Abstract
To characterize the gene expression profile of regenerated melanocytes in the narrow band UVB (NBUVB)-treated vitiligo epidermis and their precursors in the hair follicle, we present here a strategy of RNA isolation from in situ melanocytes using human frozen skin. We developed a rapid immunostaining protocol using the NKI-beteb antibody, which labels differentiated and precursor melanocytes, followed by fluorescent laser capture microdissection. This technique enabled the direct isolation, from melanocyte and adjacent keratinocyte populations, of satisfactory quality RNA that was successfully amplified and analysed by qRT-PCR. The melanocyte-specific gene transcripts TYR, DCT, TYRP1 and PMEL were significantly upregulated in our NBUVB-treated melanocyte samples as compared with the keratinocyte samples, while keratinocyte-specific genes (KRT5 and KRT14) were expressed significantly higher in the keratinocyte samples as compared with the melanocyte samples. Furthermore, in both NBUVB-treated vitiligo skin and normal skin, when bulge melanocytes were compared with epidermal melanocytes, we found significantly lower expression of melanocyte-specific genes and significantly higher expression of three melanocytic stem cell genes (SOX9, WIF1 and SFRP1), while ALCAM and ALDH1A1 transcripts did not show significant variation. We found significantly higher expression of melanocyte-specific genes in the epidermis of NBUVB-treated vitiligo, as compared to the normal skin. When comparing bulge melanocyte samples from untreated vitiligo, NBUVB-treated vitiligo and normal skin, we did not find significant differences in the expression of melanocyte-specific genes or melanocytic stem cell genes. These techniques offer valuable opportunities to study melanocytes and their precursors in vitiligo and other pigmentation disorders.
Keywords: laser capture microdissection, melanocyte, melanocyte stem cell, repigmentation, vitiligo
1 | INTRODUCTION
Vitiligo is the most common pigmentation disorder, characterized by the presence of disfiguring white spots on the skin, due to melanocyte loss, which causes social and psychological stigma among affected individuals.1,2 Progress has been made in identifying depigmentation pathways in vitiligo,3–5 although little is currently known about repigmentation pathways. No previous research strategy has characterized the precursor populations of melanocytes from the hair follicle of normal human and vitiligo skin, and no reports described the genetic signature of regenerated epidermal melanocytes in vitiligo. For a better characterization of these populations, we developed a practical human model using fluorescent immunostaining of skin biopsies from untreated and narrow band UVB (NBUVB)-treated vitiligo patients. These techniques enabled us to accurately localize melanocyte precursors and identify some of their functions.6 For a more detailed characterization of the melanocyte populations emerging in repigmenting vitiligo skin, we designed a protocol to localize melanocytes in human skin, harvest melanocyte and keratinocyte populations by laser capture and to obtain RNA from populations enriched in melanocyte and keratinocyte genes in the hair follicles and epidermis. This includes a novel application of an immunostaining technique, of short duration, which minimizes RNA degradation, and which labels the immature melanocytes in the hair follicle bulge and the mature epidermal melanocytes. The labelled melanocytes and adjacent keratinocytes from the bulge and epidermis are isolated by laser capture microdissection, and the RNA from these cells is collected. The quality of the RNA obtained from this protocol is sufficient for qRT-PCR and will be followed by whole-transcriptome RNA sequencing studies.
This approach allows accurate and detailed protein and gene expression studies of repigmentation in vitiligo skin following NBUVB treatment.
2 | MATERIALS AND METHODS
2.1 | Tissue sample collection and processing
Skin biopsies of depigmented, untreated skin from 6 vitiligo patients, and of repigmented skin from 10 vitiligo patients treated for 4 months with NBUVB were collected at the Dermatology Clinic at University of Colorado Hospital. Written, informed consent was obtained from all subjects, and the study was approved by Human Subjects Committee (COMIRB) at the University of Colorado. Normal human skin samples from 6 control subjects were either purchased from the National Disease Resource Center or obtained from the Skin Cancer Biorepository at the University of Colorado.
2.2 | Hair follicle bulge mapping
To locate the hair follicle bulge in all of our tissue samples, frozen transverse sections were immunostained with a combination of anti-K15 Lab Vision, Fremont, CA, USA and anti-desmin antibodies Dako, Carpinteria, CA, USA every 10th slide,7 as presented in Fig. S1.
2.3 | NKI-beteb antibody expression in normal human skin
For our rapid immunostaining procedure, we chose the NKI-beteb antibody (Cell Sciences, Neponset, MA, USA) which targets the premelanosomal and melanosomal melanocyte protein PMEL, identified in both mature melanocytes from the epidermis and immature melanocytes from the hair follicle outer root sheath.8 We first combined the NKI-beteb antibody with anti-K15 (that labels epithelial stem cells) and examined their co-expression in epidermis and hair follicle. Using standard immunostaining procedures, we then combined the NKI-beteb antibody with either anti-TYR or anti-DCT, as well as with an anti-K14 antibody (to label the surrounding keratinocytes), and examined the co-expression of these markers (Fig. S2). TYR is a marker of terminally differentiated melanocytes which is not expressed in the bulge,6 while DCT is a lineage marker expressed in both mature and immature melanocytes from the bulge and interfollicular epidermis.6
2.4 | Rapid fluorescent immunostaining for LCM
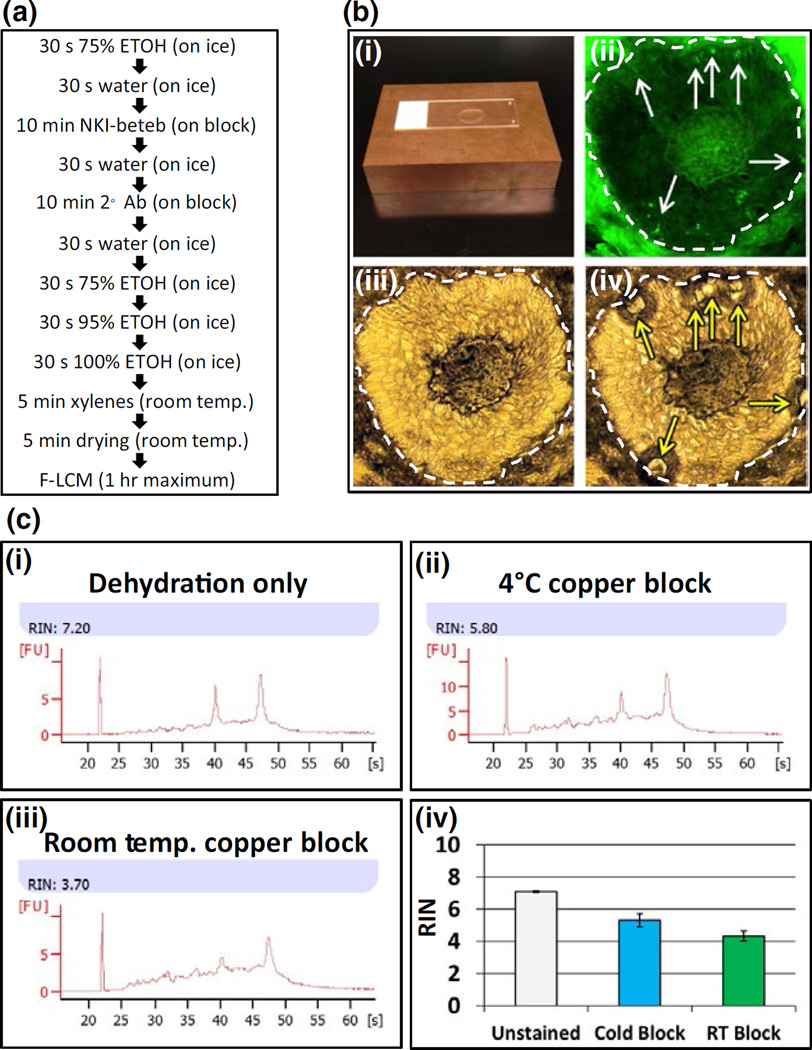
Frozen sections were immunostained using the mouse monoclonal antibody NKI-beteb (10-minute incubation). We used an Alexa Fluor® 488 goat anti-mouse IgG (H + L) secondary antibody and nuclease-free buffers from the HistoGene® LCM Frozen Section Staining Kit. To further prevent RNA degradation, we added GeneAmp® RNAse inhibitor to antibody dilutions in PBS. All materials mentioned in this section (except NKI-Beteb antibody) are from Thermo Fisher Scientific Waltham, MA, USA. The entire immunostaining procedure is presented in Fig. 1a and Supplementary Materials and Methods.
FIGURE 1.
Isolation of individual melanocytes (MCs) from epidermis and hair follicles. Panel a: Rapid immunostaining protocol to preserve RNA quality using NKI-beteb antibody. Panel b: The copper blocks used for rapid immunostaining at 4°C to preserve RNA integrity (b-i); NKI-beteb- positive MCs (fluorescent field, b-ii white arrows) laser captured after immunostaining. Bright field images show sample before (b-iii) and after (b-iv) laser capture indicating spots where tissue was captured (yellow arrows). Panels ii, iii and iv, scale bar: 50 µm. Panel c: RNA integrity of tissue scraped from slides using different staining methods. Slides stained using cold copper block (c-ii; c-iv, blue bar) showed less degradation than slides stained at room temperature (c-iii; c-iv, green bar); unstained slides showed the best RNA integrity (c-i; c-iv, white bar).
2.5 | Fluorescent laser capture microdissection
F-LCM was performed on slides within 1 hour of completing immunostaining. We used an infrared laser (Arcturus XTTM) microdissection system, under direct fluorescent microscopic visualization, in the LCM Core at the University of Colorado, using cold copper blocks9 and standard techniques detailed in the Supplementary Materials and Methods. From each sample, we captured 100 NKI-beteb- positive melanocytes from the epidermis, 100 NKI-beteb- positive cells from the hair follicle bulge (Fig. 1b) and 100 adjacent keratinocytes (NKI-beteb negative) from each of these two regions. Laser capture samples were enriched for target cells (as shown in Fig. S3), although most laser pulses could capture modest amounts of surrounding tissue. As illustrated in Fig. S3, the surrounding tissue consisted mainly of adjacent keratinocyte material when capturing melanocytes and adjacent melanocyte dendrites when capturing keratinocytes.
2.6 | RNA extraction, amplification and qRT-PCR
Total RNA was extracted from frozen LCM lysates using the ARCTURUS® PicoPure® RNA Isolation Kit Thermo Fisher Scientific Waltham, MA, USA. producing 1–5 ng of total RNA per sample of 100 cells.
Two different commercially available kits were used to amplify total RNA from captured melanocytes and keratinocytes (Supplementary Material). A total of 2 ng of cDNA produced from either amplification protocol was used for qRT-PCR using the QuantiTect SYBR® Green PCR Kit (Qiagen CA, USA) and primers designed by ourselves. Primer pairs are shown in Table S1. qRT-PCR was performed on a MX3000p real-time detection system using standard techniques. Analysis of RNA integrity is presented in Fig. 1c.
3 | RESULTS
3.1 | NKI-beteb antibody expression in human skin
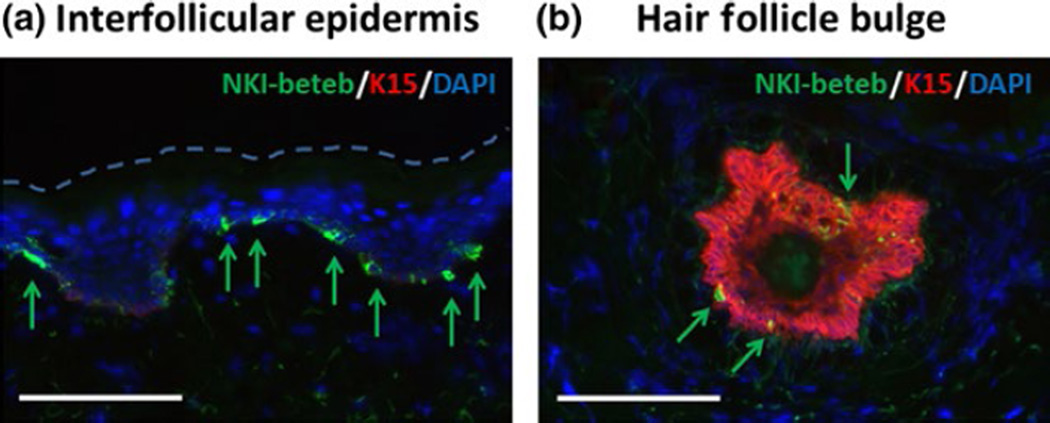
The NKI-beteb antibody detected the expression of PMEL protein in the interfollicular epidermis (Fig. 2a), infundibulum (not shown), bulge (Fig. 2b) and bulb (not shown). Further, the NKI-beteb antibody showed high concomitant localization with the anti-TYR antibody in the interfollicular epidermis (Fig. S2a), while NKI-beteb cells in the bulge were not TYR-positive (Fig. S2b). NKI-beteb antibody showed high concomitant localization with the anti-DCT antibody in both epidermis (Fig. S2c) and bulge (Fig. S2d). Moreover, our 30-minute rapid immunostaining procedure (Fig. 1a), implemented to label the melanocytes for F-LCM, produced strong NKI-beteb signal intensity (Fig. 1b).
FIGURE 2.
NKI-beteb expression in the epidermis and hair follicle bulge of normal skin. Double fluorescent immunostaining of frozen transverse sections using the NKI-beteb antibody (labels mature and immature MCs) in combination with anti-K15 (labels epithelial stem cells) in the interfollicular epidermis (a) and hair follicle bulge (b). White scale bars: 100 µm.
3.2 | RNA extraction and quality assessment
We compared the quality of RNA extracted from tissue scraped from slides subjected to two different rapid immunostaining conditions, using the cold (Fig. 1c) copper block at 4°C vs room temperature. We also evaluated the quality of RNA in the unstained tissue. Example RNA plots show intact 18s and 28s ribosomal subunit peaks. The slides stained on the cold copper block (Fig. 1c-ii,c-iv, blue bar) showed mild RNA degradation (n=3; average RIN=5.3), as compared to the unstained slides (Fig. 1c-i,c-iv, grey bar) (n=3; average RIN=7.1). The greatest RNA degradation was seen in slides stained at room temperature (Fig. 1c-iii,c-iv, green bar) (n=3; average RIN=4.3). Based on the above results, we decided to include the cold copper block for all subsequent staining procedures.
3.3 qRT-PCR analysis of amplified LCM samples
3.3.1 | Assessment of cell-type enrichment in the LCM samples
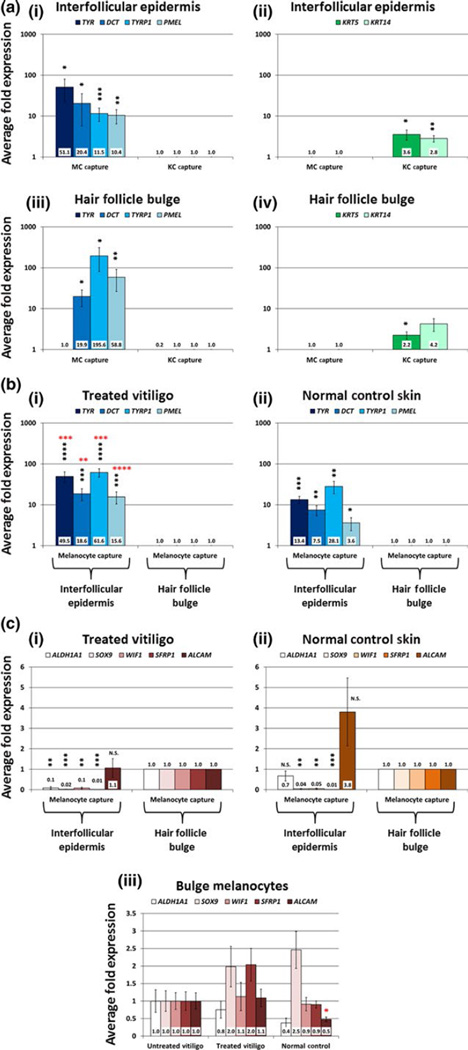
We analysed the expression values for melanocyte-specific gene transcripts (TYR, DCT, TYRP1 and PMEL) and keratinocyte-specific gene transcripts (KRT5 and KRT14) in the melanocyte samples compared with the adjacent keratinocyte samples captured from the epidermal basal layer and from the hair follicle bulge. For this initial analysis of cell-type enrichment, we captured 100 melanocytes and 100 adjacent keratinocytes from 6 NBUVB-treated vitiligo patients (Fig. S3). In the interfollicular epidermis, all four melanocyte-specific genes were significantly enriched in our melanocyte samples, as compared to the adjacent keratinocyte samples (TYR, 51-fold higher in melanocytes, P<0.05; DCT, 20-fold higher in melanocytes, P<0.05; PMEL, 12-fold higher in melanocytes, P<0.01; TYRP1, 10-fold higher in melanocytes, P<0.001) (Fig. 3a-i, represented in logarithmic scale, with the expression values in the keratinocyte samples set to 1 fold). Similarly, in the interfollicular epidermis, both keratinocyte-specific genes were significantly enriched in the keratinocyte samples as compared to the melanocyte samples (KRT5, 4-fold higher in keratinocytes, P<0.005; KRT14, 3-fold higher in keratinocytes, P<0.01) (Fig. 3a-ii, represented in logarithmic scale, with the expression values in the melanocyte capture set to 1-fold). In the hair follicle bulge, 3 of the 4 melanocyte-specific genes were significantly enriched in the melanocyte samples as compared to the keratinocyte samples (DCT, 20-fold higher in melanocytes, P<0.05; TYRP1, 196-fold higher in melanocytes, P<0.05; PMEL, 59-fold higher in melanocytes, P<0.01) (Fig. 3a-iii, with the expression values in the keratinocyte capture set to 1 fold). TYR expression in the bulge was very low in both melanocyte and keratinocyte samples and did not vary significantly between these two groups (TYR, 0.2-fold expression in keratinocytes P=0.13) (Fig. 3a-iii). Further, in the bulge keratinocyte samples, KRT5 was significantly enriched in comparison with the melanocyte samples (2-fold higher in keratinocytes, P<0.05), while KRT14 exhibited some enrichment, although non-significant (4-fold higher in keratinocytes, P=0.13) (Fig. 3a-iv; expression values in the melanocyte capture were set to 1 fold).
FIGURE 3.
qRT-PCR analysis of melanocyte (MC) and keratinocyte (KC) samples from fluorescent laser capture microdissection (F-LCM). Panel a: F-LCM samples of 100 MC and 100 KC from the interfollicular epidermis and hair follicle bulge of 6 NBUVB-treated vitiligo patients were amplified using the Ovation RNA-Seq System V2 (NuGEN Technologies) and subjected to qRT-PCR analysis. MC-specific genes (a-i, iii; blue bars) were significantly enriched in the MC samples compared to the KC samples, while KC-specific genes (a-ii, iv; green bars) exhibited significantly higher expression in the KC capture compared to MC capture in both regions tested. To illustrate enrichment for MC genes, fold changes for MC genes were set to 1 in KC samples from each region and were compared with expression values in the MC samples (blue bars). Similarly, to show enrichment of KC genes in KC samples, fold changes for KC genes were set to 1 in MC samples from each region and were compared with expression values in the KC samples (green bars). Data shown in logarithmic scale. Panel b: F-LCM samples of 100 MC from the interfollicular epidermis and hair follicle bulge of 7 NBUVB-treated vitiligo patients (b-i) and 6 normal control patients (b-ii) were amplified using the Arcturus RiboAmp HS PLUS Kit Termo Fisher Scientific and subjected to qRT-PCR analysis of MC-specific genes. Fold changes were set to 1 for MC samples from the bulge and were compared with expression values in the interfollicular epidermis within each skin type (blue bars). MC-specific genes (TYR, DCT, TYRP1 and PMEL) showed significantly higher expression (black stars) in MC samples from the interfollicular epidermis compared to MC samples from the bulge within each skin type. In the interfollicular epidermis, all MC-specific genes showed significantly higher expression (red stars) in MC samples from treated vitiligo compared to MC samples in normal control skin. Data shown in logarithmic scale. Panel c: The same F-LCM samples from 7 NBUVB-treated vitiligo patients (c-i) and 6 normal control patients (c-ii) were subjected to qRT-PCR analysis of genes associated with stem cells and the hair follicle bulge. c-i— In the NBUVB-treated vitiligo skin, 4 of the 5 tested melanocytic stem cell genes were expressed at significantly lower levels in the epidermis as compared to the bulge. ALCAM transcript’s expression did not show significant variation in the NBUVB-treated epidermis vs treated bulge. c-ii— In the normal control skin, 3 of the 5 melanocytic stem cell genes transcripts (SOX9, WIF1, SFRP1) showed significantly reduced expression levels in the interfollicular epidermis, as compared to the bulge. Both ALCAM and ALDH1A1 transcripts expression did not exhibit a significant difference between interfollicular epidermis and bulge. Fold changes were set to 1 for MC samples from the bulge and were compared with reduced expression values in the interfollicular epidermis within each skin type. All panels: *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001. c-iii— In the bulge, the expression values for all five melanocytic stem cell genes transcripts tested (ALDH1A1, SOX9, WIF1, SFRP1 and ALCAM) did not vary significantly between interfollicular epidermis of treated vitiligo, untreated vitiligo and normal control skin (one-way ANOVA, Tukey’s post hoc test: adjusted P>0.05 for all pairwise comparisons). Fold changes were set to 1-fold in untreated vitiligo, as compared to treated vitiligo and normal control skin.
3.3.2 | Testing the difference in candidate gene signatures between bulge and epidermis of NBUVB-treated vitiligo skin and normal skin
We analysed the expression of melanocyte-specific genes (TYR, DCT, TYRP1 and PMEL) in melanocyte captures (100 cells/sample) from the interfollicular epidermis compared to melanocyte captures (100 cells/sample) from the bulge of 7 NBUVB-treated vitiligo samples and 6 normal control skin samples (Fig. 3b). In both NBUVB-treated vitiligo (Fig. 3b-i) and normal control skin (Fig. 3b-ii), the expression values for all 4 melanocyte-specific genes were significantly higher (P<0.05) in the melanocyte captures from the interfollicular epidermis as compared to melanocyte samples from the bulge. The expression values, represented in logarithmic scale, were set to 1-fold in the bulge.
Next, we compared the expression levels of 5 previously reported melanocyte or melanoma stem cell genes (ALDH1A1, SOX9, WIF1, SFRP1 and ALCAM)10–15 in the hair follicle bulge and interfollicular epidermis of NBUVB-treated vitiligo and normal control skin using RNA samples of 100 melanocytes captured from each region (Fig. 3c).
In the NBUVB-treated vitiligo skin (Fig. 3c-i), 4 of the 5 tested melanocytic stem cell genes were expressed at significantly lower levels in the interfollicular epidermis as compared to the hair follicle bulge (ALDH1A1, 0.1-fold in the epidermis, P<.01; SOX9, 0.02-fold in the epidermis, P<0.001; WIF1, 0.1-fold in the epidermis, P<0.01; SFRP1, 0.01-fold in the epidermis, P<0.001). ALCAM transcript’s expression did not show significant variation in the NBUVB-treated epidermis vs treated bulge (P>0.05, 1.1-fold change in the epidermis).
In the normal control skin (Fig. 3c-ii), 3 of the 5 melanocytic stem cell genes transcripts showed significantly reduced expression levels in the interfollicular epidermis, as compared to the bulge (SOX9, 0.04-fold in the epidermis, P<0.01; WIF1, 0.05-fold in the epidermis, P<0.01; SFRP1, 0.01-fold in the epidermis, P<0.001). Both ALCAM and ALDH1A1 transcripts expression did not exhibit a significant difference between interfollicular epidermis and bulge (P>0.05) (in the epidermis ALDH1A1, 0.7-fold; ALCAM, 3.8-fold). The expression values, represented in logarithmic scale, were set to 1-fold in the bulge.
3.3.3 | Testing the difference in candidate gene signatures between untreated vitiligo, NBUVB-treated vitiligo and normal skin
- In the interfollicular epidermis, we could compare only the expression values of vitiligo NBUVB-treated vs normal skin (Fig. 3b), as there were no melanocytes to capture in the depigmented untreated vitiligo skin.a1. Expression values for all four melanocyte-specific gene transcripts (TYR, DCT, TYRP1 and PMEL) were significantly higher in the melanocyte samples captured from the interfollicular epidermis of treated vitiligo compared to normal control skin (Fig. 3b-i,b-ii, red stars) (TYR, 7.2-fold higher in treated vitiligo epidermis, P<0.001; DCT, 3.2-fold higher in treated vitiligo epidermis, P<0.01; TYRP1, 7.1-fold higher in treated vitiligo epidermis, P<0.001; PMEL, 9.5-fold higher in treated vitiligo epidermis, P<0.0001).a2. Expression values for all 5 melanocytic stem cell genes transcripts tested (Fig. 3c-i,c-ii) did not vary significantly between interfollicular epidermis of treated vitiligo and normal control skin.
- For melanocyte samples from the bulge, we compared expression values for all 3 groups (NBUVB-treated vitiligo, untreated vitiligo and normal control skin) using multiple pairwise post hoc comparisons.b1. As shown in Fig. 3c-iii, the expression values of melanocytic stem cell gene transcripts (ALDH1A1, SOX9, WIF1, SFRP1) were not significantly different between the 3 groups for melanocyte samples captured from the bulge (adjusted P>0.05; fold changes are indicated in Fig. 3c-iii, with expression values set to 1-fold for untreated vitiligo). Interestingly, there was a lower expression of borderline significance (one-way ANOVA, unadjusted P=0.05) for ALCAM in the normal skin bulge as compared to NBUVB-treated vitiligo bulge (2.3-fold lower) and untreated vitiligo bulge (2-fold lower). However, Tukey’s post hoc tests showed adjusted P>0.05 for all pairwise comparisons.b2. As shown in Fig. S4, the expression values of all melanocyte- specific gene transcripts (TYR, DCT, TYRP1 and PMEL) were not significantly different between groups (adjusted P>.05), being maximal in the treated bulge and minimal in the untreated bulge.
4 | DISCUSSION
We successfully isolated RNA for gene expression analysis from mature epidermal melanocytes and from their hair follicle bulge precursors by developing a rapid fluorescent immunostaining procedure for melanocytes followed by F-LCM assay. Our goal was to identify a strategy that can offer a better characterization of melanocyte populations in the regenerated epidermis and in the bulge of NBUVB-treated vitiligo skin. In addition, we tested the accuracy of our method in the bulge and epidermis of normal skin. We also used skin from depigmented untreated vitiligo to compare the gene signature in the bulge with that of the NBUVB-treated vitiligo and of normal skin. We showed that the resulting RNA from these enriched melanocyte samples is of satisfactory quality for qRT-PCR for two reasons: first, we found a significant enrichment of melanocyte-specific genes (TYR, DCT, TYRP1 and PMEL) in the melanocyte samples as compared with keratinocyte samples and of keratinocyte-specific genes (K15, K5) in the keratinocyte samples as compared with melanocyte samples in both epidermis and bulge of NBUVB-treated skin (Fig. 3a). Second, we found a good delineation between the epidermal mature reservoir and bulge stem cell reservoir, in regard to gene signature, findings that will be further discussed. In both NBUVB-treated vitiligo skin and normal skin, the expression of melanocyte-specific genes was significantly higher in the epidermis as compared to the bulge (Fig. 3b). These results parallel our previous immunostaining study,6 in which the bulge of both NBUVB-treated vitiligo and of normal skin showed a minimal expression of melanocyte-specific markers tested (including unremarkable TYR expression), while the epidermis in NBUVB-treated vitiligo skin, but also the normal epidermis of control skin, showed an abundant expression of these markers. Moreover, we tested five candidate melanocytic stem cell genes, looking at the expression of their transcripts in epidermal and bulge melanocyte captures. We found a significantly higher expression of three genes SOX9, WIF1 and SFRP1 in the bulge as compared to the epidermis in both NBUVB-treated (Fig. 3c-i) and normal skin (Fig. 3c-ii), an indication that we have successfully isolated RNA from stem cell-like cells (SOX9-positive, WIF1-positive and SFRP1-positive) from the hair follicle stem cell niche, populations that were previously documented by other reports on melanoma or human melanocytes.10,13–15 The above results suggest a good delineation between the melanocyte bulge stem cell reservoir enriched in melanocytic stem cell genes (SOX9, WIF1 and SFRP1) and the mature epidermal melanocyte pool, enriched in melanogenic genes (TYR, DCT, TYRP1 and PMEL). The significant expression of melanocyte-specific genes in the basal layer reflects an active melanogenesis process, accompanying melanocyte maturation.
Furthermore, we found a significantly higher expression for all four melanocyte-specific genes in epidermis of NBUVB-treated vitiligo skin, compared to normal control skin (Fig. 3b), and in general higher expression (but not significant differences) for the melanocyte-specific genes in the NBUVB-treated vitiligo bulge as compared to untreated vitiligo or to normal skin bulge (Fig. S4). Again, these gene expression results parallel our previous immunostaining findings6 that showed a higher expression of melanocyte-specific markers in the treated skin as compared with the normal skin in both epidermis and bulge, reflecting the stimulatory role of NBUVB on melanocyte maturation. Moreover, our previous immunostaining study did not show significant variation of the melanocyte-specific protein expression in the bulge of untreated vitiligo, NBUVB-treated vitiligo and normal skin, results that are paralleled at the transcription level by our current study. In addition, the current study did not show significant variation in the expression of melanocytic stem cell genes in melanocyte samples isolated from the bulge of untreated vitiligo, NBUVB-treated vitiligo or normal skin (Fig. 3c-iii). As an exception, ALCAM showed a borderline significant higher expression in the bulge of NBUVB-treated vitiligo and in untreated vitiligo as well, as compared to the bulge of normal control skin (Fig. 3c-iii). Future studies can clarify whether this is indeed a gene dysregulated in vitiligo.
Previous studies have examined the gene expression profiles associated with vitiligo and normal epidermal melanocytes using whole skin samples or cultured cell samples16,17; however, they could not relate these profiles to their anatomic localization in the skin or to specific cellular subpopulations. Other studies did utilize laser capture microdissection (of combined keratinocyte and melanocyte populations) from the bulge and supra-bulge outer root sheath and from the inner root sheath,7,18,19 or of T and B cells from lymph nodes,20 but they did not isolate specifically RNA from melanocytes in the basal layer or bulge outer root sheath. There are important advantages of the laser capture technique, embodied in our methods: a. it allows direct analysis of the RNA isolated from melanocytes and their precursors from the epidermal basal layer and hair follicle bulge; b. it minimizes the RNA degradation; c. it preserves the in situ transcription-level cellular communication signals between melanocytes and keratinocytes; d. it avoids the genetic and environmental changes in primary cell culture and cell lines grown in culture; e. it preserves the anatomic context of cells from which RNA was isolated, such as the depth and location within the hair follicle. The latter is a key aspect that could not be approached in previous human models using other methods. It also allows the comparison of various populations of cells within the natural 3D organization of the tissue, an option not available to FACS-isolated populations of cells.
The limitations of our method were a. inability to avoid a modest contamination with RNA material of neighbour cells; b. a small number of cells captured, although there are numerous previous studies that laser captured a small number of cells, followed by successful qRT-PCR and gene expression analysis21–23; c. the small sample size; and d. inability to avoid a degree of RNA degradation. Like others,23 we tried to overcome this inconvenience by designing short amplicons for qRT-PCR runs (≤150 bp in our case) and by optimizing the rapid immunostaining protocol.
We have shown for the first time that NKI-beteb was expressed specifically in melanocyte lineage cells in the normal hair follicle bulge (Fig. S2b,d), offering additional information to other studies that described NKI-beteb expression in the amelanotic and pigmented melanocytes in the hair follicle outer root sheath.9,24 Further, we have shown that the NKI-beteb antibody is an excellent tool for identifying a wide range of melanocytes in various stages of differentiation from hair follicles and epidermis. Moreover, the NKI-beteb antibody exhibited strong signal intensity in frozen tissue samples and in samples subjected to our rapid staining protocol, making this antibody a useful tool for minimizing RNA degradation.
Taken together, these data suggest that our LCM samples are significantly enriched and anatomically distinct enough to analyse a priori changes in melanocyte-specific or keratinocyte-specific gene signals associated with NBUVB treatment in human vitiligo. Currently, we are applying this technique to isolate RNA from melanocyte and keratinocyte populations in repigmented vitiligo skin and untreated skin for full characterization of the gene expression profiles using whole-transcriptome RNA sequencing, to achieve a better understanding of the repigmentation process in vitiligo. The techniques presented here offer other opportunities for researchers to study melanocyte and keratinocyte biology and can have broader implication for the study of various pigmentary disorders other than vitiligo.
Supplementary Material
Acknowledgments
This work was supported by a Research Scholar Award in Vitiligo from the American Skin Association and by UC Denver LCM Shared Resource funded by the NIH/NCATS Colorado CTSA Grant UL1 TR001082. We are grateful to our team in Dermatology Clinic at UCH for sample collection. We are grateful to Dr Vince Hearing (for the generous DCT antibody gift) and Drs Manabu Ohyama, Randall Cohrs, Mark Burgoon Yiqun Shellman and Karoline Lambert for valuable advice. We thank Adriaan van Bokhoven, Zachary Grasmick and Nicole Spoelstra from Laser Capture Microdissection Core for their valuable support with the laser capture machine. We acknowledge National Disease Research Institute for providing human skin samples.
Footnotes
AUTHORS CONTRIBUTION
Nathaniel Goldstein wrote the first draft of the manuscript, contributed to the study design, performed the experimental work, including the rapid immunostaining and sample processing for laser capture microdissection. Dr Maranke Koster supervised the histology work and contributed to the first draft of the manuscript. Laura Hoaglin performed the tissue processing, tissue cutting and some tissue staining. Michael Wright performed some qRT-PCR work needed for the first revision. Steven Robinson and Dr William Robinson provided most of the normal skin needed for the first revision. Dr Dennis Roop offered valuable advice on this project and contributed to the first draft of the manuscript. Dr David Norris contributed to all steps of this study, the study design and performed repeatedly reviews of the first draft of the manuscript and reviewed the draft for the first revision. Dr Stanca Birlea contributed to the study design, collected the skin biopsies performed the laser capture microdissection and prepared the manuscript for the first submission and first revision.
SUPPORTING INFORMATION
Additional Supporting Information may be found online in the supporting information tab for this article.
Data S1. Supplementary Materials and Methods.
Figure S1. Bulge mapping.
Figure S2. Expression of NKI-beteb in the normal human epidermis and hair follicle bulge.
Figure S3. Cartoon example of fluorescent laser capture microdissection (F-LCM) technique.
Figure S4. qRT-PCR analysis of melanocyte (MC) samples from Fluorescent Laser Capture Microdissection (F-LCM) in the bulge of untreated vitiligo, NBUVB-treated vitiligo and normal control skin.
Table S1. Primer sequences used for qRT-PCR.
Data S2. Supplementary References.
REFERENCES
- 1.Birlea SA, Spritz RA, Norris DA. Vitiligo. In: Wolff K, Goldsmith LA, Katz SI, et al., editors. Fitzpatrick’s Dermatology in General Medicine. 8th. New York, NY: McGraw-Hill; 2012. pp. 792–803. [Google Scholar]
- 2.Picardo M, Taïeb A. Epidemiology, definitions, and classification. In: Picardo M, Taïeb A, editors. Vitiligo. Heidelberg: Springer Berlin; 2010. pp. 13–23. [Google Scholar]
- 3.Mosenson JA, Eby JM, Hernandez C, et al. Exp Dermatol. 2013;22:566–569. doi: 10.1111/exd.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spritz RA. J Dermatol. 2013;40:310–318. doi: 10.1111/1346-8138.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dell’Anna ML, Ottaviani M, Bellei B, et al. J Cell Physiol. 2010;223:187–193. doi: 10.1002/jcp.22027. [DOI] [PubMed] [Google Scholar]
- 6.Goldstein NB, Koster MI, Hoaglin L, et al. J Invest Dermatol. 2015;135:2068–2076. doi: 10.1038/jid.2015.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohyama M, Terunuma A, Vogel JC. J Clin Invest. 2006;116:249–260. doi: 10.1172/JCI26043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horikawa T, Norris DA, Johnson TW, et al. J Invest Dermatol. 1996;106:28–35. doi: 10.1111/1523-1747.ep12326989. [DOI] [PubMed] [Google Scholar]
- 9.Shellman YG, Ribble D, Yi M, et al. Biotechniques. 2004;36:968–976. doi: 10.2144/04366ST01. [DOI] [PubMed] [Google Scholar]
- 10.Vidal V, Chaboissier MC, Lützkendorf S, et al. Curr Biol. 2005;15:1340–1351. doi: 10.1016/j.cub.2005.06.064. [DOI] [PubMed] [Google Scholar]
- 11.Marchitti SA, Brocker C, Stagos D, et al. Expert Opin Drug Metab Toxicol. 2008;4:697–720. doi: 10.1517/17425250802102627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Koppaka V, Thompson DC, et al. Exp Eye Res. 2012;102:105–106. doi: 10.1016/j.exer.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kadaja M, Keyes BE, Lin M, et al. Genes Dev. 2014;28:328–341. doi: 10.1101/gad.233247.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lang D, Mascarenhas JB, Shea CR. Clin Dermatol. 2013;31:166–178. doi: 10.1016/j.clindermatol.2012.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein WM, Wu BP, Zhao S, et al. Mod Pathol. 2007;20:102–107. doi: 10.1038/modpathol.3800720. [DOI] [PubMed] [Google Scholar]
- 16.Yu R, Broady R, Huang Y, et al. PLoS One. 2012;7:e51040. doi: 10.1371/journal.pone.0051040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reemann P, Reimann E, Ilmjärv S, et al. PLoS One. 2014;9:e115717. doi: 10.1371/journal.pone.0115717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu X, Lyle S, Liu Y, et al. Am J Pathol. 2003;163:969–978. doi: 10.1016/S0002-9440(10)63456-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amoh Y, Aki R, Hamada Y, et al. J Dermatol. 2012;39:33–38. doi: 10.1111/j.1346-8138.2011.01413.x. [DOI] [PubMed] [Google Scholar]
- 20.Fend F, Emmert-Buck MR, Chuaqui R, et al. Am J Pathol. 1999;154:61–66. doi: 10.1016/S0002-9440(10)65251-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dolter KE, Braman JC. Biotechniques. 2001;30:1358–1361. doi: 10.2144/01306pf01. [DOI] [PubMed] [Google Scholar]
- 22.Nakamura N, Ruebel K, Jin L, et al. Methods Mol Med. 2007;132:11–18. doi: 10.1007/978-1-59745-298-4_2. [DOI] [PubMed] [Google Scholar]
- 23.Vandewoestyne M, Goossens K, Burvenich C, et al. Anal Biochem. 2013;439:88–98. doi: 10.1016/j.ab.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 24.Anbar TS, Abdel-Raouf H, Awad SS, et al. J Eur Acad Dermatol Venereol. 2009;23:934–939. doi: 10.1111/j.1468-3083.2009.03233.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.