Abstract
We report development of the first genetically encoded bioluminescent indicator for membrane voltage called LOTUS-V. Since it is bioluminescent, imaging LOTUS-V does not require external light illumination. This allows bidirectional optogenetic control of cellular activity triggered by Channelrhodopsin2 and Halorhodopsin during voltage imaging. The other advantage of LOTUS-V is the robustness of a signal-to-background ratio (SBR) wherever it expressed, even in the specimens where autofluorescence from environment severely interferes fluorescence imaging. Through imaging of moving cardiomyocyte aggregates, we demonstrated the advantages of LOTUS-V in long-term imaging are attributable to the absence of phototoxicity, and photobleaching in bioluminescent imaging, combined with the ratiometric aspect of LOTUS-V design. Collectively LOTUS-V extends the scope of excitable cell control and simultaneous voltage phenotyping, which should enable applications in bioscience, medicine and pharmacology previously not possible.
Voltage imaging is a powerful method to observe the spatiotemporal regulation of cellular excitability1,2,3,4,5,6. Recently, various genetically encoded voltage indicators (GEVIs) have been developed5,7,8,9,10,11,12,13,14,15,16, which are advantageous for reproducible labelling of a target specimen, as well as offering compatibility with genetic control systems that target specific cell types with or without inducible expression6,17. However, strong autofluorescence from naturally existing chromophores, such as NADPH and flavin, reduces the signal-to-background ratio (SBR) of green or yellow fluorescent GEVIs (fGEVIs) in some cases, e.g. in vivo voltage imaging. In addition, excitation light irradiation causes photobleaching and phototoxicity, which often hamper long-term observation by fluorescence imaging.
Optogenetics, which enables millisecond-order fast and reversible manipulation (activation or silencing) of neural and brain function, has revolutionised contemporary neuroscience. “All-optical electrophysiology”, a combinational technique in which a voltage indicator is used with an optogenetic actuator19,20,21has been proposed5,12,15. However, all-optical electrophysiological studies are possible only with carefully selected partners with distinct spectral properties, for example simultaneous use of a near-infrared archaerhodopsin-based fluorescent GEVI (fGEVI) with a blue-light gated channelrhodopsin actuator5. The choice of partners in this approach is critical in order to prevent unintended activation of the optogenetic actuator with the excitation light required for fluorescence imaging. Incorporating additional complexity, for example using multiple optical perturbations (i.e. both activation and silencing together) during simultaneous voltage imaging remain challenging because of the unavoidable overlap between the fGEVI excitation spectrum and the composite absorption spectrum of multiple optogenetic actuators20.
To overcome these issues, we have focussed on “bioluminescent proteins”, which produce light chemically by catalyzing the oxidative reaction of substrates enabling the observation of specimens without any of the external illumination apparatus required for fluorescence imaging22. We previously developed the “Nano-lantern”22, which consists of an improved Renilla luciferase (RLuc8-S257G) fused to the yellow fluorescent protein Venus23. This allows enhancement of the bioluminescence intensity by Förster resonance energy transfer (FRET) between RLuc8-S257G and Venus. As a result, Nano-lantern emits ten times more signal than RLuc, making it possible to perform continuous detection of cancer cells in a freely moving mouse22.
Nano-lantern-based “intensiometric” indicators for biological elements including Ca2+ and ATP were also developed, and importantly, Nano-lantern (Ca2+) enabled Ca2+ imaging with simultaneous and independently-regulated optogenetic stimulation22,24. In line with this trend, we initially intended to expand the application of bioluminescent indicators to voltage imaging according to the development strategy of Nano-lantern-based “intensiometric” indicators. However, one problem with this approach is that the bioluminescence intensity is affected by the local concentration change of the bioluminescent substrate. This particularly occurs during long-term imaging as substrate addition is required to preserve bioluminescence intensity. This suggests that the approach would struggle to detect membrane voltage dynamics, due to local substrate consumption, in short or long term observation.
Thus, we attempted to develop a “bioluminescent” and “ratiometric” indicator, enabling voltage imaging free from excitation light and mitigating artefact regarding a local concentration change of the bioluminescent substrates by ratio processing. Here, we describe indicator design, functional characterization, and application to long-term voltage imaging with bidirectional optical control and an in-vitro cardiomyocyte model.
Results
Design and optimization of a FRET-based voltage sensor
To develop a ratiometric bioluminescent GEVI (bGEVI), we followed the paradigm of the fGEVIs such as the voltage sensitive fluorescent protein (VSFP) BF1.2 (ref. 10) and Mermaid2 (ref. 11). These fGEVIs are composed of a voltage-sensing domain (VSD) fused with two different fluorescent proteins functioning as a FRET pair donor and acceptor. Voltage-dependent structural change in the VSD alters the FRET efficiency between the two fluorescent proteins. We utilized the VSD from a voltage-sensing phosphatase of Ciona intestinalis with the R217Q mutation (Fig. 1a)25. As a donor, we used the bioluminescent protein NanoLuc26, which produces approximately 150-times more signal than RLuc. Following the conventional strategy for developing FRET indicators, we chose mNeonGreen27 and Venus23as FRET acceptor candidates since their absorption spectra highly overlaps with the NanoLuc emission spectrum.
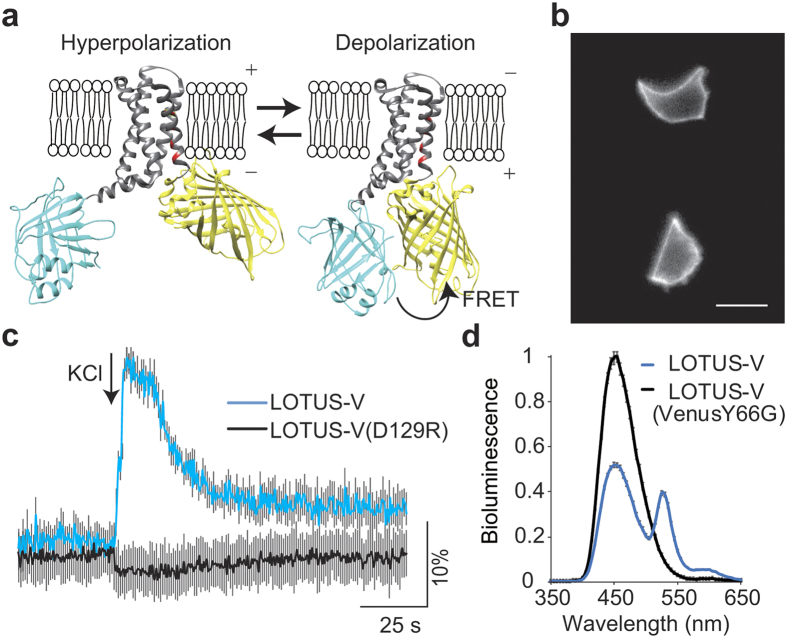
Figure 1. Development and characterization of LOTUS-V.
(a) Molecular design of LOTUS-V (gray, VSD; blue, NanoLuc; yellow, Venus; red, positively charged amino acid). The 3D structure of VSD (4G7V) and Venus (GFP; 1KYS) were obtained from the Protein Data Bank (PDB), and that of NanoLuc was predicted by I-TASSER44. Each 3D structure was arranged by UCSF Chimera 1.10.2. (b) Representative bioluminescent image of GH3 cells expressing LOTUS-V. Scale bar, 20 μm. (c) Time course of ratio change (ΔR/R0) in LOTUS-V (blue) and LOTUS-V (D129R) (black) upon 150 mM KCl stimulation. (d) Bioluminescence spectra of LOTUS-V and LOTUS-V(VenusY66G) in GH3 cells (n = 10). Gray bars indicate mean ± s.e.m. at each wavelength.
To preserve voltage sensitivity, and maximise signal, we tested multiple combinations of insertion points of NanoLuc or the acceptor within the VSD(R217Q) (Supplementary Fig. 1). Each construct was then transfected into rat pituitary epithelial-like tumor (GH3) cells (Fig. 1b), and the change in FRET signal in response to KCl-induced voltage change (depolarization) was measured to determine the largest dynamic range (Supplementary Fig. 1).
The construct in which NanoLuc was inserted between residues 103 and 104 of VSD(R217Q) and Venus was fused at the C-terminus of the VSD(R217Q) exhibited the highest FRET signal change with small deviation (ΔR/R0 = 22.6 ± 0.9% [mean ± s.e.m.], n = 5 cells) (Fig. 1c). We designated it as LOTUS-V (Luminescent Optical Tool for Universal Sensing of Voltage). The D129R mutant of the VSD(R217Q) was previously shown to be insensitive to voltage changes in physiological range28. We found that a corresponding mutation in VSD(R217Q) of LOTUS-V completely abolished the FRET change upon KCl stimulation in GH3 cells (ΔR/R0 = −2.3 ± 2.4% [mean ± s.e.m.], n = 5 cells, p = 0.0090), suggesting that the signal shown by LOTUS-V reflects a membrane voltage change (Fig. 1c).
Subsequently, we compared the bioluminescence spectrum of LOTUS-V with that of LOTUS-V(VenusY66G) lacking a chromophore of its Venus moiety (Fig. 1d)11. The donor emission peak of LOTUS-V(VenusY66G) at 455 nm, derived from direct NanoLuc bioluminescence significantly recovered in the absence of a functional FRET acceptor, suggesting FRET occurs between the NanoLuc and Venus moieties in LOTUS-V.
For further applications of LOTUS-V, we investigated whether furimazine could cause cellular toxicity using a HEK293T cell viability assay (Supplementary Fig. 2). Many dead cells were observed 12 h after continuous excitation light irradiation (22 and 100% for 210 and 830 mW/cm2, respectively), paralleling power densities in the typical range for fluorescent voltage recording16,29. Fewer dead cells were seen by weak excitation light power (52 mW/cm2) or by the treatment with 50 μM furimazine (0 and 5%, respectively). Simultaneously we identified that cell proliferation seemed relatively suppressed by 50 μM furimazine, however it is still less harmful than the excitation light for typical voltage recording.
Voltage sensitivity and kinetics of LOTUS-V
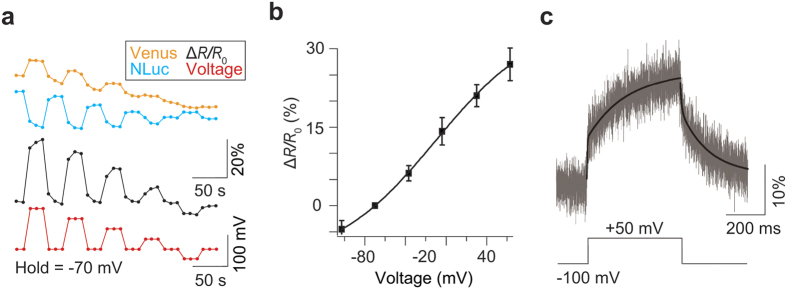
To further characterize the properties of LOTUS-V quantitatively, we performed simultaneous patch-clamp recording and photometry in HEK293T cells. The emission intensity of Venus and that of NanoLuc reciprocally changed depending on the amplitude of the applied voltage (Fig. 2a). Compared with previously reported fluorescent and FRET-based GEVIs10,11, the voltage dependency of LOTUS-V was less steep with applied voltage (Mermaid2, the value of the effective valence Z = 1.05; LOTUS-V, Z = 0.5) (equation (1)), and the ratio change for a 100-mV voltage step (from −70 to + 30 mV) was 21.0 ± 0.9% (mean ± s.e.m., n = 5 cells) (Fig. 2b). Importantly, its voltage sensitivity covered the physiological range, suggesting compatibility with various biological phenomena. Since the signal emitted from a single HEK293T cell was too weak to accurately calculate kinetics, we expressed LOTUS-V in Xenopus oocytes and conducted kinetics analysis by two-component exponential curve fitting, after data acquisition at 5 kHz with off-line time averaging of 32 sweeps and Bessel filtering at 1 kHz (Fig. 2c). Time constants of the fast component (τfast) were 3.09 ms (fraction, 37.3%) for the activation curve and 6.12 ms (fraction, 29.4%) for the deactivation curve, while time constants of the slow component (τslow) were 204 ms and 144 ms, respectively, at +50 mV from a holding potential (−100 mV). The time constant of LOTUS-V was comparable to the widely used fGEVI, ArclightQ239 (refs 13 and 14). In summary, the signal from LOTUS-V was free from external illumination and its voltage sensitivity covered the physiological range. These traits suggest its practical usefulness in further applications.
Figure 2. Electrophysiological characterization of LOTUS-V.
(a) Representative optical responses of the Venus (yellow) and NanoLuc (blue) signals (ΔL/L0), and ΔR/R0 (black) in response to stepwise voltage changes (+63, +30, +4, −37 and −107 mV, respectively) from a holding voltage (−70 mV; red) in a HEK293T cell. (b) Plot of the fractional ΔR/R0 versus membrane voltage. Error bars indicate mean ± s.d. The curved line indicates a Boltzmann fit. (c) The ΔL/L0 of Venus in response to voltage changes (+50 mV) from the holding voltage (−100 mV) in Xenopus oocytes (n = 6 cells). The black curved line indicates the result of two-component exponential curve fitting.
Voltage imaging with optogenetic actuators
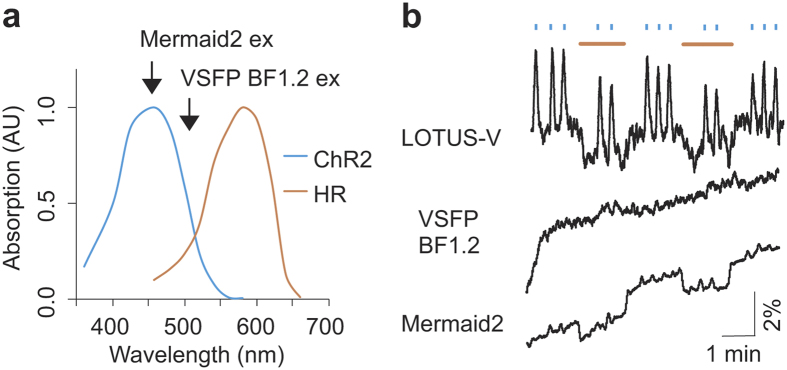
Perturbation of cellular function using optogenetic actuators can be compromised by the excitation light used for simultaneous fluorescence imaging. Therefore, the applicable combinations of optogenetic actuators and fluorescent indicators are limited. For instance, the depolarizing and hyperpolarizing optical control tools ChR2(H134R)30 and eNpHR3.0 (ref. 20) have absorption spectra ranging from 350–600 nm and 425–650 nm, respectively, giving a total spectral range of 350–650 nm for bidirectional optical control (Fig. 3a). Therefore, simultaneous voltage imaging with the current fGEVIs, including the QuasAr series5of near-infrared fGEVIs, is not possible without some effect on optogenetic actuators expressed in the same cells. In the case of bGEVIs, on the other hand, since they do not require illumination, and the emitted bioluminescence intensity (ca. 5 μW/cm2 in the case of NanoLuc)22,26is more than 10,000 times lower than the light sensitivity EC50 for activating the optogenetic actuators (ca. 105 mW/cm2for ChR2)31, suggesting that bioluminescence-driven activation of optogenetic actuators is negligible and versatile optical perturbation during LOTUS-V imaging becomes possible.
Figure 3. Voltage imaging with optogenetic actuators.
(a) Overlay of the absorption spectra of ChR2(H134R) and eNpHR3.0. Arrows show the excitation peak of VSFP BF1.2 and Mermaid2. (b) Optical response of LOTUS-V, VSFP BF1.2 and Mermaid2 in PC12 cells coexpressing eNpHR3.0 and ChR2(H134R) upon blue (438 nm, 25.5 mW/cm2) and orange (580 nm, 47 mW/cm2) light irradiation. The excitation light for VSFP BF1.2 (500 nm, 1.14 mW/cm2) and Mermaid2 (438 nm, 2.28 mW/cm2) was applied from the bottom of the imaging dish. Blue and orange bars indicate the durations of blue and orange light irradiation, respectively. The ΔR/R0 processed by the moving average (window length of 50 frames) is shown.
To validate the compatibility of live cell imaging using LOTUS-V with optical perturbation, we performed voltage imaging in pheochromocytoma (PC12) cells co-expressing LOTUS-V, ChR2(H134R) and eNpHR3.0 (Supplementary Fig. 3). For ChR2(H134R) and eNpHR3.0 activation, the wavelength and power density of blue (438 nm, 25.5 mW/cm2) and orange (580 nm, 47 mW/cm2) stimulation lights were carefully adjusted not to wrongly activate the other in line with a previous report using same optogenetic actuators20. Blue light pulses caused a rapid increase in the emission ratio of LOTUS-V, due to membrane depolarization by activated ChR2(H134R) (ΔR/R0 = 2.9 ± 0.1% [mean ± s.e.m.], n = 12 sessions). In contrast, orange light pulses caused a rapid decrease in the ratio (ΔR/R0 = −1.5 ± 0.2% [mean ± s.e.m.], n = 3 sessions) due to membrane hyperpolarization by the activation of eNpHR3.0, in the same cell. Co-irradiation with blue light during orange light pulses caused an increase in the ratio, with an attenuated maximum value compared to that obtained with blue light alone (ΔR/R0 = 2.1 ± 0.1% [mean ± s.e.m.], n = 6 sessions) (Fig. 3b and Supplementary Fig. 4a). In the absence of ChR2(H134R) and eNpHR3.0, we didn’t observe such signal changes, and thus those correctly reflected the voltage changes caused by the activity of optogenetic actuators (Supplementary Fig. 4b). These results reproduced bidirectional modification of membrane voltage electrophysiologically confirmed in hippocampal pyramidal neurons20, revealing reliability of our method. In contrast, we also performed same experiment using various fGEVIs, including QuasAr2. When we used ratiometic fGEVIs, VSFP BF1.2 and Mermaid2 for voltage imaging, upward drift of ΔR/R0 baseline was observed (0.72%/min and 0.59%/min for VSFP BF1.2 and Mermaid2, respectively; in contrast, only 0.03%/min for LOTUS-V) (Fig. 3b), which was due to photobleaching of the FRET donor caused by the continuous application of intense excitation light. For further quantitative evaluation, we corrected the baseline ratio of the Mermaid2 signal by single exponential curve fitting10. When Mermaid2 was used with ChR2(H134R) and eNpHR3.0, the emission ratio decreased less efficiently upon irradiation with orange light (ΔR/R0 = −1.0 ± 0.1% [mean ± s.e.m.], n = 3 sessions) and no obvious increase in the ratio was detected with blue light irradiation (ΔR/R0 = 0.4 ± 0.1% [mean ± s.e.m.], n = 12 sessions) (Fig. 3b). Since Mermaid2 has a larger dynamic range than LOTUS-V (48.5% ± 3.5%/100 mV vs 21.0 ± 0.9%/100 mV), it was a surprise to see that although hyperpolarization was identified as well as LOTUS-V, depolarization caused by ChR2(H134R) activation11 was harder to observe. This significant difference between LOTUS-V and Mermaid2 upon blue light irradiation (p = 3.2 × 10−5, n = 12 sessions) was due to constitutive activation of ChR2(H134R), which resulted from spectral overlap (Fig. 3a). In contrast, a green fGEVI (ArclightQ239) and red-shifted fGEVIs (FlicR1.0 and QuasAr2), which were shown to sense electrophysiologically manipulated hyperpolarization5,12,13, detected only depolarization, indicating that excitation light for these fGEVIs constitutively activated eNpHR3.0 for the same reason (Supplementary Fig. 4c–e)5,12,13. VSFP BF1.2 was not sensitive enough to detect attenuated depolarization since ArclightQ239 which was excited by shorter excitation light (Arclight, 472 nm, 1.01 mW/cm2; VSFP BF1.2, 500 nm, 1.14 mW/cm2) was able to sense it (Fig. 3b and Supplementary Fig. 4c). These results show that LOTUS-V offers improved compatibility with optogenetic actuators across a broad absorbance spectrum, and reduced upward baseline drift that would be problematic in long-term imaging.
Voltage imaging in hiPSC-CMs
Next, to prove the utility of LOTUS-V in long-term imaging and the compatibility with excitable cell types that move we chose an in-vitro cardiomyocyte model. Recently, cardiomyocytes derived from human induced pluripotent stem cells (hiPSC-CMs) have gained attention as a powerful tool for personalized drug screening in vitro. Several optical techniques have been tested for analyzing drug effects on cardiomyocytes’ beating frequency and action potential duration (APD). These include the use of chemical indicators (for Ca2+ and voltage) and fGEVIs17,18,32,33. However, techniques that rely on fluorescent excitation are hampered by photobleaching and phototoxicity, and thus long-term imaging was difficult17,18. Also, in moving specimens, motion artefact is a major concern for quantitative measurements using intensiometric indicators. Thus, the use of bioluminescent and ratiometric indicators should overcome such problems as indicated above.
To test the applicability of LOTUS-V for this purpose, we expressed it in an aggregate of hiPSC-CMs via lentivirus infection and conducted voltage imaging during contraction. As we expected, LOTUS-V showed a higher SBR than ArclightQ239 (5.23 ± 1.39 and 0.31 ± 0.09, respectively; n = 5 aggregates, p = 0.036) since bioluminescence imaging is unaffected by environmental autofluorescence, in contrast to fluorescence imaging where SBR of fGEVIs is reduced especially at low expression levels (Supplementary Fig. 5b).
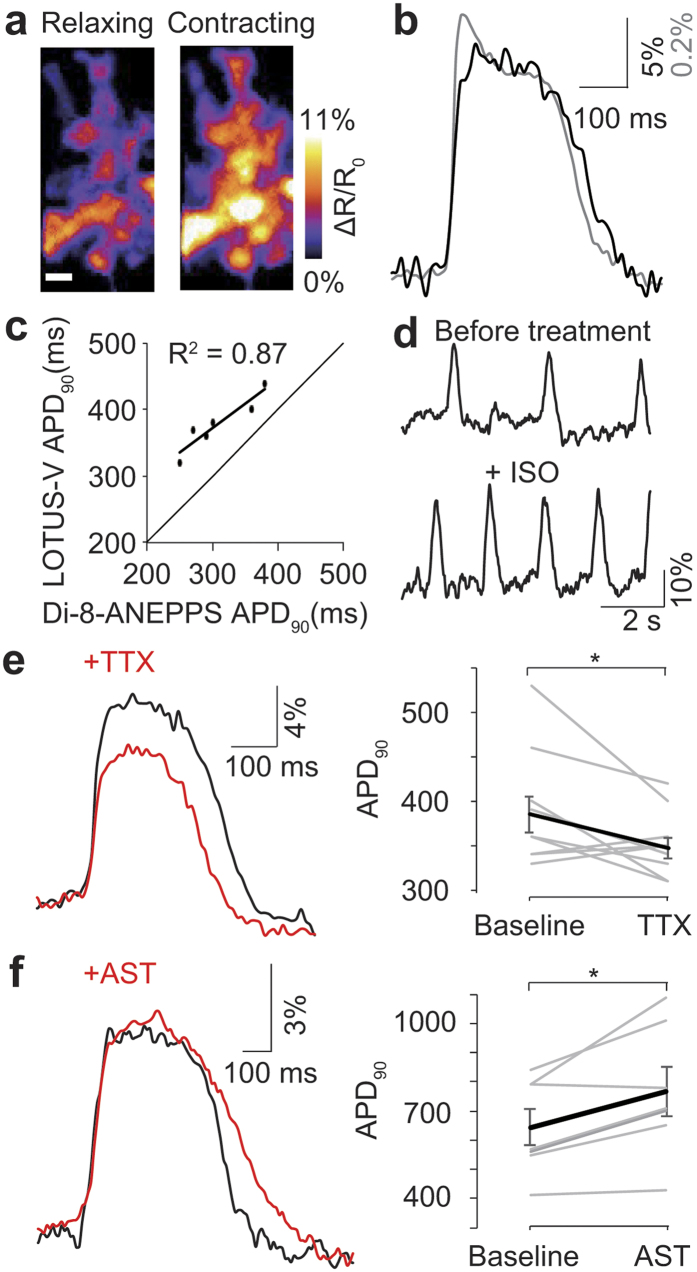
During synchronized contraction, the emission ratio (ΔR/R0) increases, reflecting the action potential of cardiomyocytes (Fig. 4a,b and Supplementary Video 1). We compared this to Di-8-ANEPPS, a commonly used membrane voltage chemical dye (Fig. 4b)17,18. Although some high frequency components were lost, overall the signal of LOTUS-V closely resembled the action potential morphology reported by Di-8-ANEPPS. APD90 measured by bioluminescence using LOTUS-V was linearly correlated with that of fluorescence recording using Di-8-ANEPPS (R2 = 0.87) (Fig. 4c).
Figure 4. Voltage imaging in hiPSC-CMs.
(a) Ratio images of hiPSC-CMs expressing LOTUS-V before and during contraction (n = 23). Scale bar, 50 μm. (b) Bioluminescence recording (black) overlaid on a fluorescence recording using Di-8-ANEPPS (gray) during hiPSC-CM contraction (n = 38). The power density of excitation light for Di-8-ANEPPS was 735 mW/cm2. (c) APD90 measured by LOTUS-V plotted against APD90 measured by Di-8-ANEPPS. The line fitted by least squares method is shown in black. (d) Time course of the ΔR/R0 emission ratio before and after treatment with 100 nM isoproterenol. (e,f) Evaluation of action potential morphology before and after treatment with 30 μM TTX (n = 17) (e), and 10 μM astemizole (n = 12 and 8, respectively) (f). The graphs indicate individual APD90 changes upon drug treatment. Wilcoxon signed-rank test was performed. *p < 0.05; Error bars indicate mean ± s.e.m.
Next we tested whether LOTUS-V could distinguish electrophysiological changes in hiPSC-CMs induced by various chemicals. Since LOTUS-V is compatible with long-term imaging, and hiPSC-CMs behave heterogeneously34, we compared the same cell populations before and after drug addition to minimize the impact of variability in the cells themselves. After addition of isoproterenol (ISO), a non-selective β-adrenergic agonist often used to treat bradycardia, the frequency of spikes increased as expected (Fig. 4d)35. Also tetrodotoxin (TTX), a sodium channel blocker, changed action potential morphology, reducing peak amplitude, and shortening duration (ΔR/R0, 13.2 ± 0.6% falls to 10.9 ± 0.4% following TTX; APD90, 387 ± 20 ms reduced to 350 ± 11 ms [mean ± s.e.m.] following TTX; p = 5.1 × 10−3 and 0.028, respectively; n = 10, Two-tailed Wilcoxon signed-rank test) (Fig. 4e)36. Finally, astemizole (AST), a hERG channel blocker, prolonged APD90 (APD90, 644 ± 61 ms to 767 ± 84 ms [mean ± s.e.m.]; p = 0.019; n = 7, Two-tailed Wilcoxon signed-rank test) (Fig. 4f)37. These results suggested that LOTUS-V is able to appraise drug effect in hiPSC-CMs, based on not only action potential frequency but also its morphological change induced by chemical addition.
It is known that the bioluminescence produced by NanoLuc decays with a half-life over 2 h26. Typically recording using LOTUS-V is done over 30 min while SBR is preserved. Longer studies require additional furimazine. To test utility in long-term imaging studies we perfused the aggregates with medium supplemented with the intact bioluminescent substrate, furimazine, and compared the result with various fGEVIs (Fig. 5 and Supplementary Fig. 6). The upstroke peak amplitude of LOTUS-V did not change significantly over more than 1 h ((ΔR/R0)0h, 7.61 ± 0.17%; (ΔR/R0)1h, 6.82 ± 0.27% [mean ± s.e.m.]; p = 0.42; decreasing rate, 10.3%; n = 5 sessions) (Fig. 5a), though the signal from VSFP BF1.2 and Mermaid2 was severely diminished following long-term imaging ((ΔR/R0)0h, 7.01 ± 0.20% and 2.07 ± 0.20%; (ΔR/R0)1h, 3.58 ± 0.73% and 0.59 ± 0.08% [mean ± s.e.m.]; p = 0.012 and 0.008; decreasing rate, 48.9% and 78.3%, respectively; n = 5 sessions) (Fig. 5b and c). Actually, the spikes in the emission ratio of Mermaid2 were difficult to distinguish even at 0 h because of strong autofluorescence in YFP channel. Although ArclightQ239 kept largely unchanged detectability of an action potential for 1 hour ((ΔF/F0)0h, −6.74 ± 0.06%; (ΔF/F0)1h, −6.62 ± 0.08% [mean ± s.e.m.]; p = 0.31; decreasing rate, 1.7%; n = 5 sessions) (Supplementary Fig. 6), there were clear regional differences in spike morphology within an aggregate at any given time (Supplementary Fig. 7b). Considering that local fluorescence intensity of EGFP alone also changed upon contraction falling or rising depending on direction of motion (Supplementary Fig. 7a), some of the differences in spike morphology of ArclightQ239 result from motion artefact, and also regional expression differences. Therefore, signal from the whole aggregate is made of inexact regional signals. In contrast, the spike morphology of LOTUS-V was almost identical irrespective of position within the sample. No ratio changes of the voltage insensitive mutant, LOTUS-V(D129R) were observed, suggesting that the signal of LOTUS-V truly reflected a membrane voltage change and was free from motion and expression artefacts (Supplementary Fig. 7c and d).
Figure 5. Long-term recording in hiPSC-CMs.
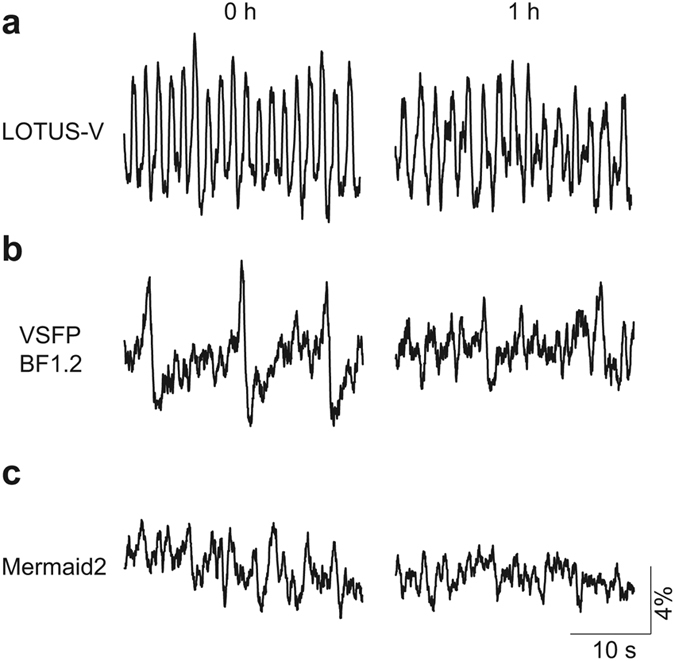
(a–c) Time course of the ΔR/R0 of LOTUS-V (a), VSFP BF1.2 (b), and Mermaid2 (c) at 0 h or 1 h. Images were taken at 30 ms/frame, and the medium was continuously replaced to provide fresh frimazine. Excitation light for VSFP BF1.2 (500 nm, 44.9 mW/cm2) and Mermaid2 (438 nm, 34.8 mW/cm2) was applied continuously for 1 h. The ratio change processed by the moving average (window length of 20 frames) is shown in all graphs.
Collectively we have shown that LOTUS-V enables not only long-term imaging, an important parameter for chronic pharmacological toxicity testing, but also high contrast imaging in samples where the autofluorescence from environment significantly lowers the SBR of fGEVIs. Furthermore, LOTUS-V offers quantitative measurement without concerns for motion artefact in moving specimens.
Discussion
Here, we propose and demonstrate utility of a new technique for membrane voltage imaging that does not require excitation light, using LOTUS-V. Compared to conventional fGEVIs, although the SBR of LOTUS-V in single cell is limited due to low signal intensity, nevertheless bioluminescence imaging using LOTUS-V has multiple advantages over conventional fluorescence imaging. Firstly, voltage imaging with LOTUS-V enables concomitant bidirectional control with paired optogenetic actuators due to the absence of external illumination requirements for imaging. This compatibility of LOTUS-V with multiple optogenetic actuators is highly beneficial for analyzing the electrical and physiological properties of cells, because it enables the advanced spatiotemporal manipulation of arbitrary targets without concerns about misactivation of the optogenetic actuators. Second, LOTUS-V enables imaging free from photobleaching and phototoxicity, which are problems for fluorescence imaging. This allows long-term monitoring of membrane voltage by continuously supplying bioluminescent substrate. This should be useful for research requiring prolonged imaging (such as drug evaluation) or light sensitive processes (e.g. circadian rhythm). Furthermore, measurements in the hiPSC-CMs aggregates by LOTUS-V showed a superior SBR to that by fGEVIs. This suggests sensitive measurements with extremely low background noise are possible even in specimens exhibiting strong autofluorescence and low expression level of the indicator.
In this study, we used various excitable cell types to demonstrate the applicability of bioluminescence recording with LOTUS-V. Quantitative investigations of indicator performance were performed in HEK293T as they lack other ion currents. Indicator performance may change depending on the cellular context in which it is used. For example, Arch(D95N) expressed in neurons showed almost same voltage dependency as that seen in HEK293T cells9,15. However although the voltage dependency of the Arclight variant, taken in HEK293T was preserved in human embryonic stem cell-derived cardiomyocytes, a lower signal amplitude was apparent (44% reduction in ΔF/F0 per 100 mV)13,17. Therefore, it is possible that although the voltage dependency of LOTUS-V might be preserved in neuronal and hiPSC-CMs, signal amplitude may change. Where quantitative data is required, calibration of LOTUS-V emissions in the cell type of interest may be needed. Further improvement, especially in response speed to voltage change, will improve detection of membrane voltage. This might be achieved by introducing mutations in the VSD or employing a rhodopsin-based VSD.
Another conceivable advantage of bioluminescent proteins is high serviceability in ex and in vivo experiments because of negligible background noise from the tissue22,38. Thus, LOTUS-V might allow optical mapping of the whole heart39, which could be combined with the techniques described in this study, such as bidirectional optical manipulation and long-term observation using perfusion systems. Since LOTUS-V is genetically encoded it may be possible to restrict it to distinct cell populations within the heart expanding the possibility of optical recording with voltage indicators and contribute to the understanding of complicated biological systems like the heart.
Methods
Gene construction
The cDNAs for the C-terminus of VSD(R217Q) were amplified by PCR from pCS4 + -Mermaid2, using sense primers containing an EcoRI site and a reverse primer containing a XhoI site. To create constructs that had a fluorescent protein following the C-terminus of VSD(R217Q), we amplified the cDNAs of mNeonGreen (Allele Biotechnology), Venus, or circularly permuted Venus by PCR using sense primers containing a XhoI site and reverse primers containing a NotI site and a stop codon. These fragments were subcloned into pcDNA3 (Invitrogen; pcDNA3-VSD_C-FPs). The cDNA for the N-terminus of VSD(R217Q) was amplified by PCR using a sense primer containing a Kozak sequence following a HindIII site and reverse primers containing a BamHI site. The cDNA for NanoLuc (Promega) was amplified by PCR using a sense primer containing a BamHI site and a reverse primer containing an EcoRI site. These fragments were subcloned into pcDNA3-VSD_C-FP to generate the cDNA for a FRET-based voltage indicator containing NanoLuc and a fluorescent protein at the N-terminus and C-terminus, respectively. Constructs with NanoLuc at the C-terminus were also created following the same procedure. The Y66G and D129R mutations inside Venus and VSD(R217Q) respectively were introduced by PCR mutagenesis using a sense primer40.
For oocyte experiments, the cDNAs of LOTUS-V (∆EcoRI) were amplified by PCR using sense primers containing a Kozak sequence following an EcoRI site and reverse primers containing a XbaI site and a stop codon. The cDNA for LOTUS-V (∆EcoRI) was then ligated between the EcoRI and XbaI site of pCS4+ -Mermaid2.
For the equal expression of ChR2(H134R) and eNpHR3.0, each synthesized oligonucleotide annealed to the cDNA for P2A peptide41 was inserted between the EcoRI and NotI site of pcDNA3. The cDNA for ChR2(H134R) (Addgene: Plasmid #20945) was also amplified by PCR using a sense primer containing a NotI site and a reverse primer containing a XbaI site and a stop codon, then subcloned into pcDNA3-P2A downstream of P2A. Subsequently, the cDNA for eNpHR3.0 (Addgene: Plasmid#26966) was amplified by PCR using a sense primer containing a HindIII site and a Kozak sequence and a reverse primer containing an EcoRI site, then each of them was subcloned into pcDNA3-P2A-ChR2(H134R) upstream of P2A.
For the lentivirus expression system, CS-CDF-LOTUS-V-PRE was created from CS-CDF-EG-PRE by replacing EGFP with LOTUS-V (ΔXhoI). At the same time, an AgeI site following the EF-1 promoter was replaced with a BamHI site. The point mutations described above were introduced by PCR mutagenesis using a sense primer following the previously described protocol40.
All of the constructs were verified by DNA sequencing and the primers are listed in Supplementary Table 1.
Characterization of bGEVIs with KCl stimulation
GH3 cells (ATCC, CCL-82.1) were cultured in Dulbecco’s Modified Eagle’s Medium: Nutrient Mixture F-12 Ham (DMEM/F12) (Invitrogen) containing heat-inactivated 15% horse serum and 2.5% fetal bovine serum at 37 °C in air with 5% CO2. One day before imaging, the cells were mounted in homemade 35-mm glass-bottom dishes coated with 0.1 mg/ml poly-D-lysine (Sigma), and transfected using Lipofectamine 2000 (Invitrogen), following the manufacturer’s protocol.
We performed the screening using an Eclipse Ti-E inverted microscope with a 40×, NA 1.30, Plan Fluor oil-immersion objective (Nikon). Bioluminescence was observed by adding 50 μM furimazine. The light from bioluminescent specimens was separated by W-VIEW GEMINI A12801-01 (Hamamatsu) equipped with a FF509-FDi01-25 × 36 dichroic mirror (Semrock) and passed through FF01-483/32-25 or FF01-525/45-25 emission filters (Semrock) for the bioluminescence from NanoLuc or fluorescent proteins, respectively. Bioluminescence was recorded by an iXon Ultra EMCCD camera (Andor Technology) with MetaMorph software (Molecular Devices), and ΔR/R0 was calculated after adding 20 μl of 150 mM KCl.
Bioluminescence spectral measurement
LOTUS-V or LOTUS-V(VenusY66G)-expressing GH3 cells cultured on plastic dishes were detached from the dish using a scraper, and suspended in DMEM (Invitrogen). The suspension was transferred to 96-well plates (1.2 × 105 cells/well), and furimazine was added to a final concentration of 5 μM. The bioluminescence spectrum was measured by a SH-9000 microplate reader (Colona Electric).
Electrophysiology and photometry
LOTUS-V-expressing HEK293T cells (RIKEN BRC Cell Bank, RCB2202) were subjected to simultaneous patch-clamp and FRET recordings at 23–30 hours post-transfection. The recording setup consisted of an IX71 inverted microscope (Olympus), RC-41LP coverslip chamber (Warner Instruments), QE-1RC temperature controller (Warner Instruments), and Axopatch 200B patch clamp amplifier (Axon Instruments). HEPES-buffered saline (15 mM HEPES, pH 7.4, containing 120 mM NaCl, 2.5 mM KCl, 1.5 mM MgSO4, 2.0 mM CaCl2, 26 mM NaHCO3, 1.1 mM NaH2PO4, and 10 mM dextrose) was used as a bath solution. The pipette solution (10 mM HEPES, pH 7.3 [adjusted with methanesulfonic acid]) contained 5 mM NaCl, 10 mM KCl, 10 mM HEPES, 130 mM KOH, 2.5 mM MgATP, 0.3 mM Na2GTP, and 1 mM EGTA. Bioluminescence was observed by adding 50 μM furimazine and recordings were performed at 30 °C. Images were acquired sequentially using an OrcaFlash 4.0 sCMOS camera (Hamamatsu).
LOTUS-V was expressed in Xenopus oocytes using a previously described protocol42 and bioluminescence was observed by adding 50 μM furimazine. Simultaneous two-electrode voltage-clamp and photometry was carried out using an OC-725C voltage-clamp amplifier (Warner Instruments) and IX70 inverted microscope (Olympus). The resistance of the intracellular glass microelectrodes filled with 2.5 M KCl ranged from 0.1 to 3.0 MΩ. The Venus signal was collected with a 20×, NA 0.70 UPlanApo objective lens (Olympus) focused onto an H5784-02 photomultiplier tube (Hamamatsu). The output of the optical signal was digitized and stored using a 1322AA/D converter (Axon Instruments) and pClamp8 software (Axon Instruments).
Voltage imaging with optogenetic actuators
PC12 cells (ATCC, CRL-1721) were grown in Dulbecco’s Modified Eagle’s Medium (Sigma) containing heat-inactivated 10% HS and 5% FBS at 37 °C in air with 5% CO2. Three days before imaging, the cells were mounted in homemade 35-mm glass-bottomed dishes coated with 0.04% polyethyleneimine and transfected using Lipofectamine 2000 (Invitrogen). After 4-6 hours of incubation, the medium was replaced with new medium containing 100 ng/ml nerve growth factor (Promega) to induce neuron-like differentiation. Images were taken with the same microscope set-up as shown in the section above, “Characterization of bGEVIs with KCl stimulation”.
To activate ChR2(H134R) and eNpHR3.0 during voltage imaging, stimulating light from a LightEngine SPECTRA (Lumencor) was applied within the dead time of the charge-coupled device camera, as we reported previously (Supplementary Fig. 2)43. Briefly, the exposure time-out signals from the EMCCD camera were used as the trigger for a WF1973 multifunction generator (NF Corporation) to generate the pulsed signals for turning on the stimulation light. The entire illumination duration was modulated by a function of the multifunction generator under the control of LabView (National Instruments). Camera binning was set at 16. Light was passed through a 438/24 nm filter or a 580/27 nm filter from above the culture dish to activate ChR2(H134R) and eNpHR3.0 (25.5 mW/cm2 and 47 mW/cm2, respectively). Irradiation from above the dish was applied by replacing the halogen lamp for the transmitted light source with the LightEngine liquid light guide, connected using a homemade adaptor.
Lentivirus production
HEK293T cells were grown in Dulbecco’s Modified Eagle’s Medium (Sigma) containing heat-inactivated 10% FBS at 37 °C in 5% CO2. Equal amounts of CS-CDF-LOTUS-V-PRE, pCAG-HIVgp, and pCMV-VSV-G-RSV-Rev were transfected by FuGENE HD transfection reagent (Roche), following the manufacturer’s protocol. Two or three days after transfection, the viruses were harvested and concentrated using a Lenti-X concentrator (Takara), following the manufacturer’s protocol. The virus titer was approximately 1.0 × 107 Infectious units (IFU)/ml, as evaluated by Lenti-X GoStix (Takara).
hiPSC-CM culture and imaging
ReproCardio2 (ReproCell) was purchased, and aggregates of hiPSC-CMs were prepared following the manufacturer’s protocol with some modifications. Briefly, the cells were mounted on 96-well round-bottomed plates for the aggregation process with lentivirus (1.0 × 105 IFU/ml) on day 0, and aggregates were transferred to 96-well flat-bottomed plates coated with ReproCoat on day 3. The cells were incubated at 37 °C, 5% CO2, and half of the culture medium was replaced with ReproCardio Culture Medium 2 containing 20% FBS and 1% Penicillin-Streptomycin on days 2 and 4–10.
Imaging was conducted on days 8–10 using an Eclipse Ti-E inverted microscope with a 20×, NA 0.7, Plan Fluor objective (Nikon), and the bioluminescence of LOTUS-V was generated by adding 50 μM furimazine. Light from the bioluminescent specimens was separated by W-VIEW GEMINI A12801-01 (Hamamatsu) equipped with a FF509-Di01-25 × 36 dichroic mirror (Semrock) and passed thorough no emission filters. Bioluminescence was recorded by an iXon3 EMCCD camera (Andor Technology) with camera binning 16 for 20–30 ms/frame acquisition with MetaMorph software (Molecular Devices). During the entire imaging period, the temperature was kept at 37 °C by an iNUG2 stage top incubator (Tokai Hit). Fresh 50 μM furimazine was provided by an MP-1000 peristaltic pump (EYELA) for long-term observation.
Data analysis
The ratio value was calculated by dividing Venus intensity by NanoLuc intensity. ∆R/R0 was calculated by subtracting the averaged basal value of ratio (R0) from individual raw ratio values at each time point and further dividing the difference by the R0. The following Boltzmann function was used for fitting R-V relationships in HEK293T cells:
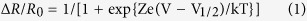 |
where e was the elementary electric charge; Z was the effective valence; k was the Boltzmann constant; T was the room temperature in kelvin; V1/2 was the voltage at which ΔR/R0 is half-activated. To analyze the voltage kinetics of LOTUS-V in Xenopus oocytes, we fitted the activation and deactivation curves of the signal from Venus to a two-component exponential equation using the nonlinear curve-fitting function in Origin 8.5.1 (OriginLab). The following equation was used:
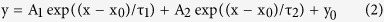 |
where y was the signal intensity at time x−x0; x0 and y0 were the initial time point and intensity, respectively; A1 and A2 were the fraction of each exponential function, representing fast and slow components; and τ1 and τ2 were time constants for the fast and slow components. We also corrected upward baseline drift of Mermaid2 signal (segments before and after optical stimulations) by a single exponential equation using the nonlinear curve-fitting function in Origin 8.5.1 for voltage imaging with optogenetic actuators.
For imaging in hiPSC-CMs, the following equation was used to calculate the decreasing rate of the ∆R/R0 from 0 h to 1 h:
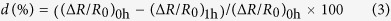 |
where d was the decreasing rate in ∆R/R0; (ΔR/R0)0h and (ΔR/R0)1h were ∆R/R0 at 0 h and 1 h, respectively. APD90 was calculated by measuring time interval at 90% repolarization.
All of the data were processed by Fiji and R-3.3.0 software. Two-tailed Wilcoxon rank sum test was performed using R-3.3.0 software for statistical analysis, unless otherwise noted.
Additional Information
Accession codes: GenBank/EMBL/DDBJ: the LOTUS-V nucleotide sequence has been deposited under the accession code LC061443.
How to cite this article: Inagaki, S. et al. Genetically encoded bioluminescent voltage indicator for multi-purpose use in wide range of bioimaging. Sci. Rep. 7, 42398; doi: 10.1038/srep42398 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by a Grant-in-aid for Scientific Research on Innovative Areas, ‘Spying minority in biological phenomena (No. 3306)’, from the Ministry of Education, Culture, Sports, Science and Technology, Japan (No. 23115003, No. 23115001), by the JST-SENTAN program, and by JSPS Core-to-Core Program, A. Advanced Research Networks to T.N.; S.I. is supported by “Program for Leading Graduate Schools” of the Ministry of Education, Culture, Sports, Science and Technology, Japan, and Grant-in-Aid for JSPS Research Fellow (16J00111). M.J.D. is supported by the Wellcome Trust, ref WT WT098519MA, and a Royal Society International Joint project grant. We thank Thomas Knöphel (Department of Medicine, Imperial college of London) for giving us pCAG-VSFP BF1.2, Robert E. Campbell (Department of Chemistry, University of Alberta) for giving us pcDNA3-FlicR1.0 and Promega Corporation for providing us the cDNA of NanoLuc. For the lentivirus expression system, CS-CDF-EG-PRE, pCAG-HIVgp, and pCMV-VSV-G-RSV-Rev developed by Hiroyuki Miyoshi were obtained from the RIKEN Bioresource Center (BRC). We also thank the Bionanophotonics Consortium (BNPC) for assistance in experiments using microscopy.
Footnotes
The authors declare no competing financial interests.
Author Contributions T.N. conceived and coordinated the project. S.I., H.T., T.M., Y.O. and T.N. designed the experiments; S.I. constructed and characterized LOTUS-V, and performed live cell imaging with support of T.M. and Y.A.; K.S. performed a cell viability assay.; Y.A. constructed the microscope system for dead time imaging; G.B. established the production system of lentivirus and performed the differentiation of iPS cells into cardiomyocytes with support of M.J.D.; H.T., Y.J., and Y.O. performed electrophysiological experiments; all authors analyzed data; S.I., M.A, T.M., and T.N. wrote the paper, with contributions from all authors. Data described can be found in the main figures and Supplementary materials.
References
- Tasaki I., Watanabe A., Sandlin R. & Carnay L. Changes in fluorescence, turbidity, and birefringence associated with nerve excitation. Proc. Natl. Acad. Sci. USA 61, 883–888 (1968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre F., Chavarha M. & Lin M. Z. Designs and sensing mechanisms of genetically encoded fluorescent voltage indicators. Curr. Opin. Chem. Biol. 27, 31–38 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterka D. S., Takahashi H. & Yuste R. Imaging Voltage in Neurons. Neuron. 69, 9–21 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohajerani M. H. et al. Spontaneous cortical activity alternates between motifs defined by regional axonal projections. Nat. Neurosci. 16, 1426–1435 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochbaum D. R. et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat. Methods. 11, 825–833 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L. et al. Transgenic Mice for Intersectional Targeting of Neural Sensors and Effectors with High Specificity and Performance. Neuron. 85, 942–958 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel M. S. & Isacoff E. Y. A Genetically Encoded Optical Probe of Membrane Voltage. Neuron. 19, 735–741 (1997). [DOI] [PubMed] [Google Scholar]
- Dimitrov D. et al. Engineering and characterization of an enhanced fluorescent protein voltage sensor. PLoS One. 2, e440 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralj J., Douglass A., Hochbaum D., Maclaurin D. & Cohen A. Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nat. Methods. 9, 90–95 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akemann W. et al. Imaging neural circuit dynamics with a voltage-sensitive fluorescent protein. J. Neurophysiol. 108, 2323–2337 (2012). [DOI] [PubMed] [Google Scholar]
- Tsutsui H. et al. Improved detection of electrical activity with a voltage probe based on a voltage-sensing phosphatase. J. Physiol. 591, 4427–4437 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelfattah A. S. et al. A Bright and Fast Red Fluorescent Protein Voltage Indicator That Reports Neuronal Activity in Organotypic Brain Slices. J. Neurosci. 36, 2458–2472 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L. et al. Single action potentials and subthreshold electrical events imaged in neurons with a fluorescent protein voltage probe. Neuron. 75, 779–785 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou P. et al. Bright and fast multicoloured voltage reporters via electrochromic FRET. Nat. Commun. 5, 4625 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y., Li J. Z. & Schnitzer M. J. Enhanced Archaerhodopsin Fluorescent Protein Voltage Indicators. PLoS One. 8, e66959 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y. et al. High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. Science. 350, 1361–1366 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyton-Mange J. S. et al. Rapid cellular phenotyping of human pluripotent stem cell-derived cardiomyocytes using a genetically encoded fluorescent voltage sensor. Stem Cell Reports. 2, 163–170 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnawi R. et al. Monitoring Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes with Genetically Encoded Calcium and Voltage Fluorescent Reporters. Stem Cell Reports. 5, 582–596 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden E. S., Zhang F., Bamberg E., Nagel G. & Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 8, 1263–1268 (2005). [DOI] [PubMed] [Google Scholar]
- Gradinaru V. et al. Molecular and Cellular Approaches for Diversifying and Extending Optogenetics. Cell. 141, 154–165 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X. et al. A high-light sensitivity optical neural silencer: development and application to optogenetic control of non-human primate cortex. Front. Syst. Neurosci. 5, 18 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K. et al. Luminescent proteins for high-speed single-cell and whole-body imaging. Nat. Commun. 3, 1262 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T. et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20, 87–90 (2002). [DOI] [PubMed] [Google Scholar]
- Takai A. et al. Expanded palette of Nano-lanterns for real-time multicolor luminescence imaging. Proc. Natl. Acad. Sci. USA 112, 4352–4356 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y., Iwasaki H., Sasaki M., Inaba K. & Okamura Y. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 435, 1239–1243 (2005). [DOI] [PubMed] [Google Scholar]
- Hall M. P. et al. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS. Chem. Biol. 7, 1848–1857 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N. C. et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods. 10, 407–409 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui H., Jinno Y., Tomita A. & Okamura Y. Optically detected structural change in the N-terminal region of the voltage-sensor domain. Biophys. J. 105, 108–15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G. et al. Genetically targeted optical electrophysiology in intact neural circuits. Cell. 154, 904–913 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulver S. R., Pashkovski S. L., Hornstein N. J., Garrity P. a. & Griffith L. C. Temporal dynamics of neuronal activation by Channelrhodopsin-2 and TRPA1 determine behavioral output in Drosophila larvae. J. Neurophysiol. 101, 3075–3088 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. Y. A user’s guide to channelrhodopsin variants: features, limitations and future developments. Exp. Physiol. 96, 19–25 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P. et al. Simultaneous voltage and calcium mapping of genetically purified human induced pluripotent stem cell-derived cardiac myocyte monolayers. Circ. Res. 110, 1556–1563 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Liao M. L. et al. Sensing Cardiac Electrical Activity With a Cardiac Myocyte–Targeted Optogenetic Voltage IndicatorNovelty and Significance. Circ. Res. 117, 401–412 (2015). [DOI] [PubMed] [Google Scholar]
- Doss M. X. et al. Maximum diastolic potential of human induced pluripotent stem cell-derived cardiomyocytes depends critically on I(Kr). PLoS One. 7, e40288 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel Y. et al. Human embryonic and induced pluripotent stem cell-derived cardiomyocytes exhibit beat rate variability and power-law behavior. Circulation. 125, 883–893 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel O. et al. Action potential characterization of human induced pluripotent stem cell-derived cardiomyocytes using automated patch-clamp technology. Assay. Drug. Dev. Technol. 12, 457–469 (2014). [DOI] [PubMed] [Google Scholar]
- Gibson J. K. et al. Human stem cell-derived cardiomyocytes detect drug-mediated changes in action potentials and ion currents. J. Pharmacol. Toxicol. Methods. 70, 255–267 (2014). [DOI] [PubMed] [Google Scholar]
- Hamada T. et al. In vivo imaging of clock gene expression in multiple tissues of freely moving mice. Nat. Commun. 7, 11705 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attin M. & Clusin W. T. Basic concepts of optical mapping techniques in cardiac electrophysiology. Biol. Res. Nurs. 11, 195–207 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawano A. & Miyawaki A. Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res. 28, E78 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H. et al. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One. 6, e18556 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldin A. L. Maintenance of Xenopus laevis and oocyte injection. Methods Enzymol. 207, 266–279 (1992). [DOI] [PubMed] [Google Scholar]
- Chang Y. F., Arai Y. & Nagai T. Optogenetic activation during detector ‘dead time’ enables compatible real-time fluorescence imaging. Neurosci. Res. 73, 341–347 (2012). [DOI] [PubMed] [Google Scholar]
- Roy A., Kucukural A. & Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.