Abstract
Lyme disease, a zoonotic disease, is the most prevalent vector-borne disease in the Northern Hemisphere. Diversity of the vector (tick) microbiome can impact pathogen transmission, yet the biotic and abiotic factors that drive microbiome diversity are largely unresolved, especially under natural, field conditions. We describe the microbiome of Ixodes pacificus ticks, the vector for Lyme disease in the western United States, and show a strong impact of host blood meal identity on tick microbiome species richness and composition. Western fence lizards, a host that is refractory to the Lyme disease pathogen, significantly reduces microbiome diversity in ticks relative to ticks that feed on a mammalian reservoir host. Host blood meal-driven reduction of tick microbiome diversity may have lifelong repercussions on I. pacificus vector competency and ultimately disease dynamics.
Animal hosted microbiome communities are now recognized as essential assemblages that can affect host health, phenotype and disease susceptibility (Gosalbes et al., 2012; Jumpstart Consortium Human Microbiome Project Data Generation Working Group, 2012; Christian et al., 2015). Further, the microbiomes of arthropod vectors of human pathogens can affect the transmission of zoonotic pathogens (Weiss and Aksoy, 2011; Hughes et al., 2014; Narasimhan et al., 2014). Laboratory experiments have found that higher tick microbiome diversity is negatively correlated with colonization success of the Lyme disease pathogen, Borrelia burgdorferi (Narasimhan et al., 2014). But it remains unclear what factors drive natural variation of the tick microbiome and whether such variation would affect pathogen transmission in the wild and therefore regulate disease risk. Prior field studies have identified microbiome differences by tick species (Hawlena et al., 2013), sex (Williams-Newkirk et al., 2014; Zhang et al., 2014) and region (Carpi et al., 2011; van Treuren et al., 2015) but have not found correlations with host blood or the immediate environment (Hawlena et al., 2013; Rynkiewicz et al., 2015).
Tick larvae must feed on an infected blood meal source in order to acquire B. burgdorferi and then molt into an infected nymph. This simple life history and feeding strategy provides an ideal system to investigate the natural drivers of microbiome diversity and its effect on pathogen transmission in a disease vector. A previous study did not find a correlation between host blood meal and Ixodes scapularis microbiomes (Rynkiewicz et al., 2015), yet Ixodes pacificus, the focal tick of this study, has a distinct natural history (Lane and Loye, 1989; Eisen et al., 2001). As a generalist, I. pacificus feeds on numerous vertebrate species that may be pathogen reservoirs, but the primary blood meal host for juvenile I. pacificus is the western fence lizard, Sceloporus occidentalis, a Borrelia-refractory host (Lane and Quistad, 1998; Kuo et al., 2000). Infected ticks that feed on S. occidentalis are cleared of their B. burgdorferi infections and are no longer infective as a result of complement proteins in the lizard's innate immune system (Kuo et al., 2000).
We collected all stages of I. pacificus, the vector of Lyme disease in the western United States, from the field and analyzed the interactions between tick microbiome diversity, host blood meal and pathogen infection using amplicon-based deep sequencing analyses. I. pacificus of all life stages were collected from the field and nymphs were tested for B. burgdorferi infection (Lane et al., 2005). Engorged larvae were collected from a reservoir host (mice) or a non-reservoir host (lizards) and molted into nymphs for microbiome analysis (Swei et al., 2012). Then 16S rRNA microbiome libraries were prepared for individually barcoded samples and sequenced on an Illumina MiSeq (San Diego, CA, USA). Sequences were quality-filtered, clustered at 97% sequence similarity and analyzed in QIIME (Caporaso et al., 2010).
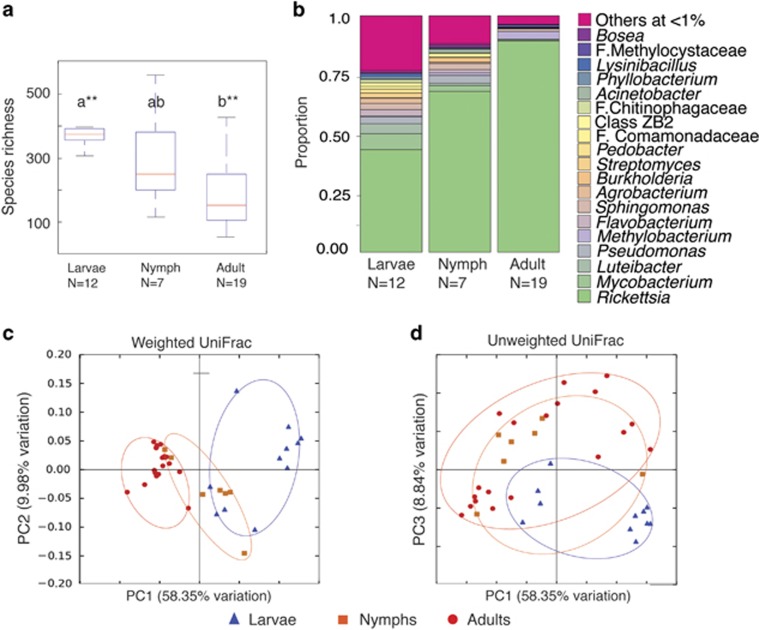
Our analysis found a significant loss of microbiome species richness and evenness as field-collected I. pacificus mature (Figures 1a and b). The loss of species richness in microbiome communities across vector life stage has not been observed in other arthropod species and may reflect unique factors that drive microbiome diversity in I. pacificus. Beta diversity was also significantly different between the life stages measured by relative abundance (weighted Unifrac distance, t-test=−18.40, P=0.001; Figure 1c) and occurrence (unweighted UniFrac distance, t-test=−7.35, P=0.001, Figure 1d). At all stages, tick microbiomes were dominated by one bacterial OTU belonging to the genus Rickettsia and identified as the endosymbiont Rickettsia G021 based on 16S sequence (Supplementary Figure 1; Hunter et al., 2015). The pathogenicity of Rickettsia G021 is unknown but it is believed to be nonpathogenic. In addition, I. pacificus is not known to harbor pathogenic Rickettsia species (Cheng et al., 2013). Our analysis found that Rickettsia was ubiquitous in I. pacificus and becomes increasingly dominant at older life stages simultaneous with loss of microbiome species richness and evenness (Supplementary Table 2). Thus, environmental exposure (for example, age) does not have a strong role in the introduction of microbes into the tick microbiome.
Figure 1.
Microbiome results of field-collected, host-seeking Ixodes pacificus life stages depicting (a) alpha diversity as measured by observed OTUs. Group significance is indicated above the bars with ** indicating P-value<0.01. (b) Life stage differences in genus-level taxonomic assignment based on 97% OTU phylogroups. Genera present at <1% were binned together into the ‘Others at <1%' group. (c) Weighted and (d) unweighted PCOA analysis of life stage beta diversity.
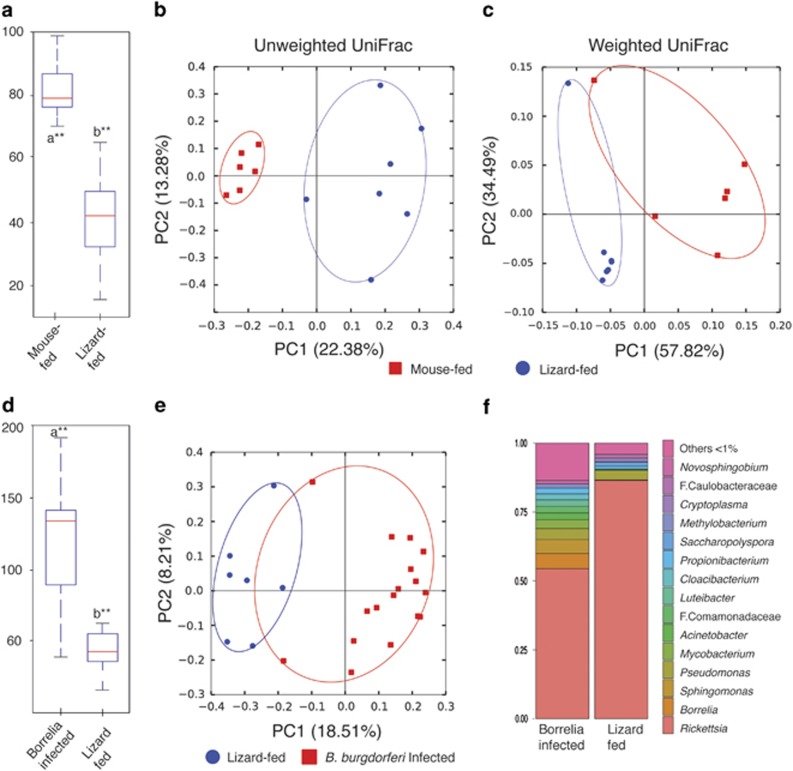
We examined I. pacificus ticks that were B. burgdorferi positive or negative and found no significant differences in microbiome species richness or composition (Supplementary Figure 2A). While B. burgdorferi positive ticks likely fed on a mammal reservoir, negative ticks may be negative because they either fed on a Borrelia-refractory lizard or an uninfected mammal. To specifically examine how host blood meal affects the nymphal tick microbiome, we evaluated the microbiome of nymphs known to have fed on mice or lizards as larvae (Figures 2a and b, Supplementary Figure 4). I. pacificus nymphs that fed on lizards as larvae were characterized by significantly lower species richness relative to mouse-fed ticks (t-test=5.41, P=0.003 Figure 2a). Microbiome community composition was distinct between lizard-fed and mouse-fed nymphs by unweighted UniFrac (t-test=−5.14, P=0.001, Figure 2b) and weighted UniFrac (t-test=−5.47, P=0.001, Figure 2c). Rickettsia comprises a significantly greater portion of the microbiome in lizard-fed ticks relative to mouse-fed ticks primarily as a function of reductions in richness of other microbiome species (P=0.002, Supplementary Figure S1). These results show that S. occidentalis reduces the diversity of other bacteria comprising the tick microbiome but does not reduce the relative abundance of Rickettsia perhaps because it is an intracellular bacteria not found in the midgut where interaction with host blood would occur (Supplementary Table 1).
Figure 2.
Host blood meal impacts on tick microbiome diversity shown by (a) alpha diversity as measured by observed OTUs, and group significance indicated by the box plots with ** indicating P-value<0.01. (b) Beta diversity analysis by unweighted UniFrac analysis and (c) Weighted UniFrac analysis of nymphal Ixodes pacificus that fed on wild lizards or mice as larvae. Microbiome summaries of nymphal ticks that fed on Sceloporus occidentalis (‘Lizard-fed') as larvae and questing nymphs that were infected with Borrelia burgdorferi (‘Borrelia infected') are shown by (d) species richness as measured by observed OTUs. Group significance is indicated by the box plots with ** indicating P-value<0.01. (e) Unweighted UniFrac analysis of lizard-fed versus Borrelia-infected ticks shown along PC1 and PC2. (f) Genus level taxonomic composition of lizard-fed versus Borrelia-infected ticks showing relative proportion of dominant taxa at the genus level.
B. burgdorferi-infected I. pacificus nymphs were distinct from lizard-fed ticks both in terms of species richness and composition (Figure 2d–f) and had a lower proportion of Rickettsia reads (Supplementary Table 1, Supplementary Figure 3). Although the source of infection is unknown, infected nymphs must have fed on a pathogen reservoir such as a small mammal. Western fence lizards host an overwhelming proportion of juvenile ticks, especially nymphs (Eisen et al., 2001), therefore, the loss of species richness from subadult to adult ticks is unique to I. pacificus (Zolnik et al., 2016) and appears to be driven by lizard blood meals. We find that when ticks feed on mice or other pathogen reservoirs, microbiome species richness is significantly higher. These results are intriguing because when microbiome diversity of I. scapularis was experimentally lowered, colonization success of Borrelia burgdorferi was impaired (Narasimhan et al., 2014). Thus, ticks with reduced microbiome richness due to feeding on lizards may exhibit decreased ability to acquire B. burgdorferi in subsequent blood meals.
We show that S. occidentalis significantly alters the composition of the tick microbiome. Laboratory studies (Narasimhan et al., 2014) suggest that reduction of tick microbiome diversity may affect future acquisition of B. burgdorferi by I. pacificus. These results demonstrate that ecological factors (that is, blood meal source) can significantly influence vector microbiomes and may fundamentally impact pathogen transmission in natural systems.
Acknowledgments
We thank Caitlin Miller and Betsabel Chicana for field and laboratory assistance. We thank Frank Cipriano for technical support for sequencing assistance. This research was supported by an NSF grant #1427772 and CSUPERB New Investigator Grant to AS. The microbiome sequence data are archived at Sequence Read Archive under the BioProject ID PRJNA352452.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpi G, Cagnacci F, Wittekindt NE, Zhao FQ, Qi J, Tomsho LP et al. (2011). Metagenomic profile of the bacterial communities associated with Ixodes ricinus ticks. PLoS One 6: e25604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D, Vigil K, Schanes P, Brown RN, Zhong J. (2013). Prevalence and burden of two rickettsial phylotypes (G021 and G022) in Ixodes pacificus from California by real-time quantitative PCR. Ticks Tick Borne Dis 4: 280–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian N, Whitaker BK, Clay K. (2015). Microbiomes: unifying animal and plant systems through the lens of community ecology theory. Front Microbiol 6: 869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen RJ, Eisen L, Lane RS. (2001). Prevalence and abundance of Ixodes pacificus immatures (Acari: Ixodidae) infesting western fence lizards (Sceloporus occidentalis in northern California: Temporal trends and environmental correlates. J Parasitol 87: 1301–1307. [DOI] [PubMed] [Google Scholar]
- Gosalbes MJ, Abellan JJ, Durban A, Perez-Cobas AE, Latorre A, Moya A. (2012). Metagenomics of human microbiome: beyond 16s rDNA. Clin Microbiol Infect 18: 47–49. [DOI] [PubMed] [Google Scholar]
- Hawlena H, Rynkiewicz E, Toh E, Alfred A, Durden LA, Hastriter MW et al. (2013). The arthropod, but not the vertebrate host or its environment dictates bacterial community composition of fleas and ticks. ISME J 7: 221–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes GL, Dodson BL, Johnson RM, Murdock CC, Tsujimoto H, Suzuki Y et al. (2014). Native microbiome impedes vertical transmission of Wolbachia in Anopheles mosquitoes. Proc Natl Acad Sci USA 111: 12498–12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter DJ, Torkelson JL, Bodnar J, Mortazavi B, Laurent T, Deason J et al. (2015). The Rickettsia endosymbiont of Ixodes pacificus contains all the genes of de novo folate biosynthesis. PLoS One 10: e0144552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumpstart Consortium Human Microbiome Project Data Generation Working Group. (2012). Evaluation of 16S rDNA-based community profiling for human microbiome research. PLoS One 7: e39315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M, Lane RS, Giclas PC. (2000). A comparative study of mammalian and reptilian alternative pathway of complement-mediated killing of the Lyme disease spirochete (Borrelia burgdorferi. J Parasitol 86: 1223–1228. [DOI] [PubMed] [Google Scholar]
- Lane RS, Loye. JE. (1989). Lyme disease in California USA interrelationship of Ixodes pacificus (Acari: Ixodidae) the western fence lizard Sceloporus occidentalis and Borrelia burgdorferi. J Med Entomol 26: 272–278. [DOI] [PubMed] [Google Scholar]
- Lane RS, Mun J, Eisen RJ, Eisen L. (2005). Western gray squirrel (Rodentia: Sciuridae): a primary reservoir host of Borrelia burgdorferi in Californian oak woodlands? J Med Entomol 42: 388–396. [DOI] [PubMed] [Google Scholar]
- Lane RS, Quistad GB. (1998). Borreliacidal factor in the blood of the western fence lizard (Sceloporous occidentalis. J Parasitol 84: 29–34. [PubMed] [Google Scholar]
- Narasimhan S, Rajeevan N, Liu L, Zhao YO, Heisign J, Pan J et al. (2014). Gut microbiota of the tick vector Ixodes scapularis modulate colonization of the Lyme disease spirochete. Cell Host Microbe 15: 58–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rynkiewicz EC, Hemmerich C, Rusch DB, Fuqua C, Clay K. (2015). Concordance of bacterial communities of two tick species and blood of their shared rodent host. Mol Ecol 24: 2566–2579. [DOI] [PubMed] [Google Scholar]
- Swei A, Briggs CJ, Lane RS, Ostfeld RS. (2012). Impacts of an introduced forest pathogen on the risk of Lyme disease in California. Vector Borne Zoonot Dis 12: 624–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Treuren W, Ponnusamy L, Brinkerhoff RJ, Gonzalez A, Parobek CM, Juliano JJ et al. (2015). Variation in the microbiota of Ixodes ticks with regard to geography, species, and sex. Appl Environ Microbiol 81: 6200–6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss B, Aksoy S. (2011). Microbiome influences on insect host vector competence. Trends Parasitol 27: 514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Newkirk AJ, Rowe LA, Mixson-Hayden TR, Dasch GA. (2014). Characterization of the bacterial communities of life stages of free living lone star ticks (Amblyomma americanum. PLoS One 9: e102130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yang Z, Lu B, Ma X, Zhang C, Xu H. (2014). The composition and transmission of microbiome in hard tick, Ixodes persulcatus, during blood meal. Ticks Tick Borne Dis 5: 864–870. [DOI] [PubMed] [Google Scholar]
- Zolnik CP, Prill RJ, Falco RC, Daniels TJ, Kolokontronis SO. (2016). Microbiome changes through ontogeny of a tick pathogen vector. Mol Ecol 25: 4963–4977. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.