Abstract
We hypothesized that brain regions showing neuronal activation after refeeding comprise major nodes in a satiety network, and tested this hypothesis with two sets of experiments. Detailed c-Fos mapping comparing fasted and refed rats was performed to identify candidate nodes of the satiety network. In addition to well-known feeding-related brain regions such as the arcuate, dorsomedial and paraventricular hypothalamic nuclei, lateral hypothalamic area, parabrachial nucleus (PB), nucleus of solitary tract and central amygdalar nucleus; other refeeding activated regions were also identified, such as the parastrial and parasubthalamic nuclei. To begin understanding the connectivity of the satiety network, the interconnectivity of PB with other refeeding-activated neuronal groups was studied following administration of anterograde or retrograde tracers into the PB. After allowing for tracer transport time, the animals were fasted and then refed before sacrifice. Refeeding-activated neurons that project to the PB were found in the agranular insular area; bed nuclei of terminal stria; anterior hypothalamic area; arcuate, paraventricular and dorsomedial hypothalamic nuclei; lateral hypothalamic area; parasubthalamic nucleus; central amygdalar nucleus; area postrema; and nucleus of solitary tract. Axons originating from PB were observed to closely associate with refeeding-activated neurons in the agranular insular area; bed nuclei of terminal stria; anterior hypothalamus; paraventricular, arcuate and dorsomedial hypothalamic nuclei; lateral hypothalamic area; central amygdalar nucleus; parasubthalamic nucleus; ventral posterior thalamic nucleus; area postrema; and nucleus of solitary tract. These data indicate that the PB has bidirectional connections with most refeeding-activated neuronal groups, suggesting that short loop feedback circuits exist in this satiety network.
Keywords: refeeding, parabrachial nucleus, anterograde tract tracing, retrograde tract tracing, c-Fos
INTRODUCTION
Energy homeostasis depends on a tightly controlled balance between food intake and energy expenditure that is regulated by a complex neuronal network in the central nervous system (Schwartz et al., 2000). Under most conditions, these two components are regulated simultaneously, but become uncoupled during refeeding after a prolonged period of fasting (Rothwell et al., 1983). When animals are refed after two days of fasting, they become rapidly satiated within two hours, yet their energy expenditure increases only 24h later (Rothwell et al., 1983; Singru et al., 2007). Mapping the neuronal groups activated by 2h of refeeding and elucidating the connections between these neuronal populations, therefore, may facilitate the identification of neuronal networks specifically involved in the development of satiety. Accordingly, we generated detailed maps of c-Fos-immunoreactive (c-Fos-IR) neurons in the brain that are associated with refeeding, mapped their patterns within the context of a defined ontology of brain parts (Swanson, 2015a,b) to inform future studies, and have begun to elucidate the connections among these cell groups, focusing on the parabrachial nucleus (PB).
The PB is a major refeeding-activated neuronal population and an important anorexic center of the brain that receives inputs from two major feeding-related sensor regions of the brain, the arcuate hypothalamic nucleus (ARH) and the nucleus of solitary tract (NTS) (Schwartz, 2000). Ablation of the orexigenic neurons of the ARH that express Agouti-related protein (AGRP), neuropeptide Y and gamma-aminobutyric acid (GABA), results in lethal anorexia due to over-excitation of anorexigenic neurons in the PB (Wu et al., 2009). This effect can be prevented by inhibition of PB cells by GABA agonists or ablation of vesicular glutamate transporter 2 in the NTS (Wu et al., 2012), implicating the PB as an important integration center for inputs from ARH and NTS and emphasizing its critical role in the inhibition of food intake. It is unknown, however, how these PB neurons are integrated into the network of refeeding-activated neuronal groups in the central nervous system. In this study, we provide detailed maps of the inputs and outputs of refeeding-activated PB neurons. Since we have shown that activation of anorexigenic, α-melanocyte stimulating hormone-synthesizing neurons of the ARH is critical for the determination of meal size during refeeding and that the neurons of the ventral and lateral parvicellular subdivisions of the hypothalamic paraventricular nucleus (PVH) mediate these effects (Singru et al., 2012), we also determined whether the ARH-PVH-PB pathway may be involved in the mediation of the effect of refeeding on PB neurons.
MATERIALS AND METHODS
Animals
The experiments were carried out on adult male Wistar rats (TOXI-COOP KKT, Budapest, Hungary) weighing 270–310 g (n=16). The animals were housed under standard environmental conditions (lights on between 06.00 and 18.00 h, temperature 22±1 °C, rat chow and water ad libitum). All experimental protocols were reviewed and approved by the Animal Welfare Committee at the Institute of Experimental Medicine of the Hungarian Academy of Sciences.
Preparation of sections immunostained for c-Fos for mapping the refeeding activated neuronal groups
Eight rats were used for the general c-Fos mapping study. The rats were fasted for 40 h, during which time they had free access to water. The weight loss of the fasted animals was less than that is allowed by the NIH guidelines (−7.26±0.90%). Fasting did not influence the water intake of the rats (2 day water intake of fed vs. fasted rats (ml): 85.0±11.7 vs. 78.7±3.7 P=0.635). After the fasting period, food was reintroduced to four of the eight animals and they were allowed to eat ad lib for 2 h. Upon completion of the refeeding interval, both control and experimental animals were anesthetized deeply with ketamine-xylazine (ketamine: 50 mg/kg body weight; xylazine: 10 mg/kg body weight) and perfused transcardially with 20 ml 0.01M phosphate buffered saline (PBS; pH 7.4), followed by 150 ml of 4% paraformaldehyde (PFA) in 0.1M phosphate buffer (PB, pH 7.4). The brains were rapidly removed and cryoprotected by immersion in 30% sucrose in PBS overnight. Coronal, 25 μm thick sections through the forebrain and brainstem were cut with a freezing microtome (Leica Microsystems, Wetzlar, Germany), and four series of sections, obtained at 100 μm intervals, were collected into antifreeze solution (30% ethylene glycol; 25% glycerol; 0.05M PB) and stored at −20°C until used for immunohistochemistry.
One series of sections from each animal was pre-treated with 0.5% H2O2 and 0.5% Triton X-100 in PBS for 20 min. Nonspecific antibody binding was reduced with 2% normal horse serum in PBS, and then the sections were incubated in rabbit anti-c-Fos antiserum (Ab5; Oncogene Science, Cambridge, MA) at 1:10000 dilution in PBS containing 2% normal horse serum and 0.2% sodium azide (antibody diluent) for 1 day at room temperature. After washing in PBS, sections were incubated in biotinylated donkey anti-rabbit IgG (Jackson ImmunoResearch, West Grove, PA) at 1:500 for 2 h at room temperature, and after further rinsing in PBS, incubated in avidin-biotin-peroxidase complex (ABC Elite Kit, Vector) at 1:1000 dilution for 1 h. Following further rinses in PBS, the peroxidase activity was visualized with Ni-DAB developer (0.05% diaminobenzidine, 0.15% nickel ammonium sulfate, 0.005% H2O2 in 0.05 M Tris buffer at pH 7.6), and the sections mounted on gelatin coated slides, air dried, counterstained with 1% cresyl violet and coverslipped with DPX mounting medium (Sigma).
Mapping refeeding-activated, c-Fos-IR neurons to the Swanson (2004) atlas
The mapping of brain regions activated after two hours of refeeding was performed on eight rats (four fasted only versus four fasted and then refed). Large mosaic images of full brain sections for these animals were taken by AxioVision software (Zeiss). The identification of nuclei was facilitated by Nissl counterstaining and was based on the rat brain atlas of Swanson (2004). A subset of these images was used to map the c-Fos-IR signal to digital plates of the Swanson (2004) rat brain atlas according to the following procedure.
Contrast enhancement, importation into Adobe Illustrator, and fiducial demarcation
First, two copies of each mosaic image, one unmodified (native) and the other first contrast-enhanced (CE) in Adobe Photoshop, were imported into Adobe Illustrator (AI), and fiber tracts and nuclei were outlined onto the CE image within an overlying layer by consulting AI files of the Nissl-stained atlas sections from the Swanson (2004) rat brain atlas.
Atlas level assignment
A third AI layer was used to assign the appropriate Swanson atlas level for each section. Midline structures in each image were considered stable fiducials and carried more weight when assigning a level to each section as compared to more lateral structures. When there was a discernable plane of section difference (e.g., dorsal and ventral), the layer included the two atlas level assignments and a horizontal line was drawn across the section to indicate where the plane of section began to differ.
Overlaying and transformation
Due to tissue processing differences between the brain documented in the Swanson (2004) rat brain atlas and the tissue sets we analyzed in this study, the datasets had to be transformed. Specifically, prior to overlaying the CE image, the AI file of the selected atlas level was transformed such that the X plane was scaled by 139% and the Y plane was scaled by 161%. The atlas level was then scaled to 70% so that it fit within the boundaries of the art board. The remaining layers within the AI file were then grouped and overlayed onto the transformed atlas level, and then scaled to 15%. The Nissl atlas image and the contrast-enhanced Mosaic image were then aligned.
Mapping of c-Fos immunoreactivity
To map the c-Fos data onto the atlas, dots were drawn in AI over every c-Fos-IR nucleus in the native mosaic image.
Annotations
Each AI file included observer comments on how a section was assigned to an atlas level, including detailed comments on plane of section, the appearance/disappearance of nuclei and fiber tracts, and a description of their size and shape.
Plane of section analysis
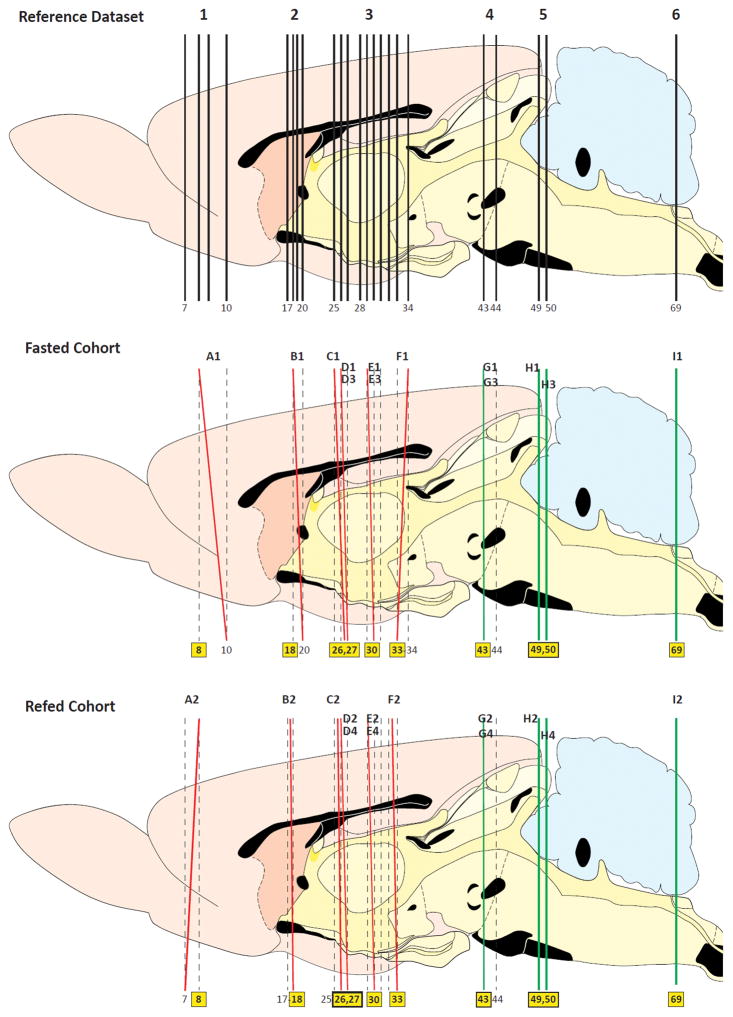
In order to compare precisely the patterns of distribution of the c-Fos-IR signal from our tissue sections in fasted and refed animals, we sought to ensure that the semi-quantitative analysis we performed was for comparable sets of sampled tissue between the two groups. To this end, we first compared the plane of section for each tissue section analyzed against the appropriate reference plane from the Swanson (2004) atlas. This allowed us to determine the common regions within each section that could be compared reliably between the two groups. The results of this analysis are shown in Figure 1. For example, since the anteriorly sampled tissue section used from the fasted and re-fed cohorts included the dorsal portion of the section that corresponded in plane to Swanson Atlas Level 8, this portion of those sections in each group were then mapped and compared (see Figure 1, red lines marking the tissue section planes ‘A1’ vs. ‘A2’). In contrast, because of plane-of-section differences between the two sampled sections, the ventral portions of the sections were not at comparable planes and hence were excluded from analysis (one mapped to Level 7 and the other to Level 10 of the Swanson (2004) reference planes). In this way, we could determine with certainty which portions of the tissue sets were suitable for accurate and reliable semi-quantitative analyses.
Figure 1.
Plane of section analysis for the images used to map c-Fos-immunoreactive signal to the Swanson (2004) atlas. The top panel shows a sagittal view of the Swanson reference atlas brain, with vertical lines marking the selected atlas levels used for our analysis. The middle and bottom panels show the plane of section of the tissue analyzed (red or green) with respect to the Swanson levels (black). Black alphanumeric designations (e.g., ‘A1’) correspond the appropriate panel in Figure 2 that shows the detailed map at that plane. Yellow boxes highlight the atlas levels assigned for the particular section analyzed. If a section’s plane fell between two atlas levels (i.e., there was a dorsoventral difference in plane), the plane of section is drawn as a red line. In such cases, only a portion of the section was used; the level for the portion chosen is indicated by the yellow box assignment. That portion was then used to compare against a comparable portion of a section from the other cohort. Levels completely in plane with the reference atlas are drawn as green lines.
Tabulation
The data summarized in Table 2 reflect c-Fos-IR counts obtained from those selected regions of the dataset that were mapped to the Swanson (2004) atlas (i.e., the tabulated counts are those directly performed from the maps). The topographic arrangement of brain regions listed in Table 2 follows the latest nomenclature standards of the Swanson atlas (Swanson, 2015a; see Table 2 Legend for details).
Table 2.
c-Fos-immunoreactivity (Fos-ir) in brains from fasted and refed rats mapped to Swanson (2004)
| Cell Group or Region (Abbreviation, Swanson Atlas Level) | Density of Fos-ir(1)
|
|
|---|---|---|
|
| ||
| Fasted | Refed | |
| Cerebrum (CH) | ||
| Cerebral Nuclei | ||
| Pallidum | ||
| Globus pallidus (GP) | ||
| Medial globus pallidus (GPi, 27) | (+) | (+) |
| Lateral globus pallidus (GPe, 27) | (−) | (−) |
| Substantia innominata (SI, 27) | (−) | (−) |
| Medial septal complex (MSC) | ||
| Medial septal nucleus (MS, 18) | ++++ | +++* |
| Bed nuclei of terminal stria (BST) | ||
| Anteromedial area (BSTam, 18) | +++ | ++++* |
| Striatum | ||
| Lateral septal nucleus (LS) | ||
| rostral (rostroventral) part (LSr, 18) | + | +* |
| ventral part (LSv, 18) | ++++ | +++ |
| Caudoputamen (CP, 27) | (−) | (−) |
| (CP, 30) | (−) | (+) |
| Central amygdalar nucleus (CEA) | ||
| Medial part (CEAm, 27) | (−) | (+++) |
| Lateral part (CEAl, 27) | (−) | (++) |
| (CEAl, 30) | (−) | (−) |
| Intercalated amygdalar nuclei (IA, 27) | (−) | (−) |
| (IA, 30) | (−) | (−) |
| Medial amygdalar nucleus (MEA) | ||
| Anteroventral part (MEAav, 27) | (+) | (++) |
| Anterodorsal part (MEAad, 27) | (+) | (+) |
| Posterodorsal part, sublayer c (MEApd.c, 30) | (−) | (++) |
| Cerebral Cortex (CTX) | ||
| Cortical Plate (CTXpl) | ||
| Limbic region | ||
| Olfactory region (OLF) | ||
| Piriform area (PIR) | ||
| molecular layer (PIR1, 27) | (−) | (+) |
| (PIR1, 30) | (−) | (+) |
| pyramidal layer (PIR2, 27) | (−) | (−) |
| (PIR2, 30) | (−) | (+) |
| polymorph layer (PIR3, 27) | (−) | (−) |
| (PIR3, 30) | (−) | (++) |
| Cortical amygdalar complex | ||
| Cortical amygdalar area (COA) | ||
| Anterior part (COAa, 27) | (+) | (++) |
| Posterior part (COAp, 30) | ||
| Medial zone (COApm, 30) | ||
| layer 1 | (−) | (+) |
| layer 3 | (+) | (+) |
| Lateral zone (COApl, 30) | ||
| layer 1 | (−) | (−) |
| layer 2 | (−) | (+) |
| layer 3 | (+) | (+) |
| Piriform-amygdalar area (PAA, 30) | ||
| layer 1 | (−) | (+++) |
| layer 3 | (−) | (−) |
| Retrohippocampal region (RHP) | ||
| Entorhinal area (ENT) | ||
| Lateral part (ENTl) | ||
| layer 1 (ENTl1) | (−) | (−) |
| layer 2 (ENTl2) | (−) | (−) |
| layer 3 (ENTl3) | (−) | (−) |
| layer 4 (ENTl4) | (−) | (−) |
| layer 5 (ENTl5) | (−) | (−) |
| layer 6 (ENTl6) | (−) | (−) |
| Cingulate region (CNG) | ||
| Prelimbic area (PL) | ||
| layer 1 (PL1, 8) | - | +* |
| layer 2 (PL2, 8) | - | ++* |
| layer 3 (PL3, 8) | - | +* |
| layer 5 (PL5, 8) | - | ++++* |
| layer 6a (PL6a, 8) | - | +++* |
| Anterior cingulate area, dorsal part (ACAd) | ||
| layer 1 (ACAd1, 8) | - | +* |
| layer 5 (ACAd5, 8) | + | + |
| layer 6a (ACAd6a, 8) | + | + |
| layer 6b (ACAd6b, 8) | - | - |
| Frontal region (FRO) | ||
| Somatomotor areas (MO) | ||
| Secondary somatomotor areas (MOs) | ||
| layer 1 (MOs1, 8) | - | +* |
| layers 2/3 (MOs2/3, 8) | - | +* |
| layer 5 (MOs5, 8) | + | + |
| layer 6a (MOs6a, 8) | ++* | + |
| layer 6b (MOs6b, 8) | - | - |
| Temporal region (TE) | ||
| Perirhinal area (PERI) | ||
| layer 1 (PERI1) | (−) | (−) |
| layer 3 (PERI3) | (−) | (−) |
| layer 5 (PERI5) | (+) | (−) |
| Ectorhinal area (ECT) | ||
| layer 1 (ECT1) | (−) | (+) |
| layer 3 (ECT3) | (−) | (+) |
| layer 5 (ECT6) | (−) | (+) |
| Temporal association areas (TEa) | ||
| layer 1 (TEa1) | (−) | (−) |
| layer 3 (TEa2) | (−) | (−) |
| layer 4 (TEa4) | (−) | (+) |
| layer 5 (TEa5) | (−) | (+) |
| Cortical subplate (CTXsp) | ||
| Basolateral amygdalar complex | ||
| Basomedial amygdalar nucleus (BMA) | ||
| Anterior part (BMAa, 27) | (−) | (+) |
| Posterior part (BMAp, 27) (2) | (−) | (−) |
| (BMAp, 30) | (−) | (+) |
| Basolateral amygdalar nucleus (BLA, 30) | ||
| Anterior part (BLAa, 27) | (−) | (−) |
| Posterior part (BLAp, 30) | (−) | (−) |
| Lateral amygdalar nucleus (LA, 27) | (−) | (−) |
| (LA, 30) | (−) | (−) |
| Posterior amygdalar nucleus (PA, 30) | (+) | (−) |
| (PA, 33) | - | - |
| Endopiriform nucleus (EP) | ||
| Ventral part (EPv, 27) | (−) | (−) |
| (EPv, 30) | (−) | (−) |
| Dorsal part (EPd, 27) | (−) | (−) |
| (EPd, 30) | (−) | (−) |
| Interbrain (IB) | ||
| Hypothalamus (HY) | ||
| Periventricular hypothalamic zone | ||
| Terminal lamina (lam) | ||
| Median preoptic nucleus (MEPO, 18) | + | + |
| Internuclear area, hypothalamic periventricular part (I, 30) | ++* | + |
| Anterodorsal preoptic nucleus (ADP, 18) | ++++ | ++++ |
| Medial preoptic area (MPO, 18) | ++* | +++ |
| Parastrial nucleus (PS, 18) | ++ | ++++* |
| Subparaventricular zone (SBPV, 26) | +* | + |
| (SBPV, 27) | +* | + |
| Paraventricular hypothalamic nucleus, magnocellular division (PVHm) | ||
| Posterior magnocellular part (PVHpm) | ||
| lateral zone (PVHpml, 26) | + | +++* |
| Paraventricular hypothalamic nucleus, parvicellular division (PVHp) | ||
| Periventricular part (PVHpv, 27) | + | + |
| Medial parvicellular part, dorsal zone (PVHmpd, 27) | + | !!!! |
| [Lateral parvicellular part] (PVHlp, 27) | ++++ | !!!! |
| [Forniceal part] (PVHf, 27) | +* | ++++ |
| Supraoptic nucleus (SOg) | ||
| retrochiasmatic part (SOr) | - | - |
| Neurohypophysis | ||
| Infundibulum (INF) | ||
| Median eminence (ME, 30) | - | + |
| Arcuate hypothalamic nucleus (ARH, 30) | - | ++++ |
| Periventricular hypothalamic nucleus intermediate part (PVi) | - | ++ |
| Dorsomedial hypothalamic nucleus (DMH) | ||
| Anterior part (DMHa, 30) | + | + |
| Posterior part (DMHp, 30) | + | ++ |
| Ventral part (DMHv, 30) | ++* | !!!!! |
| Posterior hypothalamic nucleus (PH, 30) | ++* | +* |
| (PH, 33) | !!!! | !!!! |
| Periventricular hypothalamic nucleus, posterior part (PVp, 33) | +++* | ++ |
| Medial hypothalamic zone | ||
| Anterior hypothalamic nucleus (AHN) | ||
| Central part (AHNc, 27) | +* | ++* |
| Posterior part (AHNp, 27) | ++* | +++ |
| Dorsal part (AHNd, 27) | - | + |
| Ventromedial hypothalamic nucleus (VMH) | ||
| Ventrolateral part (VMHvl, 27) | + | + |
| (VMHvl, 30) | ++* | ++ |
| Central part (VMHc, 27) | +* | ++* |
| (VMHc, 30) | +* | ++ |
| Dorsomedial part (VMHdm, 27) | +* | +* |
| Ventral premammillary nucleus (PMv, 33) | +++* | - |
| Dorsal premammillary nucleus (PMd, 33) | + | ++* |
| Mammillary body (MB) | ||
| Tuberomammillary nucleus (TM) | ||
| ventral part (TMv, 33) | + | + |
| Medial mammillary nucleus (MMg) | ||
| median part (MMe, 33) | - | - |
| Supramammillary nucleus (SUM) | ||
| lateral part (SUMl, 33) | +++* | + |
| Lateral hypothalamic zone | ||
| Lateral preoptic area (LPO, 18) | +* | ++++ |
| Lateral hypothalamic area (LHA) | ||
| Anterior group (LHAag) | ||
| Anterior region (LHAa) | ||
| intermediate zone (LHAai, 26) | +++ | ++++ |
| dorsal zone (LHAad, 26) | + | ++++ |
| Middle group (LHAmg) | ||
| Medial tier (LHAm) | ||
| Juxtaventromedial region (LHAjv) | ||
| Ventral zone (LHAjvv, 27) (3) | ++ | ++ |
| (LHAjvv, 30) | ++++* | +++ |
| Dorsal zone (LHAjvd, 27) (3) | + | ++* |
| Juxtaparaventricular region (LHAjp, 26) | + | ++++ |
| Juxtadorsomedial region (LHAjd, 27) | + | + |
| (LHAjd, 30) | ++++* | ++++ |
| Perifornical tier (LHApf) | ||
| Subfornical region (LHAsf) | ||
| Anterior zone (LHAsfa, 27) | +++* | !!!!* |
| Posterior zone (LHAsfp, 30) | ++ | +++* |
| Premammillary zone (LHAsfpm, 33) | ++++ | +++* |
| Suprafornical region (LHAs, 30) | ++++* | ++ |
| Lateral tier (LHAl) | ||
| Tuberal nucleus (TU) | ||
| Subventromedial part (TUsv, 27) | - | ++++ |
| (TUsv, 33) | - | - |
| Intermediate part (TUi, 27) (3) | +* | - |
| (TUi, 33) | - | + |
| Terete part (TUte, 33) | - | + |
| Lateral part (TUl, 33) | + | - |
| Ventral region (LHAv) | ||
| Medial zone (LHAvm, 30) | ++++* | ++++* |
| Lateral zone (LHAvl, 30) | + | + |
| (LHAvl, 33) | + | + |
| Parvicellular region (LHApc, 27) | + | + |
| Magnocellular nucleus (LHAma, 30) | + | - |
| Dorsal region (LHAd, 27) | !!!! | !!!! |
| (LHAd, 30) | ++++ | ++++ |
| Posterior group (LHApg) | ||
| Posterior region (LHAp, 33) | !!!!* | !!!!* |
| Parasubthalamic nucleus (PSTN, 33) | ++ | !!!!* |
| Subthalamic nucleus (STN, 30) | - | - |
| (STN, 33) | +++ | !!!! |
| Thalamus (TH) | ||
| Ventral part of thalamus (VNT) | ||
| Zona incerta, general (ZIg) | ||
| Zona incerta (ZI, 26) | +++* | ++++ |
| (ZI, 27) | +++* | !!!!! |
| (ZI, 30) | ++++ | +++* |
| (ZI, 33) | +++* | ++* |
| Fields of Forel (FF, 33) | ++++ | + |
| Dorsal part of thalamus (DOR) | ||
| Midline thalamic nuclei (MTN) | ||
| Nucleus reuniens (RE) | ||
| Rostral division (REr, 26) (4) | ++++* | +++ |
| Caudal division (REca, 27) (4) | ++ | +++ |
| Paraventricular thalamic nucleus (PVT, 25) | ++ | ++++ |
| Paratenial nucleus (PT, 25) | ++ | ++++ |
| Anterior thalamic nuclei (ATN) | ||
| Anteromedial thalamic nucleus (AM, 25) | ||
| Dorsal part (AMd) | + | - |
| Intralaminar thalamic nuclei (ILM) | ||
| Central medial thalamic nucleus (CM, 25) | + | ++ |
| (CM, 33) | + | - |
| Medial thalamic nuclei (MED) | ||
| Perireuniens nucleus (PR, 27) | +* | +* |
| (PR, 30) | - | + |
| Submedial thalamic nucleus (SMT, 30) | - | - |
| Mediodorsal nucleus of the thalamus (MD) | ||
| Medial part (MDm, 25) | - | +* |
| Ventral thalamic nuclei | ||
| Ventral medial thalamic nucleus (VM, 30) | - | - |
| Ventral posterior thalamic nucleus (VP) | ||
| Ventral posteromedial thalamic nucleus, general (VPMg) | ||
| Parvicellular part (VPMpc, 33) | - | +* |
| Ventral posterolateral thalamic nucleus, general (VPLg) | ||
| Parvicellular part (VPLpc, 33) | - | - |
| Subparafascicular nucleus (SPF) | ||
| Magnocellular part (SPFm, 33) | +* | ++* |
| Midbrain (MB) | ||
| Tegmentum (TG) | ||
| Interpeduncular nucleus (IPN) | ||
| apical subnucleus (IPNa, 43) | +++ | + |
| dorsomedial subnucleus (IPNd, 43) | + | ++ |
| lateral subnucleus (IPNl, 43) | ||
| dorsal part (IPNld) | - | - |
| intermediate part (IPNli) | + | - |
| ventral part (IPNlv, 43) | + | - |
| intermediate subnucleus (IPNi, 43) | + | - |
| central subnucleus (IPNc, 43) | + | - |
| Central linear raphe nucleus (CLI, 43) | + | - |
| Superior central raphé nucleus (CS) | ||
| medial part (CSm, 43) | + | - |
| Dorsal raphe nucleus (DR, 43) | + | - |
| (DR, 49) | !!!! | +++ |
| Ventral tegmental area (VTA, 43) | - | + |
| Midbrain reticular nucleus (MRN) | ||
| Magnocellular part (MRNm, 43) | ++++* | +++ |
| Parvicellular part (MRNp, 43) | ++* | - |
| Retrorubral area (RR, 43) | ++++* | + |
| Periaqueductal gray (PAG) | ||
| Medial division (PAGm, 43) | - | + |
| Ventrolateral division (PAGvl, 43) | !!!! | !!!! |
| Dorsolateral division (PAGdl, 43) | - | - |
| Dorsal division (PAGd, 43) | ++ | ++ |
| Midbrain nucleus of trigeminal nerve (MEV, 49) | + | ++ |
| (MEV, 50) | + | !!!! |
| (MEV, 69) | - | - |
| Nucleus sagulum (SAG) | - | + |
| Parabigeminal nucleus (PBG, 43) | - | - |
| Nucleus of brachium of inferior colliculus (NB) | +++* | !!!! |
| Tectum (TC) | ||
| Superior colliculus (SC) | ||
| zonal layer (SCzo, 43) | - | + |
| superficial gray layer (SCsg, 43) | - | - |
| sublayer a of intermediate gray layer (SCig.a, 43) | - | ++* |
| sublayer b of intermediate gray layer (SCig.b, 43) | ++* | ++++* |
| sublayer c of intermediate gray layer (SCig.c, 43) | ++* | +++* |
| Inferior colliculus (IC) | - | - |
| Central nucleus (ICc, 49) | !!!!!* | - |
| (ICc, 50) | - | - |
| Dorsal nucleus (ICd, 49) | !!!! | - |
| (ICd, 50) | - | - |
| External nucleus (ICe, 43) | +++* | ++++ |
| Rhombic Brain (RB) | ||
| Hindbrain (HB) | ||
| Pons (P) | ||
| Motor nucleus of trigeminal nerve (V) | ||
| Magnocellular part (Vma, 49) | + | !!!!* |
| (Vma, 50) | +++ | !!!!!* |
| Parvicellular part (Vpc, 49) | +* | - |
| (Vpc, 50) | - | + |
| Pontine nuclei, general (PGg) | ||
| Pontine nuclei (PG, 43) | !!!!! | + |
| Tegmental reticular nucleus (TRN, 49) (5) | ++ | + |
| (TRN, 50) (5) | +++ | ++ |
| Magnus raphe nucleus (RM, 49) | + | + |
| (RM, 50) | + | + |
| Pontine raphe nucleus (RPO, 49) | +++ | +++* |
| (RPO, 50) | +++* | +* |
| Pontine reticular nucleus, Caudal part (PRNc, 49) | !!!!* | ++++ |
| (PRNc, 50) | !!!!* | ++++* |
| Pedunculopontine nucleus (PPN, 43) | +++ | ++* |
| Supratrigeminal nucleus (SUT, 49)Pontine central gray, general (PCGg) | ||
| Pontine central gray (PCG, 49) | !!!! | ++++* |
| (PCG, 50) | +++* | ++* |
| Nucleus incertus (NI) | ||
| compact part (NIc, 50) | ++++ | ++++ |
| diffuse part (NId, 50) | ++++ | ++* |
| Lateral tegmental nucleus (LTN, 49) | + | +++* |
| Dorsal tegmental nucleus (DTN, 49) | +++* | + |
| (DTN, 50) | !!!!* | ++++* |
| Sublaterodorsal nucleus (SLD, 49) (5) | ++++* | + |
| (SLD, 50) | ++* | + |
| Laterodorsal tegmental nucleus (LDT, 49) | ++++ | +++ |
| (LDT, 50) | !!!!* | +++* |
| Barrington’s nucleus (B, 49) | + | +* |
| (B, 50) | ++++* | !!!!* |
| Locus ceruleus (LC, 49) | - | ++++ |
| (LC, 50) | ++ | ++++ |
| Parabrachial Nucleus (PB) | ||
| Medial division (PBm) | ||
| Ventral medial part (PBmv, 50) | +* | ++++* |
| Medial medial part (PBmm, 49) | + | !!!! |
| (PBmm, 50) | + | ++++* |
| Kölliker-Fuse subnucleus (KF, 49) | +* | !!!!!* |
| Lateral division | ||
| Ventral lateral part (PBlv, 49) | + | !!!!* |
| (PBlv, 50) | - | !!!!* |
| Central lateral part (PBlc, 49) | ++ | !!!!* |
| (PBlc, 50) | ++* | !!!!!* |
| External lateral part (PBle, 49) | - | ++* |
| (PBle, 50) | - | !!!!* |
| Dorsal lateral part (PBld, 49) | +* | +++* |
| Principal sensory nucleus of trigeminal nerve (PSV, 49) | !!!!* | ++++* |
| (PSV, 50) | !!!!* | +++* |
| Nucleus of lateral lemniscus (NLL) | ||
| ventral part (NLLv, 43) | + | + |
| Superior olivary complex (SOC) | ||
| medial part (SOCm, 49) (5) | +* | - |
| (SOCm, 50) (5) | + | + |
| lateral part (SOCl, 50) (5) | ++* | +++* |
| periolivary region (POR, 49) (5) | !!!!* | !!!!* |
| (POR, 50) (5) | !!!!* | !!!!* |
| Nucleus of trapezoid body (NTB, 49) | + | !!!! |
| (NTB, 50) | + | +++* |
| Medulla (MY) | ||
| Hypoglossal nucleus (XII, 69) | - | + |
| Ambiguus nucleus, dorsal division (AMBd, 69) | - | - |
| Ambiguus nucleus, ventral division (AMDv, 69) | - | + |
| Dorsal motor nucleus of vagus nerve (DMX, 69) | - | +* |
| Obscurus raphe nucleus (RO, 69) | - | - |
| Pallidal raphe nucleus (RPA) | + | - |
| Parvicellular reticular nucleus (PARN, 69) | + | + |
| Medullary reticular nucleus (MDRN) | ||
| Ventral part (MDRNv, 69) | + | +* |
| Inferior olivary complex (IO) | ||
| Medial accessory olive (IOma, 69) | + | + |
| Lateral reticular nucleus (LRN, 69) | ||
| Magnocellular part (LRNm) | ++ | + |
| Cochlear nuclei (CN) | ||
| Ventral cochlear nucleus, anterior part (VCOa, 49) | !!!!* | ++* |
| (VCOa, 50) | ++* | + |
| Subpeduncular granular region (CNspg, 49) | - | - |
| (CNspg, 50) | !!!!* | ++* |
| Parvicellular part (LRNp) | - | + |
| Paramedian reticular nucleus (PMR, 69) | - | - |
| Parasolitary nucleus (PAS, 69) | - | +* |
| Nucleus of solitary tract (NTS) | ||
| Medial part (NTSm) | ||
| Caudal zone (NTSm, 69) | - | !!!!! |
| Gelatinous part (NTSge, 69) | - | +++* |
| Lateral part (NTSl, 69) | - | ++* |
| Commissural part (NTSco, 69) | - | ++++* |
| Area postrema (AP, 69) | ++++ | !!!! |
| Dorsal column nuclei (DCN, 69) | ||
| Gracile nucleus, general (GRg) | ||
| gracile nucleus, principal part (GR) | + | + |
| Cuneate nucleus (CU, 69) | - | - |
| External cuneate nucleus (ECU, 69) | ++ | ++ |
| Paratrigeminal nucleus (PAT, 69) | - | +++* |
| Spinal nucleus of the trigeminal nerve (SPV) | ||
| Interpolar part (SPVI, 69) | + | ++* |
| Caudal part (SPVC, 69) | - | + |
|
|
||
| Density of Fos-ir(1) | ||
|
|
||
| Fos Scattered in White Matter Tracts (Abbreviation, Atlas Level) | Fasted | Refed |
|
| ||
| Cerebrospinal Axis | ||
| Spinal tract of the trigeminal nerve (sptV, 69) | ++ | +++ |
| Tegmentum | ||
| Brachium of inferior colliculus (bic, 43) | +++* | !!!! |
| Pons | ||
| Middle cerebellar peduncle (mcp, 43) | +++ | - |
| Ventral spinocerebellar tract (sctv, 49) | !!!! | !!!!* |
| (sctv, 50) | !!!!* | +++ |
| Pyramid (py, 49) | ++ | !!!!! |
| (py, 50) | +++ | !!!!* |
This tabular, hierarchical arrangement for anatomical regions is based (with minor variations) on the provisional topographic histological groupings provided in Table C, Rat CNS Gray Matter Regionalization, BM4 Beta 3.1 and Table D, Rat CNS White Matter Tracts, BM4 Beta 1, available at the time of this writing in the online material from Swanson LW, 2015a (white papers dated 6/25/15 for Table C and 4/5/15 for Table D). This order standardizes topographic ordering based on embryological/developmental sequence, with rostral before caudal, then medial before lateral, and then ventral before dorsal. The names of the structures have evolved from Swanson (2004) to become standardized by Swanson (2015a). Therefore, all newer, standard terms for each brain part are used whenever possible (these are denoted by underlining; for example, ‘paratrigeminal nucleus’). If the older term from Swanson (2004) is retained, it is denoted in brackets (e.g., ‘[Lateral parvicellular part]’ of the paraventricular hypothalamic nucleus). Note, however, that all symbols of the structures themselves (e.g., ‘GPi’) are from Swanson LW (2004) and correspond to the symbols within the maps presented in Figure 2.
Explanations of symbols
Scale (c-Fos-IR cells): 0: – | 1–5 cells: + | 6–10 cells: ++ | 11–20 cells: +++ | 20–30 cells: ++++ | 31–99 cells: !!!! | >100 cells: !!!!!;
Other superscripted notations:
The scoring represents combined bilateral counts at each mapped level (i.e., the counts were performed on the maps shown in Figure 2, not on the tissue itself). Asterisks (*) denote a bilateral difference in the same subject (i.e., the combined bilateral count reflects mainly signal on one side, as it was not distributed uniformly between sides). Scoring symbols marked in parentheses (e.g., ‘(+++)’) represent unilateral counts, since in some cases, only one side of the tissue shared a common plane of section and therefore was the only side mapped.
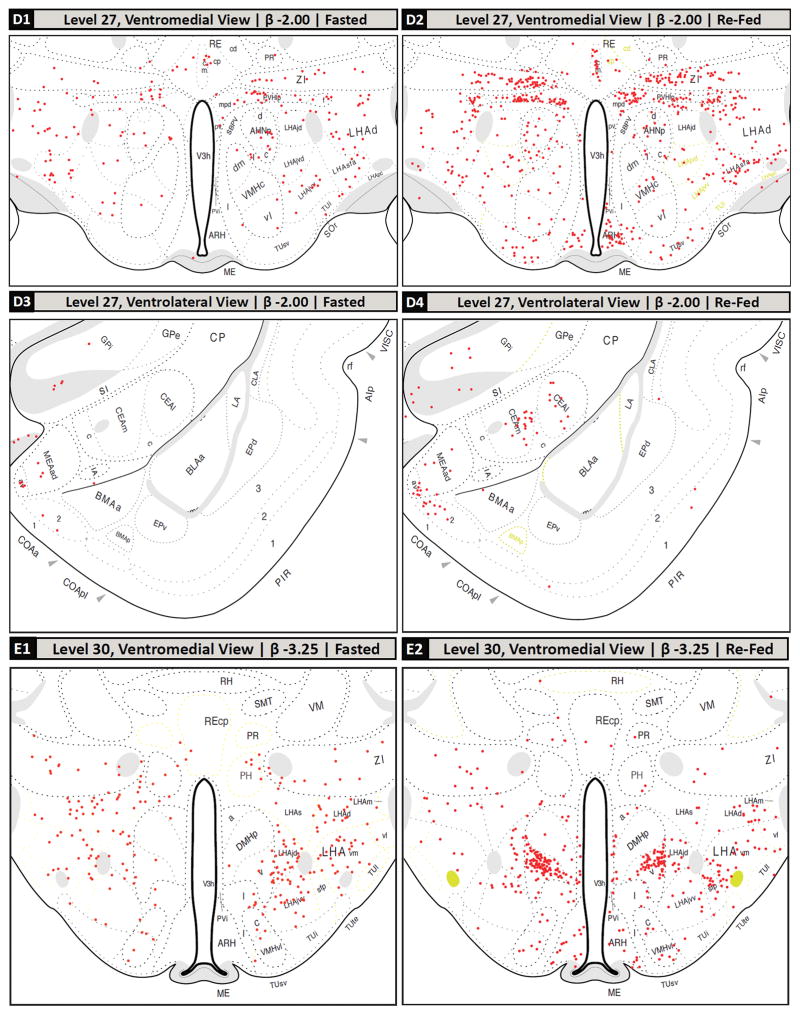
For Level 27, the boundaries for the BMAp were indistinct in Nissl-stained tissue analyzed for the refed cohort, and were inferred (this uncertainty is denoted by pale yellow colored boundaries; see Figure 2, Panel D4).
For Level 27, the boundaries for the LHAjvd, LHAjvv, and TUi were inferred, as they were indistinct in Nissl-stained sections (this uncertainty is denoted by pale yellow colored boundaries; see Figure 2, Panels D2, E1 and E2).
For the reuniens nucleus, boundaries for the subdivisions were inferred, as they were indistinct in the Nissl-stained sections (this uncertainty is denoted by pale yellow colored boundaries; see Figure 2, Panels C1, C2 and D2). Therefore, c-Fos+ counts represent those found in the structure’s main divisions (rostral vs. caudal) rather than from specific subdivisions.
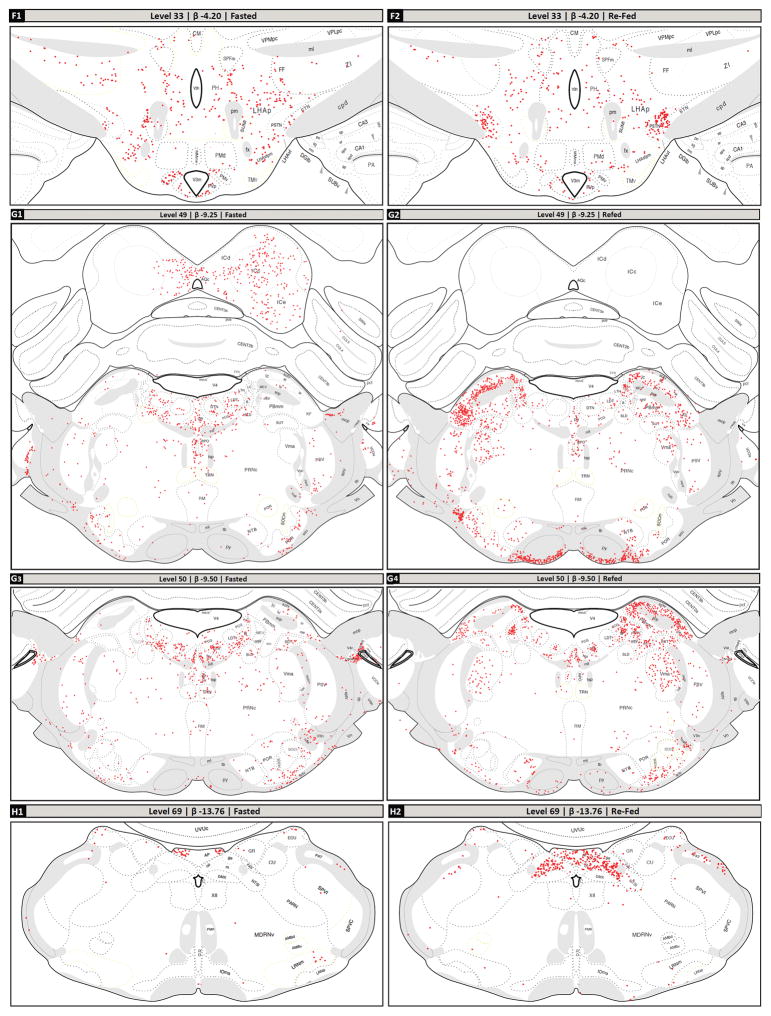
For Levels 49 and 50, the boundaries for the SLD, TRN, SOCm, SOCl, and POR were often indistinct in Nissl-stained tissue analyzed for one or both cohorts, and were therefore inferred (this uncertainty is denoted by pale yellow colored boundaries; see Figure 2, Panels H1–H4).
Analyses of the number of c-Fos-IR neurons in refeeding-activated brain areas
The refeeding induced changes of the number of c-Fos-IR neurons was measured with ImageJ software (http://rsbweb.nih.gov/ij/) using automatic particle analysis. The color images were converted to black and white, then the monochrome images were thresholded. The threshold range was set to differentiate the c-Fos containing nuclei from the background. Finally, the numbers of immunolabeled nuclei were counted by the automatic particle analyses. Each nuclei were analyzed in three to five sections in each brain. The data presented as mean ± SEM of the number of c-Fos-IR nuclei/section.
Retrograde tract tracing experiments
The retrograde tracer, cholera toxin β subunit (CTB, List Biological Laboratories, Campbell, CA), was injected by iontophoresis into the PB in three animals. Rats were anesthetized with ketamine-xylazine (ketamine: 50 mg/kg; xylazine: 10 mg/kg, i.p.) and their head positioned in a stereotaxic apparatus with the Bregma and Lambda landmarks in the horizontal plane. Through a burr hole in the skull, a glass micropipette (20 μm outer tip diameter) filled with 0.5% CTB was lowered into the brain at stereotaxic coordinates corresponding to the PB (anterior–posterior, −9.3 mm from the Bregma; lateral, −2.0 mm; and dorsoventral, −7.0 mm from the surface of the skull), based on the atlas of Paxinos and Watson (1998). The tracer was deposited by iontophoresis for 10 min (6 μA positive current, pulsed on-off at 7s intervals) using a constant-current source (Stoelting, Wood Dale, IL). Rats were allowed to survive for 10 days, than fasted for 40 h. The weight of the animals was monitored during the recovery period. All of the animals gained weight during the recovery period compared to their preoperative weight. After the fasting period, the rats were refed for 2 h and the brains fixed by perfusion as described above.
Anterograde tract tracing experiments
The anterograde tracer, Phaseolus vulgaris leuco-agglutinin (PHA-L; Vector Laboratories, Burlingame, CA), was injected into the PB of five rats. Animals were anesthetized intraperitoneally with ketamine-xylazine and their head mounted in a stereotaxic apparatus as described above. Using the same stereotaxic coordinates as described in the retrograde tract tracing experiment section, 2.5% PHA-L in 0.01M PB at pH 8.0 was injected by a glass micropipette into the PB using iontophoresis (6 μA positive current, pulsed on-off at 7 s intervals) for 15–20 min. Following a 10 day transport time, animals were fasted for two days and then refed for 2 h. The weight of the animals was monitored during the recovery period. All of the animals gained weight during the recovery period compared to their preoperative weight. Perfusion of the animals with fixative, sectioning of the tissue, and preparation and the pretreatment of the sections were performed as described above.
Immunohistochemical identification of the CTB injection sites
CTB injection sites were identified in single immunolabeled sections. One of the four series of sections was pre-treated sequentially, first with 0.5% H2O2 and 0.5% Triton X-100 in PBS for 15 min, and then 2% normal horse serum in PBS for 20 min. Sections were then incubated in a mixture of goat anti-CTB serum (List Biological Labs) at 1:10,000 dilution for 2 days at 4°C. Following washes in PBS, the sections were immersed in biotinylated donkey anti-sheep IgG at 1:500 (Jackson) and incubated for 2 h at room temperature. After rinsing with PBS, the sections were incubated in avidin-biotin complex (ABC Elite, Vector Laboratories) at 1:1,000 dilution for 1 h. Following rinses in PBS, peroxidase activity was visualized by NiDAB developer consisting of 0.05% diaminobenzidine (DAB), 0.15% nickel ammonium sulfate (Ni) and 0.005% H2O2 in 0.05M Tris buffer (pH 7.6). The sections were mounted on gelatin coated slides, air dried, counterstained with 1% cresyl violet and coverslipped with DPX mounting medium (Sigma).
Identification of the sources of the refeeding-activated inputs of the PB using triple-labelling immunofluorescence for CTB, POMC and c-Fos
To map the refeeding-activated inputs of the PB and to determine whether the pro-opiomelanocortin (POMC) neurons of the ARH are also among the refeeding activated inputs of this nucleus, triple-labeling immunofluorescence was performed for POMC, CTB and c-Fos on sections from animals with CTB injection positioned into the PB. After the pre-treatment, the sections were incubated in a mixture of goat anti-CTB serum at 1:10,000 dilution and rabbit anti-c-Fos serum at 1:2,000 dilution for 2 days at 4 °C. Following washes in PBS, the sections were immersed in biotinylated donkey anti-sheep IgG (1:500, Jackson) and incubated for 2 h at room temperature. After rinsing with PBS, sections were incubated in ABC solution at 1:1,000 dilution for 1 h, rinsed in PBS and the immunoreaction amplified with biotinylated tyramide using the TSA amplification kit (Perkin Elmer Life and Analytical Sciences, Waltham, MA) according to the manufacturer’s instructions. After further washes, the sections were incubated in a cocktail of streptavidin-conjugated FITC at 1:250 dilution (Vector) and Alexa 555-conjugated anti rabbit IgG at 1:500 dilution, and then incubated in rabbit antiserum against POMC (Phoenix Pharmaceuticals, Inc.) at 1:2,000 dilution for 2 days at 4 °C. Following washes in PBS, the sections were incubated in Cy5-conjugated anti-rabbit IgG for 2 h at room temperature, mounted onto glass slides, air dried, and coverslipped with Vectashield mounting medium (Vector).
Immunohistochemistry for identification of the PHA-L injection sites
To determine the location of the PHA-L injection sites, single labeling immunocytochemistry was performed on one series of sections incubated in rabbit anti-PHA-L serum (Vector) at 1:10,000 dilution in antibody diluent for 1 day at room temperature. After washing in PBS, the sections were incubated in biotinylated donkey anti-rabbit IgG (Jackson ImmunoResearch, West Grove, PA) at 1:500 for 2 h at room temperature, rinsed in PBS, and incubated in ABC solution at 1:1,000 dilution for 1 h. Following rinses in PBS, peroxidase activity was visualized with Ni-DAB developer. Sections were mounted on gelatin coated slides, air dried, counterstained with 1% cresyl violet and coverslipped with DPX mounting medium.
Identification of the refeeding-activated targets of the PB using triple-labeling immunofluorescence for PHA-L, c-Fos and the neuronal marker HuC/HuD
To identify the brain regions where refeeding-activated neurons are contacted by PB neurons, triple-labeling immunofluorescence for PHA-L, c-Fos and the neuronal marker, HuC/HuD, was performed on sections of all animals (n=5) with a PHA-L injection site confined within the area of the PB. Following pre-treatment as described above, the sections were incubated in anti-PHA-L serum at 1:5,000 for 2 days at room temperature, followed by biotinylated donkey anti-rabbit IgG (Jackson Immunoresearch) diluted to 1:500 and then ABC (1:1,000) for 1 h after rinses in PBS. The immunoreaction was amplified with biotinylated tyramide as above, and after further washes, the sections were incubated in streptavidin-conjugated Alexa 555 (Vector) at a 1:500 dilution. Then, the sections were incubated in a mixture of rabbit antiserum against c-Fos at 1:2,000 dilution and mouse antiserum against HuC/HuD (Molecular Probes) at 1:500 for 2 days at 4°C. After washes in PBS, the sections were immersed in a cocktail of fluorescein isothiocyanate (FITC)-conjugated donkey anti-rabbit IgG (Jackson) at 1:250 dilution and Cy5-conjugated donkey anti-mouse IgG (Jackson) at 1:100 dilution and incubated for 2 h at room temperature. The sections were then mounted onto glass slides and coverslipped with Vectashield Mounting Medium.
Image and data analysis
The DAB- and Ni-DAB-labeled immunocytochemical preparations were examined and imaged using a Zeiss AxioImager M1 bright field microscope equipped with an AxioCam MR digital camera (Carl Zeiss AG, Göttingen, Germany). The fluorescent preparations were examined with a Nikon A1R Confocal System (Nikon Instruments Ltd.). Confocal images were taken using line by line sequential scanning with laser excitation lines 488 nm for FITC, 561.6 nm for Alexa 555 and 641.8 for Cy5, dichroic mirror 405/488/561/640, emission filters 525/50 for FITC, 595/50 for Alexa 555 and 700/50 for Cy5 fluorophores were used. For 20x and 60x oil lenses, pinhole sizes were set to obtain optical slices of 2 and 1 μm thickness, respectively, and the series of optical sections were recorded with 2.0 and 1.0 μm Z step. To enhance visibility of triple-labeled PHA-L/c-Fos/HuC/HuD and CTB/c-Fos/POMC cell bodies, consecutive optical sections (from 3 to 10) were projected into one image with ImageJ image analysis software (public domain at http://rsb.info.nih.gov/ij/download/src/). Adobe Photoshop 7.0 (Adobe Systems Incorporated, San Jose, CA) was used to create composite images and to modify brightness and contrast of the images. Line drawings representing the distribution of c-Fos-IR of fasted and refed rats were made using Corel Draw 11 (Corel Corporation, Ottawa, Canada).
Antibody characterizations
Table 1 summarizes the primary antisera used in this study. The antibody directed against the protein product of the immediate-early gene, c-Fos (rabbit anti-c-Fos polyclonal antibody, PC-38 (Ab-5), Merck Biosciences, Nottingham, Notts, UK), was prepared against a peptide mapping at residues 4–17 of the human c-Fos protein. This antiserum stains a single band at ~50–55 kDa as observed by Western blot analysis of fibroblast-like BHK 21 C13 cells (Archer et al., 1999).
Table 1.
Description of the used antibodies
| Structure of the immunogen | Manufacturing | Dilution | |
|---|---|---|---|
| c-Fos | Synthetic peptide (SGFNADYEASSSRC) corresponding to amino acids 4–17 of human c-Fos | Merck Millipore, rabbit, polyclonal, #cat. PC38 RRID: AB_213663 |
1:2000 1:10000 |
| CTB | Purified CTB isolated from Vibrio cholerae | List Biological Laboratories, goat polyclonal, #cat. 703 RRID: AB_10013220 |
1:10000 |
| PHAL | Purified PHAL | Vector Laboratories, goat polyclonal, #cat. AS-2224 RRID: AB_10000080 |
1:5000 1:10000 |
| POMC | Amino acids of 27–52 of porcine POMC precursor | Phoenix Pharmaceuticals, rabbit, polyclonal, #cat. H-029-30 RRID: AB_2307442 |
1:2000 |
| HuCD | Recognizes human HuC/HuD neuronal protein and the expressed product of the ELAVL3 gene | Molecular Probes, mouse, monoclonal, #cat. A-21271 RRID: AB_221448 |
1:500 |
According to descriptions of the manufacturers, the PHA-L antiserum was produced by hyperimmunization of goat with purified PHA-L, and specific antibodies to PHA-L were isolated by affinity chromatography on PHA-L-agarose columns, whereas the CTB antiserum forms an immunoprecipitated band against a 0.5 mg/ml solution of CTB. Because PHA-L and CTB are normally not present in the brain, the specificities of PHA-L and CTB antisera were verified by the lack of any labeling in brain sections from animals that were not injected with PHA-L and CTB.
The anti-POMC serum (Phoenix Pharmaceuticals, Inc., Belmont, CA; catalog No. H-029-30, lot R163-5) was raised in rabbit against the porcine POMC precursor (AA 27–52). The specificity of this antiserum was confirmed by dual immunofluorescent labeling of rat and mouse hypothalamic sections using a sheep α-MSH antiserum (Wittmann et al., 2013).
Neuronal protein HuC/HuD (HuC/D) monoclonal (clone 16A11) antibody labeled neuronal cell nuclei and perikarya, recognizing an epitope within the carboxy-terminal domain of HuD (Marusich et al., 1994). Staining pattern of cellular morphology and distribution is the same as previously described (Lin et al., 2002; Murphy et al., 2007). The staining pattern of biotin-conjugated HuC/D monoclonal antibody is identical to that of the unconjugated antibody.
RESULTS
Brain regions displaying marked activation in association with refeeding
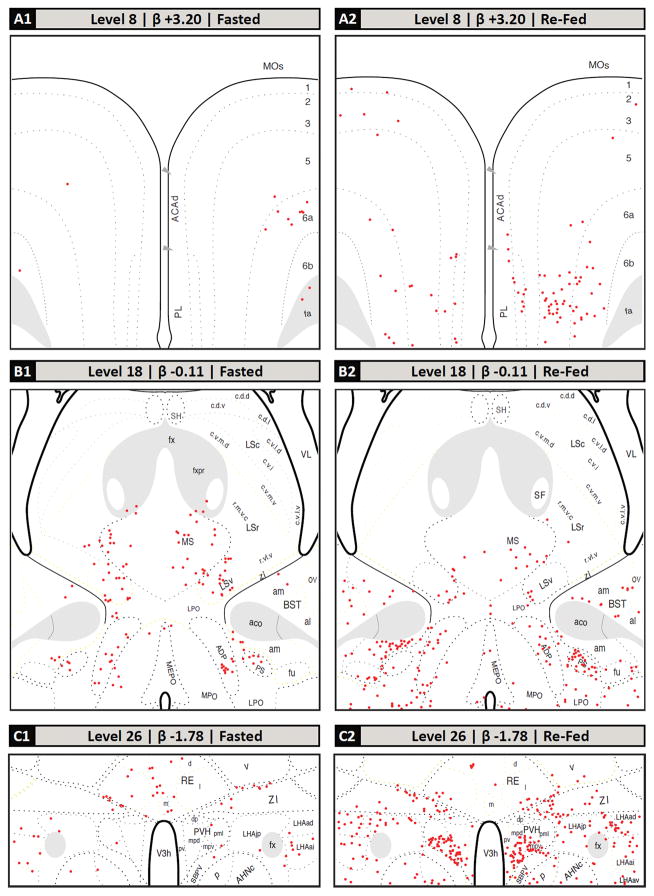
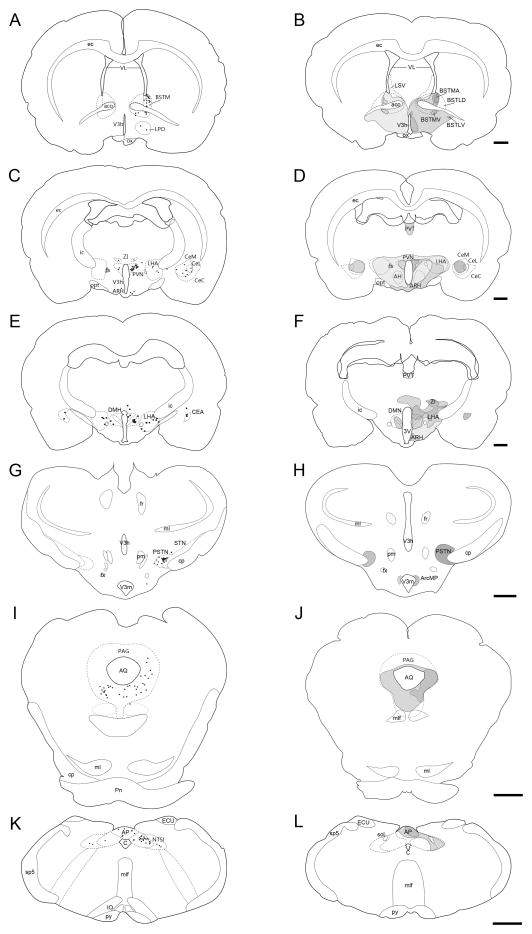
Following the screening of the tissue sets based on plane of section differences, the remaining common regions between the two groups’ sections were mapped. The mapped brain regions displaying c-Fos-IR neurons in fasted and refed rats are detailed in Figure 2 and summarized in Table 2. In the forebrain, the greatest increases in the density of c-Fos-IR cells in refed animals compared to fasted animals were detected in the prelimbic area (Fig. 2, Panels A1 and A2); the anteromedial area of the bed nuclei of terminal stria, parastrial nucleus, and lateral preoptic area (Fig. 2, Panels B1 and B2); the medial, lateral and forniceal parts of the parvicellular subdivision of the paraventricular hypothalamic nucleus (as well as the posterior part of its magnocellular division; Fig. 2, Panels C1, C2, D1 and D2); caudal portions of the piriform cortex and piriform-amygdalar transition (Fig. 2, Panels D3, D4); the medial and lateral parts of the central amygdalar nucleus (Fig. 2, Panels D3, D4); the arcuate and dorsomedial hypothalamic nuclei (Fig. 2, Panels D1, D2, E1 and E2); the anterior, juxtaparaventricular, and subfornical sub-regions of the lateral hypothalamic area (Fig. 2, Panels C1, C2, D1, D2, E1 and E2); parts of the tuberal nucleus (Fig. 2, Panels D1, D2, E1 and E2); and the parasubthalamic and subthalamic nuclei (Fig. 2, Panels F1 and F2). Additionally, thalamic areas displaying marked increases in c-Fos-IR cells in refed versus fasted rats included portions paratenial nucleus, and zona incerta (Fig. 2, Panels C1, C2, D1, D2, E1 and E2), while c-Fos-immunoreactivity was decreased in the nucleus reuniens.
Figure 2.
Selected regions of c-Fos-immunoreactive (c-Fos-IR) signal in fasted and refed rats mapped to the Swanson (2004) atlas. (A–H): c-Fos-immunoreactivity plotted to Swanson (2004) atlas levels (red dots; each dot signified one c-Fos-IR cell). The levels selected are based on the plane of section analysis summarized in Figure 1. Dotted lines within each atlas plate that are rendered in faint yellow indicate areas where the Nissl image did not provide clear boundaries for the boundary to be assigned with certainty. These should be considered as inferred boundaries based on the remaining data obtained for that section. (A1, A2): dorsal portion of Level 8; (B1, B2): middle portion of Level 18; (C1, C2): lower middle portion of the Level 26; (D1, D2): ventromedial portion of Level 27; (D3, D4): ventrolateral portion of Level 27; (E1, E2): ventromedial portion of Level 30; (F1, F2): ventral portion of Level 33; (G1, G2): full view of Level 49, except for outer portions of the cerebellum; (G3, G4): full view of Level 50, except for outer portions of the cerebellum; (H1, H2): full view of Level 69, except for overlying cerebellum. Note that these mapped data are tabulated in Table 2 according to more recent nomenclature standards (see legend of Table 2 for details).
In the hindbrains of refed animals, a large number of c-Fos-IR neurons was observed in the the magnocellular part of the motor nucleus of the trigeminal nerve (Vma), the supratrigeminal nucleus, and the ventral medial (PBmv), medial medial (PBmm), ventral lateral (PBlv), central lateral (PBlc), external lateral (PBle), and dorsal lateral (PBld) parts of the PB (Fig. 2, Panels G1–G4). Increased c-Fos-IR was also observed in refed animals in the medial, gelatinous, lateral and commissural subdivisions of the NTS and the area postrema (Fig. 2, Panels H1 and H2). Table 2 shows the quantitative analyses of the number of c-Fos-IR nuclei in the refeeding activated brain regions relative to their fasted counterparts.
Origins of the refeeding-activated neuronal inputs of the PB
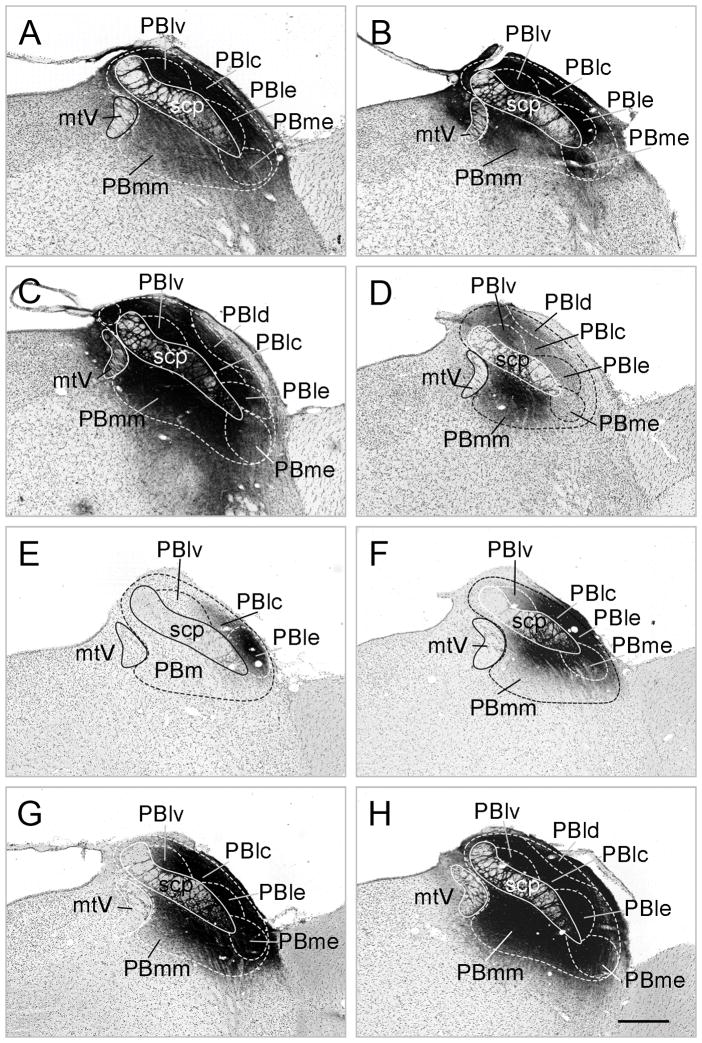
The CTB injection site in all three cases spread throughout the entire PB including the medial and lateral divisions, but the injection sites were centered in different parts of the nucleus. In animal A, the center of the injection site was in the lateral part of PB, but the medial part was also covered by the tracer. In animal B, there was intense CTB signal in the medial and lateral parts of the PB. In animal C, the middle of the injection was in the medial part of PB and the tracer spread to the lateral part of the PB. The number of CTB containing neurons in the refeeding-activated areas was higher in cases where the injection site was centered in the medial part of the PB (Animals B and C) compared to the animal where the injection was centered into the lateral part of PB (Fig. 3), although the distribution of CTB containing cells were similar in all cases.
Figure 3.
Localization of CTB (A–C) and PHA-L (D–H) injection sites using the nomenclature of Paxinos and Watson (1998). Scale bar: 200 μm
In refed, CTB-injected animals, the greatest number of refeeding-activated neurons that project to the PB (CTB- and c-Fos-IR) were observed in the PVH, particularly in ventral and lateral parvicellular subdivisions (Fig. 5B), the PSTN (Fig. 5D) and in the medial, intermediate and commissural parts of the NTS. A moderate number of double-labeled neurons were observed in the bed nuclei of terminal stria, primarily their medial portion (Fig. 5A), the medial part of the central amygdalar nucleus (Fig. 5C), lateral hypothalamic area and area postrema (Fig. 5F). Scattered c-Fos and CTB-containing neurons were detected in the agranular insular area, anterior hypothalamic area, arcuate and dorsomedial hypothalamic nuclei, and zona incerta.
Figure 5.
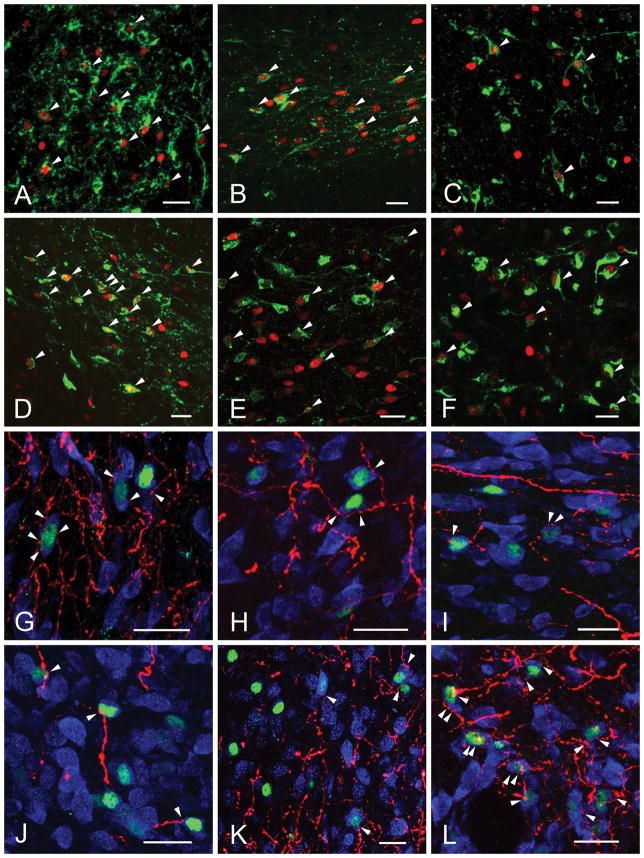
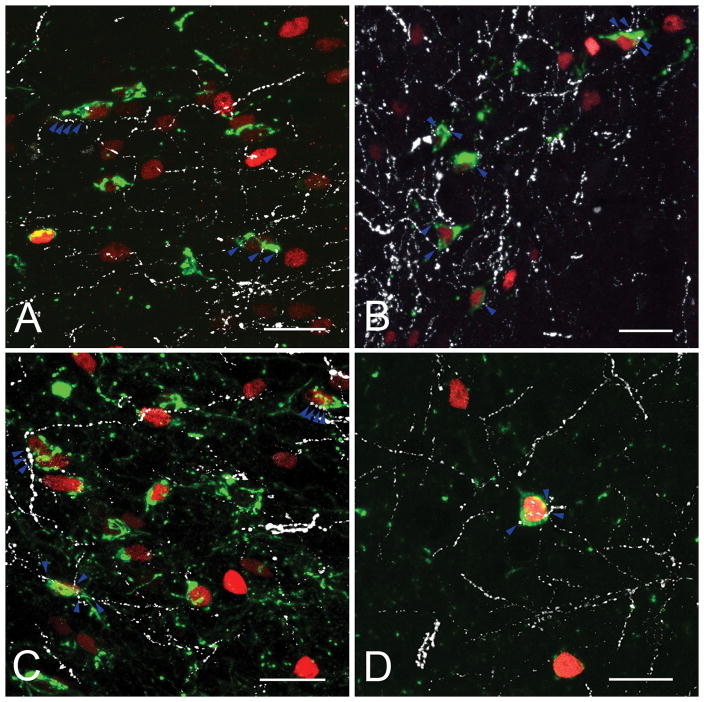
Confocal images illustrate the refeeding-activated neurons (green) (A–F) and close association of PHA-L-immunoreactive (PHA-L-ir) axons (red) (G–L). The majority of refeeding-activated neurons are contacted by PHA-L-ir axon varicosities (arrowheads). The cytoplasm of neurons is labeled with HUC/D-immunoreactivity (blue). Bed nuclei of terminal stria (A, G); ventral (B, H) and lateral (I) portions of the paraventricular hypothalamic nucleus; central nucleus of amygdala (C,K); parasubthalamic nucleus (D, L); nucleus of solitary tract (E); area postrema (F) and dorsomedial hypothalamic nucleus (J).
Identification of the direct and indirect inputs of the PB from refeeding-activated POMC neurons
Triple-labeling immunocytochemistry for c-Fos, CTB and POMC was used to determine whether the PB receives direct or indirect refeeding-activated inputs from POMC neurons in the arcuate nucleus. A large number of POMC neurons in the arcuate nucleus contained c-Fos-immunoreactivity in their nuclei after refeeding, but only a very small portion of these cells contained also CTB-immunoreactivity (1 or 2 cells/section), but were seen to heavily innervate refeeding-activated, neurons in the ventral and lateral parvicellular subdivisions of the hypothalamic paraventricular nucleus and parasubthalamic nucleus (Fig. 4).
Figure 4.
Schematic drawings, based on nomenclature of Paxinos & Watson (1998), illustrating the distribution of CTB-containing c-Fos-immunoreactive (c-Fos-IR) neurons in a representative brain injected with CTB into the PB. (A–F) The c-Fos-IR and CTB containing neurons are visualized by black dots. (G–L) The localization of PHA-L immunoreactive (-ir) fibers is represented on schematic drawings. The darkness of the area corresponds to the density of PHA-L-ir fibers. Most efferent regions of PB overlap with refeeding activated areas. Scale bar: 1 mm.
Identification of the refeeding-activated targets of the PB
Of the five animals (D-H) where the PHA-L injection sites were centered in the PB, the localization of injection sites was somewhat different (Fig. 3). The center of the PHA-L injection site in animal D was localized in the medial part of the nucleus and spread only to a small portion of the ventral part of the lateral PB (Fig. 3D). In animal E, the PHA-L injection site was centered in the external part of the lateral PB (Fig. 3E). In animal F, PHA-L entered the central and external part of lateral PB and spread into a small part of the medial PB close to the superior cerebellar peduncle (Fig. 3F). In animals G and H, PHA-L spread into all parts of PB (Fig. 3G,H).
PHA-L-IR fibers were found in refeeding-activated areas only in animals D, G and H, where the injection centered or spread into the medial part of the PB. In animal F, there were only few PHA-L-IR fibers in the central nucleus of amygdala, whereas in animal E, PHA-L-IR axons were not observed in any of the refeeding-activated areas. In animals D, G and H, the pattern of the PHA-L-IR axons was similar with a large number of PHA-L-IR fibers observed in close association with refeeding-activated neurons in the bed nucleus of the stria terminalis (Fig. 5L) and central nucleus of the amygdala, mainly in the medial parts of both nuclei, and the PSTN. Fewer, but still numerous fibers were observed in association with c-Fos-IR neurons in the anterior hypothalamus, ventral and lateral parvocellular subdivisions of the PVH (Fig. 5H,I), arcuate nucleus, hypothalamic dorsomedial nucleus (Fig. 5J), lateral hypothalamus, zona incerta, and anterior hypothalamus. Only scattered PHA-L-IR fibers were found in association with c-Fos containing neurons in the agranular insular cortex and NTS.
DISCUSSION
During the last decades, an enormous effort has been made to elucidate the central mechanisms governing appetite control and energy homeostasis. While several brain regions have now been implicated, little information is available about how these neuronal groups are interconnected and function together as a network. To better understand the satiety-related neuronal network, as a first step, we took advantage of an experimental model that we believe has made it possible to separate loci involved in satiety versus energy expenditure, and generated a detailed series of maps for the c-Fos-IR, activated neuronal groups associated with refeeding. We used an explicitly defined plane-of-section analysis together with a rigorous hierarchical anatomical framework based on a defined ontology of brain parts (Swanson, 2004; 2015a,b). The use of this analysis and ontology will allow investigators to integrate their own datasets within a common spatial framework to inform future connectomic, neuroinformatic, and functional analyses (see Bard and Rhee, 2004; Khan, 2013).
In agreement with previous reports (Timofeeva et al., 2002; Fekete et al., 2012), c-Fos-IR neurons were observed in two well-known, major metabolic sensor areas in the brain, the NTS and hypothalamic arcuate nucleus (ARH). The NTS is known to receive feeding-related inputs from the periphery through the vagus nerve, conveying information about the amount and content of the consumed food in response to distension of the esophagus and stomach, and also from the chemosensors in the wall of gastrointestinal tract (Hayes and Covasa, 2006; Grill and Hayes, 2009). Distension of the esophagus and stomach results in neuronal activation in the caudal NTS, particularly the medial and commissural parts (Sabbatini et al., 2004), regions that overlap with the distribution of the refeeding-activated neurons identified in our study. These sub-regions of the NTS are also known to receive satiety information from amylin-sensitive neurons in the area postrema and to also express leptin receptors (Grill and Hayes, 2012).
The ARH contains POMC neurons that are known to have potent anorectic effects, and are regulated primarily by circulating levels of peripheral hormones like leptin, insulin and ghrelin, rather than through the vagus nerve (Schwartz et al., 2000). These neurons play a critical role in the determination of meal size during refeeding, as inhibition of melanocortin signaling markedly increases the amount of food consumed during the first two hours of refeeding (Singru et al., 2007).
In addition to these known metabolic sensor areas, neuronal activation associated with refeeding was also observed in second order feeding-related neuronal groups including the PVH, DMH, LHA, PB and limbic structures with a known role in the control of food intake, including the central nucleus of the amygdala (CEA). A number of other neuronal groups that have not been described previously as anorectic brain centers were also observed to show c-Fos-immunoreactivity including the parasubthalamic (PSTN) nucleus. The PSTN is primarily known as a pre-autonomic nucleus with depressor effects on circulation (Ciriello, 2007), but is of particular interest in that it may be involved with conditioned taste aversion (Yasoshima et al., 2006) and shows c-Fos activation in association with anorexia induced by a diet deficient in an amino acid (Watts et al., 1999; Zhu et al., 2012).
Connections of PB with other refeeding-activated areas
To begin to untangle the complicated circuitry involved in appetite regulation, we focused on the c-Fos-IR cells in the PB. The PB is a central relay nucleus in rodent taste pathways (Cho et al., 2002) and has crucial role in the control of food intake as chronic activation of PB leads to anorexia (Wu et al., 2012). Retrograde tract tracing experiments revealed that the largest number of refeeding-activated neurons that project to the PB reside in the NTS, PVH and PSTN. The NTS relays vagal and humoral inputs toward the PB using glutamate as the major, anorexic neurotransmitter (Wu et al., 2012), indicating its important role in the mediation of refeeding-related peripheral signals toward the PB. The PVH, especially, the ventral and lateral parvicellular subdivisions, contain a large number of PB projecting, refeeding-activated neurons that are glutamatergic and densely innervated by POMC neurons in the arcuate nucleus (Singru et al., 2012). Since only few refeeding-activated POMC neurons project directly to the PB, neurons in the ventral and lateral parvocellular subdivisions of the PVH appear to serve as the major relay centers between the anorexigenic POMC neurons and the PB. Ventral and lateral parvicellular PVH neurons also project to the NTS and activate neurons in the medial and commissural sub-nuclei in response to the activation of the melanocortin receptors in the PVH (Singru et al., 2012). Therefore, it is likely that the ascending NTS-PB anorexigenic pathway and the descending ARH-PVH-PB/NTS pathway interact at multiple levels that result in multi-nodal integration of hypothalamic and brainstem signals.
The third major source of the refeeding-related input of the PB is the PSTN. The majority of PSTN neurons are glutamatergic (Ronald M. Lechan, personal observation), further supporting the observation that the refeeding-induced activation of PSTN neurons results in neuronal activation of PB. In addition, anorexigenic stimuli have been shown to induce CRH expression in the PSTN (Zhu et al., 2012), and CRH neurons in the PSTN are known to project to the PB (Magableh and Lundy, 2014). In addition to glutamate, therefore, CRH may also be involved in the inhibition of food intake in the PSTN-PB pathway. Since, like the PVH, the PSTN is densely innervated by POMC-containing axons, the PSTN may also serve as a relay station between ARH POMC neurons and the PB similar to that observed for the ARH-PVH-PB pathway. The PB was also observed to receive refeeding-activated inputs from limbic structures, including the central amygdalar and the bed nuclei of terminal stria. Activation of neurons in these brain areas is known to reduce appetite (Carter et al., 2013; Jennings et al., 2013), but also to be involved in the integration of motivation, stress, and learning (Petrovich and Gallagher, 2007; Roman et al., 2012). Carter et al., (2013) recently demonstrated a major anorexigenic pathway between the PB and the laterocapsular division of the central amydalar nucleus, mediated by calcitonin gene-related peptide (CGRP)-expressing neurons in the outer external lateral subdivision of the PB. This region of the PB, however, may primarily responsible for anorexia associated with illness or exposure to toxins (Carter et al., 2013). Anterograde tract tracing, however, also identified massive projections from the PB to the medial part of the central amygdalar nucleus, primarily originating from the medial PB, indicative of a second major projection from the PB to this nucleus and the existence of a medial PB to medial part of the central amygdalar nucleus anorexigenic pathway.
A major projection from the PB was also observed to refeeding-activated neurons of the bed nuclei of terminal stria (BST). Although recent studies were unable to demonstrate that activation of the CGRP expressing PB-BST pathway has an effect on food intake when stimulated optogenetically (Carter et al., 2013), these data do not exclude the possibility that projection of refeeding activated PB neurons to the BST is involved in the control of food intake. CGRP input from the PB may represent only a fraction of the total PB-BST input. Therefore, further, studies are needed to determine the role of the medial PB-BST projection, observed in this study, on appetite regulation.
The third major connection of the PB to refeeding-activated areas is the PSTN. Currently, no information is available about the physiological function of this pathway. We hypothesize on the basis of the anatomical data presented in the study, however, that the PSTN is another, major, integrating center in the regulation of food intake, as it receives inputs from multiple feeding-related centers such as the PB and ARH. Further studies using optogenetic activation of this center are necessary to support this hypothesis.
Since the PB is known to receive inhibitory inputs from AGRP/NPY/GABA neurons in the ARH (Wu et al., 2009), inhibitory pathways may also contribute to the refeeding-induced activation of the PB neurons. It is likely, therefore, that PB neurons have wider refeeding related inputs than recognized in the current study. Unfortunately, markers to allow immunocytochemical detection of inhibited neurons are not currently available. Further studies are needed to understand whether, in addition to the regulation of satiety, the described connections of the PB are involved in the control of vegetative functions such as blood pressure, heart rate, and respiration.
In summary, these data provide a detailed series of maps of known and unknown regions in the brain that may be involved in the regulation of satiety, and serve to facilitate the identification of novel feeding pathways not previously recognized. Most importantly, they demonstrate that the PB is highly interconnected with a large number of refeeding-activated neuronal groups in the brain through bidirectional connections, suggesting that the network of refeeding-activated neurons regulated by the PB is not uni-directionally connected, but rather is composed of a series of feedback loops that simultaneously facilitate or restrain information flow in multiple regions of the brain.
Figure 6.
Confocal images of triple-immunolabeled sections illustrating POMC-ir innervation (white) of refeeding activated (c-Fos-IR, red) CTB-containing (green) neurons in the lateral PVH (A), ventral PVH (B), PSTN (C) and CeA (D). Blue arrowheads point to the axon varicosities on the surface of the c-Fos-IR neurons. Scale bars = 25 μm.
Acknowledgments
This work was supported by Grants from the Hungarian Science Foundation (OTKA K109710), Seventh EU Research Framework Programme (Health-F2-2010-259772), Lendület Award of the Hungarian Academy of Sciences and the Dr. Gerald J. and Dorothy R. Friedman New York Foundation for Medical Research. Work performed at the UTEP Systems Neuroscience Laboratory was funded by a grant from the U.S. National Institutes of Health (GM109817) and a Grand Challenges Award from the UTEP Office of Research and Sponsored Projects. A.M. was supported by a Frank B. Cotton Trust Scholarship, a National Science Foundation STEM GK-12 Fellowship, and the UTEP PERSIST Program Grant awarded by the Howard Hughes Medical Institute.
The authors are grateful for Vera Maruzs and Agnes Simon for the expert technical assistant and wish to thank to the Nikon Microscopy Center at IEM, Nikon Austria GmbH and Auro-Science Consulting Ltd. for kindly providing microscopic support.
Footnotes
Disclosure summary: The authors have nothing to disclose.
Conflict of interest statement: There is no identified conflict, and the authors have nothing to disclose.
Role of authors:
Györgyi Zséli: Data acquisition and analyses
Barbara Vida: Data analyses
Anais Martinez: Spatial analysis and mapping of the distribution of c-Fos-immunoreactivity, manuscript preparation
Arshad M. Khan: Supervision of the general c-Fos mapping to the Swanson atlas, preparation of the final maps of the distribution of c-Fos-immunoreactivity, semi-quantitative analysis and tabulation of the c-Fos data, manuscript preparation
Ronald M. Lechan: Experimental design, data interpretation, manuscript preparation
Csaba Fekete: Supervision of the work, experimental design, data interpretation, manuscript preparation.
References
- Archer S, Li TT, Evans AT, Britland ST, Morgan H. Cell reactions to dielectrophoretic manipulation. Biochemical and biophysical research communications. 1999;257(3):687–698. doi: 10.1006/bbrc.1999.0445. [DOI] [PubMed] [Google Scholar]
- Bard JBL, Rhee SY. Ontologies in biology: design, applications and future. Nat Rev Gen. 2004;5(3):213–222. doi: 10.1038/nrg1295. [DOI] [PubMed] [Google Scholar]
- Carter ME, Soden ME, Zweifel LS, Palmiter RD. Genetic identification of a neural circuit that suppresses appetite. Nature. 2013;503(7474):111–114. doi: 10.1038/nature12596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YK, Li CS, Smith DV. Gustatory projections from the nucleus of the solitary tract to the parabrachial nuclei in the hamster. Chem Sens. 2002;27(1):81–90. doi: 10.1093/chemse/27.1.81. [DOI] [PubMed] [Google Scholar]
- Ciriello J. Cardiovascular depressor responses to stimulation of the parasubthalamic nucleus. FASEB J. 2007;21(5):A474–A474. [Google Scholar]
- Fekete C, Zseli G, Singru PS, Kadar A, Wittmann G, Fuzesi T, El-Bermani W, Lechan RM. Activation of anorexigenic pro-opiomelanocortin neurones during refeeding is independent of vagal and brainstem inputs. J Neuroendocrinol. 2012;24(11):1423–1431. doi: 10.1111/j.1365-2826.2012.02354.x. [DOI] [PubMed] [Google Scholar]
- Grill HJ, Hayes MR. The nucleus tractus solitarius: a portal for visceral afferent signal processing, energy status assessment and integration of their combined effects on food intake. Intl J Obes. 2009;33(Suppl 1):S11–15. doi: 10.1038/ijo.2009.10. [DOI] [PubMed] [Google Scholar]
- Grill HJ, Hayes MR. Hindbrain neurons as an essential hub in the neuroanatomically distributed control of energy balance. Cell Metab. 2012;16(3):296–309. doi: 10.1016/j.cmet.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes MR, Covasa M. Gastric distension enhances CCK-induced Fos-like immunoreactivity in the dorsal hindbrain by activating 5-HT3 receptors. Brain Res. 2006;1088(1):120–130. doi: 10.1016/j.brainres.2006.03.018. [DOI] [PubMed] [Google Scholar]
- Jennings JH, Rizzi G, Stamatakis AM, Ung RL, Stuber GD. The inhibitory circuit architecture of the lateral hypothalamus orchestrates feeding. Science. 2013;341(6153):1517–1521. doi: 10.1126/science.1241812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AM. Controlling feeding behavior by chemical or gene-directed targeting in the brain: what’s so spatial about our methods? Front Neurosci. 2013;7(Article 182):1–49. doi: 10.3389/fnins.2013.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Gao N, Hu HZ, Liu S, Gao C, Kim G, Ren J, Xia Y, Peck OC, Wood JD. Immunoreactivity of Hu proteins facilitates identification of myenteric neurones in guinea-pig small intestine. Neurogastroenterol Motil. 2002;14:197–204. doi: 10.1046/j.1365-2982.2002.00317.x. [DOI] [PubMed] [Google Scholar]
- Magableh A, Lundy R. Somatostatin and corticotrophin releasing hormone cell types are a major source of descending input from the forebrain to the parabrachial nucleus in mice. Chem Sens. 2014;39(8):673–82. doi: 10.1093/chemse/bju038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusich MF, Furneaux HM, Henion PD, Weston JA. Hu neuronal proteins are expressed in proliferating neurogenic cells. J Neurobiol. 1994;25:143–155. doi: 10.1002/neu.480250206. [DOI] [PubMed] [Google Scholar]
- Murphy EMA, Defontgalland D, Costa M, Brookes SJH, Wattchow DA. Quantification of subclasses of human colonic myenteric neurons by immunoreactivity to Hu, choline acetyltransferase and nitric oxide synthase. Neurogastroenterol Motil. 2007;19:126–134. doi: 10.1111/j.1365-2982.2006.00843.x. [DOI] [PubMed] [Google Scholar]
- Nilaweera KN, Archer ZA, Campbell G, Mayer CD, Balik A, Ross AW, Mercer JG, Ebling FJ, Morgan PJ, Barrett P. Photoperiod regulates genes encoding melanocortin 3 and serotonin receptors and secretogranins in the dorsomedial posterior arcuate of the Siberian hamster. J Neuroendocrinol. 2009;21(2):123–131. doi: 10.1111/j.1365-2826.2008.01810.x. [DOI] [PubMed] [Google Scholar]
- Petrovich GD, Gallagher M. Control of food consumption by learned cues: a forebrain-hypothalamic network. Physiol Behav. 2007;91(4):397–403. doi: 10.1016/j.physbeh.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman CW, Lezak KR, Kocho-Schellenberg M, Garret MA, Braas K, May V, Hammack SE. Excitotoxic lesions of the bed nucleus of the stria terminalis (BNST) attenuate the effects of repeated stress on weight gain: evidence for the recruitment of BNST activity by repeated, but not acute, stress. Behav Brain Res. 2012;227(1):300–304. doi: 10.1016/j.bbr.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell NJ, Saville ME, Stock MJ. Role of insulin in thermogenic responses to refeeding in 3-day-fasted rats. Am J Physiol. 1983;245(2):E160–165. doi: 10.1152/ajpendo.1983.245.2.E160. [DOI] [PubMed] [Google Scholar]
- Sabbatini M, Molinari C, Grossini E, Mary DA, Vacca G, Cannas M. The pattern of c-Fos immunoreactivity in the hindbrain of the rat following stomach distension. Exp Brain Res. 2004;157(3):315–323. doi: 10.1007/s00221-004-1845-x. [DOI] [PubMed] [Google Scholar]
- Schwartz MW, Woods SC, Porte D, Jr, Seeley RJ, Baskin DG. Central nervous system control of food intake. Nature. 2000;404(6778):661–671. doi: 10.1038/35007534. [DOI] [PubMed] [Google Scholar]
- Singru PS, Sanchez E, Fekete C, Lechan RM. Importance of melanocortin signaling in refeeding-induced neuronal activation and satiety. Endocrinology. 2007;148(2):638–646. doi: 10.1210/en.2006-1233. [DOI] [PubMed] [Google Scholar]
- Singru PS, Wittmann G, Farkas E, Zseli G, Fekete C, Lechan RM. Refeeding-activated glutamatergic neurons in the hypothalamic paraventricular nucleus (PVN) mediate effects of melanocortin signaling in the nucleus tractus solitarius (NTS) Endocrinol. 2012;153(8):3804–3814. doi: 10.1210/en.2012-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachniak TJ, Ghosh A, Sternson SM. Chemogenetic synaptic silencing of neural circuits localizes a hypothalamus-->midbrain pathway for feeding behavior. Neuron. 2014;82(4):797–808. doi: 10.1016/j.neuron.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson LW. Cerebral hemisphere regulation of motivated behavior. Brain Res. 2000;886(1–2):113–164. doi: 10.1016/s0006-8993(00)02905-x. [DOI] [PubMed] [Google Scholar]
- Swanson LW. Brain Maps: Structure of the Rat Brain. 3. Amsterdam: Elsevier; 2004. [Google Scholar]
- Swanson LW. Brain Maps 4 beta version. 2015a accessible at larrywswanson.com.
- Swanson LW. Brain maps online: Toward open access atlases and a pan-mammalian nomenclature. J Comp Neurol. 2015b;523(15):2272–2276. doi: 10.1002/cne.23788. [DOI] [PubMed] [Google Scholar]
- Timofeeva E, Picard F, Duclos M, Deshaies Y, Richard D. Neuronal activation and corticotropin-releasing hormone expression in the brain of obese (fa/fa) and lean (fa/?) Zucker rats in response to refeeding. Eur J Neurosci. 2002;15(6):1013–1029. doi: 10.1046/j.1460-9568.2002.01942.x. [DOI] [PubMed] [Google Scholar]
- Watts AG, Sanchez-Watts G, Kelly AB. Distinct patterns of neuropeptide gene expression in the lateral hypothalamic area and arcuate nucleus are associated with dehydration-induced anorexia. J Neurosci. 1999;19(14):6111–6121. doi: 10.1523/JNEUROSCI.19-14-06111.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann G, Hrabovszky E, Lechan RM. Distinct glutamatergic and GABAergic subsets of hypothalamic pro-opiomelanocortin neurons revealed by in situ hybridization in male rats and mice. J Comp Neurol. 2013;521(14):3287–3302. doi: 10.1002/cne.23350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Boyle MP, Palmiter RD. Loss of GABAergic signaling by AgRP neurons to the parabrachial nucleus leads to starvation. Cell. 2009;137(7):1225–1234. doi: 10.1016/j.cell.2009.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Clark MS, Palmiter RD. Deciphering a neuronal circuit that mediates appetite. Nature. 2012;483(7391):594–597. doi: 10.1038/nature10899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasoshima Y, Scott TR, Yamamoto T. Memory-dependent c-Fos expression in the nucleus accumbens and extended amygdala following the expression of a conditioned taste aversive in the rat. Neuroscience. 2006;141(1):35–45. doi: 10.1016/j.neuroscience.2006.03.019. [DOI] [PubMed] [Google Scholar]
- Zhu X, Krasnow SM, Roth-Carter QR, Levasseur PR, Braun TP, Grossberg AJ, Marks DL. Hypothalamic signaling in anorexia induced by indispensable amino acid deficiency. Am J Physiol Endocrinol Metab. 2012;303(12):E1446–1458. doi: 10.1152/ajpendo.00427.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]