Abstract
Several cosmopolitan marine picoeukaryotic phytoplankton are B1 auxotrophs requiring exogenous vitamin B1 or precursor to survive. From genomic evidence, representatives of picoeukaryotic phytoplankton (Ostreococcus and Micromonas spp.) were predicted to use known thiazole and pyrimidine B1 precursors to meet their B1 demands, however, recent culture-based experiments could not confirm this assumption. We hypothesized these phytoplankton strains could grow on precursors alone, but required a thiazole-related precursor other the well-known and extensively tested 4-methyl-5-thiazoleethanol. This hypothesis was tested using bioassays and co-cultures of picoeukaryotic phytoplankton and bacteria. We found that specific B1-synthesizing proteobacteria and phytoplankton are sources of a yet-to-be chemically identified thiazole-related precursor(s) that, along with pyrimidine B1 precursor 4-amino-5-hydroxymethyl-2-methylpyrimidine, can support growth of Ostreococcus spp. (also Micromonas spp.) without B1. We additionally found that the B1-synthesizing plankton do not require contact with picoeukaryotic phytoplankton cells to produce thiazole-related precursor(s). Experiments with wild-type and genetically engineered Ostreococcus lines revealed that the thiazole kinase, ThiM, is required for growth on precursors, and that thiazole-related precursor(s) accumulate to appreciable levels in the euphotic ocean. Overall, our results point to thiazole-related B1 precursors as important micronutrients promoting the survival of abundant phytoplankton influencing surface ocean production and biogeochemical cycling.
Introduction
Vitamin B1 (thiamine, called B1 herein) is an essential micronutrient for all cells, specifically in the form of thiamine diphosphate (TDP; also called thiamine pyrophosphate), a cofactor for enzymes with critical functions in intracellular carbon and amino-acid metabolism, for example, oxidoreductases, dehydrogenases and decarboxylases (Frank et al., 2007; Jurgenson et al., 2009). B1 auxotrophic cells are unable to carry out de novo synthesis of B1 (specifically TDP), and inherently depend on exogenous supply of the vitamin or precursor compounds to survive.
B1 auxotrophy occurs across diverse marine bacterioplankton and phytoplankton lineages (Provasoli and Carlucci, 1974; Croft et al., 2006; Sañudo-Wilhelmy et al., 2014), and recently genome and culture-based studies revealed that representatives of environmentally abundant SAR11 and SAR86 bacterioplankton clades (Dupont et al., 2011; Carini et al., 2014), bloom-forming phytoplankton and cosmopolitan picoeukaryotic phytoplankton (Tang et al., 2010; Bertrand and Allen, 2012; McRose et al., 2014; Paerl et al., 2015) are B1 auxotrophs. These findings directly link availability of B1 and/or precursors to elemental cycling (for example, C, N, P, S), water quality and productivity in the upper ocean.
The de novo B1 biosynthesis pathway is composed of thiazole and pyrimidine branches that converge to yield thiamine monophosphate (TMP) and ultimately TDP (Jurgenson et al., 2009). In B1 auxotrophic organisms, only portions of the complete B1 biosynthesis pathway are present, preventing de novo TDP synthesis; however, remnant partial pathways in B1 auxotrophs can function in salvaging TDP from exogenous precursor(s)—enabling cells to grow on precursor(s) alone. For example, in SAR11 bacterioplankton clade affiliate Pelagibacter ubique HTCC1062 and bloom-forming phytoplankton Emiliania huxleyi, ThiC is missing (resulting in B1 auxotrophy) an enzyme that generates precursor exogenous 4-amino-5-hydroxymethyl-2-methylpyrimidine (HMP). However, when exogenous HMP is available these organisms can meet their B1 demands by feeding HMP into their partial pyrimidine B1 biosynthesis pathway to make TDP (Carini et al., 2014; McRose et al., 2014). Early research demonstrated that marine B1 auxotrophic plankton can use pyrimidine and/or thiazole B1 moieties to meet their B1 demands (Lwoff, 1947; Droop, 1957; Burkholder, 1963). Recent genome surveys highlight that diverse marine bacterioplankton and phytoplankton lack parts of the pyrimidine and/or thiazole branches of the B1 biosynthesis pathway (McRose et al., 2014; Sañudo-Wilhelmy et al., 2014)—suggesting they can use B1 precursors to meet B1 demands.
Ostreococcus and Micromonas spp. are B1 auxotrophic picoeukaryotic phytoplankton with incomplete pyrimidine and thiazole pathways. They are representatives of keystone picoeukaryotic phytoplankton in the upper ocean that are widespread (Countway and Caron, 2006; Demir-Hilton et al., 2011; Worden et al., 2012), exhibit high transcriptional activity (Ottesen et al., 2013; Aylward et al., 2015) and can contribute >70% of total carbon fixation (Worden et al., 2004). Based on genome sequences, several Ostreococcus and Micromonas spp. were predicted to meet B1 demands by using exogenous pyrimidine precursor HMP and 4-methyl-5-thiazoleethanol (HET; McRose et al., 2014; Paerl et al., 2015), the latter being a well-studied thiazole B1 precursor useful for B1 auxotrophic phytoplankton and non-marine model organisms (Droop, 1958; Burkholder, 1963; Jurgenson et al., 2009). However, in culture, they could not meet their B1 demands using HMP and HET (McRose et al., 2014; Paerl et al., 2015), underscoring the need for additional physiological testing of genomic inferences regarding B1 metabolism in these environmentally important phytoplankton.
We hypothesized these picoeukaryotic phytoplankton could grow on precursors alone, using HMP and a thiazole-related precursor different from the well-known thiazole precursor HET. We tested this hypothesis in co-culture and bioassay experiments where first multiple B1-synthesizing plankton were identified as sources of B1 and/or precursors, then examining if the plankton were sources of a thiazole-related B1 precursor useful for B1 auxotrophic picoeukaryotic phytoplankton (primarily Ostreococcus spp.). Using genetic engineering of Ostreococcus tauri RCC745, we investigated if the known thiazole kinase, ThiM, typically thought to phosphorylate HET, has a role in its use of B1 precursors. Finally, using bioassays with Ostreococcus wild-type and mutant lines, we tested if thiazole-related precursor(s) available to picoeukaryotic phytoplankton occur at appreciable levels in the mixed layer of coastal and pelagic ocean waters.
Materials and methods
Experimental reagents
High-performance liquid chromatography grade (>98% pure) compounds were used in experiments (Spectrum Inc. (Gardena, CA, USA) and Alfa Aesar (Ward Hill, MA, USA)). Purified (>90% based on nuclear magnetic resonance) 4-amino-5-aminomethyl-2-methylprimidine (HMP) was purchased from VitasMLab Ltd (Apeldoorn, The Netherlands) or Enamine Ltd (Kiev, Ukraine), and HET was manufactured by Alfa Aesar. Aliquots of thiamine, HMP and HET were freshly prepared in autoclaved Milli Q water, (0.22 μm pore-sized) filter-sterilized, and kept in the dark on ice before use in experiments.
Culture growth conditions and monitoring
All cultures except for those containing O. tauri RCC745 and co-cultures were grown on charcoal-treated seawater with F/2 medium components (Guillard, 1975) plus Se and citrate as previously described, with B1 omitted to induce limitation (Paerl et al., 2015). Light and temperature conditions were as described in Paerl et al. (2015). Cultures containing O. tauri RCC745 were grown under 25 μE m−2 s−1 white light and at 20 °C on artificial seawater supplemented with trace metals and vitamins as previously described (Lozano et al., 2014). Synechococcus sp. WH8102 was grown under lower light intensity (~40 μE m−2 s−1) at ~20 °C. All cultures were handled using aseptic technique in a laminar flow hood. B1 was omitted from media to induce limitation. Axenicity of phytoplankton cultures was confirmed by 4-6-diamidino-2-phenylindole staining and epifluorescence microscopy (Porter and Feig, 1980) as well as culture inoculations into marine broth 2216 (ZoBell, 1941). O. tauri RCC745 cultures were non-axenic and determined to contain only a single strain related to Marinobacter lipolyticus (97.55% 16 S ribosomal RNA (rRNA) gene sequence identity) based on plating of the culture upon Marine Agar 2216 (Difco, Detroit, MI, USA) and subsequent PCR amplification (27 F, 1492 R primers; Lane, 1991) and sequencing of (10) colonies. Colony frequency per volume plated was comparable to bacterial abundances in the O. tauri RCC745 cultures (see below). The 16 S rRNA gene sequence for the single strain (Marinobacter sp. OTB1) is available via GenBank accession ID: KT828547.
Phytoplankton growth was monitored using in vivo (chlorophyll a) fluorescence (Paerl et al., 2015) or phycoerythrin fluorescence using an AU-10 fluorometer (Turner Designs, Sunnyvale, CA, USA). Synechococcus sp. WH8102 and Phaeodactylum tricornutum CCMP632 cell abundances were quantified from formaldehyde fixed (2% final concentration) subsamples using an Influx flow cytometer (Becton Dickinson, San Jose, CA, USA) or epifluorescence microscopy (using a Nikon C1 microscope equipped with a UV light source). Abundances of O. tauri RCC745 and co-occurring Marinobacter sp. OTB1 were determined on an Accuri C6 flow cytometer (Becton Dickinson) using SYBR Green I staining (Corellou et al., 2005). Based on flow cytometry analysis, 1–4% of total cells in O. tauri RCC745 cultures were bacterial cells. Cultures treated with an antibiotic cocktail (penicillin G 50 μg ml−1, streptomycin 200 μg ml−1) did not contain detectable bacteria using flow cytometry, or plating (100 μl of culture).
Establishment of bacteria, O. lucimarinus CCE9901 co-cultures
Multiple heterotrophic bacterioplankton isolates were used in co-culture experiments with O. lucimarinus CCE9901 (and related phytoplankton). Scripps bacterial isolates (SBIs) were isolated by plating surface seawater off the Scripps Pier, La Jolla, CA, USA on (1%) agar enriched marine broth 2216 (ZoBell, 1941). Taxonomy of SBIs was determined by 16S rRNA gene sequencing as described above. Isolation details for all other bacterioplankton is described elsewhere (Bidle and Azam, 2001; Long and Azam, 2001; Miller and Belas, 2004; Supplementary Table S1).
Before the start of co-culture experiments, bacterial strains were grown to stationary phase on marine broth 2216, centrifuged (6 min at 6000 × g) and washed three times, and ultimately resuspended in B1-deplete F/2 medium (described above) plus NH4+ (50 μM). Washed-resuspended cells were added to B1-limited O. lucimarinus CCE9901 cultures at a 1:1000 dilution. Co-cultures were maintained on F/2+NH4+ medium and labile organic carbon (pyruvate, maltose and glucose, 100 μM each) was added to a subset. Co-cultures were transferred twice (1:10 dilution of culture to fresh medium) following increases in in vivo fluorescence, before use in experiments.
Collection of filtrate and seawater for bioassays
Synechococcus sp. WH8102, P. tricornutum CCMP632 and heterotrophic bacterioplankton cultures were maintained in exponential growth on F/2 –B1 medium, then transferred to duplicate flasks for filtrate collection at different growth stages (bacterioplankton cultures were provided with labile organic carbon). Specifically, Synechococcus WH8102 cultures were maintained in exponential growth for more than five transfers (1:10 dilution) in the B1-deplete F/2 medium, then used in experiments. P. tricornutum and proteobacterial cultures (following centrifugation and washing) were first transferred two or more times (1:10 000 dilution) before use. Filtrates were collected by gently filtering (<5 in. Hg vacuum) cultures onto 0.22 μm pore-sized filters, and slowly (drop wise, collecting any few remaining cells) filtering the collected liquid through a 0.22 μm pore-sized polyethersulfone syringe filter (Pall, Port Washington, NY, USA) before immediate storage at −20 °C.
Surface seawater collected off the Scripps Pier was incubated and sub-sampled over diel changes in light. A time zero seawater sample was processed and the remaining collected seawater was incubated at in-situ temperature (21 °C) under 12 h of ~70 μE m−2 s−1 white light (T5 28 W fluorescent bulbs) and 12 h of darkness. Near-surface seawater (3 m) from the north Mediterranean (Service d'Observation du Laboratoire Arago (SOLA) site, Banyuls-sur-Mer, France) was collected and filtered for bioassays with O. tauri RCC745 bioassays. Filtered seawater samples were collected (filtered) as was done for obtaining culture filtrates.
Archived seawater samples from oligotrophic open-ocean sites were collected using trace-metal clean sampling techniques, filtered through sterile 0.22 μm pore-size filters and immediately stored and kept at −20 °C (personal communication R Bundy, Scripps Institution of Oceanography). Seawater from 10 m at Line P station 27 was collected on 10 June 2008. N. Atlantic seawater, near the Bermuda Atlantic Time-series Study (BATS) station (30° 3.47' N, 58° 6.20' W), was collected from 2 m on 22 November 2011.
O. lucimarinus CCE9901 and O. tauri RCC745 bioassays
B1 and utilizable related micronutrients (for example, precursors) were detected in seawater samples by bioassays with B1-limited O. lucimarinus CCE9901 or O. tauri RCC745 cells. O. lucimarinus CCE9901 in vivo fluorescence changed linearly with cell abundances under culture conditions until high cell densities (Supplementary Figure S1); hence, fluorescence was used to track biomass of the bioassay organism. The upper quantification limit of the bioassays was conservatively set at ~170 AU, whereas the lower quantification limit was ~1.45 AU (fluorescence of the medium alone; Supplementary Figure S1). Flow cytometry was used to determine O. tauri RCC745 cell yields in bioassays.
To start bioassays, B1-limited cells were transferred (1:10 dilution for O. lucimarinus CCE9901; 1:1000 for O. tauri RCC745) to tubes containing seawater or filtrate sample, plus medium nutrients except B1. For O. lucimarinus CCE9901 bioassays F/2 medium nutrients were added—for O. tauri RCC745 bioassays only phosphate (10 μM), nitrate (1 mM) and antibiotics (described above) were added. Culture filtrates were assayed in the same manner as seawater samples, but were diluted with B1-deplete sterile seawater. Dilution of seawater or filtrate samples was considered in final calculations of yields in bioassays. Triplicate positive and negative controls were run in parallel with sample tubes. For positive controls, 1 nM B1 was added to tubes with or without seawater sample or filtrate; maximum yields in positive controls were comparable to those of the bioassay organism grown on F/2 –B1 medium plus 1 nM B1. For negative controls, cell culture was transferred to B1-deplete culture medium, without seawater or filtrate; no significant growth was observed in negative controls.
ThiM phylogenetic tree construction
A ThiM phylogenetic tree was constructed using MUSCLE (Edgar, 2004) aligned ThiM sequences obtained from Uniprot or JGI genome portals (ID's provided). A 111 amino-acid section of the ThiM sequence was used to construct the tree in ARB (Ludwig et al., 2004), and bootstrap values were obtained using 1000 replicate trees.
Generation of O. tauri RCC745 ΔThiM deletion mutant
Ostreococcus and Micromonas spp. cannot use thiazole B1 precursor HET, yet possess ThiM (McRose et al., 2014; Paerl et al., 2015), a kinase known to phosphorylate HET to 4-methyl-5-thiazoleethanol phosphate (HET-P) in model organisms (Moulin et al., 2013; Yazdani et al., 2013). To test involvement of ThiM in use of thiazole-related precursor(s), we genetically engineered a (ΔThiM) deletion mutant in the strain O. tauri RCC745, originally isolated from coastal Mediterranean seawater off of southern France (Derelle et al., 2006). The thiM gene was deleted by homologous recombination according to Lozano et al. (2014). The transgene used in the methodology consisted of the pH4:KanMx:Tef selection gene flanked by 1 kb of sequences complementary to thiM-bordering sequence (lacking any thiM sequence) in wild-type O. tauri RCC745 so that upon recombination of the transgene at the ThiM locus, homologous recombinants would lack the ThiM coding sequence (Supplementary Figure S2). The primers used to amplify the 5′ and 3′ thiM flanking regions were Thim 5′F AAGGCCATGAGCCGCTCCGCCAAGGCGGGC and Thim 5′R ACATACCCCGTCGCCGACGATCGCG, and Thim 3′F ATTCGCATTACGATTTTTGTAGGG and Thim 3′R ACTGCTGATTGACGATGTCCGCCAGACCAGTC, respectively. The transgene was introduced into Ostreococcus cells by electroporation and transformants were selected on G148 plates as described previously (Corellou et al., 2009). Homologous recombinants were identified by PCR amplification using the primers described in Supplementary Figure S2. The ΔThiM mutant line is available per request to F-Y Bouget.
Results
Diverse bacterioplankton sustain O. lucimarinus CCE9901 in B1-deplete co-cultures
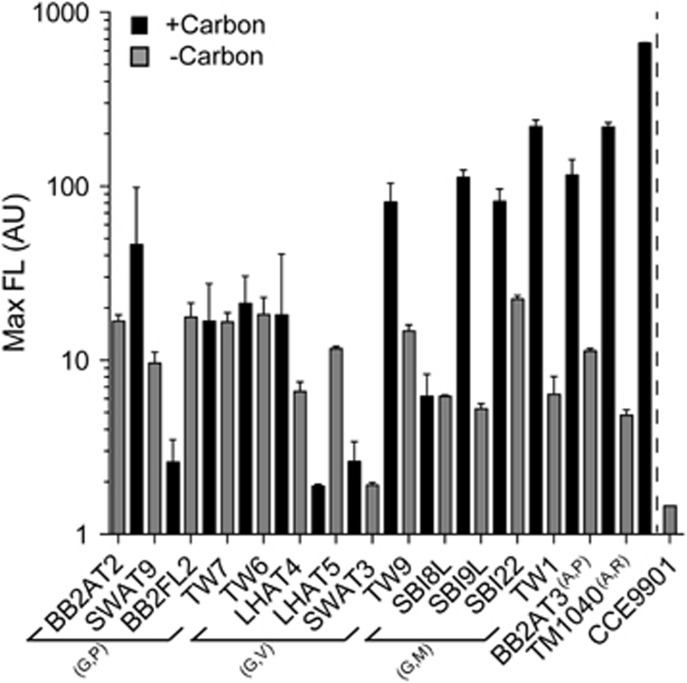
Before testing for provision of thiazole-related precursor(s), we first examined if diverse proteobacterial isolates (largely obtained from waters off La Jolla, CA, USA where O. lucimarinus CCE9901 was also isolated) would supply micronutrients that maintained growth of initially axenic cultures of O. lucimarinus CCE9901 in B1-deplete medium. After a single 1:10 transfer to B1-deplete medium, axenic O. lucimarinus CCE9901 cultures rapidly became growth-limited and ultimately cells were diluted following another transfer; in contrast, yields of O. lucimarinus CCE9901 co-cultured with bacterioplankton strains on the same medium were maintained above the limit of detection in all pairings (Figure 1). Also in a majority of co-cultures (9 of 15), the addition of labile organic carbon (glucose, maltose, pyruvate) increased O. lucimarinus CCE9901 maximum yields, indicating carbon availability limited supply of B1 and/or related micronutrients by the bacteria (via increased cell yields or per cell production) in these co-cultures (Figure 1). Growth of bacterioplankton was not directly determined in co-cultures, but inferred based on maintained yields of B1-limited O. lucimarinus CCE9901 following multiple transfers to B1-deplete medium (five 1:10 dilutions; see Materials and methods section).
Figure 1.
Multiple bacterioplankton provide compounds that sustain the B1 auxotrophic phytoplankton O. lucimarinus CCE9901 in co-cultures maintained on B1-deplete medium. The average maximum chlorophyll a fluorescence (Max FL; indicating phytoplankton biomass) is plotted for co-cultures containing a single proteobacterial strain (listed in Supplementary Table S1) and were either not amended (gray columns) or amended (adjacent black columns) with labile organic carbon (noted as carbon in the legend). The phytoplankton alone does not grow on the B1-deplete medium—as a result fluorescence of B1-limited axenic cultures are quickly diluted down to levels of the medium blank (~1.45 AU) after a single 1:10 transfer to the B1-deplete medium (gray bar separated by a dashed line). The Max FL of B1-limited co-cultures was maintained above the blank in all cases, indicating the bacteria sustained growth of CCE9901 in the B1-deplete medium. Yields of the phytoplankton (Max FL) often increased because of carbon addition. The carbon added to co-cultures with SBI8L, SBI9L, SBI22, TW1 and TW7 was citrate (50 μM); for all others a combination of glucose, maltose and pyruvate (50 μM each) was added. Co-cultures were maintained over five transfers (1:10 dilution each time) and the plotted data are averages and respective standard deviations (error bars) for each of the final three transfers. Bacterioplankton used in experiments are listed along the x axis and arranged (some bracketed) according to their phylum and genus affiliation. Letters is parentheses next to brackets, or as superscripts next to strain names, indicate their respective affiliated phyla (G, Gammaproteobacteria; A, Alphaproteobacteria) and genus (P, Pseudoalteromonas; V, Vibrio; M, Marinobacter; (A,)P, Phaeobacter; R, Rugeria).
O. lucimarinus CCE9901 uses a thiazole-related precursor (and HMP) to grow without exogenous B1
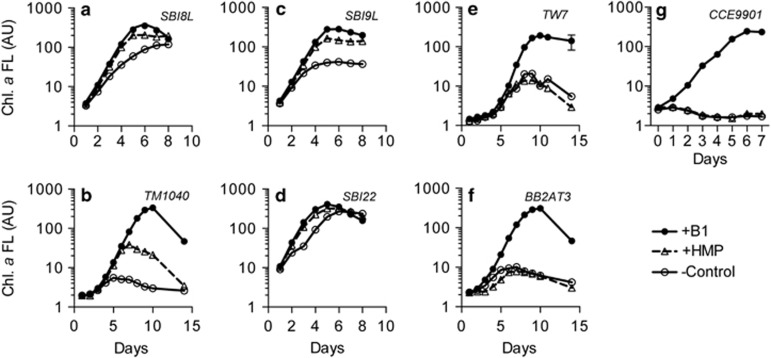
Grown in axenic cultures, O. lucimarinus CCE9901 cannot use B1 precursors HMP or HET to meet its B1 demands despite possessing genes encoding for parts of the B1 biosynthesis pathway (Paerl et al., 2015). We tested if a subset of the B1-synthesizing bacterioplankton strains that provided useful compounds for O. lucimarinus CCE9901 to meet its B1 demands in our initial experiment (Figure 1) could provide specifically a thiazole-related precursor that could be used in conjunction with known pyrimidine B1 precursor HMP. Strikingly, in four of six co-cultures of O. lucimarinus CCE9901 and bacterioplankton (Figure 2a–d), growth of B1-limited O. lucimarinus CCE9901 was stimulated after additions of HMP (1 nM), contrasting with axenic O. lucimarinus CCE9901 (Figure 2g). Another Mamiellales-affiliated picoeukaryote, Micromonas pusilla CCMP487, which cannot meet its B1 demands by using HMP and/or HET in axenic cultures (Paerl et al., 2015), was also stimulated by HMP addition in B1-deplete co-cultures with the bacterium Marinomonas sp. SBI22 (Supplementary Figure S3), as observed in co-cultures of O. lucimarinus CCE9901 and Marinomonas sp. SBI22 (Figure 2d). Moreover, growth of B1-limited O. tauri RCC745 was stimulated following HMP addition in B1-limited co-cultures with Marinobacter sp. OTB1, but not when antibiotics were added to prevent Marinobacter sp. OTB1 growth (Supplementary Figure S4). The positive growth responses by Ostreococcus and Micromonas spp. to HMP addition to co-cultures strongly suggested the bacterioplankton provided a precursor (or precursors) related to the thiazole B1 moiety that is useful (in conjunction with HMP) by the picoeukaryotic phytoplankton to synthesize TDP.
Figure 2.
HMP addition (1 nM) stimulates growth of O. lucimarinus CCE9901 in co-cultures with individual B1-synthesizing bacterioplankton (a–f). In co-cultures with TW7 (e) or BB2AT3 (f), HMP addition did not stimulate growth of O. lucimarinus CCE9901 in co-cultures. Alone, O. lucimarinus CCE9901 does not grow on exogenous HMP (g). Presented chlorophyll a fluorescence data are averages and respective s.d. (error bars) for triplicate co-cultures. All co-cultures were maintained on F/2 –B1 +NH4+ medium plus citrate, except for TM1040 (b) and BB2AT3 (f) co-cultures. Taxonomic information for all strains is in Supplementary Table S1.
Production of a thiazole-like B1 precursor(s) by axenic bacterioplankton and phytoplankton cultures
Addition of HMP possibly stimulated B1 production by bacterioplankton in co-culture experiments (Figures 2a–d). Therefore, we tested if axenically grown B1-synthesizing plankton produced thiazole-related precursor(s). This was determined via O. lucimarinus CCE9901 bioassays where filtrates (medium from axenic cultures of B1-synthesizing strains, collected using low vacuum filtration to remove cells; see Materials and methods section) were provided to the bioassay organism. Filtrates from two previously tested B1-synthesizing bacterioplankton were tested (Figures 2d and e), but also two phytoplankton strains as some phytoplankton are known to provide compounds that support B1 auxotrophic plankton (Carlucci and Bowes, 1970; Carini et al., 2014). Further, the two tested phytoplankton, Synechococcus sp. WH8102 and P. tricornutum CCMP632 (Figures 3a and d), are also extensively studied model organisms (Brahamsha, 1996; Bowler et al., 2008) representing cosmopolitan B1-synthesizing phytoplankton (cyanobacteria and diatoms) in the euphotic ocean (Partensky et al., 1999; Malviya et al., 2016).
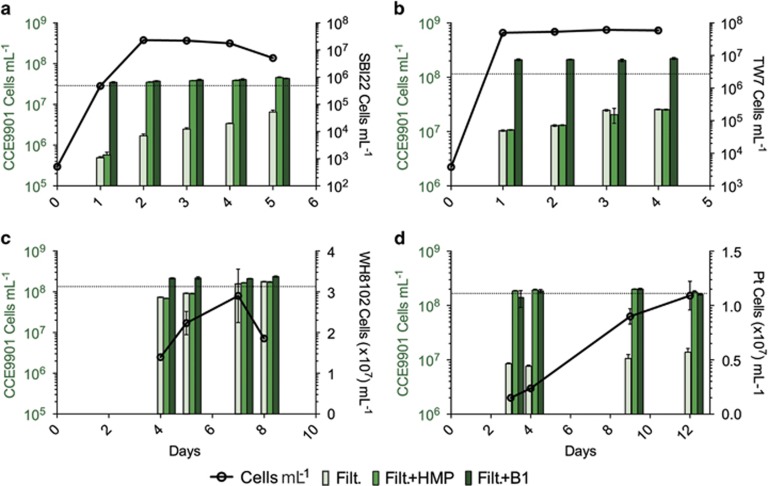
Figure 3.
Select axenic bacterioplankton and phytoplankton cultures provide relatively high amounts of a thiazole-related B1 precursor(s) useful for O. lucimarinus CCE9901. Filtrates for use in O. lucimarinus CCE9901 bioassays and samples for determining cell abundances were collected from bacterioplankton (a, Marinomonas sp. SBI22; b, Pseudoalteromonas sp. TW7) and phytoplankton (c, Synechococcus sp. WH8102; d, P. tricornutum CCMP632 (Pt)) cultures after multiple days of growth (days of growth denoted along the x axis). Black circles connected by lines represent cell abundances of B1-synthesizing cultures on filtrate sampling days (x axis value), and the respective scales for the abundance data are on the right y axes of panels in black text. Grouped columns represent maximum yields of the bioassay organism (O. lucimarinus CCE9901) grown on filtrate and modified filtrate (light green=filtrate alone (Filt.); green=filtrate plus HMP (Filt. +HMP); dark green=filtrate plus B1 (Filt. +B1) from respective filtrate collection days (on x axis; also corresponding with the cell abundance data). Scales on left y axes of panels in green text are associated with CCE9901 maximum yield data (columns). The higher yields of O. lucimarinus CCE9901 grown on Filt +HMP (green) versus Filt. (light green), specifically filtrates from Marinomonas sp. SBI22 and diatom P. tricornutum CCMP632, indicates a thiazole-related precursor(s) was more abundant relative to B1 and/or useful pyrimidine precursor (panels a and d). No detection of the thiazole-related precursor indicates the absence of secretion or equivalent availability of B1 or complementary pyrimidine precursor (for example, HMP; panels b and c). Fluorescence and cell abundance data for Synechococcus sp. WH8102, P. tricornutum CCMP632 cultures are in the Supplementary Information (Supplementary Figure S5). HMP or B1 were at added at a final concentration of 1 nM. Data points and columns represent the means and s.d. from duplicate cultures.
Thiazole-related precursor(s) were detected in select filtrates based on increased yields of CCE9901 in bioassays when grown on filtrate plus HMP versus filtrate alone (Figure 3). In regards to tested filtrates from heterotrophic bacterioplankton, Marinomonas sp. SBI22 filtrate amended with HMP supported notably higher yields in CCE9901 bioassays, while Pseudoalteromonas sp. TW7 filtrate did not, suggesting Marinomonas sp. SBI22 produces relatively high amounts of exogenous thiazole-related precursor(s) while grown in isolation (Figures 3a and b). These strain-specific bioassay results are in line with the prior observation that HMP stimulated growth of CCE9901 when grown in B1-limited co-culture with SBI22, but not with TW7 (Figures 2d and e). Detection of thiazole-related precursor(s) in phytoplankton filtrates also varied between tested strains. Thiazole-related precursor(s) were not detectable in Synechococcus sp. WH8102 filtrate as yields of our bioassay organism grown on the filtrate were comparable to that of our B1 replete treatment (B1+filtrate; Figure 3c), suggesting B1 and/or thiazole-related precursor(s) plus HMP were simultaneously available. In contrast, P. tricornutum CCMP632 filtrates contained high relative amounts of the thiazole-related precursor(s) (Figure 3d).
Previously annotated thiazole (HET) transporters (ThiU, ThiW; Rodionov et al., 2008) could also be involved in efflux of thiazole precursor from cells. Searches for these proteins (using BLASTP) against genomes of plankton clearly providing thiazole-related precursor(s) in our experiments (Figures 2a, b and d; Figures 3a and d), namely Marinomonas spp. SBI8L and SBI22, Rugeria sp. TM1040, and P. tricornutum CCMP632, failed to yield any significant hits—ruling out any contribution by these transporters to efflux of precursor (Supplementary Table S2). Similarly, no homolog of either transporter was found in Ostreococcus or Micromonas spp. genomes (using NCBI BLASTP-based searches).
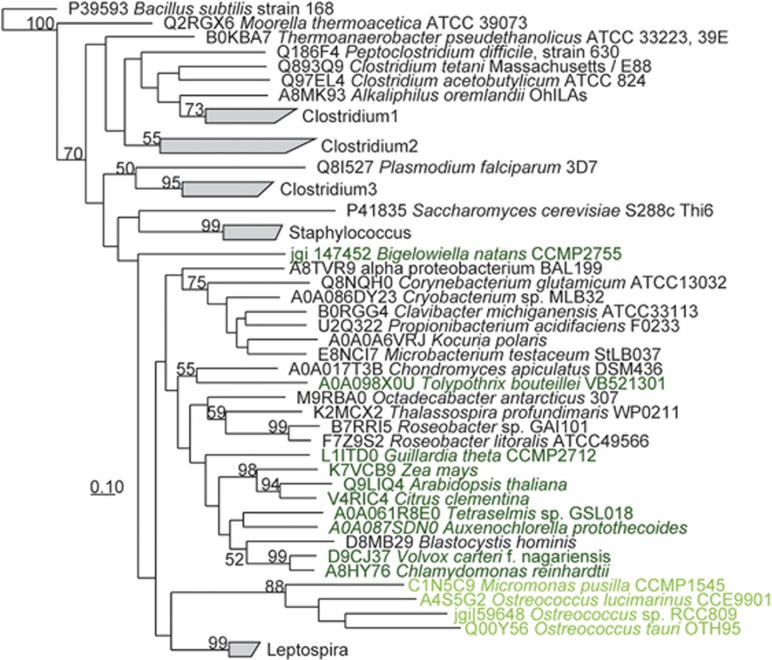
Picoeukaryotic phytoplankton require thiazole kinase (ThiM) to grow on B1 precursor(s)
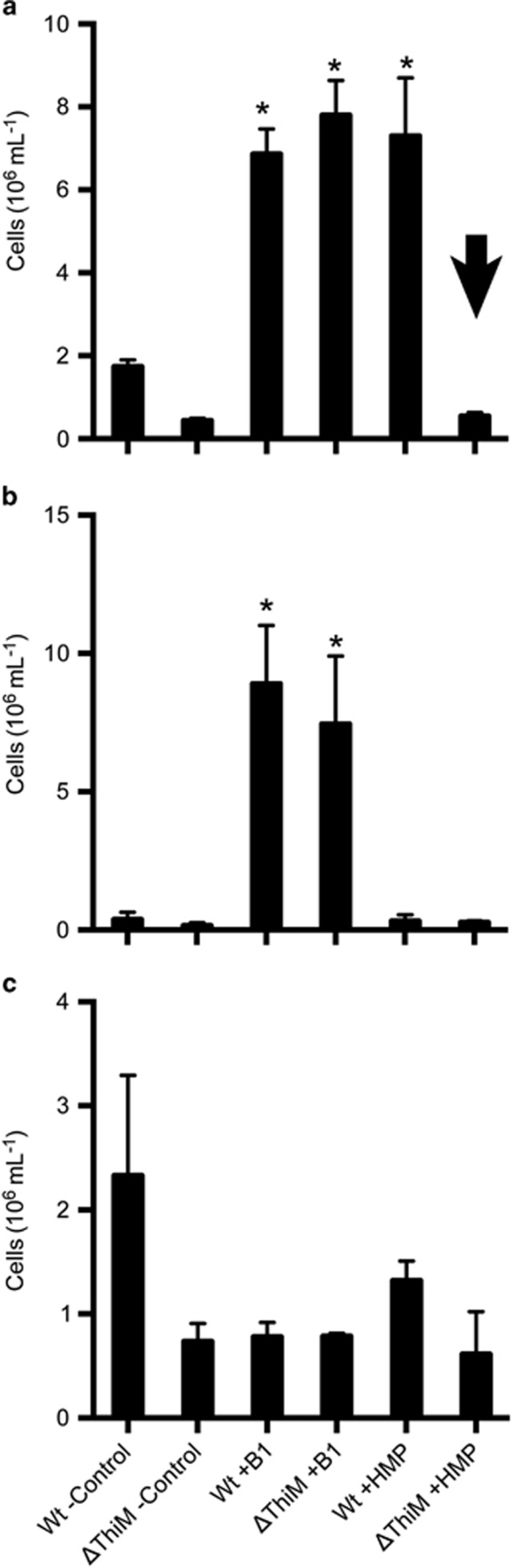
ThiM is a kinase normally thought to phosphorylate HET to make HET-P in model organisms as part of de novo biosynthesis or salvaging B1 from exogenous HET (Jurgenson et al., 2009; Moulin et al., 2013; Yazdani et al., 2013). The protein occurs in select bacteria, phytoplankton and terrestrial plants with notable amino-acid sequence divergence, and in regards to Ostreococcus and Micromonas spp., their ThiM sequences appear closely related likely indicating functional similarity (Figure 4). Knowing that HET is not used by Ostreococcus and Micromonas spp. (McRose et al., 2014; Paerl et al., 2015), we hypothesized ThIM is integral to the use of the thiazole-related precursor(s) observed in our previous experiments (Figures 2 and 3). To test this hypothesis, we generated an O. tauri RCC745 mutant line lacking ThiM (ΔThiM) (Supplementary Figure S2; also see Materials and methods section) and examined if its use of precursors mirrored that of wild-type strains noted in initial co-culture experiments (Supplementary Figures 2a–d). We found that in co-cultures with bacterium Marinobacter sp. OTB1, HMP addition did not stimulate growth of the ΔThiM mutant, with or without antibiotic addition (Figures 5a and b), which permitted or inhibited growth of Marinobacter sp. OTB1 (Figure 5c). In contrast, HMP addition stimulated growth of the wild-type line only in the treatment with active Marinobacter OTB1 cells (no antibiotics) (Figure 5a). Across treatments, the O. tauri RCC745 ΔThiM mutant grew only after addition of the complete B1 molecule (Figures 5a and b).
Figure 4.
ThiM amino-acid sequence phylogeny highlights Ostreococcus and Micromonas sequences are closely related relative to sequences from bacteria, terrestrial plants and other phytoplankton. Uniprot or JGI genome portal ID's for sequences are provided. Bootstrap values >50% are included to the left of nodes or collapsed branches. The sequences in collapsed branches are listed in Supplementary Table S1. Dark green text highlights terrestrial plant and phytoplankton sequences. Light green text highlights Mamiellales (picoeukaryotic phytoplankton) affiliates.
Figure 5.
O. tauri RCC745 requires ThiM to salvage B1 from thiazole-related precursor(s) provided by co-cultured bacterioplankton Marinobacter sp. OTB1 and supplemented HMP. The cell abundances of O. tauri RCC745 wild-type (Wt) and ΔThiM mutant (ΔThiM) lines are presented from day 3 of co-culture incubations without (a) or with (b) antibiotics (penicillin G and streptomycin). Bacterioplankton OTB1 growth was detectable only in co-cultures not treated with antibiotics (c). An arrow points to the lack of O. tauri RCC745 ΔThiM mutant growth in co-cultures supplemented with HMP and not treated with antibiotics (enabling bacterioplankton OTB1 cells to grow (c)). In contrast, wild-type RCC745 grew under the same co-culture conditions (a); underscoring the need for ThiM in salvaging B1 from added HMP and thiazole-related precursor(s) provided by OTB1 cells. Increased growth of the RCC745 wild-type versus ThiM mutant in (a) also suggests OTB1 cells provide precursors, not intact B1, useful for wild-type RCC745 to meet its B1 demands. Additions of B1 stimulated growth of RCC745 and ΔThiM mutant lines (a, b) confirming their B1 growth limitation and viability. Averages (columns) and s.d. (error bars) of triplicate co-cultures are presented. B1, HMP (1 nM each) and/or antibiotics were added on day 0. Negative controls (- Control) received no vitamin addition. Asterisks denote values significantly higher than respective negative controls in (a, b; t-test, n=3, P<0.05).
Thiazole-related B1 precursor(s) are present in the upper ocean mixed layer
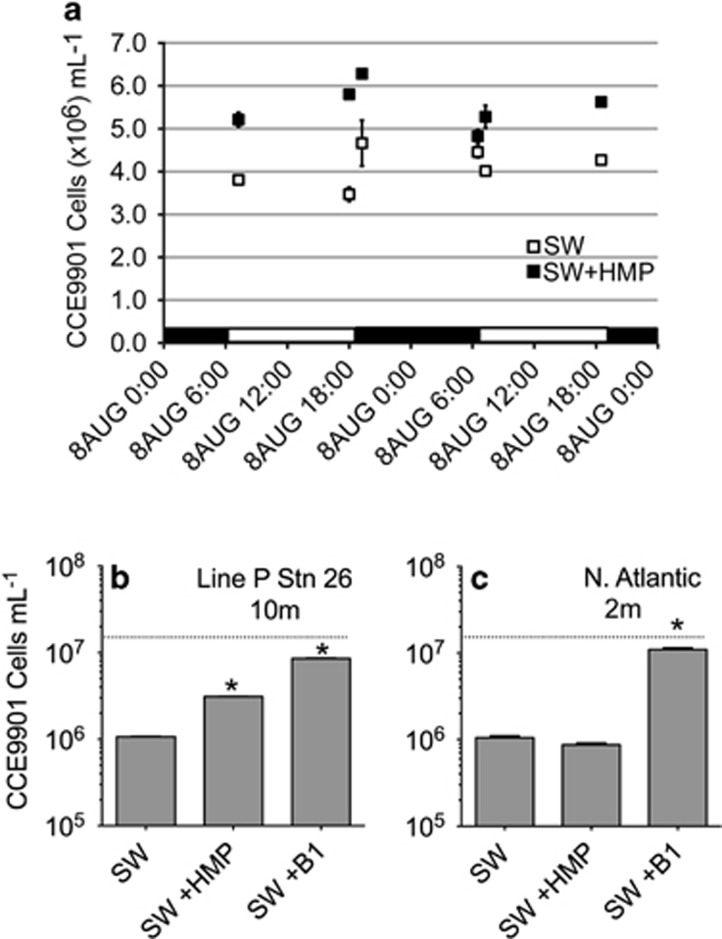
Applying the same O. lucimarinus CCE9901 bioassay approach used to examine culture filtrates (Figure 3), we tested if thiazole-related B1 precursor(s) was present in seawater samples from the mixed layer of the coastal and open-ocean (Figure 6). Thiazole-related B1 precursor(s) was detected in seawater collected from the mixed layer off the Scripps Pier and the northeast Pacific Ocean (furthest offshore station along monitoring transect Line P; Figures 6a and b), but not from the oligotrophic N. Atlantic open-ocean (Figure 6c). In addition, temporal changes in the availability of thiazole-related precursors were assessed during a ~24-h incubation of freshly collected Scripps Pier seawater (Figure 6a). Via the bioassay approach, excess of thiazole-related precursor(s) relative to B1 or other utilizable B1-related micronutrients was determinable (for example, differences in the yield of our bioassay organism O. lucimarinus CCE9901 grown on seawater samples alone versus seawater samples amended with HMP) and changed by up to a factor of 6 across incubation samplings while not obviously fluctuating across diel changes in light conditions (Figure 6a). Altogether, the thiazole-related B1 precursor(s) was available in seawater samples with its relative abundance fluctuating over time and across coastal and open-ocean domains (Figure 6).
Figure 6.
Based on bioassays with O. lucimarinus CCE9901, a thiazole-related B1 precursor (or precursors) is abundant in coastal and open-ocean near-surface seawater. Data in (a) are from an incubation of surface seawater off La Jolla, CA, USA. SW+HMP data represent maximum O. lucimarinus CCE9901 yields from seawater plus HMP, and SW data represent yields from seawater alone. SW+HMP maximum yields were significantly higher than unamended samples, indicating the presence of a thiazole-related B1 precursor(s), at all time points except 13th August 6:30 (t-test, n=3, P<0.05). Black and white bars along the x axis indicate the light and dark periods of the incubation. Positive controls (+1 nM B1) yielded ~1.6–1.9 x 107 cells ml−1 at all time points, but are not plotted. Similar to results in (a), HMP addition to bioassays of 10 m N. Pacific, Line P station 26 seawater led to increased maximum yields (b). In contrast, HMP addition did not stimulate yields in bioassays of 2 m seawater from near the open-ocean N. Atlantic BATS station (c). Dashed horizontal lines in (b, c) represent the upper quantification limit of the CCE9901 bioassay; this threshold was not plotted in (a), as all values were below this threshold. Asterisks in (b, c) denote data that were significantly higher (t-test, n=3, P<0.05) than respective unamended SW controls. Additions of B1 or HMP were at a final concentration of 1 nM.
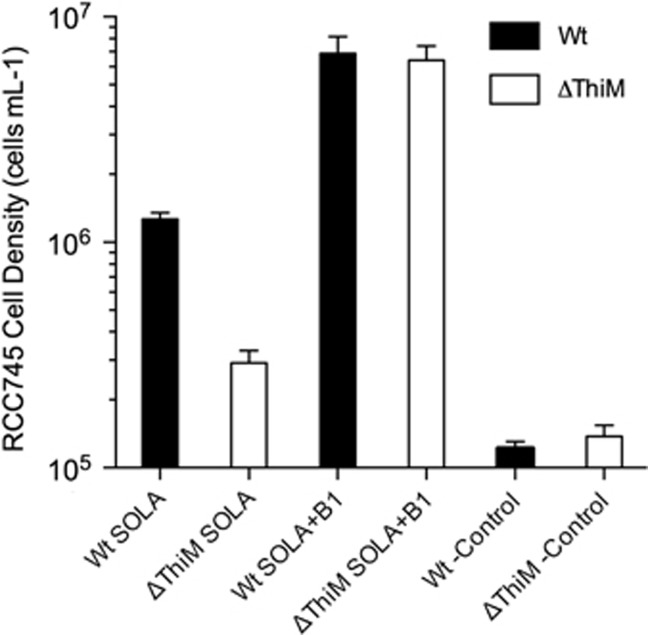
Finally, we used the O. tauri RCC745 wild-type and ΔThiM mutant lines in bioassays to examine the relative abundance of useful precursors versus B1 in near-surface seawater from a coastal Mediterranean monitoring site (SOLA). Maximum yields in bioassays with B1-limited RCC745 ΔThiM mutant (which can use only B1) were notably lower than yields in bioassays with the wild-type RCC745, accounting for only 23% the wild-type bioassay yields (Figure 7)—suggesting precursors were more available than intact B1 in the bioassayed SOLA seawater.
Figure 7.
Bioassays with O. tauri RCC745 indicate B1 precursors are more abundant relative to B1 in coastal seawater from the northwestern Mediterranean Sea (SOLA site). Wild-type (Wt) and ΔThiM O. tauri RCC745 (ΔThiM) cells deprived of B1 for 7 days were inoculated (10 cells ml−1) in filter-sterilized SOLA seawater from 3 m. After 7 days, maximum yields of the wild-type were significantly higher than those of the ΔThiM mutant; in contrast, yields of the wild-type and ΔThiM mutant grown on SOLA water supplemented with B1 were identical. The means and s.d. of biological triplicates are shown. Yields from negative control cultures (-Control), cells added to vitamin deplete medium, are also shown.
Discussion
The ecophysiological significance of Ostreococcus and Micromonas spp. using only precursors to meet their B1 demands
Here we report that key picoeukaryotic phytoplankton taxa, Ostreococcus and Micromonas spp., can meet their B1 requirements using solely precursors (Figures 2 and 3, and Supplementary Figure S3). Previous results suggested they required the B1 molecule to grow (McRose et al., 2014; Paerl et al., 2015), and could face growth limitation in B1-scarce euphotic waters based on their B1 growth kinetics (Paerl et al., 2015). It is now apparent that these phytoplankton are likely to minimize such limitation by using HMP, a precursor recently highlighted as being abundant in the upper ocean (Carini et al., 2014), and a yet-to-be identified thiazole-related precursor that can also be abundant (Figures 6 and 7).
From a broad marine microbiological perspective, our results elevate the ecological significance of thiazole-related precursor(s) by revealing they: sustain cosmopolitan marine picoeukaryotic phytoplankton (Figures 2 and 3, and Supplementary Figure S3), are provided by diverse B1-synthesizing plankton (Figure 3), and are present at significant concentrations in the euphotic ocean (Figures 6 and 7). These results expand prior knowledge regarding the ecological importance of thiazole-related B1 precursors to marine phytoplankton, which was limited to early demonstrations of HET being used to sustain primarily neritic B1 auxotrophic strains (Lwoff, 1947; Droop, 1958; Provasoli and Carlucci, 1974).
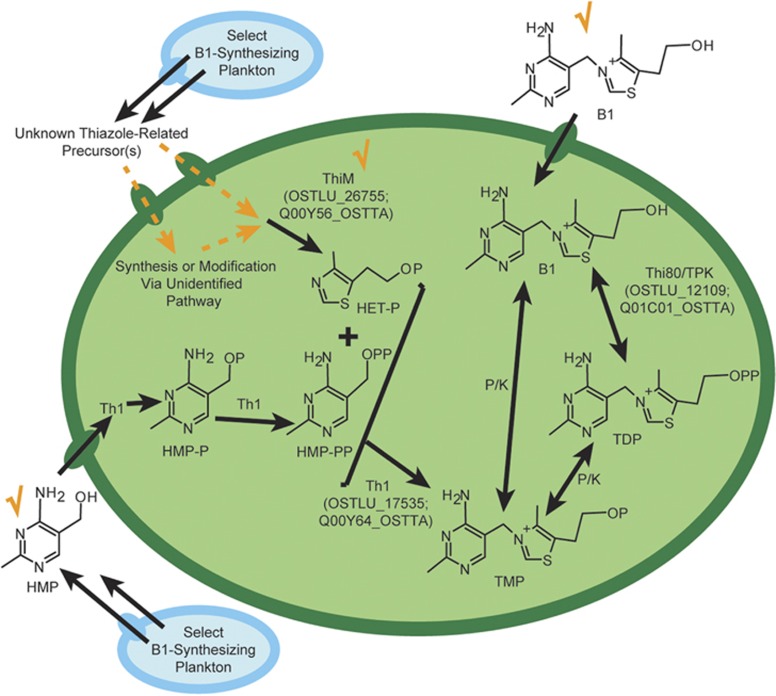
Taking into account their ability to use B1 precursors, we present an updated model of B1 biosynthesis and salvaging pathways in O. lucimarinus CCE9901 and O. tauri RCC745 (Figure 8). Considering pyrimidine and thiazole-related precursors are provided by B1-synthesizing plankton and accumulate in euphotic waters (Figures 2, 3, 6 and 7) (Carini et al., 2014), we hypothesize partial B1 pathways in these picoeukaryotic phytoplankton are the result of co-evolution with B1-synthesizing strains, enabling them to use both B1 precursors and intact B1 compared with being strict B1 auxotrophs.
Figure 8.
An updated view of B1 biosynthesis and salvaging pathways in O. lucimarinus CCE9901 and O. tauri RCC745 incorporating their use of B1 precursors. Orange checkmarks denote that ThiM and HMP are utilizable in salvaging TDP by Ostreococcus (large green oval; demonstrated in this study), and that exogenous B1 can also support the phytoplankton (Figures 2–4; Paerl et al., 2015). Dashed orange lines denote uptake and biosynthesis pathways hypothesized to be involved in the utilization of a thiazole-related B1 precursor(s) to generate TMP from HET-P and HMP-PP by Th1 (Pourcel et al., 2013). A limited set of proteins associated with canonical thiazole B1 biosynthesis are present in these phytoplankton (Supplementary Table S3), but they do not generate HET, and hence components to scavenging of thiazole-like precursor upstream of ThiM are unclear. Select B1-synthesizing plankton (represented by blue ovals) are sources of thiazole-related precursor(s) (Figures 2–4, and S3) and HMP (Carini et al., 2014). Kinases and phosphatases (K/P) are presumed present in the Ostreococcus spp. but their identities are unknown. Smaller ovals (blue or green) represent membrane transporters; putative B1 or HMP-associated transporters and symporters are present in Ostreococcus spp. but their transport capacities are not well-characterized. Closely related phytoplankton strains (Ostreococcus sp. RCC809, Micromonas pusilla CCMP1545 and Micromonas sp. CCMP487) are expected to similarly salvage B1 from precursors as they possess ThiM (Figure 4) or exhibit a similar growth response to HMP in co-cultures with bacteria (Supplementary Figure S3). Other B1-related compounds specifically TDP, TMP and HET are not utilizable by axenic O. lucimarinus CCE9901 cultures; however, in co-culture with bacteria, TDP becomes accessible presumably via dephosphorylation to B1 (Paerl et al., 2015; data not shown). Uniprot ID's for B1 biosynthesis proteins found in both Ostreococcus strains are provided in parentheses, and the amino-acid sequence similarity between Ostreococcus B1 biosynthesis proteins and Swiss-Prot reviewed reference sequences are in Supplementary Table S3. B1, thiamine; HMP-PP, 4-amino-5-hydroxymethyl-2-methylpyrimidine diphosphate; Th1; thiamine phosphate synthase; Thi80, thiamine diphosphokinase; P (in chemical compounds), PO4.
We speculate the newly detected thiazole-related precursor(s) is either an unrecognized substrate for ThiM, or a compound modified to generate HET upstream of ThiM (Figure 8). HET-P is not anticipated to be the precursor, as ThiM phosphorylates HET to make HET-P, nor are known low molecular weight thiazole-related B1 precursors, for example, dehydroglycine (Jurgenson et al., 2009), as multiple enzymes involved in the thiazole branch of canonical B1 biosynthesis are not present in the phytoplankton (for example, ThiG/Thi4, ThiS; Supplementary Table S3). Possibly the useful thiazole-like precursor(s) is a partly degraded (for example, oxidized) thiazole as B1 and thiazole precursors degrade in alkaline solutions in vitro (Dwivedi and Arnold, 1973). Chemical identification of the newly detected thiazole-related precursor(s) is important future work requiring substantial analytical efforts, for example, development of an isolation method, bioassay testing of iterative isolated fractions and nuclear magnetic resonance-based analysis of purified fractions, and will build on the ecophysiological results presented here.
Newly realized facets of B1-related micronutrient flux from marine microbes
Release or excretion of vitamins is considered to be an important input to the upper ocean (Sañudo-Wilhelmy et al., 2014), but few details are known about the flux of B1 and/or related micronutrients from marine plankton—beyond that axenic B1-synthesizing marine phytoplankton and bacteria can provide compounds that sustain B1 auxotrophic plankton (Burkholder, 1963; Carlucci and Bowes, 1970), including at least one specific precursor HMP (Carini et al., 2014).
Here we do not disentangle release versus excretion (respectively interpreted here as passive loss via diffusion or leakage, versus, active export, for example, via specific protein-mediated processes) by B1-synthesing plankton, but do demonstrate that through either process multiple micronutrients related to B1 are fluxed from cells to their surroundings. Further, the types of micronutrients that become available appear to vary between strains. This is evident based on the growth of our bioassay organism on filtrates with and without HMP addition (Figure 3 and Supplementary Figure S3) and the inability to detect thiazole-related precursor(s) in some filtrates (Figure 3). Filtrates were collected using gentle filtration (to prevent cell lysis) and culture growth curves did not exhibit signs of viral lysis (Figure 3), hence flux of B1 or precursors from cells in our experiments is attributed to release or excretion, as has been suggested in previous vitamin-focused studies (Carlucci and Bowes, 1970; Bonnet et al., 2010; Carini et al., 2014). We speculate that the higher availability of thiazole-related precursor(s) observed in seawater samples (Figures 6 and 7) is due to relatively high release or excretion of thiazole-related precursor(s) by select plankton, for example, Marinomonas spp. SBI8L, SBI9L and SBI22, Marinobacter sp. OTB1, and P. tricornutum CCMP632 (Figures 2a–d, Figures 3a and d). Efficient acquisition of exogenous B1 and/or pyrimidines (for example, HMP) may also contribute to relatively high net flux of thiazole-related precursor(s). The strains that provided relatively high amounts of the thiazole-related precursor(s) in our experiments (SBI8L, SBI22, TM1040 and CCMP632; Figures 2 and 3) all possess B1 or pyrimidine-associated uptake transporters (Supplementary Table S2), potentially enabling them to efficiently acquire exogenous B1 and pyrimidine precursor while leaving a higher external concentration of thiazole-related precursor(s). Variability in the type and quantity of B1-related micronutrients released or excreted by plankton carries environmental significance as their use differs between B1 auxotrophic marine plankton (Burkholder and Burkholder, 1958; Droop, 1958; Carini et al., 2014; McRose et al., 2014; Paerl et al., 2015).
Shifting focus to released or excreted B1 precursors
Results presented here, and in recent studies, highlight that B1 precursors being released or excreted from plankton, not just B1 or B1 degradation products, are vital micronutrients that sustain key plankton in the upper ocean. HET, a B1 degradation product used in all previous tests of marine plankton thiazole B1 precursor utilization (Droop, 1958; Burkholder, 1963; Provasoli and Carlucci, 1974; Turner, 1979) is not useful for Ostreococus and Micromonas spp. (McRose et al., 2014; Paerl et al., 2015). Similarly, the also frequently tested pyrimidine, and B1 degradation product, 4-amino-5-aminomethyl-2-methylpyrimidine (Burkholder, 1963; Carlucci and Silbernagel, 1966; Provasoli and Carlucci, 1974; Turner, 1979) is not utilizable by SAR11 bacterioplankton clade affiliate Pelagibacter ubique HTCC1062 (Carini et al., 2014). In contrast, thiazole-related precursor(s) (other than HET) and HMP that can be released or excreted (Figure 3) (Carini et al., 2014), are relatively abundant in seawater (Figures 6 and 7; Carini et al., 2014) and useful for prevalent picophytoplankton (Figures 2–4, and Supplementary Figure S3; McRose et al., 2014; Paerl et al., 2015) and bacterioplankton (for example, SAR11 representatives; Carini et al., 2014) in the upper ocean. A spectrum of interactions involving these precursors is expected to occur in the upper ocean, more than previously realized. In particular, competitive interactions, especially for HMP, are likely widespread and in need of assessment as it is now known that multiple biogeochemically important plankton require or can utilize HMP (alone or with a thiazole-related precursor) to meet their B1 demands, including SAR11 clade representatives (Carini et al., 2014), Proteobacteria (lacking thiC; Sañudo-Wilhelmy et al., 2014), and both cosmopolitan and bloom-forming eukaryotic phytoplankton (for example, Ostreococcus, Micromonas and Pelagomonas spp., and Emiliania huxleyi; Figures 2, 3 and 5) (McRose et al., 2014; Paerl et al., 2015).
Conclusion
Our results provide valuable new information about thiamine cycling and B1 auxotrophy in marine phytoplankton and collectively underscore the importance of thiazole-related precursor(s) in sustaining environmentally abundant B1 auxotrophic picoeukaryotic phytoplankton. The results also highlight released or excreted B1 precursors as vital micronutrients in the upper ocean, in line with findings of recent studies (Carini et al., 2014; McRose et al., 2014). Multiple lines of evidence now demonstrate B1 precursors as vital micronutrients for numerically dominant marine plankton—future work is needed to better resolve how availability of precursors fluctuates relative to B1 and relates to environmentally important conditions, for example, changes in productivity or plankton community composition.
Acknowledgments
This research is funded in part by the Gordon and Betty Moore Foundation through grants # GBMF2758 and GMBF4827 (to FA), the French Agence Nationale de la recherche (Photo-Phyto, ANR-14-CE02-0018) and the PIA EMBRC France (F-YB). Thanks are extended to E Bertrand and A Allen for providing CCMP632, B Brahamsha for discussion of experimental results, K Barbeau and lab members S Hogle and R Bundy for providing Rugeria sp. TM1040 and archived seawater, E Daniels and Y Zhou for culture monitoring, J Freeze and L Intertaglia for 16S rRNA gene sequencing. RWP recognizes E Bertrand, L Riemann, J Smith and H Paerl for helpful comments on the manuscript.
Author contributions
RWP, F-YB, BP and FA designed the research; RWP, F-YB, J-CL, PS and VV performed experiments; RWP wrote the manuscript and all authors edited the manuscript.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Aylward FO, Eppley JM, Smith JM, Chavez FP, Scholin CA, DeLong EF. (2015). Microbial community transcriptional networks are conserved in three domains at ocean basin scales. Proc Natl Acad Sci USA 112: 5443–5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand EM, Allen AE. (2012). Influence of vitamin B auxotrophy on nitrogen metabolism in eukaryotic phytoplankton. Front Microbiol 3: 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidle KD, Azam F. (2001). Bacterial control of silicon regeneration from diatom detritus: significance of bacterial ectohydrolases and species identity. Limnol Oceanogr 46: 1606–1623. [Google Scholar]
- Bonnet S, Webb EA, Panzeca C, Karl DM, Capone DG, Wilhelmy SS. (2010). Vitamin B12 excretion by cultures of the marine cyanobacteria Crocosphaera and Synechococcus. Limnol Oceanogr 5: 1959–1964. [Google Scholar]
- Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A et al. (2008). The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456: 239–244. [DOI] [PubMed] [Google Scholar]
- Brahamsha B. (1996). A genetic manipulation system for oceanic cyanobacteria of the genus Synechococcus. Appl Environ Microbiol 62: 1747–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkholder PR. (1963) Some nutritional relationships among microbes of sea sediments and water. In: Oppenheimer CH (ed), Symposium on Marine Microbiology. Charles C Thomas: Springfield, IL, USA, pp 133–150. [Google Scholar]
- Burkholder PR, Burkholder LM. (1958). Studies on B vitamins in relation to productivity of the Bahia Fosforescente, Puerto Rico. Bull Mar Sci Gulf Caribbean 8: 201–223. [Google Scholar]
- Carini P, Campbell EO, Morré J, Sañudo-Wilhelmy SA, Cameron Thrash J, Bennett SE et al. (2014). Discovery of a SAR11 growth requirement for thiamin's pyrimidine precursor and its distribution in the Sargasso Sea. ISME J 8: 1727–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlucci AF, Bowes PM. (1970). Production of vitamin B12, thiamine and biotin by phytoplankton. J Phycol 6: 351–357. [Google Scholar]
- Carlucci AF, Silbernagel SB. (1966). Bioassay of seawater II. Methods for the determination of concentrations of dissolved vitamin B1 in seawater. Can J Microbiol 12: 1079–1089. [DOI] [PubMed] [Google Scholar]
- Corellou F, Camasses A, Ligat L, Peaucellier G, Bouget F-Y. (2005). Atypical regulation of a green lineage-specific B-type cyclin-dependent kinase. Plant Physiol 138: 1627–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corellou F, Schwartz C, Motta J-P, Batoul Djouani-Tahri El, Sanchez F, Bouget F-Y. (2009). Clocks in the green lineage: comparative functional analysis of the circadian architecture of the picoeukaryote Ostreococcus. Plant Cell 21: 3436–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Countway PD, Caron DA. (2006). Abundance and distribution of Ostreococcus sp. in the San Pedro Channel, California, as revealed by quantitative PCR. Appl Environ Microbiol 72: 2496–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft MT, Warren MJ, Smith AG. (2006). Algae need their vitamins. Eukary Cell 5: 1175–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demir-Hilton E, Sudek S, Cuvelier ML, Gentemann CL, Zehr JP, Worden AZ. (2011). Global distribution patterns of distinct clades of the photosynthetic picoeukaryote Ostreococcus. ISME J 5: 1095–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derelle E, Ferraz C, Rombauts S, Rouzé P, Worden AZ, Robbens S et al. (2006). Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci USA 103: 11647–11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droop MR. (1957). Auxotrophy and organic compounds in the nutrition of marine phytoplankton. J Gen Microbiol 16: 286–293. [DOI] [PubMed] [Google Scholar]
- Droop MR. (1958). Requirement for thiamine among some marine and supra-littoral protista. J Marine Biol Assoc UK 37: 323–329. [Google Scholar]
- Dupont CL, Rusch DB, Yooseph S, Lombardo M-J, Alexander Richter R, Valas R et al. (2011). Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J 6: 1186–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi BK, Arnold RG. (1973). Chemistry of thiamine degradation in food products and model systems: a review. J Agric Food Chem 21: 54–60. [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform 5: 113–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank RAW, Leeper FJ, Luisi BF. (2007). Structure, mechanism and catalytic duality of thiamine-dependent enzymes. Cell Mol Life Sci 64: 892–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillard RRL. (1975) Culture of phytoplankton for feeding marine invertebrates. In: Smith WL, Chanley MH (eds). Culture of Marine Invertebrate Animals. Springer: Boston, MA, USA, pp 29–60. [Google Scholar]
- Jurgenson CT, Begley TP, Ealick SE. (2009). The structural and biochemical foundations of thiamin biosynthesis. Annu Rev Biochem 78: 569–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DJ. (1991). 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds.), Nucleic Acid Techniques in Bacterial Systematics Wiley: NY, USA, pp 115–175.
- Long RA, Azam F. (2001). Antagonistic interactions among marine pelagic bacteria. Appl Environ Microbiol 67: 4975–4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano J-C, Schatt P, Botebol H, Vergé V, Lesuisse E, Blain S et al. (2014). Efficient gene targeting and removal of foreign DNA by homologous recombination in the picoeukaryote Ostreococcus. Plant J 78: 1073–1083. [DOI] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lwoff A. (1947). Some aspects of the problem of growth factors for protozoa. Annu Rev Microbiol 1: 101–114. [Google Scholar]
- Malviya S, Scalco E, Audic S, Vincent F, Veluchamy A, Poulain J et al. (2016). Insights into global diatom distribution and diversity in the world's ocean. Proc Natl Acad Sci USA 113: E1516–E1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRose D, Guo J, Monier A, Sudek S, Wilken S, Yan S et al. (2014). Alternatives to vitamin B1 uptake revealed with discovery of riboswitches in multiple marine eukaryotic lineages. ISME J 8: 2517–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller TR, Belas R. (2004). Dimethylsulfoniopropionate metabolism by Pfiesteria-associated Roseobacter spp. Appl Environ Microbiol 70: 3383–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulin M, Nguyen GTDT, Scaife MA, Smith AG, Fitzpatrick TB. (2013). Analysis of Chlamydomonas thiamin metabolism in vivo reveals riboswitch plasticity. Proc Natl Acad Sci USA 110: 14622–14627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottesen EA, Young CR, Eppley JM, Ryan JP, Chavez FP, Scholin CA et al. (2013). Pattern and synchrony of gene expression among sympatric marine microbial populations. Proc Natl Acad Sci USA 110: E488–E497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paerl RW, Bertrand EM, Allen AE, Palenik B, Azam F. (2015). Vitamin B1 ecophysiology of marine picoeukaryotic algae: strain-specific differences and a new role for bacteria in vitamin cycling. Limnol Oceanogr 60: 215–228. [Google Scholar]
- Partensky F, Blanchot J, Vaulot D. (1999). Differential distribution and ecology of Prochlorococcus and Synechococcus in oceanic waters: a review. Bulletin de l'Institut Oceanographique, Monaco, special 19: 457–475. [Google Scholar]
- Porter KG, Feig YS. (1980). Use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr 25: 943–948. [Google Scholar]
- Pourcel L, Moulin M, Fitzpatrick TB. (2013). Examining strategies to facilitate vitamin B1 biofortification of plants by genetic engineering. Front Plant Sci 4: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provasoli L, Carlucci AF. (1974) Vitamins and growth regulators. In: Stewart WDP (ed). Algal physiology and biochemistry. Blackwell: Oxford, pp 741–787. [Google Scholar]
- Rodionov DA, Hebbeln P, Eudes A, Beek ter J, Rodionova IA, Erkens GB et al. (2008). A novel class of modular transporters for vitamins in prokaryotes. J Bacteriol 191: 42–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sañudo-Wilhelmy SA, Gómez-Consarnau L, Suffridge C, Webb EA. (2014). The role of B vitamins in marine biogeochemistry. Annu Rev Marine Sci 6: 339–367. [DOI] [PubMed] [Google Scholar]
- Tang YZ, Koch F, Gobler CJ. (2010). Most harmful algal bloom species are vitamin B1 and B12 auxotrophs. Proc Natl Acad Sci USA 107: 20756–20761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner MF. (1979). Nutrition of some marine microalgae with special reference to vitamin requirements and utilization of nitrogen and carbon sources. J Marine Biol Assoc UK 59: 535–552. [Google Scholar]
- Worden AZ, Janouskovec J, McRose D, Engman A, Welsh RM, Malfatti S et al. (2012). Global distribution of a wild alga revealed by targeted metagenomics. Curr Biol 22: R675–R677. [DOI] [PubMed] [Google Scholar]
- Worden AZ, Nolan JK, Palenik B. (2004). Assessing the dynamics and ecology of marine picophytoplankton: the importance of the eukaryotic component. Limnol Oceanogr 49: 168–179. [Google Scholar]
- Yazdani M, Zallot R, Tunc-Ozdemir M, de Crécy-Lagard V, Shintani DK, Hanson AD. (2013). Identification of the thiamin salvage enzyme thiazole kinase in Arabidopsis and maize. Phytochem 94: 68–73. [DOI] [PubMed] [Google Scholar]
- ZoBell CE. (1941). Studies on marine bacteria. I. The cultural requirements of heterotrophic aerobes. J Mar Res 4: 42–75. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.