Abstract
Bacterial genome sizes have previously been shown to exhibit a bimodal distribution. This phenomenon has prompted discussion regarding the evolutionary forces driving genome size in bacteria and its ecological significance. We investigated the level of inherent redundancy in the public database and the effect it has on the shape of the apparent bimodal distribution. Our study reveals that there is a significant bias in the genome sequencing efforts towards a certain group of species, and that correcting the bias using species nomenclature and clustering of the 16S rRNA gene, results in a unimodal rather than the previously published bimodal distribution. The true genome size distribution and its wider ecological implications will soon emerge as we are currently witnessing rapid growth in the number of sequenced genomes from diverse environmental niches across a range of habitats at an unprecedented rate.
Significant progress has been made in understanding interactions between ecology and genome evolution in prokaryotes. A number of recent studies have focussed on the evolution of bacterial genome sizes (Kempes et al., 2016), indicating that the interaction between an organism and its ecological niche, for example, resource availability and environmental stability, selects the genome size of the species (Konstantinidis and Tiedje, 2004; Bentkowski et al., 2015). The exact mechanisms driving the genome sizes are still not fully resolved (Sabath et al., 2013, Kempes et al., 2016). It has, however, been speculated that species living in invariant niches tend to have small genomes, as stability acts to reduce genome size due the metabolic burden of replicating DNA with no adaptive value (Giovannoni et al., 2005, 2014) such as in obligatory and intracellular pathogens or mutualists (Moran, 2003; Klasson and Andersson, 2004; Moya et al., 2009). Due to their metabolic diversity, species with large genomes are potentially able to tackle a wider range of environmental conditions (Schneiker et al., 2007) and tend to be more ecologically successful where resources are scarce but diverse, and where there is little penalty for slow growth (Konstantinidis and Tiedje, 2004). The effect by which these two opposing evolutionary forces exert on the overall distribution of genome sizes was first observed by Koonin and Wolf in 2008, where it was reported that bacterial genome sizes show a bimodal distribution (Koonin and Wolf, 2008). The authors speculated that the observation of two distinct groups of bacteria, those with 'small' and those with 'large' genomes, directly reflects the balance between the opposing trends of genome expansion through gene duplication, horizontal gene transfer and replication, and genome contraction caused by genome streamlining and degradation (Koonin and Wolf, 2008). The observed bimodality in the database was the first empirical evidence to show the two forces at work in bacterial genomes, and the bimodalilty in the distribution has since attracted numerous citations in both peer-reviewed articles (Lane, 2011; Mock and Kirkham, 2012; Giovannoni et al., 2014; Morán et al., 2015) and textbooks (Bergman, 2011; Koonin, 2011; Kirchman, 2012; Saitou, 2014; Seshasayee, 2015).
A substantial proportion of complete bacterial genomes in the public domain belong to human pathogens and very closely related genomes representing variations within the species (Tatusova et al., 2014). As first reported by Graur, 2014, it has been suggested that this fact might introduce a bias to the bimodal distribution seen in the previous analyses. No formal treatment, however, has been carried out in the peer-reviewed literature to examine the extent of database bias and how it may affect bacterial genome size bimodality. The distribution of the bacterial genome size has broad and far-reaching implications in our understanding of prokaryotes and this in turn necessitates reassessment of the distribution and the extent to which the bias distorts the apparent bimodality. Here we present our finding that the bias in the database has profound influence in shaping the overall distribution of bacterial genome size.
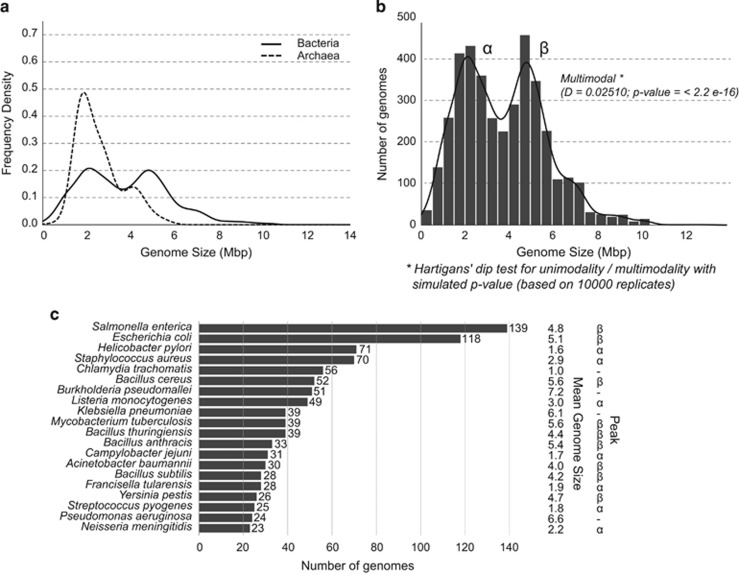
Having obtained a total of 3923 complete bacterial genomes from Ensembl Bacteria database, which is the most comprehensive source of complete bacterial genomes (see Supplementary Information for detailed methods), the distribution of genome sizes was first evaluated and compared against the distribution from Koonin and Wolf, 2008. Despite that almost six times more genomes have been archived since 2007, the current dataset exhibited a remarkably similar bimodal distribution with its distinctive bimodal peaks around 2 and 5 Mbp (Figure 1a). Hartigans' dip test (Hartigan and Hartigan, 1985) was used to confirm that it features significant bimodality with a P-value of 2.2e−16 (Figure 1b), where P-values <0.05 indicate significant bimodality (or multimodality) and P-values >0.10 indicate unimodality (Freeman and Dale, 2013).
Figure 1.
(a) Distribution of genome sizes in bacteria and archaea: the curves were generated by Gaussian–kernel smoothing of the individual data points. The figure has a very similar pattern to the figure generated by Koonin and Wolf, 2008. The distribution of archaea was included for comparison only. (b) Distribution of genome sizes in bacteria on a different scale: the distribution shows clear-cut bimodality. Hartigans' dip test for unimodality/multimodality with simulated P-value with 10 000 Monte Carlo replicates: D=0.02510, P<2.2e−16, where values <0.05 indicate significant bi- or multimodality and values >0.10 indicate unimodality (Freeman and Dale, 2013). (c) Number of genomes from the top 20 most redundant species in the database with mean genome size and peak in which they belong. (Peak α: 1.5–3 Mbp, Peak β: 4–5.5 Mbp). The top 20 most redundant species belonged to 971 genomes representing almost 25% of the entire dataset. Most of them (18 species in total) formed part of the peaks (α and β), including the top 4 species, namely Salmonella enterica, Escherichia coli, Helicobacter pylori and Staphylococcus aureus.
The level of redundancy in the dataset was next assessed by counting the number of genomes, which shared the same species classification. The entire dataset of 3923 genomes represented 1706 groups of species with a unique species classification based on names. As shown in Figure 1c, there was a significant amount of bias in the genome sequencing efforts towards a certain group of species, most of which belonged to well-characterised human pathogens. In fact, almost 25% of the entire genome dataset was composed of just 20 species (971 genomes). We also found that most of these highly redundant species belonged to the peaks in the bimodal distribution. Notably, the two most redundant species, namely Salmonella enterica, Escherichia coli belonged to peak β and Helicobacter pylori, Staphylococcus aureus belonged to peak α.
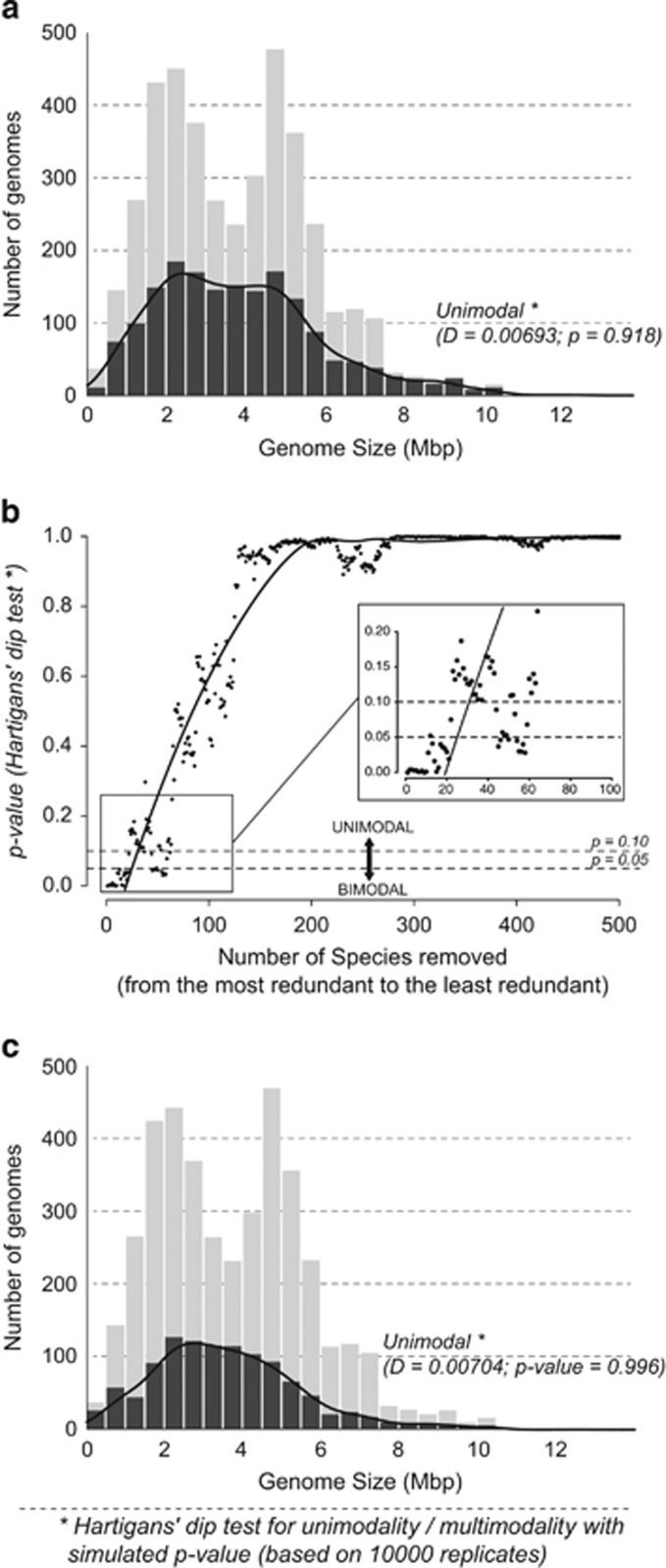
Having observed the bias in the dataset, we assessed how much impact this has on the modality of the distribution by removing the redundant genomes from the dataset (Figure 2a). The resulting distribution exhibited much less pronounced peaks, and as confirmed by Hartigans' dip test, the distribution was non-significant for bimodality (P=0.91). The influence these redundant species has on the distribution became more apparent (Figure 2b) as we evaluated the modality of the distribution by progressively removing species from the dataset (from the most redundant to the least). There is a sharp incline towards unimodality as redundant species were gradually excluded (Figure 2b). In fact, the distribution became more or less unimodal after the top 60 redundant species were removed from the dataset of 1706 species.
Figure 2.
(a) Distribution of genome sizes in bacteria after removing redundant genomes. The grey area indicates 2217 redundant genomes (out of 3923 genomes in total). The distribution indicates unimodality (Hartigans' dip test: D=0.0069289, P=0.908). (b) Effect of removing 500 most redundant species from the database on the modality of distribution measured by Hartigans' dip test. After removing around 60 most redundant species, the distribution becomes mostly unimodal. (c) Distribution of genome sizes in bacteria after removing redundant and very closely related genomes using 16S rRNA (2841 genomes). The distribution shows a clear-cut unimodal distribution (Hartigans' dip test: D=0.0070418, P=0.996).
One of the issues we faced with our approach was that a large number of genomes in the dataset had disorganized and inconsistent taxonomic classification. For instance, there were genomes using different naming convention such as ones with square brackets or strain identifier attached to their species name (for example, ‘[Clostridium]-cellulolyticum', ‘Francisella sp. TX077308'). This meant that removing redundant genomes using a text-based approach was only able to partially extirpate the bias. Also, using this approach could not resolve the bias arising from very closely related genomes representing variations within the species but with different species classification. A more suitable approach was to use a biomarker gene directly extracted from each genome to cluster the dataset into units of redundant or very closely related species. For this purpose, we chose 16S rRNA gene as it had been demonstrated that 16S rRNA sequence on an individual strain with another exhibiting a similarity score of 97% or above represents the same species (Stackebrandt and Goebel, 1994; Tindall et al., 2010). The clustering resulted in 1081 groups of species or very closely related species, and as Figure 2c shows, the resulting distribution from the dataset indicated a unimodal distribution (P=0.99, Hartigans' dip test).
Our results revealed that there is a significant amount of inherent redundancy in the public database with a strong bias towards a certain group of species, and they have strong influence in driving bacterial genome size distribution into bimodal. While it is plausible that bacterial genome size is heavily influenced by the specialist or generalist lifestyle, it is not immediately apparent whether or not this should lead to any particular distribution. To a great degree, it is still too early to make any conclusions as to whether the true distribution exhibits certain modality as the majority of genomes sequenced so far have only focussed on culturable species, in particular human pathogens and closely related species. Some interesting observations with a potential link to the nature of distribution have been emerging in recent years. For example, (i) the bimodality in flow cytometric analysis of bacterial DNA content has been implicated with the bimodal genome size distribution (Schattenhofer et al., 2011; Morán et al., 2015); (ii) there may be other factors such as physical cell space constraints having a role in genome size selection (Kempes et al., 2016) and (iii) perhaps most intriguingly, numerous studies from metagenomics are indicating that species with small genomes are more common than previously thought (Giovannoni et al., 2014; Morán et al., 2015). With the rise of single-cell genomics and improved bioinformatic assembly methods coupled with the continual reduction in genome sequencing, we are currently witnessing rapid growth in the number of sequenced genomes. Consequently, the true nature of the distribution together with its ecological implications will become more apparent as we gather more sequenced genomes from diverse niches across a wide range of habitats.
Acknowledgments
HSG acknowledges the support of NERC NBAF-W (NEC04916).
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Bentkowski P, Van Oosterhout C, Mock T. (2015). A model of genome size evolution for prokaryotes in stable and fluctuating environments. Genome Biol Evol 7: 2344–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman NH. (2011) Bacillus anthracis and Anthrax. John Wiley and Sons. [Google Scholar]
- Freeman JB, Dale R. (2013). Assessing bimodality to detect the presence of a dual cognitive process. Behav Res Methods 45: 83–97. [DOI] [PubMed] [Google Scholar]
- Giovannoni SJ, Cameron Thrash J, Temperton B. (2014). Implications of streamlining theory for microbial ecology. ISME J 8: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D et al. (2005). Genome streamlining in a cosmopolitan oceanic bacterium. Science 309: 1242–1245. [DOI] [PubMed] [Google Scholar]
- Graur D. (2014). ‘Take Another Good Look at the Data': The Bimodal Distribution that Wasn't, Retrieved from http://judgestarling.tumblr.com/post/84095742522/take-another-good-look-at-the-data-the-bimodal.
- Hartigan JA, Hartigan PM. (1985). The dip test of unimodality. Ann Stat 13: 70–84. [Google Scholar]
- Kempes CP, Wang L, Amend JP, Doyle J, Hoehler T. (2016). Evolutionary tradeoffs in cellular composition across diverse bacteria. ISME J 10: 2145–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchman DL. (2012) Processes in Microbial Ecology. Oxford University Press: Oxford, UK. [Google Scholar]
- Klasson L, Andersson SGE. (2004). Evolution of minimal-gene-sets in host-dependent bacteria. Trends Microbiol 12: 37–43. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. (2004). Trends between gene content and genome size in prokaryotic species with larger genomes. Proc Natl Acad Sci USA 101: 3160–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI. (2008). Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res 36: 6688–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. (2011) The Logic of Chance: The Nature and Origin of Biological Evolution. FT Press: New Jersey, USA. [Google Scholar]
- Lane N. (2011). Energetics and genetics across the prokaryote-eukaryote divide. Biol Direct 6: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mock T, Kirkham A. (2012). What can we learn from genomics approaches in marine ecology? from sequences to eco-systems biology!. Mar Ecol 33: 131–148. [Google Scholar]
- Morán XAG, Alonso-Sáez L, Nogueira E, Ducklow HW, González N, López-Urrutia Á et al. (2015). More, smaller bacteria in response to ocean's warming? Proc R Soc B 282: 20150371 Available at http://dx.doi.org/10.1098/rspb.2015.0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA. (2003). Tracing the evolution of gene loss in obligate bacterial symbionts. Curr Opin Microbiol 6: 512–518. [DOI] [PubMed] [Google Scholar]
- Moya A, Gil R, Latorre A, Pereto J, Pilar Garcillan-Barcia M, De La Cruz F. (2009). Toward minimal bacterial cells: evolution vs design. FEMS Microbiol Rev 33: 225–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabath N, Ferrada E, Barve A, Wagner A. (2013). Growth temperature and genome size in bacteria are negatively correlated, suggesting genomic streamlining during thermal adaptation. Genome Biol Evol 5: 966–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N. (2014) Introduction to Evolutionary Genomics. Springer. [Google Scholar]
- Schattenhofer M, Wulf J, Kostadinov I, Glöckner FO, Zubkov MV, Fuchs BM. (2011). Phylogenetic characterisation of picoplanktonic populations with high and low nucleic acid content in the North Atlantic Ocean. Syst Appl Microbiol 34: 470–475. [DOI] [PubMed] [Google Scholar]
- Schneiker S, Perlova O, Kaiser O, Gerth K, Alici A, Altmeyer MO et al. (2007). Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat Biotechnol 25: 1281–1289. [DOI] [PubMed] [Google Scholar]
- Seshasayee ASN. (2015) Bacterial Genomics: Genome Organization and Gene Expression Tools. Cambridge University Press. [Google Scholar]
- Stackebrandt E, Goebel BM. (1994). Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44: 846–849. [Google Scholar]
- Tatusova T, Ciufo S, Federhen S, Fedorov B, McVeigh R, O'Neill K et al. (2014). Update on RefSeq microbial genomes resources. Nucleic Acids Res 43: D599–D605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tindall BJ, Rosselló-Móra R, Busse HJ, Ludwig W, Kämpfer P. (2010). Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol 60: 249–266. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.