Abstract
Symbiodinium, the dinoflagellate photosymbiont of corals, is posited to become more susceptible to viral infections when heat-stressed. To investigate this hypothesis, we mined transcriptome data of a thermosensitive and a thermotolerant type C1 Symbiodinium population at ambient (27 °C) and elevated (32°C) temperatures. We uncovered hundreds of transcripts from nucleocytoplasmic large double-stranded DNA viruses (NCLDVs) and the genome of a novel positive-sense single-stranded RNA virus (+ssRNAV). In the transcriptome of the thermosensitive population only, +ssRNAV transcripts had remarkable expression levels in the top 0.03% of all transcripts at 27 °C, but at 32 °C, expression levels of +ssRNAV transcripts decreased, while expression levels of anti-viral transcripts increased. In both transcriptomes, expression of NCLDV transcripts increased at 32 °C, but thermal induction of NCLDV transcripts involved in DNA manipulation was restricted to the thermosensitive population. Our findings reveal that viruses infecting Symbiodinium are affected by heat stress and may contribute to Symbiodinium thermal sensitivity.
Tropical reef-building corals form a multipartite symbiosis with the photosynthetic dinoflagellate Symbiodinium and a diverse microbial community that includes bacteria, archaea, fungi, protists and viruses; collectively termed the coral holobiont (Rohwer et al., 2002). The Symbiodinium–coral symbiosis can be disrupted by heat stress, which results in the loss of Symbiodinium cells from coral tissues, i.e., coral bleaching (Hoegh-Guldberg, 1999). Heat stress has been alleged to promote lytic viral infections of Symbiodinium, as virus-like particles have been found in heat-stressed Symbiodinium from the temperate sea anemone Anemonia viridis (Wilson et al., 2001) and from the corals Pavona danai, Acropora formosa and Stylophora pistillata (Wilson et al., 2005; Davy et al., 2006). Additionally, following heat shock at 31 °C, the expression of a protein with homology to a eukaryotic viral protein increased >100-fold in a Symbiodinium-enriched fraction of Stylophora pistillata tissue (Weston et al., 2012).
Using transcriptome data generated by Levin et al. (2016) for two Symbiodinium type C1 populations cultured at 27 °C and 32 °C (n=4), we explore the effect of heat stress on viruses associated with Symbiodinium and Symbiodinium anti-viral responses. The Symbiodinium populations were originally isolated from the coral Acropora tenuis at South Molle Island (SM) and Magnetic Island (MI) (Great Barrier Reef, Australia). The thermosensitive SM population was found to suffer physiological damage in culture and to bleach in hospite at 32 °C, whereas the thermotolerant MI population was unaffected (Howells et al., 2012; Levin et al., 2016).
As many viral RNAs are polyadenylated (Wilson et al., 2000; Priet et al., 2015), they were retained in the poly(A)+ purified Symbiodinium RNA samples used for RNA-Seq (Levin et al., 2016). Viral transcripts in each de novo transcriptome were identified through a robust BLASTx bit score approach adapted from Boschetti et al. (2012), which calculates the difference between the highest viral and the highest non-viral bit score to determine if a transcript is from a virus or the host, followed by GC content, trinucleotide frequency and codon usage analyses (Supplementary Materials and Methods, Supplementary Figure 1 and Supplementary Datasets 1 and 2). Symbiodinium anti-viral transcripts were identified by searching the transcriptomes for transcripts encoding anti-viral gene types found in corals and for transcripts with anti-viral Gene Ontology (Supplementary Materials and Methods).
The thermosensitive SM and thermotolerant MI transcriptomes contain 306 and 238 viral transcripts (Supplementary Discussion), as well as 62 and 65 anti-viral transcripts, respectively (NCBI GEO accession: GSE77911). In both transcriptomes, viral transcripts show homology to genes from NCLDVs Mimiviridae and Phycodnaviridae and the major capsid protein (MCP) gene from the dinoflagellate-specific +ssRNAV Alvernaviridae (Dinornavirus), which is in agreement with previous sequencing and transmission electron microscopy findings (Wilson et al., 2005; Correa et al., 2013, 2016). The +ssRNAV transcripts in the thermosensitive SM transcriptome share 94% nucleotide (nt) identity (TR74740|c13_g1_i1, 5202 nt; TR74740|c13_g1_i2, 2154 nt). The +ssRNAV transcript in the thermotolerant MI transcriptome is much shorter (TR97578|c0_g1_i1, 475 nt) but shares 100% nt identity with TR74740|c13_g1_i1. Successful PCR amplification of the MCP genes from complementary DNA reverse-transcribed from RNA—but not from genomic DNA—of both Symbiodinium populations supports that they are from +ssRNAVs (Supplementary Materials and Methods and Supplementary Figure 2). However, phylogenetic analysis of the translated MCP gene sequences revealed that they are highly divergent from the Dinornavirus MCP gene and previously identified partial-length Dinornavirus-like MCP genes (Supplementary Discussion, Supplementary Table 1 and Supplementary Figure 3).
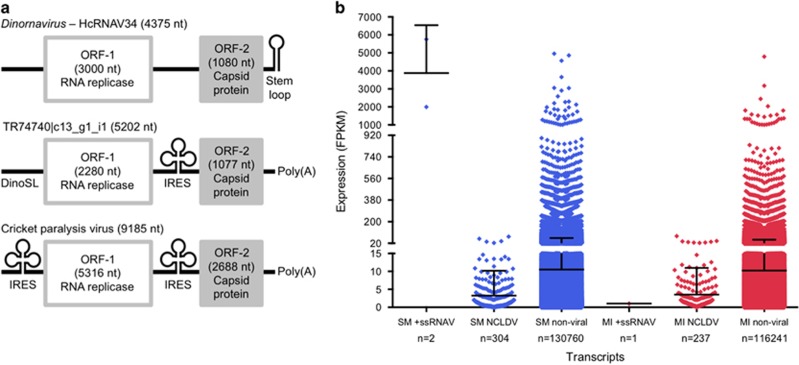
Both +ssRNAV transcripts in the thermosensitive SM transcriptome contain a putative viral internal ribosomal entry site (Supplementary Figure 4), which is related to the internal ribosomal entry site of the +ssRNA cricket paralysis virus, directly upstream from the MCP gene. The longer +ssRNAV transcript, TR74740|c13_g1_i1, also encodes an unannotated open reading frame (ORF) determined to be a +ssRNAV RNA replicase (RNA-dependent RNA-polymerase) polyprotein based on protein structure modeling (Supplementary Figure 5). The RNA replicase gene precedes the internal ribosomal entry site and MCP gene, giving the full transcript a markedly similar arrangement to the Dinornavirus and cricket paralysis virus complete genomes (Wilson et al., 2000; Nagasaki et al., 2005). Thus, we conclude TR74740|c13_g1_i1 to be the RNA genome of a novel +ssRNAV, making this the first discovered genome of any virus infecting Symbiodinium (Figure 1a). Surprisingly, the conserved dinoflagellate spliced leader, which is present on >95% of Symbiodinium mRNAs (Zhang et al., 2013), is at the 5′ end of TR74740|c13_g1_i1. Although, if the +ssRNAV is dinoflagellate-specific like Dinornavirus, incorporation of the dinoflagellate spliced leader in the viral RNA genome is likely a case of molecular mimicry, a well-documented viral strategy to evade host immune responses that detect foreign nucleic acids (Elde and Malik, 2009), or for efficient cap-dependent translation by host polysomes (Zeiner et al., 2003).
Figure 1.
Genome of the novel +ssRNAV and its expression in Symbiodinium transcriptomes. (a) Genome model of the +ssRNAV infecting Symbiodinium (TR74740|c13_g1_i1, GenBank accession: KX538960) and its similarities to the RNA genomes of the dinoflagellate-specific Dinornavirus (Heterocapsa circularisquama RNA virus strain 34, NCBI accession: AB218608) and cricket paralysis virus (NCBI accession: AF218039). (b) TMM-normalized FPKM at 27 °C (averaged across replicates for each transcript on day −1, n=8) for the +ssRNAV transcripts (TR74740|c13_g1_i1, 5757 FPKM; TR74740|c13_g1_i2, 1996 FPKM; TR97578|c0_g1_i1, 1 FPKM), NCLDV transcripts and non-viral transcripts in the thermosensitive SM and thermotolerant MI transcriptomes. Black lines mark the mean FPKM+s.d. for each subset of transcripts.
Viral transcripts had similar expression levels to many non-viral transcripts at 27 °C, with the exception of the two +ssRNAV transcripts in the thermosensitive SM transcriptome (Figure 1b). Both +ssRNAV transcripts in the thermosensitive SM transcriptome maintained average expression levels >1300 fragments per kb of transcript per million mapped reads (FPKM) on all sampling time points at 27 °C, whereas the +ssRNAV transcript in the thermotolerant MI transcriptome had an average expression level <2 FPKM on all sampling time points. The vastly dissimilar expression levels of +ssRNAV transcripts between the transcriptomes suggest that the thermosensitive SM Symbiodinium population was experiencing a severe viral infection.
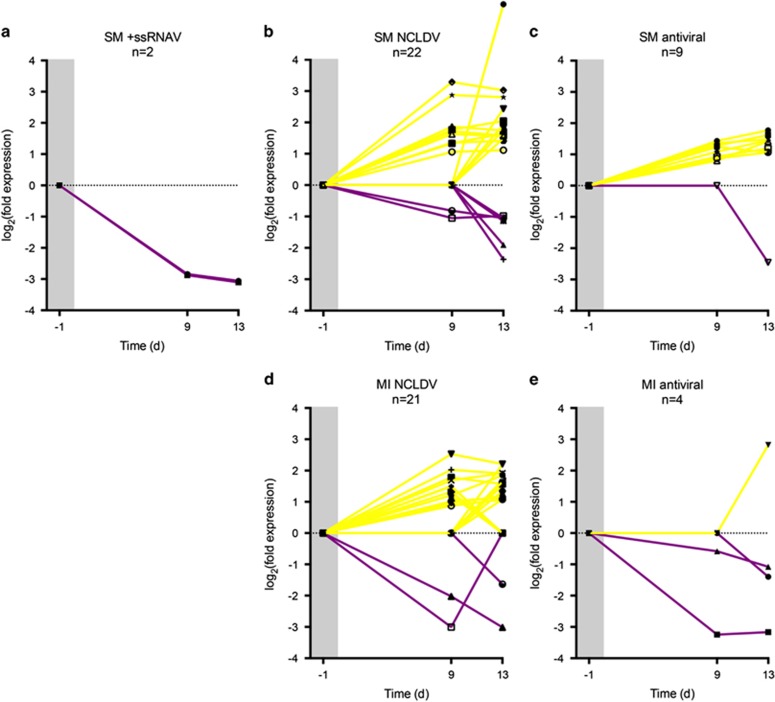
Differential expression analysis (Supplementary Materials and Methods) confirmed that no viral or anti-viral transcripts were differentially expressed between experimental groups of either population on day −1 (preheating; all samples acclimated at 27 °C) (Figures 2a–e). In both transcriptomes, upregulation of NCLDV transcripts was induced at 32 °C and increased from day 9 to day 13 (Figures 2b and d, and Supplementary Tables 2–9). However, the upregulated NCLDV transcripts in the thermosensitive SM transcriptome encoded for a greater diversity of genes (F-box and FNIP repeat-containing proteins, resolvase, transposase, ankyrin repeat protein) compared to those in the thermotolerant MI transcriptome (only F-box and FNIP repeat-containing proteins). Viral F-box and FNIP repeat-containing proteins and ankyrin repeat proteins have possible roles in degrading proteins through protein–protein interactions, countering host defences and exploiting the host's ubiquitin-proteasome system to create an appropriate cellular environment for viral replication (Suhre, 2005; Sonnberg et al., 2008; Fischer et al., 2010), whereas resolvases and transposases directly participate in DNA manipulation (Iyer et al., 2001; Schroeder et al., 2009).
Figure 2.
Viral infections and anti-viral responses of Symbiodinium under heat stress. Differential expression of (a) thermosensitive SM transcriptome +ssRNAV transcripts, (b) thermosensitive SM transcriptome NCLDV transcripts, (c) thermosensitive SM transcriptome anti-viral transcripts, (d) thermotolerant MI transcriptome NCLDV transcripts and (e) thermotolerant MI transcriptome anti-viral transcripts at 32 °C. Only transcripts with ⩾2-fold expression between 27 °C and 32 °C treatments on at least one sampling time point were analyzed. Transcripts were considered to have no differential expression on a sampling time point where the false discovery rate (FDR) was >0.001. Upregulated transcripts at 32 °C are shown in yellow. Downregulated transcripts at 32 °C are shown in purple. The gray regions represent the preheating sampling time point on day −1 when all replicates (n=8) were still at 27 °C. On day 0, replicates were ramped to 32 °C (n=4) or maintained at 27 °C (n=4) for the duration of the study (Supplementary Materials and Methods). Differential expression and transcript annotation results are detailed in Supplementary Tables 2–9.
In the thermosensitive SM transcriptome, heat stress resulted in downregulation of the highly expressed +ssRNAV transcripts and upregulation of anti-viral transcripts (Figures 2a and c and Supplementary Tables 2–3 and 6–7). Conversely, the thermotolerant MI transcriptome showed no differential expression of its lowly expressed +ssRNAV transcript and downregulation of anti-viral transcripts at 32 °C (Figure 2e, Supplementary Tables 8–9). Symbiodinium anti-viral responses may therefore become activated from increased DNA manipulation by NCLDVs or initial upregulation of highly expressed +ssRNAV transcripts at 32 °C that was mitigated by the sampling time points on days 9 and 13.
Our study exemplifies how RNA-Seq data can be used to gain valuable insight into resident viruses. Our results indicate that only the thermosensitive SM Symbiodinium population experienced an extreme +ssRNAV infection and thermally induced NCLDV DNA manipulation. Thus, viral infections may factor into Symbiodinium thermal sensitivity, and consequently, coral bleaching.
Acknowledgments
The Australian Institute of Marine Science supplied the Symbiodinium strains (aims-aten-C1-WSY, aims-aten-C1-MI) used in this study. Yi Jin Liew provided assistance with the bit score analysis. Rhys T Graham provided technical support for MATLAB. The Centre for Marine Bio-Innovation at the University of New South Wales, King Abddullah University of Science and Technology (KAUST), The Joyce W Vickery Scientific Research Fund Grant awarded to Rachel A Levin from the Linnean Society of New South Wales, and Future Fellowship No FT100100088 awarded to Madeleine JH van Oppen from the Australian Research Council contributed financial support. Raw, processed and annotated sequencing data are available through NCBI GEO (accession: GSE77911). The novel +ssRNAV genome (TR74740|c13_g1_i1) and highly related, partial +ssRNAV genome (TR74740|c13_g1_i2) discovered in this study have been deposited in GenBank (accession nos: KX538960 and KX787934, poly(A) tails were trimmed from RNA-Seq reads prior to de novo assembly of transcripts).
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Boschetti C, Carr A, Crisp A, Eyres I, Wang-Koh Y, Lubzens E et al. (2012). Biochemical diversification through foreign gene expression in Bdelloid Rotifers. PLoS Genet 8: e1003035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa AM, Ainsworth TD, Rosales SM, Thurber AR, Butler CR, Thurber RLV. (2016). Viral outbreak in corals associated with an in situ bleaching event: atypical herpes-like viruses and a new megavirus infecting Symbiodinium. Front Microbiol 7: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa AM, Welsh RM, Thurber RLV. (2013). Unique nucleocytoplasmic dsDNA and +ssRNA viruses are associated with the dinoflagellate endosymbionts of corals. ISME J 7: 13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davy S, Burchett S, Dale A, Davies P, Davy J, Muncke C et al. (2006). Viruses: agents of coral disease? Dis Aquat Organ 69: 101–110. [DOI] [PubMed] [Google Scholar]
- Elde NC, Malik HS. (2009). The evolutionary conundrum of pathogen mimicry. Nat Rev Microbiol 7: 787–797. [DOI] [PubMed] [Google Scholar]
- Fischer MG, Allen MJ, Wilson WH, Suttle CA. (2010). Giant virus with a remarkable complement of genes infects marine zooplankton. Proc Natl Acad Sci USA 107: 19508–19513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoegh-Guldberg O. (1999). Climate change, coral bleaching and the future of the world's coral reefs. Mar Freshwater Res 50: 839–866. [Google Scholar]
- Howells EJ, Beltran VH, Larsen NW, Bay LK, Willis BL, van Oppen MJH. (2012). Coral thermal tolerance shaped by local adaptation of photosymbionts. Nat Clim Change 2: 116–120. [Google Scholar]
- Iyer LM, Aravind L, Koonin EV. (2001). Common origin of four diverse families of large eukaryotic DNA viruses. J Virol 75: 11720–11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin RA, Beltran VH, Hill R, Kjelleberg S, McDougald D, Steinberg PD et al. (2016). Sex, scavengers, and chaperones: transcriptome secrets of divergent Symbiodinium thermal tolerances. Mol Biol Evol 33: 2201–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki K, Shirai Y, Takao Y, Mizumoto H, Nishida K, Tomaru Y. (2005). Comparison of genome sequences of single-stranded RNA viruses infecting the bivalve-killing dinoflagellate Heterocapsa circularisquama. Appl Environ Microbiol 71: 8888–8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priet S, Lartigue A, Debart F, Claverie J-M, Abergel C. (2015). mRNA maturation in giant viruses: variation on a theme. Nucleic Acids Res 7: 3776–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwer F, Seguritan V, Azam F, Knowlton N. (2002). Diversity and distribution of coral-associated bacteria. Mar Ecol Prog Ser 243: 1–10. [Google Scholar]
- Schroeder DC, Park Y, Yoon H-M, Lee YS, Kang SW, Meints RH et al. (2009). Genomic analysis of the smallest giant virus—Feldmannia sp. virus 158. Virology 384: 223–232. [DOI] [PubMed] [Google Scholar]
- Sonnberg S, Seet BT, Pawson T, Fleming SB, Mercer AA. (2008). Poxvirus ankyrin repeat proteins are a unique class of F-box proteins that associate with cellular SCF1 ubiquitin ligase complexes. Proc Natl Acad Sci USA 105: 10955–10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhre K. (2005). Gene and genome duplication in Acanthamoeba polyphaga Mimivirus. J Virol 79: 14095–14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston AJ, Dunlap WC, Shick JM, Klueter A, Iglic K, Vukelic A et al. (2012). A profile of an endosymbiont-enriched fraction of the coral Stylophora pistillata reveals proteins relevant to microbial–host interactions. Mol Cell Proteomics 11: M111.015487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JE, Powell MJ, Hoover SE, Sarnow P. (2000). Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol Cell Biol 20: 4990–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson W, Dale A, Davy J, Davy S. (2005). An enemy within? Observations of virus-like particles in reef corals. Coral Reefs 24: 145–148. [Google Scholar]
- Wilson WH, Francis I, Ryan K, Davy SK. (2001). Temperature induction of viruses in symbiotic dinoflagellates. Aquat Microb Ecol 25: 99–102. [Google Scholar]
- Zeiner GM, Sturm NR, Campbell DA. (2003). The Leishmania tarentolae spliced leader contains determinants for association with polysomes. J Biol Chem 278: 38269–38275. [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhuang Y, Gill J, Lin S. (2013). Proof that dinoflagellate spliced leader (DinoSL) is a useful hook for fishing dinoflagellate transcripts from mixed microbial samples: Symbiodinium kawagutii as a case study. Protist 164: 510–527. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.