., 2014). In the dark, the phosphorylation percentage of Thr78 increases (Reiland et al., 2009; Kim et al., 2016). As Thr78 is located in the region crucial for Rubisco interaction (Zhang and Portis, 1999; Kim et al., 2016), it has been suggested that Thr78 phosphorylation inhibits Rubisco activation (van de Loo and Salvucci, 1996; Stotz et al., 2011; Boex-Fontvieille et al., 2014). However, the importance of Thr78 phosphorylation for the Rubisco activation requires further investigation as the Thr78 is not conserved and replaced by Ile in maize and rice (Baginsky, 2016).
In addition to Rubisco, three other enzymes involved in Calvin cycle have been reported as phosphoproteins. Phosphoglycerate kinase (PGK) enzyme catalyzing the transfer of phosphate group from ATP to 3PG is phosphorylated in Arabidopsis, rice, and maize plants (Reiland et al., 2009; Facette et al., 2013; Roitinger et al., 2015; Baginsky, 2016). The two latter species share the identical phosphorylation site VGAVSpSPK whereas in Arabidopsis PGK is phosphorylated in a domain much closer to the N-terminus. The kinase responsible for phosphorylation is unknown, but the phosphorylation motif suggests proline-directed kinase as a possible candidate (Baginsky, 2016). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) possesses several phosphorylation sites, but as the sites differ significantly between different organisms, it is plausible that phosphorylation is not a major determinant of GAPDH activity in chloroplasts (Baginsky, 2016). Moreover, the main transketolase isoform in Arabidopsis (TKL1) is phosphorylated in a Ca2+ dependent manner at Ser428, and phosphorylation affects enzyme activity (Rocha et al., 2014). Although Ser428 is conserved in higher plants, Ser428 has been found phosphorylated only in Arabidopsis plants (Hou et al., 2015; Baginsky, 2016). It is intriguing that PGK and GAPDH are also targets of Lys acetylation and S-glutathionylation (Finkemeier et al., 2011; Zaffagnini et al., 2012a; Chardonnet et al., 2015; Shen et al., 2015). The enzymatic activity of GAPDH and PGK is increased upon deacetylation, but the functional importance of S-glutathionylation of GAPDH and PGK remains to be elucidated (Finkemeier et al., 2011; Shen et al., 2015). These examples indicate that further studies are urgently needed in order to fully understand the dynamic regulation of Calvin cycle enzymes and to pinpoint the responsible enzymes involved (Friso and van Wijk, 2015; Baginsky, 2016).
Furthermore, a number of other enzymes involved in carbon assimilation have been shown to be post-translationally modified. For instance, fructose 1,6-bisphosphate aldolase (FBA) is trimethylated at a conserved Lys residue close to the C-terminus of the protein, however, without any effect on catalytic activity or the oligomeric state of the enzyme (Mininno et al., 2012; Ma et al., 2016). In poplar trees sedoheptulose-bisphosphatase (SBPase), RA, ribose-5-phosphate isomerase (RPI), phosphoribulokinase (PRK), GAPDH, triosephosphate isomerase (TPI), and PGK were S-nitrosylated during short-term oxidative stress induced by NO treatment (Vanzo et al., 2014, 2016), but the functional importance has not been described yet (Lindermayr et al., 2005; Abat et al., 2008; Romero-Puertas et al., 2008; Abat and Deswal, 2009).
Starch Metabolism
Starch synthesis and degradation occur in a coordinated manner on a diurnal basis. In leaves, starch is synthesized during the day and degraded in darkness (Kötting et al., 2010). Reversible protein phosphorylation plays an important role also in the regulation of starch metabolism (Tetlow et al., 2004a, 2008; Grimaud et al., 2008; Reiland et al., 2009), and five different phosphoproteins (phosphoglucose isomerase, phosphoglucomutase, starch synthase and two subunits of ADP-glucose pyrophosphorylase) involved in starch biosynthesis have been identified in Arabidopsis leaves (Geigenberger, 2011). Interestingly, starch synthase has been reported to be phosphorylated in a light dependent manner, i.e., exclusively at the end of the dark period (Reiland et al., 2009). Analyses of amyloplasts and chloroplasts from Triticum aestivum (wheat) have shown that some isoforms of starch-branching enzymes (SBE) are catalytically activated by phosphorylation and deactivated by dephosphorylation of one or more of their Ser residues (Tetlow et al., 2004b). Additionally, phosphorylation is apparently involved in the formation of protein complexes composed of starch synthase, SBE isoforms as well as other enzymes with undefined role(s) (Tetlow et al., 2004b; Kötting et al., 2010). It has been speculated that the physical association of the enzymes could alter their activities thus improving the efficiency of starch polymer construction (Kötting et al., 2010; Geigenberger, 2011). Moreover, numerous enzymes involved in starch metabolism, such as glucan water dikinase (GWD, also termed SEX1), starch excess4 (SEX4), β-amylase 1 (BAM1), ADP-glucose pyrophosphorylase, ADP-Glc transporter and class II SBE (Mikkelsen et al., 2005; Balmer et al., 2006; Sokolov et al., 2006; Valerio et al., 2011; Tuncel et al., 2014) are redox activated by thioredoxin. However, it is worth noting that redox modification of starch biosynthesis enzymes in response to light (and other environmental stimuli; reviewed in Kötting et al., 2010; Geigenberger, 2011) is not the only determinant of starch accumulation in plants, but most probably other (PTM-dependent) regulatory mechanisms will be identified in the future (Li et al., 2012).
Conclusion
Recently developed new experimental tools, i.e., PTM-specific antibodies and stains as well as enrichment techniques and high quality equipment for mass spectrometry have enabled identification of a range of PTMs in chloroplast proteins. Detailed knowledge about the effects of protein phosphorylation and redox regulation on the photosynthetic reactions already exists, but the regulation of most metabolic pathways in the chloroplast is poorly understood. Because a specific amino acid residue may be targeted by different PTM types (e.g., Lys methylation or Lys acetylation), and because different PTMs may have either antagonistic or cooperative effects, it will be important to reveal the entire PTM code of a protein(s) in order to understand the physiological significance of PTM-mediated regulation in a given metabolic pathway. Future studies are likely to reveal novel modification types as well as molecular mechanisms of PTM-dependent regulation of various metabolic pathways in chloroplasts.
Author Contributions
PM, MG, and MK have made substantial intellectual contribution to the work, participated in writing and revised the paper. MK and MG have drawn the figures. All authors have approved the paper for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
- PS
photosystem
- psa
genes encoding subunits of Photosystem II
- psb
genes encoding subunits of Photosystem II
- PTM
post-translational modification
- RA
Rubisco activase
- RB
RNA binding protein
- rps
genes encoding ribosome subunits
- trn
genes encoding chloroplast transferRNAs
Footnotes
Funding. This study was financially supported by Academy of Finland (307335 “Centre of Excellence in Molecular Biology of Primary Producers”) and The Doctoral Programme in Molecular Life Sciences at the University of Turku.
References
- Abat J. K., Deswal R. (2009). Differential modulation of S-nitrosoproteome of Brassica juncea by low temperature: change in S-nitrosylation of rubisco is responsible for the inactivation of its carboxylase activity. Proteomics 9 4368–4380. 10.1002/pmic.200800985 [DOI] [PubMed] [Google Scholar]
- Abat J. K., Mattoo A. K., Deswal R. (2008). S-nitrosylated proteins of a medicinal CAM plant Kalanchoe pinnata- ribulose-1,5-bisphosphate carboxylase/oxygenase activity targeted for inhibition. FEBS J. 275 2862–2872. 10.1111/j.1742-4658.2008.06425.x [DOI] [PubMed] [Google Scholar]
- Aggarwal K. K., Saluja D., Sachar R. C. (1993). Phosphorylation of rubisco in Cicer arietinum: non-phosphoprotein nature of rubisco in Nicotiana tabacum. Phytochemistry 34 329–335. 10.1016/0031-9422(93)80004-C [DOI] [Google Scholar]
- Alban C., Tardif M., Mininno M., Brugiere S., Gilgen A., Ma S., et al. (2014). Uncovering the protein lysine and arginine methylation network in Arabidopsis chloroplasts. PLoS ONE 9:e95512 10.1371/journal.pone.0095512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alergand T., Peled-Zehavi H., Katz Y., Danon A. (2006). The chloroplast protein disulfide isomerase RB60 reacts with a regulatory disulfide of the RNA-binding protein RB47. Plant Cell Physiol. 47 540–548. [DOI] [PubMed] [Google Scholar]
- Aro E. M., Virgin I., Andersson B. (1993). Photoinhibition of photosystem II. Inactivation, protein damage and turnover. Biochim. Biophys. Acta 1143 113–134. [DOI] [PubMed] [Google Scholar]
- Baena-González E., Baginsky S., Mulo P., Summer H., Aro E.-M., Link G. (2001). Chloroplast transcription at different light intensities. Glutathione-mediated phosphorylation of the major RNA polymerase involved in redox-regulated organellar gene expression. Plant Physiol. 127 1044–1052. 10.1104/pp.010168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baginsky S. (2016). Protein phosphorylation in chloroplasts - a survey of phosphorylation targets. J. Exp. Bot. 67 3873–3882. 10.1093/jxb/erw098 [DOI] [PubMed] [Google Scholar]
- Baginsky S., Tiller K., Link G. (1997). Transcription factor phosphorylation by a protein kinase associated with chloroplast RNA polymerase from mustard (Sinapis alba). Plant Mol. Biol. 34 181–189. 10.1023/A:1005802909902 [DOI] [PubMed] [Google Scholar]
- Baginsky S., Tiller K., Pfannschmidt T., Link G. (1999). PTK, the chloroplast RNA polymerase-associated protein kinase from mustard (Sinapis alba), mediates redox control of plastid in vitro transcription. Plant Mol. Biol. 39 1013–1023. 10.1023/A:1006177807844 [DOI] [PubMed] [Google Scholar]
- Balmer Y., Vensel W. H., Hurkman W. J., Buchanan B. B. (2006). Thioredoxin target proteins in chloroplast thylakoid membranes. Antioxid. Redox Signal. 8 1829–1834. 10.1089/ars.2006.8.1829 [DOI] [PubMed] [Google Scholar]
- Barroso J. B., Valderrama R., Corpas F. J. (2013). Immunolocalization of S-nitrosoglutathione, S-nitrosoglutathione reductase and tyrosine nitration in pea leaf organelles. Acta Physiol. Plant 35 2635–2640. 10.1007/s11738-013-1291-0 [DOI] [Google Scholar]
- Baudouin E. (2011). The language of nitric oxide signalling. Plant Biol. 13 233–242. 10.1111/j.1438-8677.2010.00403.x [DOI] [PubMed] [Google Scholar]
- Bellafiore S., Barneche F., Peltier G., Rochaix J. D. (2005). State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature 433 892–895. [DOI] [PubMed] [Google Scholar]
- Bienvenut W. V., Espagne C., Martinez A., Majeran W., Valot B., Zivy M., et al. (2011). Dynamics of post-translational modifications and protein stability in the stroma of Chlamydomonas reinhardtii chloroplasts. Proteomics 11 1734–1750. 10.1002/pmic.201000634 [DOI] [PubMed] [Google Scholar]
- Boex-Fontvieille E., Daventure M., Jossier M., Hodges M., Zivy M., Tcherkez G. (2014). Phosphorylation pattern of rubisco activase in Arabidopsis leaves. Plant Biol. 16 550–557. 10.1111/plb.12100 [DOI] [PubMed] [Google Scholar]
- Bonardi V., Pesaresi P., Becker T., Schleiff E., Wagner R., Pfannschmidt T., et al. (2005). Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature 437 1179–1182. [DOI] [PubMed] [Google Scholar]
- Buchanan B. B. (1980). Role of light in the regulation of chloroplast enzymes. Ann. Rev. Plant Physiol. 31 341–371. 10.1146/annurev.pp.31.060180.002013 [DOI] [Google Scholar]
- Buchanan B. B., Balmer Y. (2005). Redox regulation: a broadening horizon. Annu. Rev. Plant Biol. 56 187–220. 10.1146/annurev.arplant.56.032604.144246 [DOI] [PubMed] [Google Scholar]
- Buchanan B. B., Wolosiuk R. A. (1976). Photosynthetic regulatory protein found in animal and bacterial cells. Nature 264 669–670. 10.1038/264669a0 [DOI] [PubMed] [Google Scholar]
- Budde R. J., Randall D. D. (1990). Light as a signal influencing the phosphorylation status of plant proteins. Plant Physiol. 94 1501–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Gao Y., Wang Y., Li C. M., Zhao Y. B., Han Z. H., et al. (2011). Differential expression and modification of proteins during ontogenesis in Malus domestica. Proteomics 11 4688–4701. 10.1002/pmic.201100132 [DOI] [PubMed] [Google Scholar]
- Carroll A. J. (2013). The Arabidopsis cytosolic ribosomal proteome: from form to function. Front. Plant Sci. 4:32 10.3389/fpls.2013.00032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecconi D., Orzetti S., Vandelle E., Rinalducci S., Zolla L., Delledonne M. (2009). Protein nitration during defense response in Arabidopsis thaliana. Electrophoresis 30 2460–2468. 10.1002/elps.200800826 [DOI] [PubMed] [Google Scholar]
- Chardonnet S., Sakr S., Cassier-Chauvat C., Le Marechal P., Chauvat F., Lemaire S. D., et al. (2015). First proteomic study of S-glutathionylation in cyanobacteria. J. Proteome Res. 14 59–71. 10.1021/pr500625a [DOI] [PubMed] [Google Scholar]
- Chen W., Gaikwad A., Mukherjee S. K., Choudhary N. R., Kumar D., Tewari K. K. (1996). A 43 kDa DNA binding protein from the pea chloroplast interacts with and stimulates the cognate DNA polymerase. Nucleic Acids Res. 24 3953–3961. 10.1093/nar/24.20.3953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Zhang W., Zhang B., Zhou J., Wang Y., Yang Q., et al. (2011). Phosphoproteins regulated by heat stress in rice leaves. Proteome Sci. 9:37 10.1186/1477-5956-9-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrestensen C. A., Starke D. W., Mieyal J. J. (2000). Acute cadmium exposure inactivates thioltransferase (glutaredoxin), inhibits intracellular reduction of protein-glutathionyl-mixed disulfides, and initiates apoptosis. J. Biol. Chem. 275 26556–26565. 10.1074/jbc.M004097200 [DOI] [PubMed] [Google Scholar]
- Clark D., Durner J., Navarre D. A., Klessig D. F. (2000). Nitric oxide inhibition of tobacco catalase and ascorbate peroxidase. Mol. Plant Microbe Interact. 13 1380–1384. 10.1094/MPMI.2000.13.12.1380 [DOI] [PubMed] [Google Scholar]
- Clarke S. G. (2013). Protein methylation at the surface and buried deep: thinking outside the histone box. Trends Biochem. Sci. 38 243–252. 10.1016/j.tibs.2013.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland W. W., Andrews T. J., Gutteridge S., Hartman F. C., Lorimer G. H. (1998). Mechanism of rubisco: the carbamate as general base. Chem. Rev. 98 549–562. [DOI] [PubMed] [Google Scholar]
- Corpas F. J., del Rio L. A., Barroso J. B. (2007). Need of biomarkers of nitrosative stress in plants. Trends Plant Sci. 12 436–438. [DOI] [PubMed] [Google Scholar]
- Danon A., Mayfield S. P. (1994). ADP-dependent phosphorylation regulates RNA-binding in vitro: implications in light-modulated translation. EMBO J. 13 2227–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depege N., Bellafiore S., Rochaix J. D. (2003). Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science 299 1572–1575. 10.1126/science.1081397 [DOI] [PubMed] [Google Scholar]
- Dinh T. V., Bienvenut W. V., Linster E., Feldman-Salit A., Jung V. A., Meinnel T., et al. (2015). Molecular identification and functional characterization of the first Nα-acetyltransferase in plastids by global acetylome profiling. Proteomics 15 2426–2435. 10.1002/pmic.201500025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirk L. M., Trievel R. C., Houtz R. L. (2006). 7 non-histone protein lysine methyltransferases: structure and catalytic roles. Enzymes 24 179–228. 10.1016/S1874-6047(06)80009-0 [DOI] [PubMed] [Google Scholar]
- Elrouby N., Coupland G. (2010). Proteome-wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proc. Natl. Acad. Sci. U.S.A. 107 17415–17420. 10.1073/pnas.1005452107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facette M. R., Shen Z., Bjornsdottir F. R., Briggs S. P., Smith L. G. (2013). Parallel proteomic and phosphoproteomic analyses of successive stages of maize leaf development. Plant Cell 25 2798–2812. 10.1105/tpc.113.112227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares A., Rossignol M., Peltier J. B. (2011). Proteomics investigation of endogenous S-nitrosylation in Arabidopsis. Biochem. Biophys. Res. Commun. 416 331–336. 10.1016/j.bbrc.2011.11.036 [DOI] [PubMed] [Google Scholar]
- Finkemeier I., Laxa M., Miguet L., Howden A. J., Sweetlove L. J. (2011). Proteins of diverse function and subcellular location are lysine acetylated in Arabidopsis. Plant Physiol. 155 1779–1790. 10.1104/pp.110.171595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratelli M., Gianazza E., Ghezzi P. (2004). Redox proteomics: identification and functional role of glutathionylated proteins. Expert Rev. Proteomics 1 365–376. 10.1586/14789450.1.3.365 [DOI] [PubMed] [Google Scholar]
- Friso G., van Wijk K. J. (2015). Posttranslational protein modifications in plant metabolism. Plant Physiol. 169 1469–1487. 10.1104/pp.15.01378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fristedt R., Vener A. V. (2011). High light induced disassembly of photosystem II supercomplexes in Arabidopsis requires STN7-dependent phosphorylation of CP29. PLoS ONE 6:e24565 10.1371/journal.pone.0024565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fristedt R., Willig A., Granath P., Crevecoeur M., Rochaix J. D., Vener A. V. (2009). Phosphorylation of photosystem II controls functional macroscopic folding of photosynthetic membranes in Arabidopsis. Plant Cell 21 3950–3964. 10.1105/tpc.109.069435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaikwad A., Tewari K. K., Kumar D., Chen W., Mukherjee S. K. (1999). Isolation and characterisation of the cDNA encoding a glycosylated accessory protein of pea chloroplast DNA polymerase. Nucleic Acids Res. 27 3120–3129. 10.1093/nar/27.15.3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaikwad A., Van Hop D., Mukherjee S. K. (2000). Carboxy terminal region of a chloroplast DNA polymerase accessory factor stimulates DNA polymerase activity. Indian J. Biochem. Biophys. 37 424–432. [PubMed] [Google Scholar]
- Galetskiy D., Lohscheider J. N., Kononikhin A. S., Popov I. A., Nikolaev E. N., Adamska I. (2011a). Mass spectrometric characterization of photooxidative protein modifications in Arabidopsis thaliana thylakoid membranes. Rapid Commun. Mass Spectrom. 25 184–190. 10.1002/rcm.4855 [DOI] [PubMed] [Google Scholar]
- Galetskiy D., Lohscheider J. N., Kononikhin A. S., Popov I. A., Nikolaev E. N., Adamska I. (2011b). Phosphorylation and nitration levels of photosynthetic proteins are conversely regulated by light stress. Plant Mol. Biol. 77 461–473. 10.1007/s11103-011-9824-7 [DOI] [PubMed] [Google Scholar]
- Gao X., Hong H., Li W. C., Yang L., Huang J., Xiao Y. L., et al. (2016). Downregulation of Rubisco activity by non-enzymatic acetylation of RbcL. Mol. Plant. 9 1018–1027. 10.1016/j.molp.2016.03.012 [DOI] [PubMed] [Google Scholar]
- Geigenberger P. (2011). Regulation of starch biosynthesis in response to a fluctuating environment. Plant Physiol. 155 1566–1577. 10.1104/pp.110.170399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green B. R. (2011). Chloroplast genomes of photosynthetic eukaryotes. Plant J. 66 34–44. 10.1111/j.1365-313X.2011.04541.x [DOI] [PubMed] [Google Scholar]
- Grimaud F., Rogniaux H., James M. G., Myers A. M., Planchot V. (2008). Proteome and phosphoproteome analysis of starch granule-associated proteins from normal maize and mutants affected in starch biosynthesis. J. Exp. Bot. 59 3395–3406. 10.1093/jxb/ern198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guitton C., Dorne A. M., Mache R. (1984). In organello and in vitro phosphorylation of chloroplast ribosomal proteins. Biochem. Biophys. Res. Commun. 121 297–303. [DOI] [PubMed] [Google Scholar]
- Guitton C., Mache R. (1987). Phosphorylation in vitro of the large subunit of the ribulose-1,5-bisphosphate carboxylase and of the glyceraldehyde-3-phosphate dehydrogenase. Eur. J. Biochem. 166 249–254. 10.1111/j.1432-1033.1987.tb13509.x [DOI] [PubMed] [Google Scholar]
- Hayes R., Kudla J., Schuster G., Gabay L., Maliga P., Gruissem W. (1996). Chloroplast mRNA 3′-end processing by a high molecular weight protein complex is regulated by nuclear encoded RNA binding proteins. EMBO J. 15 1132–1141. [PMC free article] [PubMed] [Google Scholar]
- Hodges M., Jossier M., Boex-Fontvieille E., Tcherkez G. (2013). Protein phosphorylation and photorespiration. Plant Biol. 15 694–706. 10.1111/j.1438-8677.2012.00719.x [DOI] [PubMed] [Google Scholar]
- Hoshiyasu S., Kohzuma K., Yoshida K., Fujiwara M., Fukao Y., Yokota A., et al. (2013). Potential involvement of N-terminal acetylation in the quantitative regulation of the epsilon subunit of chloroplast ATP synthase under drought stress. Biosci. Biotechnol. Biochem. 77 998–1007. [DOI] [PubMed] [Google Scholar]
- Hou Y., Qiu J., Tong X., Wei X., Nallamilli B. R., Wu W., et al. (2015). A comprehensive quantitative phosphoproteome analysis of rice in response to bacterial blight. BMC Plant Biol. 15:163 10.1186/s12870-015-0541-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtz R. L., Poneleit L., Jones S. B., Royer M., Stults J. T. (1992). Posttranslational modifications in the amino- terminal region of the large subunit of ribulose- 1,5-bisphosphate carboxylase/oxygenase from several plant species. Plant Physiol. 98 1170–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang C. S., Shemorry A., Varshavsky A. (2010). N-terminal acetylation of cellular proteins creates specific degradation signals. Science 327 973–977. 10.1126/science.1183147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Iwabuchi M., Ogawa K. (2003). The sugar-metabolic enzymes aldolase and triose-phosphate isomerase are targets of glutathionylation in Arabidopsis thaliana: detection using biotinylated glutathione. Plant Cell Physiol. 44 655–660. 10.1093/pcp/pcg098 [DOI] [PubMed] [Google Scholar]
- Jensen R. G., Bassham J. A. (1968). Photosynthesis by isolated chloroplasts III. Light activation of the carboxylation reaction. Biochim. Biophys. Acta 153 227–234. 10.1016/0005-2728(68)90164-3 [DOI] [PubMed] [Google Scholar]
- Jin S. H., Hong J., Li X. Q., Jiang D. A. (2006). Antisense inhibition of Rubisco activase increases Rubisco content and alters the proportion of Rubisco activase in stroma and thylakoids in chloroplasts of rice leaves. Ann. Bot. 97 739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamp R. M., Srinivasa B. R., von Knoblauch K., Subramanian A. R. (1987). Occurrence of a methylated protein in chloroplast ribosomes. Biochemistry 26 5866–5870. 10.1021/bi00392a043 [DOI] [Google Scholar]
- Kim S. Y., Bender K. W., Walker B. J., Zielinski R. E., Spalding M. H., Ort D. R., et al. (2016). The plastid casein kinase 2 phosphorylates Rubisco activase at the Thr-78 site but is not essential for regulation of Rubisco activation state. Front. Plant Sci. 7:404 10.3389/fpls.2016.00404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatt P., Lamas S. (2000). Regulation of protein function by S-glutathiolation in response to oxidative and nitrosative stress. Eur. J. Biochem. 267 4928–4944. [DOI] [PubMed] [Google Scholar]
- Knight S., Andersson I., Branden C. I. (1990). Crystallographic analysis of ribulose 1,5-bisphosphate carboxylase from spinach at 2.4 A resolution. Subunit interactions and active site. J. Mol. Biol. 215 113–160. [DOI] [PubMed] [Google Scholar]
- Kötting O., Kossmann J., Zeeman S. C., Lloyd J. R. (2010). Regulation of starch metabolism: the age of enlightenment? Curr. Opin. Plant Biol. 13 321–329. 10.1016/j.pbi.2010.01.003 [DOI] [PubMed] [Google Scholar]
- Lehtimäki N., Koskela M. M., Mulo P. (2015). Posttranslational modifications of chloroplast proteins: an emerging field. Plant Physiol. 168 768–775. 10.1104/pp.15.00117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemeille S., Turkina M. V., Vener A. V., Rochaix J. D. (2010). Stt7-dependent phosphorylation during state transitions in the green alga Chlamydomonas reinhardtii. Mol. Cell. Proteomics 9 1281–1295. 10.1074/mcp.M000020-MCP201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemeille S., Willig A., Depege-Fargeix N., Delessert C., Bassi R., Rochaix J. D. (2009). Analysis of the chloroplast protein kinase Stt7 during state transitions. PLoS Biol. 7:e45 10.1371/journal.pbio.1000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Almagro G., Munoz F. J., Baroja-Fernandez E., Bahaji A., Montero M., et al. (2012). Post-translational redox modification of ADP-glucose pyrophosphorylase in response to light is not a major determinant of fine regulation of transitory starch accumulation in Arabidopsis leaves. Plant Cell Physio. 53 433–444. 10.1093/pcp/pcr193 [DOI] [PubMed] [Google Scholar]
- Liere K., Link G. (1994). Structure and expression characteristics of the chloroplast DNA region containing the split gene for tRNA(gly) (UCC) from mustard (Sinapis alba L.). Curr. Genet. 26 557–563. 10.1007/BF00309950 [DOI] [PubMed] [Google Scholar]
- Lindermayr C., Saalbach G., Durner J. (2005). Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol. 137 921–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link G. (2003). Redox regulation of chloroplast transcription. Antioxid. Redox. Signal. 5 79–87. 10.1089/152308603321223568 [DOI] [PubMed] [Google Scholar]
- Lisitsky I., Schuster G. (1995). Phosphorylation of a chloroplast RNA-binding protein changes its affinity to RNA. Nucleic Acids Res. 23 2506–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohrig K., Muller B., Davydova J., Leister D., Wolters D. A. (2009). Phosphorylation site mapping of soluble proteins: Bioinformatical filtering reveals potential plastidic phosphoproteins in Arabidopsis thaliana. Planta 229 1123–1134. 10.1007/s00425-009-0901-y [DOI] [PubMed] [Google Scholar]
- López-Torrejón G., Guerra D., Catala R., Salinas J., del Pozo J. C. (2013). Identification of SUMO targets by a novel proteomic approach in plants. J. Integr. Plant Biol. 55 96–107. 10.1111/jipb.12012 [DOI] [PubMed] [Google Scholar]
- Lozano-Juste J., Colom-Moreno R., Leon J. (2011). In vivo protein tyrosine nitration in Arabidopsis thaliana. J. Exp. Bot. 62 3501–3517. 10.1093/jxb/err042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loza-Tavera H., Vargas-Suarez M., Diaz-Mireles E., Torres-Marquez M. E., Gonzalez de la Vara L. E., Moreno-Sanchez R., et al. (2006). Phosphorylation of the spinach chloroplast 24 kDa RNA-binding protein (24RNP) increases its binding to petD and psbA 3′ untranslated regions. Biochimie 88 1217–1228. [DOI] [PubMed] [Google Scholar]
- Ma S., Martin-Laffon J., Mininno M., Gigarel O., Brugiere S., Bastien O., et al. (2016). Molecular evolution of the substrate specificity of chloroplastic aldolases/Rubisco lysine methyltransferases in plants. Mol. Plant 9 569–581. 10.1016/j.molp.2016.01.003 [DOI] [PubMed] [Google Scholar]
- Majeran W., Friso G., Asakura Y., Qu X., Huang M., Ponnala L., et al. (2012). Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves: a new conceptual framework for nucleoid functions. Plant Physiol. 158 156–189. 10.1104/pp.111.188474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus Y., Altman-Gueta H., Finkler A., Gurevitz M. (2003). Dual role of cysteine 172 in redox regulation of ribulose 1,5-bisphosphate carboxylase/oxygenase activity and degradation. J. Bacteriol. 185 1509–1517. 10.1128/JB.185.5.1509-1517.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoleni M., Figuet S., Martin-Laffon J., Mininno M., Gilgen A., Leroux M., et al. (2015). Dual targeting of the protein methyltransferase PrmA contributes to both chloroplastic and mitochondrial ribosomal protein L11 methylation in Arabidopsis. Plant Cell Physiol. 56 1697–1710. 10.1093/pcp/pcv098 [DOI] [PubMed] [Google Scholar]
- Melonek J., Oetke S., Krupinska K. (2016). Multifunctionality of plastid nucleoids as revealed by proteome analyses. Biochim. Biophys. Acta 1864 1016–1038. 10.1016/j.bbapap.2016.03.009 [DOI] [PubMed] [Google Scholar]
- Michel H., Griffin P. R., Shabanowitz J., Hunt D. F., Bennett J. (1991). Tandem mass spectrometry identifies sites of three post-translational modifications of spinach light-harvesting chlorophyll protein II. Proteolytic cleavage, acetylation, and phosphorylation. J. Biol. Chem. 266 17584–17591. [PubMed] [Google Scholar]
- Michelet L., Zaffagnini M., Morisse S., Sparla F., Perez-Perez M. E., Francia F., et al. (2013). Redox regulation of the Calvin-Benson cycle: something old, something new. Front. Plant Sci. 4:470 10.3389/fpls.2013.00470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen R., Mutenda K. E., Mant A., Schurmann P., Blennow A. (2005). Alpha-glucan, water dikinase (GWD): a plastidic enzyme with redox-regulated and coordinated catalytic activity and binding affinity. Proc. Natl. Acad. Sci. U.S.A. 102 1785–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mininno M., Brugiere S., Pautre V., Gilgen A., Ma S., Ferro M., et al. (2012). Characterization of chloroplastic fructose 1,6-bisphosphate aldolases as lysine-methylated proteins in plants. J. Biol. Chem. 287 21034–21044. 10.1074/jbc.M112.359976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Jin J. B., Hasegawa P. M. (2007). Sumoylation, a post-translational regulatory process in plants. Curr. Opin. Plant Biol. 10 495–502. [DOI] [PubMed] [Google Scholar]
- Mulligan R. M., Houtz R. L., Tolbert N. E. (1988). Reaction-intermediate analogue binding by ribulose bisphosphate carboxylase/oxygenase causes specific changes in proteolytic sensitivity: the amino-terminal residue of the large subunit is acetylated proline. Proc. Natl. Acad. Sci. U.S.A. 85 1513–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulo P., Sakurai I., Aro E. M. (2012). Strategies for psbA gene expression in cyanobacteria, green algae and higher plants: from transcription to PSII repair. Biochim. Biophys. Acta 1817 247–257. 10.1016/j.bbabio.2011.04.011 [DOI] [PubMed] [Google Scholar]
- Mulo P., Sirpio S., Suorsa M., Aro E. M. (2008). Auxiliary proteins involved in the assembly and sustenance of photosystem II. Photosynth. Res. 98 489–501. 10.1007/s11120-008-9320-3 [DOI] [PubMed] [Google Scholar]
- Nickelsen J., Link G. (1993). The 54 kDa RNA-binding protein from mustard chloroplasts mediates endonucleolytic transcript 3′ end formation in vitro. Plant J. 3 537–544. 10.1046/j.1365-313X.1993.03040537.x [DOI] [PubMed] [Google Scholar]
- Ogrzewalla K., Piotrowski M., Reinbothe S., Link G. (2002). The plastid transcription kinase from mustard (Sinapis alba L.). A nuclear-encoded CK2-type chloroplast enzyme with redox-sensitive function. Eur. J. Biochem. 269 3329–3337. 10.1046/j.1432-1033.2002.03017_269_13.x [DOI] [PubMed] [Google Scholar]
- Pedersen T. A., Kirk M., Bassham J. A. (1966). Inhibition of photophosphorylation and photosynthetic carbon cycle reactions by fatty acids and esters. Biochim. Biophys. Acta 112 189–203. 10.1016/0926-6585(66)90320-7 [DOI] [PubMed] [Google Scholar]
- Posno M., van Noort M., Debise R., Groot G. S. (1984). Isolation, characterization, phosphorylation and site of synthesis of Spinacia chloroplast ribosomal proteins. Curr. Genet. 8 147–154. 10.1007/BF00420227 [DOI] [PubMed] [Google Scholar]
- Pribil M., Pesaresi P., Hertle A., Barbato R., Leister D. (2010). Role of plastid protein phosphatase TAP38 in LHCII dephosphorylation and thylakoid electron flow. PLoS Biol. 8:e1000288 10.1371/journal.pbio.1000288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiland S., Finazzi G., Endler A., Willig A., Baerenfaller K., Grossmann J., et al. (2011). Comparative phosphoproteome profiling reveals a function of the STN8 kinase in fine-tuning of cyclic electron flow (CEF). Proc. Natl. Acad. Sci. U.S.A. 108 12955–12960. 10.1073/pnas.1104734108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiland S., Messerli G., Baerenfaller K., Gerrits B., Endler A., Grossmann J., et al. (2009). Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol. 150 889–903. 10.1104/pp.109.138677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rintamäki E., Martinsuo P., Pursiheimo S., Aro E. M. (2000). Cooperative regulation of light-harvesting complex II phosphorylation via the plastoquinol and ferredoxin-thioredoxin system in chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 97 11644–11649. 10.1073/pnas.180054297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha A. G., Mehlmer N., Stael S., Mair A., Parvin N., Chigri F., et al. (2014). Phosphorylation of Arabidopsis transketolase at Ser428 provides a potential paradigm for the metabolic control of chloroplast carbon metabolism. Biochem. J. 458 313–322. 10.1042/BJ20130631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix J. D. (2013). Redox regulation of thylakoid protein kinases and photosynthetic gene expression. Antioxid. Redox Signal. 18 2184–2201. 10.1089/ars.2012.5110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix J. D. (2014). Regulation and dynamics of the light-harvesting system. Annu. Rev. Plant Biol. 65 287–309. 10.1146/annurev-arplant-050213-040226 [DOI] [PubMed] [Google Scholar]
- Roitinger E., Hofer M., Kocher T., Pichler P., Novatchkova M., Yang J., et al. (2015). Quantitative phosphoproteomics of the ataxia telangiectasia-mutated (ATM) and ataxia telangiectasia-mutated and rad3-related (ATR) dependent DNA damage response in Arabidopsis thaliana. Mol. Cell. Proteomics 14 556–571. 10.1074/mcp.M114.040352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Puertas M. C., Campostrini N., Matte A., Righetti P. G., Perazzolli M., Zolla L., et al. (2008). Proteomic analysis of S-nitrosylated proteins in Arabidopsis thaliana undergoing hypersensitive response. Proteomics 8 1459–1469. 10.1002/pmic.200700536 [DOI] [PubMed] [Google Scholar]
- Rouhier N., Villarejo A., Srivastava M., Gelhaye E., Keech O., Droux M., et al. (2005). Identification of plant glutaredoxin targets. Antioxid. Redox Signal. 7 919–929. 10.1089/ars.2005.7.919 [DOI] [PubMed] [Google Scholar]
- Rowland E., Kim J., Bhuiyan N. H., van Wijk K. J. (2015). The Arabidopsis chloroplast stromal N-terminome: complexities of amino-terminal protein maturation and stability. Plant Physiol. 169 1881–1896. 10.1104/pp.15.01214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakr S., Dutheil J., Saenkham P., Bottin H., Leplat C., Ortega-Ramos M., et al. (2013). The activity of the Synechocystis PCC6803 AbrB2 regulator of hydrogen production can be post-translationally controlled through glutathionylation. Int. J. Hydrogen Energy 38 13547–13555. [Google Scholar]
- Salinas P., Fuentes D., Vidal E., Jordana X., Echeverria M., Holuigue L. (2006). An extensive survey of CK2 alpha and beta subunits in Arabidopsis: multiple isoforms exhibit differential subcellular localization. Plant Cell Physiol. 47 1295–1308. 10.1093/pcp/pcj100 [DOI] [PubMed] [Google Scholar]
- Samol I., Shapiguzov A., Ingelsson B., Fucile G., Crevecoeur M., Vener A. V., et al. (2012). Identification of a photosystem II phosphatase involved in light acclimation in Arabidopsis. Plant Cell 24 2596–2609. 10.1105/tpc.112.095703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J., Herfurth E., Subramanian A. R. (1992). Purification and characterization of seven chloroplast ribosomal proteins: evidence that organelle ribosomal protein genes are functional and that NH2-terminal processing occurs via multiple pathways in chloroplasts. Plant Mol. Biol. 20 459–465. 10.1007/BF00040605 [DOI] [PubMed] [Google Scholar]
- Schweer J., Turkeri H., Link B., Link G. (2010). AtSIG6, a plastid sigma factor from Arabidopsis, reveals functional impact of cpCK2 phosphorylation. Plant J. 62 192–202. 10.1111/j.1365-313X.2010.04138.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D. C., Monda J. K., Bennett E. J., Harper J. W., Schulman B. A. (2011). N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science 334 674–678. 10.1126/science.1209307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiguzov A., Ingelsson B., Samol I., Andres C., Kessler F., Rochaix J. D., et al. (2010). The PPH1 phosphatase is specifically involved in LHCII dephosphorylation and state transitions in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 107 4782–4787. 10.1073/pnas.0913810107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Wei W., Zhou D. X. (2015). Histone acetylation enzymes coordinate metabolism and gene expression. Trends Plant Sci. 20 614–621. 10.1016/j.tplants.2015.07.005 [DOI] [PubMed] [Google Scholar]
- Shiina T., Tsunoyama Y., Nakahira Y., Khan M. S. (2005). Plastid RNA polymerases, promoters, and transcription regulators in higher plants. Int. Rev. Cytol. 244 1–68. [DOI] [PubMed] [Google Scholar]
- Shimizu M., Kato H., Ogawa T., Kurachi A., Nakagawa Y., Kobayashi H. (2010). Sigma factor phosphorylation in the photosynthetic control of photosystem stoichiometry. Proc. Natl. Acad. Sci. U.S.A. 107 10760–10764. 10.1073/pnas.0911692107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolov L. N., Dominguez-Solis J. R., Allary A. L., Buchanan B. B., Luan S. (2006). A redox-regulated chloroplast protein phosphatase binds to starch diurnally and functions in its accumulation. Proc. Natl. Acad. Sci. U.S.A. 103 9732–9737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza J. M., Peluffo G., Radi R. (2008). Protein tyrosine nitration- functional alteration or just a biomarker? Free Radic. Biol. Med. 45 357–366. 10.1016/j.freeradbiomed.2008.04.010 [DOI] [PubMed] [Google Scholar]
- Steiner S., Schroter Y., Pfalz J., Pfannschmidt T. (2011). Identification of essential subunits in the plastid-encoded RNA polymerase complex reveals building blocks for proper plastid development. Plant Physiol. 157 1043–1055. 10.1104/pp.111.184515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotz M., Mueller-Cajar O., Ciniawsky S., Wendler P., Hartl F. U., Bracher A., et al. (2011). Structure of green-type Rubisco activase from tobacco. Nat. Struct. Mol. Biol. 6 1366–1370. 10.1038/nsmb.2171 [DOI] [PubMed] [Google Scholar]
- Sugiura M. (1992). “The chloroplast genome,” in 10 Years Plant Molecular Biology, eds Schilperoort R. A., Dure L. (Berlin: Springer; ), 149–168. 10.1007/978-94-011-2656-4_10 [DOI] [PubMed] [Google Scholar]
- Takahashi S., Yamasaki H. (2002). Reversible inhibition of photophosphorylation in chloroplasts by nitric oxide. FEBS Lett. 512 145–148. [DOI] [PubMed] [Google Scholar]
- Tetlow I. J., Beisel K. G., Cameron S., Makhmoudova A., Liu F., Bresolin N. S., et al. (2008). Analysis of protein complexes in wheat amyloplasts reveals functional interactions among starch biosynthetic enzymes. Plant Physiol. 146 1878–1891. 10.1104/pp.108.116244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetlow I. J., Emes M. J. (2014). A review of starch-branching enzymes and their role in amylopectin biosynthesis. IUBMB Life 66 546–558. 10.1002/iub.1297 [DOI] [PubMed] [Google Scholar]
- Tetlow I. J., Morell M. K., Emes M. J. (2004a). Recent developments in understanding the regulation of starch metabolism in higher plants. J. Exp. Bot. 55 2131–2145. 10.1093/jxb/erh248 [DOI] [PubMed] [Google Scholar]
- Tetlow I. J., Wait R., Lu Z., Akkasaeng R., Bowsher C. G., Esposito S., et al. (2004b). Protein phosphorylation in amyloplasts regulates starch branching enzyme activity and protein-protein interactions. Plant Cell 16 694–708. 10.1105/tpc.017400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikkanen M., Aro E. M. (2012). Thylakoid protein phosphorylation in dynamic regulation of photosystem II in higher plants. Biochim. Biophys. Acta 1817 232–238. 10.1016/j.bbabio.2011.05.005 [DOI] [PubMed] [Google Scholar]
- Tikkanen M., Nurmi M., Kangasjärvi S., Aro E. M. (2008). Core protein phosphorylation facilitates the repair of photodamaged photosystem II at high light. Biochim. Biophys. Acta 1777 1432–1437. 10.1016/j.bbabio.2008.08.004 [DOI] [PubMed] [Google Scholar]
- Trebitsh T., Levitan A., Sofer A., Danon A. (2000). Translation of chloroplast psbA mRNA is modulated in the light by counteracting oxidizing and reducing activities. Mol. Cell. Biol. 20 1116–1123. 10.1128/MCB.20.4.1116-1123.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuncel A., Cakir B., Hwang S. K., Okita T. W. (2014). The role of the large subunit in redox regulation of the rice endosperm ADP-glucose pyrophosphorylase. FEBS J. 281 4951–4963. 10.1111/febs.13041 [DOI] [PubMed] [Google Scholar]
- Tyystjärvi E. (2013). Photoinhibition of photosystem II. Int. Rev. Cell. Mol. Biol. 300 243–303. 10.1016/B978-0-12-405210-9.00007-2 [DOI] [PubMed] [Google Scholar]
- Valerio C., Costa A., Marri L., Issakidis-Bourguet E., Pupillo P., Trost P., et al. (2011). Thioredoxin-regulated beta-amylase (BAM1) triggers diurnal starch degradation in guard cells, and in mesophyll cells under osmotic stress. J. Exp. Bot. 62 545–555. 10.1093/jxb/erq288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Loo F. J., Salvucci M. E. (1996). Activation of ribulose-1,5-biphosphate carboxylase/oxygenase (Rubisco) involves Rubisco activase Trp16. Biochemistry 35 8143–8148. 10.1021/bi9604901 [DOI] [PubMed] [Google Scholar]
- Vanzo E., Ghirardo A., Merl-Pham J., Lindermayr C., Heller W., Hauck S. M., et al. (2014). S-nitroso-proteome in poplar leaves in response to acute ozone stress. PLoS ONE 9:e106886 10.1371/journal.pone.0106886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanzo E., Merl-Pham J., Velikova V., Ghirardo A., Lindermayr C., Hauck S. M., et al. (2016). Modulation of protein S-nitrosylation by isoprene emission in poplar. Plant Physiol. 170 1945–1961. 10.1104/pp.15.01842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas-Suarez M., Castro-Sanchez A., Toledo-Ortiz G., Gonzalez de la Vara L. E., Garcia E., Loza-Tavera H. (2013). Protein phosphorylation regulates in vitro spinach chloroplast petD mRNA 3′-untranslated region stability, processing, and degradation. Biochimie 95 400–409. 10.1016/j.biochi.2012.10.012 [DOI] [PubMed] [Google Scholar]
- Vener A. V., van Kan P. J., Rich P. R., Ohad I., Andersson B. (1997). Plastoquinol at the quinol oxidation site of reduced cytochrome bf mediates signal transduction between light and protein phosphorylation: thylakoid protein kinase deactivation by a single-turnover flash. Proc. Natl. Acad. Sci. U.S.A. 94 1585–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra R. D. (2012). The expanding universe of ubiquitin and ubiquitin-like modifiers. Plant Physiol. 160 2–14. 10.1104/pp.112.200667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarejo A., Buren S., Larsson S., Dejardin A., Monne M., Rudhe C., et al. (2005). Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat. Cell Biol. 7 1224–1231. 10.1038/ncb1330 [DOI] [PubMed] [Google Scholar]
- Wagner L. E., II, Betzenhauser M. J., Yule D. I. (2006). ATP binding to a unique site in the type-1 S2- inositol 1,4,5-trisphosphate receptor defines susceptibility to phosphorylation by protein kinase A. J. Biol. Chem. 281 17410–17419. [DOI] [PubMed] [Google Scholar]
- Wang Y., Wang Y., Zhao Y. B., Chen D. M., Han Z. H., Zhang X. Z. (2014). Protein phosphorylation differs significantly among ontogenetic phases in malus seedlings. Proteome Sci. 12 31 10.1186/1477-5956-12-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolosiuk R. A., Buchanan B. B. (1976). Studies on the regulation of chloroplast NADP-linked glyceraldehyde-3-phosphate dehydrogenase. J. Biol. Chem. 251 6456–6461. [PubMed] [Google Scholar]
- Wu X., Oh M. H., Schwarz E. M., Larue C. T., Sivaguru M., Imai B. S., et al. (2011). Lysine acetylation is a widespread protein modification for diverse proteins in Arabidopsis. Plant Physiol. 155 1769–1778. 10.1104/pp.110.165852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K., Beligni M. V., Prieto S., Haynes P. A., McDonald W. H., Yates J. R., III, et al. (2003). Proteomic characterization of the Chlamydomonas reinhardtii chloroplast ribosome. Identification of proteins unique to the 70 S ribosome. J. Biol. Chem. 278 33774–33785. 10.1074/jbc.M301934200 [DOI] [PubMed] [Google Scholar]
- Yamaguchi K., Subramanian A. R. (2000). The plastid ribosomal proteins. Identification of all the proteins in the 50 S subunit of an organelle ribosome (chloroplast). J. Biol. Chem. 275 28466–28482. 10.1074/jbc.M005012200 [DOI] [PubMed] [Google Scholar]
- Yamaguchi K., Subramanian A. R. (2003). Proteomic identification of all plastid-specific ribosomal proteins in higher plant chloroplast 30 S ribosomal subunit. Eur. J. Biochem. 270 190–205. 10.1046/j.1432-1033.2003.03359.x [DOI] [PubMed] [Google Scholar]
- Yamaguchi K., von Knoblauch K., Subramanian A. R. (2000). The plastid ribosomal proteins. Identification of all the proteins in the 30 S subunit of an organelle ribosome (chloroplast). J. Biol. Chem. 275 28455–28465. 10.1074/jbc.M004350200 [DOI] [PubMed] [Google Scholar]
- Zaffagnini M., Bedhomme M., Groni H., Marchand C. H., Puppo C., Gontero B., et al. (2012a). Glutathionylation in the photosynthetic model organism Chlamydomonas reinhardtii: a proteomic survey. Mol. Cell. Proteomics 11 M111.014142. 10.1074/mcp.M111.014142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffagnini M., Bedhomme M., Marchand C. H., Morisse S., Trost P., Lemaire S. D. (2012b). Redox regulation in photosynthetic organisms: focus on glutathionylation. Antioxid. Redox Signal. 16 567–586. 10.1089/ars.2011.4255 [DOI] [PubMed] [Google Scholar]
- Zhang N., Portis A. R. (1999). Mechanism of light regulation of Rubisco: a specific role for the larger Rubisco activase isoform involving reductive activation by thioredoxin-f. Proc. Natl. Acad. Sci. U.S.A. 96 9438–9443. 10.1073/pnas.96.16.9438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B., Rutschow H., Friso G., Rudella A., Emanuelsson O., Sun Q., et al. (2008). Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS ONE 3:e1994 10.1371/journal.pone.0001994 [DOI] [PMC free article] [PubMed] [Google Scholar]