Abstract
Capsaicin (CAP) reduces body weight mainly through activation of transient receptor potential vanilloid 1 (TRPV1) cation channel. However, recent evidence indicates that the gut microbiota influences many physiological processes in host and might provoke obesity. This study determined whether the anti-obesity effect of CAP is related to the changes in gut microbiota. C57BL/6 mice were fed either with high-fat diet (HFD) or HFD with CAP (HFD-CAP) for 9 weeks. We observed a significantly reduced weight gain and improved glucose tolerance in HFD-CAP-fed mice compared with HFD-fed mice. 16S rRNA gene sequencing results showed a decrease of phylum Proteobacteria in HFD-CAP-fed mice. In addition, HFD-CAP-fed mice showed a higher abundance of Akkermansia muciniphila, a mucin-degrading bacterium with beneficial effects on host metabolism. Further studies found that CAP directly up-regulates the expression of Mucin 2 gene Muc2 and antimicrobial protein gene Reg3g in the intestine. These data suggest that the anti-obesity effect of CAP is associated with a modest modulation of the gut microbiota.
Keywords: gut, micriobiome, capsaicin, Akkermansia, mucin
Introduction
Obesity is a global health problem that increases the risk of many physical and mental conditions, such as metabolic syndrome (Ogden et al., 2007). Diet regulation and exercise are the main treatments to obesity. Several molecules from diet have been shown to reduce body weight. CAP is an ingredient of chili peppers, which are consumed worldwide as vegetables and flavorings (Yang et al., 2010). The anti-obesity effect of CAP has been extensively reported in both human and animal studies (Derbenev and Zsombok, 2016), but the mechanisms are very complicated, which include regulation of whole body metabolism or reduction of food intake. Ludy et al. (2012) reported a modest correlation of CAP consumption and body weight reduction. CAP activates the TRPV1 cation channel and enhances catecholamine secretion from adrenal medulla to increase thermogenesis and to reduce weight gain and adipogenesis (Kawabata et al., 2006; Hachiya et al., 2007). CAP can also enhance thermogenesis by activating gastrointestinal TRPV1 in mice (Kawabata et al., 2009). Lee et al. (2015) found that CAP can reduce food intake, which is dependent on TRPV1. The anti-obesity effect of CAP via TRPV1 activation has been extensively studied, but the impact of CAP on gut microbiota has not been well studied.
Recently, growing evidences showed that dysbiosis of gut microbiota plays a significant role in the development of obesity and metabolic syndromes (Sonnenburg and Backhed, 2016; Wang and Jia, 2016). The gut microbiota has numerous physiological and pathological interactions with the host, such as host energy balance regulation, glucose metabolism, and the chronic inflammatory state (Sonnenburg and Backhed, 2016). Profound changes in the composition of the gut microbiota have been uncovered in obese mice and humans. HFD increased the levels of phyla Firmicutes and Proteobacteria and decreased the abundance of Bifidobacterium spp. and Lactobacillus gasseri, which are known to have beneficial effects on hosts (Wang and Jia, 2016).
At the species level, the abundance of Akkermansia muciniphila has been shown to inversely correlate with obesity and type I diabetes in both mice and humans (Everard et al., 2013; Shin et al., 2014; Dao et al., 2016). A. muciniphila is a mucin-degrading bacterium that can use gastric mucin as the sole carbon and nitrogen source (Derrien et al., 2004, 2011; Collado et al., 2007; Derrien, 2007; van Passel et al., 2011). The oral administration of A. muciniphila prevents HFD-induced obesity by altering adipose tissue metabolism and gut barrier function (Everard et al., 2013). The anti-obesity effect of a polyphenol-rich cranberry was associated with increased Akkermansia spp. population in the mice gut (Anhê et al., 2015). Therefore, these experiments showed that gut microbiota dysbiosis may stimulate the development of obesity and the modulation of gut microbiome may reduce body weight (Derrien et al., 2016; Gomez-Gallego et al., 2016).
Recently, Baboota et al. (2014) detected a significant gut microbial alteration in CAP treated mice by qPCR, but the detailed changes in microbiome are lacking. In this study, we explored the effects of CAP on gut microbiota in HFD-fed mice by high throughput sequencing and determined whether the anti-obesity effect is related to the modulation of the gut microbiota.
Materials and Methods
Animals and Diets
C57BL/6J male mice (Animal Center, Third Military Medical University) were bred, no more than six mice per cage in the animal facility of Third Military Medical University. Animals were housed in a controlled environment (12 h per day/night cycle and lights off at 19:00) with free access to food and water. All animal protocols were approved by the institute of Animal Care and Use Committee (Approval number: SYXC-2014-00203). The animal experiments were performed in accordance with the Guidelines for the Care and Use of Laboratory Animals at Third Military Medical University.
After 1 week of acclimation (week 0) on a normal chow diet, 5-week-old mice (n = 12) were randomly divided into two groups of six and fed the following diets (Luo et al., 2012): (1) a HFD containing 45% fat and (2) a HFD with 0.01% CAP (a gift from Prof. Zhiming Zhu, Daping Hospital, Third Military Medical University) treatment in food (HFD-CAP). Animals were given unrestricted access to food and water.
Body weight and average food intake were monitored every day in the 1st week (week 1) after treatment and once a week after week 1. During week 9, mice feces were collected and stored at −80°C until DNA extraction, and oral glucose tolerance test (OGTT) was assessed. At last, animals were anesthetized with amobarbital and then sacrificed by cardiac puncture. Gut tissues were collected for histological staining or tissue RNA extraction.
For RT-qPCR experiment, sixteen 6-week C57BL/6J male mice were randomly divided into four groups and all were fed orally with CAP (0.4 mg in 200 μL PBS and 0.4 mg is computed according to the daily consumption of 4 g of chow per mice multiplied by 0.01%), At the four time points (0, 30, 60, and 90 min), one group of mice (n = 4) were anesthetized with amobarbital, and the jejunum and proximal colon tissue samples were collected for RNA extraction.
Glucose Homeostasis
Mice were fasted overnight (17:00–9:00), and an OGTT was performed after gavage with glucose (1 g/kg body weight, volume: ∼200 μL) (Anhê et al., 2015). Blood glucose concentrations were measured with glucometer (Johnson, USA) before (0 min) and after (15, 30, 60, 90, and 120 min) glucose challenge (Ayala et al., 2010; Anhê et al., 2015).
Gut Microbiota Analysis
Fecal bacterial DNA was extracted using QIAamp DNA Stool Mini Kit (QIAGEN, German) from approximately 100 mg feces according to the manufacturer’s protocol. For each sample, the V4 hypervariable region of the bacterial 16S rRNA gene was amplified using primers 515F (5′-GTGCCAGCMGC CGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAA T-3′) (Caporaso et al., 2011), purified, and then sent to Novogene Corporation for the generation of sequencing libraries by using TruSeq DNA PCR-Free Sample Preparation Kit (Illumina, USA) and sequenced on the Illumina Hiseq 2500 platform.
After de-multiplexing of the paired-end 250 bp raw reads, Trimmomatic (v0.32) (Bolger et al., 2014) was used to clean low-quality reads and remove adapter sequences. Paired ends were merged using FLASH (v1.2.7) (Magoc and Salzberg, 2011) and filtered under specific filtering conditions to obtain high-quality clean tags according to the QIIME (v1.91) (Caporaso et al., 2010) quality control process. The OTU picking processing adopted an open-reference OTU picking protocol by searching reads against the Greengenes (gg_13_8) (DeSantis et al., 2006) database (similarity ≥ 0.97) according the QIIME tutorial1 and the diversity analyses followed the same tutorial. LEFSe (Version 1.0) (Segata et al., 2011) was used to discover the metagenomic biomarker for both organisms and pathways. PICURSt (Version 1.0.0-dev) (Langille et al., 2013) and HUManN (Version 0.99) (Abubucker et al., 2012) were used to predict functional profiling of microbial communities using 16S rRNA amplicon sequencing result following the PICURSt tutorial2,3 .
Determination of absolute abundance of A. muciniphila followed the method from Everard et al. (2013). Briefly, A. muciniphila specific primers (forward: 5′-CAGCACGTGAAG GTGGGGAC-3′, reverse: 5′-CCTTGCGGTTGGCTTCAGAT-3′) were used to detect A. muciniphila through real time PCR. A standard curve (performed in triplicate) was made by fivefold serial dilutions of genomic DNA of A. muciniphila (ATCC BAA-835). And the cycle threshold of each sample was compared with the standard curve.
Bacterial Strains and Growth Curve
Akkermansia muciniphila strain ATCC BAA-835 was purchased from the ATCC. Strains were cultured in BHI medium in tubes at 37°C in anaerobic chamber (Whitley A35 Workstation 2.5, Don Whitley Scientific, UK). To acquire the growth curve of A. muciniphila, 3 mL cell cultures were grown with different concentrations (2, 20, and 200 μg/mL) of CAP or with equal volume (2 μL for 1 mL medium) of ethanol. Mucin (from porcine stomach, Type II, Sigma, USA, #SLBH5886V) was added to the final concentration of 4 g/L when needed. CAP (Sigma, USA, #MKBS2249V) was dissolved in ethanol. Cell density was measured by serial 10-fold dilution and plating on BHI agar plates (1% agar). Each experiment was repeated three times.
Gene Expression
Total RNA was extracted from the jejunum and proximal colon using TriPure (Roche, Switzerland). Genomic DNA removal and cDNA synthesis were performed using TransScript II One-Step gDNA Removal and cDNA Synthesis SuperMix (Transgen, China). Real-time PCR was performed using SYBR Premix Ex Taq II (Takara, Japan) with 1:5 diluted cDNA products.
Target genes including Muc2 and Klf4 (a goblet cell marker), and Reg3g (an antimicrobial marker), gene were detected with hypoxanthine guanine phosphoribosyl transferase (Hprt) as the housekeeping gene (Anhê et al., 2015). Data were calculated according to the 2-ΔΔCt method.
Goblet Cell Staining
The distal ileum and proximal colon were stained with PAS, (Sigma, USA) as previously described (Shin et al., 2014). The goblet cells were stained pink to red and nuclei were blue.
Statistical Analysis
Weight, glycemia, goblet cell count and gene expression level data are expressed as mean ± SEM. For different abundance taxon analyses in STAMP (Parks et al., 2014), Welch’s t-test was used for the comparison of two groups. One way ANOVA followed by Newman–Keuls post hoc was used for real time PCR results tests. Student’s t-test was used for other cases.
Results
Anti-obesity Effect of Capsaicin in Mice Fed with High-Fat Diet
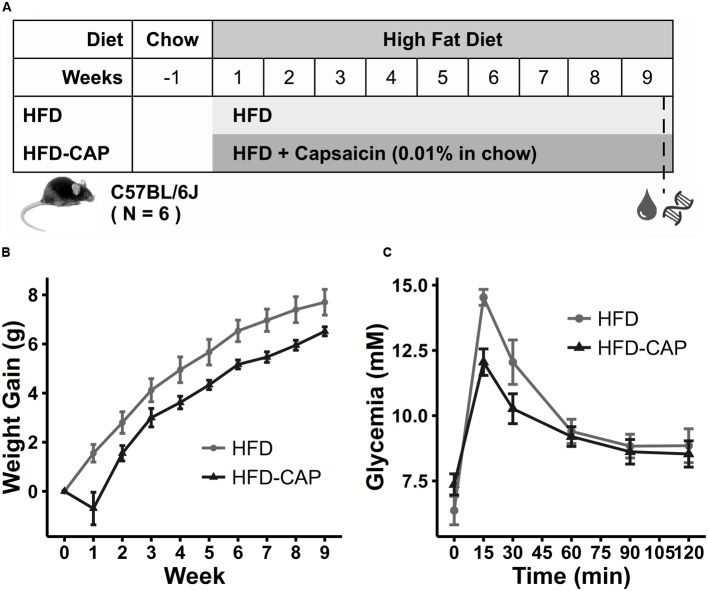
To assess the effect of CAP on obesity, 5-week-old male C57BL/6 mice were fed either with HFD or HFD-CAP diet for 9 weeks (Figure 1A). The HFD-CAP-fed mice had a lower (P < 0.05) body weight gain (6.52 ± 0.45 g) than HFD-fed mice (7.7 ± 1.29 g) (Figure 1B; Supplementary Figure S1A), which was associated with reduction of food intake (Supplementary Figure S1B). The sharp reduction of weight at week 1 could be explained by the reduction of food intake during the early days after suddenly change of food containing spicy CAP. Next, we used a glucose tolerance test to examine glucose homeostasis in CAP-treated and non-treated mice. Compared with HFD-fed mice, HFD-CAP-fed mice showed a smaller (P < 0.05) AUC (Figure 1C; Supplementary Figure S1C), which indicated an improvement in glucose tolerance. These findings confirmed that dietary CAP helped prevent obesity and improved glucose homeostasis under HFD conditions.
FIGURE 1.
(A) Groups information. Five-week-old male mice (n = 12) were randomly divided into two groups of six and fed the following diets: (1) a HFD containing 45% fat and (2) a HFD with 0.01% capsaicin treatment in food (HFD-CAP). (B) Gained body weight within 9 weeks. (C) Oral glucose tolerance test (OGTT) was performed after gavage with glucose (1 g/kg body weight). Blood glucose concentrations were measured with glucometer before (0 min) and after (15, 30, 60, 90, and 120 min) glucose challenge.
Capsaicin Altered the Gut Microbiota and Increased the Abundance of Akkermansia
Fecal DNA of all the 12 mice at week 9 were extracted and sequenced by 16S rRNA gene amplicons sequencing. The average effective tags per sample were 54 thousand (ranging from 47 thousand to 63 thousand tags/sample).
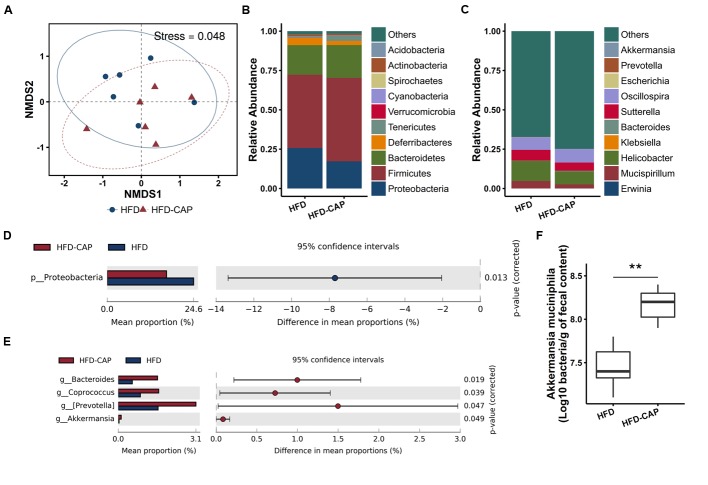
The NMDS plot of the microbial compositions of the 12 samples revealed that the HFD and HFD-CAP group have similar microbiome composition (Figure 2A). However, at the phylum level, among the top 10 abundant phyla, the average proportion of Acidobacteria, Bacteroidetes, and Firmicutes was increased in HFD-CAP group, while the abundance level of Deferribacteres and Proteobacteria was decreased (Figure 2B). According to Welch’s t-tests by STAMP (Parks et al., 2014), only the decrease (from 24.63 ± 4.18% to 16.92 ± 3.83%) of Proteobacteria abundance is significant (P = 0.013) (Figure 2D).
FIGURE 2.
Microbiome composition. (A) NMDS for ordination plot of normalized counts at the OTU level (stress = 0.048). (B,C) Taxonomy composition at the phylum and genus levels. The top 10 phyla/genera were reported for each group, and all others are grouped into “others.” (D,E) Taxa that differ significantly between HFD-CAP and HFD at the phylum and genus level. (F) Absolute abundance of Akkermansia muciniphila in fecal content by qPCR. ∗∗P < 0.01.
At the genus level, among the top 10 abundant genera, the average proportion of Helicobacter and Mucispirillum was decreased in the HFD-CAP group (Figure 2C). According to Welch’s t-tests by STAMP, the abundance levels of Bacteroides (from 0.56 ± 0.35% to 1.55 ± 0.66%), Coprococcus (from 0.88% ± 0.32% to 1.60 ± 0.57%), Prevotella (from 1.58 ± 0.84% to 3.07 ± 1.18%), and Akkermansia (from 0.03 ± 0.04% to 0.12 ± 0.07%) increased significantly (Figure 2E). Real-time PCR also confirmed the increase (p < 0.01) of absolute abundance of A. muciniphila (Figure 2F) in HFD-CAP group.
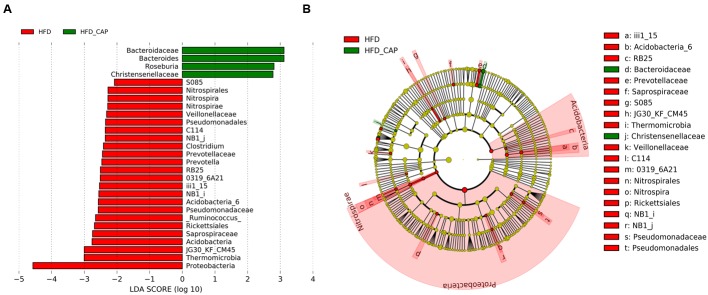
We compared the fecal microbiota in HFD and HFD-CAP groups using LEfSe (Segata et al., 2011) to identify the specific bacterial taxa associated with treatment of CAP. A cladogram representative of the structure of the fecal microbiota was shown in Figure 3B. The greatest differences in multiple levels of taxa between the two communities were displayed (Figure 3A). These data indicated the significantly decreased phylum Proteobacteria can be one of the biomarkers of HFD group. Family Bacteroidaceae and its genus Bacteroides can be the biomarkers of HFD-CAP group.
FIGURE 3.
LEfSe identified the most differentially abundant taxa at the genus level between HFD-CAP and HFD group. (A) HFD-CAP-enriched taxa are indicated with a positive LDA score (green) and taxa enriched in HFD with a negative score (red). Only taxa meeting an LDA significant threshold of >2 are shown. (B) Taxonomic cladogram obtained from LEfSe analysis of 16S sequences HFD-enriched taxa (Red); taxa enriched in HFD-CAP (Green). The brightness of each dot is proportional to its effect size. Each circle’s diameter is proportional to the taxon’s abundance.
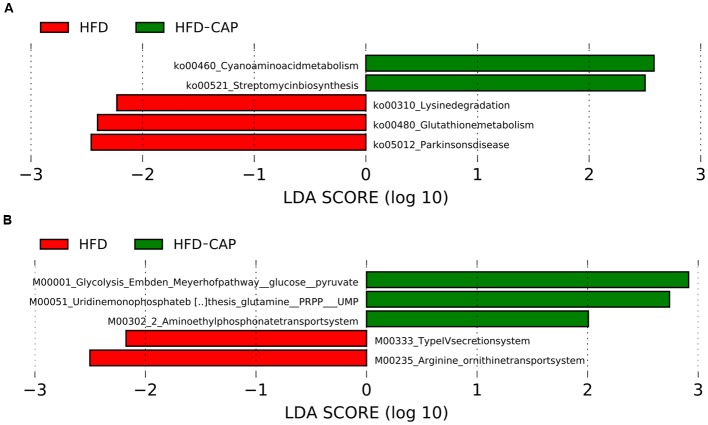
The changes in microbial taxa are associated with changes in functional gene abundances, as predicted from 16S rRNA data analysis using the PICRUSt (phylogenetic investigation of communities by reconstruction of unobserved state) software (Langille et al., 2013). LEFSe identified that three KEGG pathways, including lysine degradation, Glutathione metabolism, and Parkinsons disease, were enriched in HFD group, while two pathways, including cyanoamino acid metabolism and streptomycin biosynthesis, are enriched in HFD-CAP group (Figure 4A). At the module level (Figure 4B), the type IV secretion system modules and arginine/ornithine transport system are enriched in HFD group. Glycolysis (Embden-Meyerhof pathway), uridine monophosphate biosynthesis, and aminoethyl phosphonate transport system are enriched in HFD-CAP group.
FIGURE 4.
LEfSe identified the most differentially abundant PICRUSt-predicated KEGG pathway between HFD-CAP and HFD group. HFD_CAP-enriched pathway (A) and module (B) are indicated with a positive LDA score (green), and pathway (A) and module (B) enriched in HFD with a negative score (red). Only taxa meeting an LDA significant threshold of >2 are shown.
Capsaicin Is Toxic to A. muciniphila, But Mucin Promotes A. muciniphila Growth In vitro
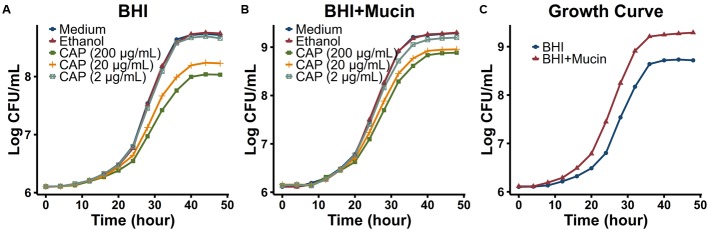
To test whether CAP directly stimulated the growth of A. muciniphila in vitro, the growth curve of A. muciniphila strain ATCC BAA-835 was monitored in both BHI medium and BHI medium with mucin (from porcine stomach, Type II) (Figure 5). High concentration of CAP (20 or 200 μg/mL) inhibited the growth of A. muciniphila, whereas low concentration of CAP (2 μg/mL) did not promote the growth of A. muciniphila in vitro directly (Figures 5A,B). However, the addition of mucin directly promoted the growth of A. muciniphila in BHI medium (Figures 5B,C). When mucin was provided, A. muciniphila grew faster at log phase and plateaued at a much higher cell density. Thus, mucin could promote A. muciniphila growth.
FIGURE 5.
Growth curves of Akkermansia muciniphila in different conditions. A. muciniphila strain ATCC BAA-835 was cultured in BHI (A) or BHI+Mucin (B) medium with different concentrations (2, 20, and 200 μg/mL) of CAP (dissolved in ethanol) or with equal volume (2 μL for 1 mL medium) of ethanol. (C) Growth curves of A. muciniphila in BHI and BHI+Mucin medium without CAP or ethanol.
Capsaicin Increases Muc2 and Reg3g Gene Expression in the Intestine
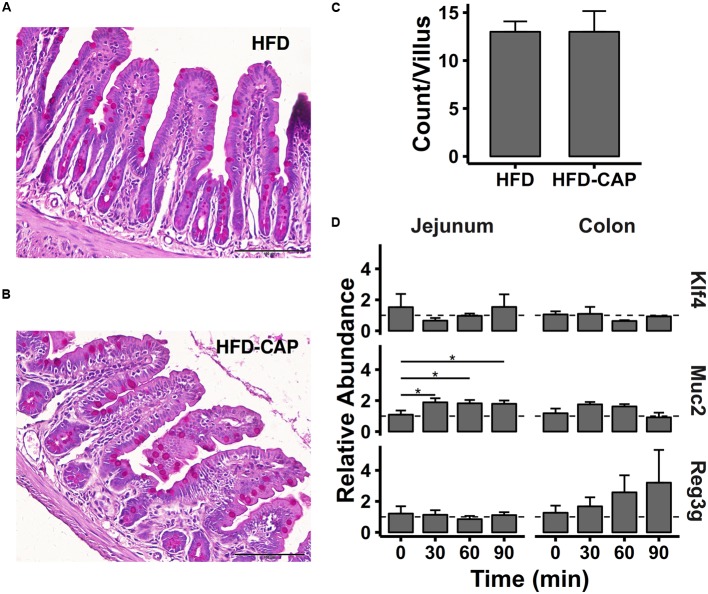
The addition of mucin directly stimulated A. muciniphila growth, and mucin is secreted by goblet cells in vivo. Thus, we examined the number of goblet cells in jejunum and colon in HFD and HFD-CAP mice at week 9. PAS staining was used to stain goblet cells (Figures 6A,B), and PAS-positive goblet cells per villus were counted. An average of 13 goblet cells was seen in HFD-CAP mice, which did not show a significant difference from that in HFD mice (Figure 6C).
FIGURE 6.
(A,B) Representative red photomicrographs showing ileal section stained with periodic acid-Schiff (PAS), and goblet cells were stained pink to red and nuclei were blue. (C) Count number of PAS-positive goblet cells per villus. (D) Gene expression level of Klf4, Muc2, and Reg3g (n = 4). ∗P < 0.05.
To further explore the effect of CAP on goblet cells, sixteen 6-week C57BL/6J male mice were randomly divided into four groups (n = 4) and all were fed orally with CAP (0.4 mg in 200 μL PBS, see Materials and Methods). Gene expressions at four time points (0, 30, 60, and 90 min) in jejunum and proximal colon tissue samples were detected by qPCR. We tested the Klf4 gene, a marker of goblet cells that regulated goblet cells’ differentiation. CAP did not directly increase Klf4 mRNA expression in jejunum or colon within 90 min after the oral administration of CAP (Figure 6D).
We next examined the expression of the major goblet cell mucin production gene (Muc2) after CAP treatment. Mucin 2 mRNA expression was increased twofold at 30 min after CAP administration and lasted for 1.5 h in the jejunum (P < 0.05).
Last, we measured the mRNA expression of Reg3g, an antimicrobial protein that is predominantly expressed in the gastrointestinal tract (Wang et al., 2016). Reg3g has bactericidal activity and restricts bacterial colonization of mucosal surfaces. As is shown in Figure 6D, according the mean expression abundance values, Reg3g mRNA expression is steadily up-regulated in the colon after CAP administration and increased to about threefold at 90 min. However, the differences were not statistically significant.
Discussion
Capsaicin is a worldwide consumed ingredient from chili peppers, and its anti-obesity function has been extensively studied through activation of TRPV1 channel (Yang et al., 2010; Ludy et al., 2012). Here, we found that the beneficial effects of CAP treatment on reducing body weight were associated with a modest modulation of gut microbiota. At the genus level, the proportion of Bacteroides, Coprococcus, Prevotella, and Akkermansia increased significantly. Recent studies indicated that obese individuals are associated with less Bacteroides (Qin et al., 2012; Johansson et al., 2013; Sonnenburg and Backhed, 2016; Wang and Jia, 2016), and the increased abundance of Bacteroides may be beneficial to the health. Genus Coprococcus was reported to be positively correlated with the GIP, (which is also known as glucose-dependent insulinotropic peptide) that induces insulin secretion (Thorens, 1995). Thus, the increase of Coprococcus might contribute to the improvement of glucose tolerance.
We observed an increase in the relative abundance of A. muciniphila at HFD-CAP mice. Recently, Baboota et al. (2014) used qPCR to detect the changes of seven gut microbes in HFD-CAP fed mice and found that the relative abundance of A. muciniphila increased. Interestingly, an increase of Akkermansia has also been reported in other microbiome studies. Anhê et al. (2015) found that a polyphenol-rich cranberry extract increased the relative abundance of Akkermansia, and the author claimed that the increase in Akkermansia population might prevent the production of the negative metabolic phenotype that is associated with obesity. Greer et al. (2016) found that A. muciniphila mediates negative effects of IFNγ on glucose metabolism. Kemperman et al. (2013) reported that complex polyphenols from black tea modulated the human gut microbial ecosystem, with an increase of Akkermansia. Roopchand et al. (2015) showed that dietary polyphenols promoted the growth of A. muciniphila and attenuate HFD-induced metabolic syndrome (Roopchand et al., 2015). Moreover, the antidiabetic drug metformin also has been found to modulate the gut microbiome and lined with an increased abundance of this bacterium (Shin et al., 2014). All these studies pointed out that a better health condition by dietary or pharmaceutical interventions is associated with increased A. muciniphila.
The probiotic effects of A. muciniphila have been studied extensively in animals and clinical studies, and the potential mechanisms were reported from cell biology studies (Derrien et al., 2011; Everard et al., 2013; Lukovac et al., 2014; Li et al., 2016). Therefore, we infer that the alteration of gut microbiota and increased A. muciniphila might contribute to the anti-obesity effects of CAP.
However, the mechanisms by which dietary or pharmaceutical interventions, including CAP, reshaped the gut microbiota and increased the relative abundance of A. muciniphila are still not unclear. Among all the current studies, the mechanism that directly stimulates the growth of A. muciniphila in vivo is lacking and a proposed mechanism is increased mucus production. We found that CAP inhibits the growth of A. muciniphila in high concentration and did not promote its growth at lower concentrations (Figures 5A,B). Since A. muciniphila is a mucin-degrading bacterium, A. muciniphila grows faster and plateaued at a higher cell density with the addition of mucin in the culture medium (Figure 5C). This result is consistent with previous reports (Derrien et al., 2004). Van den Abbeele et al. (2011) also showed that the abundance of A. muciniphila is positively correlated with the levels of mucins in the cecum. Therefore, we infer that CAP might stimulate mucin secretion in the gut to promote A. muciniphila growth.
Mucin is secreted by goblet cell, but we did not found a significant increase of goblet cell numbers in the colon after 9 weeks of CAP intervention, and CAP did not directly stimulate the expression of Kruppel-like factor (Klf4), the key factor that regulates goblet cell differentiation, in 90 min. However, an increased Muc2 mRNA expression in the colon was observed after CAP treatment for 30 min. Interestingly, Karmouty-Quintana et al. (2007) showed that CAP administration induces mucus secretion in rat airways through the activation of sensory nerves. Therefore, we infer that CAP might stimulate mucin production and promote the growth of A. muciniphila in vivo, though the molecular mechanism needs further investigation.
More interestingly, we observed a steady increase of antimicrobial peptide Reg3g mRNA expression after CAP intervention. Antimicrobial peptides are produced and secreted by intestinal epithelial cells or Paneth cells and play a key role in maintaining the homeostasis in gut microbiota (Bevins and Salzman, 2011). Reg3g has bactericidal activity and keeps gut bacteria from the intestinal epithelial surface (Wang et al., 2016). Therefore, CAP might induce the antimicrobial defense to modulate the gut microbial composition. Further investigations are needed to better understand how CAP regulate gut microbiome and stimulate A. muciniphila.
Another potential explanation for the relative increase of A. muciniphila could be a decreased food intake in HFD-CAP fed mice. We observed a reduced food intake in HFD-CAP fed mice (Supplementary Figure S1B), which is consistent with previous studies (Cui and Himms-Hagen, 1992). Contrary to many other species, A. muciniphila can survive when host mucin is the only carbon and nitrogen source (Derrien et al., 2004, 2011), while most other microbes are dependent on food intake. Recently, Holmes et al. (2017) found that a reduced nutrient intake is associated with an increase in the abundance of Akkermansia. Thus, the decreased food intake might result in a higher relative abundance of A. muciniphila (Figures 2E,F).
Conclusion
In summary, we found that CAP treatment reduces body weight and improves glucose homeostasis in HFD mice. This effect was associated with a modest modulation of gut microbiome. The changes in the gut microbiome and the increase in Akkermansia population might play a key role in this protective effect. Thus, we propose that CAP may prevent obesity through modulation of the gut microbiota.
Availability Of Data And Material
The sequence data supporting the results of this article are available in the NCBI Sequence Read Archive under SRA accession number SRP082249.
Ethics Statement
Animal experiments were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals and were approved by the Animal Care and Use Committee of the Third Military Medical University (Approval number: SYXC-2014-00203).
Author Contributions
S. Le and FH conceived the study. WS, MS, XZ, HZ, YY, and S. Lu performed the experiments. WS, YT, and GL analyzed the sequence data. ML and WJ provided intellectual support. S. Le and WS wrote the paper. All authors read and approve the final manuscript for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Zhiming Zhu from Daping Hospital, Third Military Medical University for providing capsaicin for animal experiments and advices on mice raising.
Abbreviations
- ATCC
American type culture collection
- AUC
area under the curve
- CAP
capsaicin
- GIP
gastric inhibitory polypeptide
- HFD
high-fat diet
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- Klf4
Kruppel-like factor 4
- Muc2
mucin 2 gene
- NMDS
non-metric multi-dimensional scaling
- OTU
operational taxonomic unit
- PAS
Periodic Acid-Schiff
- Reg3g
regenerating islet-derived 3 gamma
- TRPV1
transient receptor potential vanilloid 1.
Funding. This work was supported by the National Natural Science Foundation of China (grant No. 31570173 to FH).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00272/full#supplementary-material
(A) Weight gain of mice at week 9 (∗P < 0.05). (B) Average food intake (g/day/mice). (C) Area under the curve (AUC) of the oral glucose tolerance test (OGTT) (∗P < 0.05).
References
- Abubucker S., Segata N., Goll J., Schubert A. M., Izard J., Cantarel B. L., et al. (2012). Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput. Biol. 8:e1002358 10.1371/journal.pcbi.1002358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anhê F. F., Roy D., Pilon G., Dudonné S., Matamoros S., Varin T. V., et al. (2015). A polyphenol-rich cranberry extract protects from diet-induced obesity, insulin resistance and intestinal inflammation in association with increased Akkermansia spp. population in the gut microbiota of mice. Gut 64 872–883. 10.1136/gutjnl-2014-307142 [DOI] [PubMed] [Google Scholar]
- Ayala J. E., Samuel V. T., Morton G. J., Obici S., Croniger C. M., Shulman G. I., et al. (2010). Standard operating procedures for describing and performing metabolic tests of glucose homeostasis in mice. Dis. Model Mech. 3 525–534. 10.1242/dmm.006239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baboota R. K., Murtaza N., Jagtap S., Singh D. P., Karmase A., Kaur J., et al. (2014). Capsaicin-induced transcriptional changes in hypothalamus and alterations in gut microbial count in high fat diet fed mice. J. Nutr. Biochem. 25 893–902. 10.1016/j.jnutbio.2014.04.004 [DOI] [PubMed] [Google Scholar]
- Bevins C. L., Salzman N. H. (2011). Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol. 9 356–368. 10.1038/nrmicro2546 [DOI] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Lozupone C. A., Turnbaugh P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl. 1), 4516–4522. 10.1073/pnas.1000080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M. C., Derrien M., Isolauri E., de Vos W. M., Salminen S. (2007). Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl. Environ. Microbiol. 73 7767–7770. 10.1128/AEM.01477-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Himms-Hagen J. (1992). Long-term decrease in body fat and in brown adipose tissue in capsaicin-desensitized rats. Am. J. Physiol. 262(4 Pt 2), R568–R573. [DOI] [PubMed] [Google Scholar]
- Dao M. C., Everard A., Aron-Wisnewsky J., Sokolovska N., Prifti E., Verger E. O., et al. (2016). Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut 65 426–436. 10.1136/gutjnl-2014-308778 [DOI] [PubMed] [Google Scholar]
- Derbenev A. V., Zsombok A. (2016). Potential therapeutic value of TRPV1 and TRPA1 in diabetes mellitus and obesity. Semin. Immunopathol. 38 397–406. 10.1007/s00281-015-0529-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M. (2007). Mucin Utilisation and Host Interactions of the Novel Intestinal Microbe Akkermansia muciniphila. Ph.D. thesis, Wageningen University, Wageningen, The Netherlands. [Google Scholar]
- Derrien M., Belzer C., de Vos W. M. (2016). Akkermansia muciniphila and its role in regulating host functions. Microb. Pathog. 10.1016/j.micpath.2016.02.005 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Derrien M., Van Baarlen P., Hooiveld G., Norin E., Müller M., de Vos W. M. (2011). Modulation of mucosal immune response, tolerance, and proliferation in mice colonized by the mucin-degrader Akkermansia muciniphila. Front. Microbiol. 2:166 10.3389/fmicb.2011.00166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M., Vaughan E. E., Plugge C. M., de Vos W. M. (2004). Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 54(Pt 5), 1469–1476. 10.1099/ijs.0.02873-0 [DOI] [PubMed] [Google Scholar]
- DeSantis T. Z., Hugenholtz P., Larsen N., Rojas M., Brodie E. L., Keller K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72 5069–5072. 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A., Belzer C., Geurts L., Ouwerkerk J. P., Druart C., Bindels L. B., et al. (2013). Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. U.S.A. 110 9066–9071. 10.1073/pnas.1219451110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gallego C., Pohl S., Salminen S., De Vos W. M., Kneifel W. (2016). Akkermansia muciniphila: a novel functional microbe with probiotic properties. Benef. Microbes 7 571–584. 10.3920/BM2016.0009 [DOI] [PubMed] [Google Scholar]
- Greer R. L., Dong X., Moraes A. C., Zielke R. A., Fernandes G. R., Peremyslova E., et al. (2016). Akkermansia muciniphila mediates negative effects of IFNgamma on glucose metabolism. Nat. Commun. 7:13329 10.1038/ncomms13329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachiya S., Kawabata F., Ohnuki K., Inoue N., Yoneda H., Yazawa S., et al. (2007). Effects of CH-19 Sweet, a non-pungent cultivar of red pepper, on sympathetic nervous activity, body temperature, heart rate, and blood pressure in humans. Biosci. Biotechnol. Biochem. 71 671–676. 10.1271/bbb.60359 [DOI] [PubMed] [Google Scholar]
- Holmes A. J., Chew Y. V., Colakoglu F., Cliff J. B., Klaassens E., Read M. N., et al. (2017). Diet-microbiome interactions in health are controlled by intestinal nitrogen source constraints. Cell Metab. 25 140–151. 10.1016/j.cmet.2016.10.021 [DOI] [PubMed] [Google Scholar]
- Johansson M. E. V., Sjövall H., Hansson G. C. (2013). The gastrointestinal mucus system in health and disease. Nat. Rev. Gastroenterol. Hepatol. 10 352–361. 10.1038/nrgastro.2013.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmouty-Quintana H., Cannet C., Sugar R., Fozard J. R., Page C. P., Beckmann N. (2007). Capsaicin-induced mucus secretion in rat airways assessed in vivo and non-invasively by magnetic resonance imaging. Br. J. Pharmacol. 150 1022–1030. 10.1038/sj.bjp.0707168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata F., Inoue N., Masamoto Y., Matsumura S., Kimura W., Kadowaki M., et al. (2009). Non-pungent capsaicin analogs (capsinoids) increase metabolic rate and enhance thermogenesis via gastrointestinal TRPV1 in mice. Biosci. Biotechnol. Biochem. 73 2690–2697. 10.1271/bbb.90555 [DOI] [PubMed] [Google Scholar]
- Kawabata F., Inoue N., Yazawa S., Kawada T., Inoue K., Fushiki T. (2006). Effects of CH-19 sweet, a non-pungent cultivar of red pepper, in decreasing the body weight and suppressing body fat accumulation by sympathetic nerve activation in humans. Biosci. Biotechnol. Biochem. 70 2824–2835. 10.1271/bbb.60206 [DOI] [PubMed] [Google Scholar]
- Kemperman R. A., Gross G., Mondot S., Possemiers S., Marzorati M., Van de Wiele T., et al. (2013). Impact of polyphenols from black tea and red wine/grape juice on a gut model microbiome. Food Res. Int. 53 659–669. 10.1016/j.foodres.2013.01.034 [DOI] [Google Scholar]
- Langille M. G., Zaneveld J., Caporaso J. G., McDonald D., Knights D., Reyes J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31 814–821. 10.1038/nbt.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E., Jung D. Y., Kim J. H., Patel P. R., Hu X., Lee Y., et al. (2015). Transient receptor potential vanilloid type-1 channel regulates diet-induced obesity, insulin resistance, and leptin resistance. FASEB J. 29 3182–3192. 10.1096/fj.14-268300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Lin S., Vanhoutte P. M., Woo C. W., Xu A. (2016). Akkermansia muciniphila protects against atherosclerosis by preventing metabolic endotoxemia-induced inflammation in Apoe-/- mice. Circulation 133 2434–2446. 10.1161/CIRCULATIONAHA.115.019645 [DOI] [PubMed] [Google Scholar]
- Ludy M. J., Moore G. E., Mattes R. D. (2012). The effects of capsaicin and capsiate on energy balance: critical review and meta-analyses of studies in humans. Chem. Senses 37 103–121. 10.1093/chemse/bjr100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukovac S., Belzer C., Pellis L., Keijser B. J., de Vos W. M., Montijn R. C., et al. (2014). Differential modulation by Akkermansia muciniphila and Faecalibacterium prausnitzii of host peripheral lipid metabolism and histone acetylation in mouse gut organoids. MBio 5:e1438-14 10.1128/mBio.01438-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z., Ma L., Zhao Z., He H., Yang D., Feng X., et al. (2012). TRPV1 activation improves exercise endurance and energy metabolism through PGC-1α upregulation in mice. Cell Res. 22 551–564. 10.1038/cr.2011.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoc T., Salzberg S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27 2957–2963. 10.1093/bioinformatics/btr507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden C. L., Yanovski S. Z., Carroll M. D., Flegal K. M. (2007). The epidemiology of obesity. Gastroenterology 132 2087–2102. 10.1053/j.gastro.2007.03.052 [DOI] [PubMed] [Google Scholar]
- Parks D. H., Tyson G. W., Hugenholtz P., Beiko R. G. (2014). STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30 3123–3124. 10.1093/bioinformatics/btu494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Li Y., Cai Z., Li S., Zhu J., Zhang F., et al. (2012). A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490 55–60. 10.1038/nature11450 [DOI] [PubMed] [Google Scholar]
- Roopchand D. E., Carmody R. N., Kuhn P., Moskal K., Turnbaugh P. J., Raskin I., et al. (2015). Dietary polyphenols promote growth of the gut bacterium Akkermansia muciniphila and attenuate high fat diet-induced metabolic syndrome. Diabetes 64 2847–2858. 10.2337/db14-1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin N.-R., Lee J.-C., Lee H.-Y., Kim M.-S., Whon T. W., Lee M.-S., et al. (2014). An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 63 727–735. 10.1136/gutjnl-2012-303839 [DOI] [PubMed] [Google Scholar]
- Sonnenburg J. L., Backhed F. (2016). Diet-microbiota interactions as moderators of human metabolism. Nature 535 56–64. 10.1038/nature18846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorens B. (1995). Glucagon-like peptide-1 and control of insulin secretion. Diabete Metab. 21 311–318. [PubMed] [Google Scholar]
- Van den Abbeele P., Gerard P., Rabot S., Bruneau A., El Aidy S., Derrien M., et al. (2011). Arabinoxylans and inulin differentially modulate the mucosal and luminal gut microbiota and mucin-degradation in humanized rats. Environ. Microbiol. 13 2667–2680. 10.1111/j.1462-2920.2011.02533.x [DOI] [PubMed] [Google Scholar]
- van Passel M. W. J., Kant R., Zoetendal E. G., Plugge C. M., Derrien M., Malfatti S. A., et al. (2011). The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PLoS ONE 6:e16876 10.1371/journal.pone.0016876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Jia H. (2016). Metagenome-wide association studies: fine-mining the microbiome. Nat. Rev. Microbiol. 14 508–522. 10.1038/nrmicro.2016.83 [DOI] [PubMed] [Google Scholar]
- Wang L., Fouts D. E., Starkel P., Hartmann P., Chen P., Llorente C., et al. (2016). Intestinal REG3 lectins protect against alcoholic steatohepatitis by reducing mucosa-associated microbiota and preventing bacterial translocation. Cell Host Microbe 19 227–239. 10.1016/j.chom.2016.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Luo Z., Ma S., Wong W. T., Ma L., Zhong J., et al. (2010). Activation of TRPV1 by dietary capsaicin improves endothelium-dependent vasorelaxation and prevents hypertension. Cell Metab. 12 130–141. 10.1016/j.cmet.2010.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Weight gain of mice at week 9 (∗P < 0.05). (B) Average food intake (g/day/mice). (C) Area under the curve (AUC) of the oral glucose tolerance test (OGTT) (∗P < 0.05).