Abstract
By phylogenetic analysis, Mycobacterium kansasii is closely related to Mycobacterium tuberculosis. Yet, although both organisms cause pulmonary disease, M. tuberculosis is a global health menace, whereas M. kansasii is an opportunistic pathogen. To illuminate the differences between these organisms, we have sequenced the genome of M. kansasii ATCC 12478 and its plasmid (pMK12478) and conducted side-by-side in vitro and in vivo investigations of these two organisms. The M. kansasii genome is 6,432,277 bp, more than 2 Mb longer than that of M. tuberculosis H37Rv, and the plasmid contains 144,951 bp. Pairwise comparisons reveal conserved and discordant genes and genomic regions. A notable example of genomic conservation is the virulence locus ESX-1, which is intact and functional in the low-virulence M. kansasii, potentially mediating phagosomal disruption. Differences between these organisms include a decreased predicted metabolic capacity, an increased proportion of toxin–antitoxin genes, and the acquisition of M. tuberculosis-specific genes in the pathogen since their common ancestor. Consistent with their distinct epidemiologic profiles, following infection of C57BL/6 mice, M. kansasii counts increased by less than 10-fold over 6 weeks, whereas M. tuberculosis counts increased by over 10,000-fold in just 3 weeks. Together, these data suggest that M. kansasii can serve as an image of the environmental ancestor of M. tuberculosis before its emergence as a professional pathogen, and can be used as a model organism to study the switch from an environmental opportunistic pathogen to a professional host-restricted pathogen.
Keywords: mycobacteria, phylogeny, comparative genomics, virulence, Mycobacterium kansasii
Introduction
First identified in 1953 as the “yellow bacillus” (Pollak and Buhler 1953), Mycobacterium kansasii is an acid-fast bacterium that can cause a pulmonary disease in immunocompromised individuals and those with underlying pulmonary conditions such as chronic obstructive pulmonary disease and silicosis (Lillo et al. 1990; Corbett et al. 1999; Canueto-Quintero et al. 2003; Griffith et al. 2007). This disease resembles that caused by Mycobacterium tuberculosis in that patients experience similar symptoms (chest pain, productive cough, and weight loss), with comparable radiographic features (Evans, Colville, et al. 1996; Evans, Crisp, et al. 1996; Ellis 2004) and both infections can be treated with standard antituberculosis agents (Griffith et al. 2007). However, although tuberculosis (TB) is a global pandemic (World Health Organization 2013), M. kansasii infections are uncommon in the general population (Good and Snider 1982; Marras et al. 2007; Cassidy et al. 2009), and human-to-human M. kansasii transmission, if any, has rarely been documented (Davies 1994; Ricketts et al. 2014). The absence of transmission of M. kansasii infection in humans marks an evolutionary dead-end for this environmental organism, a scenario also described in Legionella, an accidental pathogen, and animal–human zoonotic pathogens such as Campylobacter jejuni and Salmonella enterica (Sokurenko et al. 2006; Sanchez-Buso et al. 2014).
Unlike M. kansasii, which is frequently found in aquatic environments (McSwiggan and Collins 1974; Joynson 1979; Kaustova et al. 1981; Thomson et al. 2013), the tubercle bacillus M. tuberculosis has no identified environmental reservoir. Rather, both phylogeographic and paleo-DNA studies present M. tuberculosis as a human-adapted pathogen that originated in Africa and accompanied the migrations of modern humans throughout the world (Wirth et al. 2008; Gagneux 2012; Comas et al. 2013). Despite these clear epidemiologic differences, the organisms share many similarities. Similar to M. tuberculosis, M. kansasii can grow at 37 °C with growth seen after 2–3 weeks on Löwenstein–Jensen medium (Roberts 1981). Mycobacterium kansasii is also positive for urease production, thiophene-2-carboxylic hydrazide resistance, and nitrate reduction (Roberts 1981; Tsukamura 1985), phenocopying biochemical characteristics long used for the laboratory identification of M. tuberculosis. In contrast, M. kansasii is a photochromogenic bacterium that produces carotenoid pigments against UV damage, a feature common to environmental organisms but lacking in M. tuberculosis (Tsukamura 1964; Robledo et al. 2011). Additionally, M. kansasii can utilize a much wider array of carbon and nitrogen sources to support growth than M. tuberculosis (Tsukamura et al. 1969; Tsukamura 1983), potentially capitalizing on a broader pool of nutrient sources in the environment.
As more complete mycobacterial genomes become available, comparative genomic studies provide the opportunity to identify testable differences between related, but biologically distinct species. For example, the smooth tubercle bacilli, exemplified by Mycobacterium canettii, are considered to be the most closely related species to M. tuberculosis, yet a number of genetic differences have been documented between these organisms (Gutierrez et al. 2005; Fabre et al. 2010; Supply et al. 2013). The observation that M. canettii causes TB in apparently immunocompetent hosts, along with its lack of a known environmental reservoir, together suggests that the biology of M. canettii may be more similar to M. tuberculosis than their putative environmental ancestor. To elucidate the speciation of M. tuberculosis on a larger evolutionary scale, we have compared M. tuberculosis with M. kansasii. Previous analysis, using multilocus sequence analysis, has identified M. kansasii as the environmental mycobacterium most closely related to the M. tuberculosis complex organisms (Veyrier et al. 2009). In this study, we determined the genome of M. kansasii and conducted comprehensive comparative genomic analyses between M. kansasii and M. tuberculosis. By apposing their genomes with phenotypic characterization, we present a novel perspective on the emergence of M. tuberculosis as a “professional” pathogen.
Materials and Methods
Mycobacterium kansasii and M. tuberculosis Strains
Mycobacterium kansasii ATCC 12478 (Hauduroy) is a clinical isolate now a type strain; belongs to subtype I (Richter et al. 1999). Mycobacterium tuberculosis H37Rv is a well-characterized virulent laboratory strain; M. tuberculosis:ΔRD1 and M. tuberculosis:Δesat-6 were gifts from Dr David Sherman (University of Washington). Mycobacterium kansasii clinical strains (62359, 62759, 62349, and 63327) were collected from the McGill University Health Centre between 2006 and 2010.
Genome Sequencing and Annotation
The first draft of M. kansasii genome was assembled following shotgun sequencing using the Genome Sequencer with 33 × coverage (Veyrier et al. 2009). In this study, we prepared ultrapure, intact genomic DNA for the Pacific Biosciences (PacBio) sequencing platform to generate a gapless genome. DNA was extracted using the PowerSoil DNA isolation kit (MoBio, Carlsbad, CA, USA). Library preparation and sequencing reaction were carried out as described previously at Genome Quebec (Labrie et al. 2014). The sequenced genome was assembled using HGAP. Genome annotation was performed by the National Center for Biotechnology Information (NCBI) Prokaryotic Genome Annotation Pipeline using GeneMarkS (Besemer et al. 2001); features annotated include coding sequences (CDS), ribosomal RNA (rRNA), transfer RNA (tRNA), noncoding RNA (ncRNA), and repeat regions. Additional analysis was carried out by Rapid Annotation by Subsystems Technology (RAST) to organize genes into functional groups or pathways (Aziz et al. 2008).
Strain Subtyping and Plasmid Detection
Mycobacterium kansasii ATCC 12478 and four clinical strains were subtyped using polymerase chain reaction (PCR)-restriction fragment length polymorphism and 16S–23S ITS sequencing as described in Alcaide et al. (1997) and Richter et al. (1999). The presence of plasmid was detected by PCR. Standard PCR reaction was carried out with an annealing temperature of 55 °C with forward (CTACCGCGACTACAACACCA) and reverse (GGGGTGAACGTGAGGTCATA) primers amplifying a 213-bp region in the conjugative transfer relaxase gene (MKAN_29815) found to be conserved in mycobacterial plasmids (Ummels et al. 2014).
Amino Acid Sequence-Based Phylogeny
The core genome was built by identifying proteins that were bidirectional best hits between pairs of genomes with at least 40% identity in protein sequence and less than 20% difference in size (Vieira-Silva et al. 2011). The intersection of the pairwise list of bidirectional best-hits provided the core genome of the clade. Each family of the core genome was then aligned using the protein sequence with MUSCLE 3.52 (Edgar 2004). The software BMGE was used to identify and extract suitable regions from multiple alignments using the BLOSUM62 matrix (Criscuolo and Gribaldo 2010). The resulting alignment had 91,530 distinct alignment patterns and was used to build a phylogenetic tree of the clade with RaxML 7.2.7 using the model PROTGAMMALG, with 100 bootstrap replicates (Stamatakis 2014). The resulting tree was visualized with FigTree (http://tree.bio.ed.ac.uk/software/figtree/, last accessed March 4, 2015).
Genomes used in this analysis included: M. abscessus, M. abscessus subspecies bolletii, M. sp. MCS, M. sp. KMS, M. sp. JLS, Mycobacterium gilvum Spyr1, M. gilvum PYR-GCK, Mycobacterium vanbaalenii PYR-1, Mycobacterium vaccae, Mycobacterium chubuense, Mycobacterium phlei, Mycobacterium hassiacum, Mycobacterium rhodesiae NBB3, Mycobacterium tusciae, Mycobacterium mageritense, Mycobacterium fortuitum, Mycobacterium smegmatis MC2155, Mycobacterium neoaurum VKM Ac-1815D, Mycobacterium thermoresistable, M. sp. JDM601, Mycobacterium xenopi, Mycobacterium indicus pranii MTCC 9506, Mycobacterium intracellulare MOTT-64, M. intracellulare ATCC 13950, Mycobacterium yongonense 05-1390, M. sp. MOTT36Y, Mycobacterium colombiense, Mycobacterium avium 104, M. avium sp. paratuberculosis K-10, Mycobacterium parascrofulaceum, Mycobacterium leprae TN, M. kansasii ATCC 12478, Mycobacterium gastri, Mycobacterium liflandii 128FXT, Mycobacterium ulcerans Agy99, Mycobacterium marinum M, M. canettii CIPT 140010059, M. tuberculosis H37Rv, Mycobacterium bovis AF2122/97, and Mycobacterium africanum GM041182.
Related species in the same order (Actinomycetales) were used as an outgroup; RHEQ: Rhodococcus equi; NOFA, Nocardia farcinica.
Genome Comparison between M. kansasii and M. tuberculosis
Genes present in M. tuberculosis but absent in other mycobacteria, including M. kansasii, have been described (Veyrier et al. 2009). To generate the list of genes potentially have been lost in M. tuberculosis since its divergence from other mycobacteria, the CDS of M. tuberculosis were compared with other mycobacteria including M. abscessus, M. gilvum, M. intracellulare, M. sp. JLS, M. vanbaalenii, M. smegmatis, M. sp. KMS, M. sp. MCS, M. avium 104, M. avium subsp. paratuberculosis, M. ulcerans, and M. kansasii, using criteria described in Veyrier et al. (2009). The resulting gene list was categorized into Clusters of Orthologous Groups (COG) functional categories (Tatusov et al. 2000).
In Vitro Comparisons
The growth of M. tuberculosis H37Rv and M. kansasii in 7H9 supplemented with ADC enrichment and 0.2% Tween-80 at 37 °C with rolling was monitored with a spectrophotometer at OD600 in triplicates every 24 h for 4 days. Bacteria were also streaked out on 7H10 agar plates for morphological observation. To verify the presence of a functional ESX-1 secretion system, culture supernatants for M. tuberculosis, M. tuberculosis:ΔRD1, M. tuberculosis:Δesat-6, and M. kansasii were prepared according to Chen et al. (2012) and probed with 1:5,000 monoclonal anti-ESAT-6 antibody (11G4, Abcam). The abundance of each protein was measured and quantified using ImageJ (Schneider et al. 2012).
Phagosomal Perturbation Assay
Detection of phagosomal rupture was performed as described using a single-cell fluorescence resonance energy transfer (FRET)-based assay (Ray et al. 2010; Simeone et al. 2012). Briefly, M. tuberculosis, M. tuberculosis:ΔRD1, and M. kansasii were used to infect phorbol-myristate-acetate (PMA)-differentiated THP-1 macrophages. THP-1 cells were incubated with CCF-4-AM (Invitrogen), an esterified, lipophilic form of the CCF-4 substrate that can readily enter into cells, where it is converted by endogenous cytoplasmic esterases into CCF-4 that is negatively charged. CCF-4 is retained in the cytosol and emits green fluorescence (535 nm) upon stimulation at 405 nm, due to FRET from the 7-hydroxycoumarin donor to the fluoroscein acceptor. When in contact with endogenous beta-lactamases produced by mycobacteria, as occurs when the phagosomal compartment is disrupted, CCF-4 is cleaved, emitting at 450 nm when excited at 405 nm. All experimental steps were performed in the presence of probenecid upon CCF-4-AM treatment to avoid the export of the fluorescent reporter. The 450/535 nm intensity ratio measurements were obtained and analyzed by algorithms provided by the Metamorph software, as previously reported (Ray et al. 2010).
In Vivo Comparisons
Eight-week-old female C57BL/6 mice purchased from Jackson Laboratories were aerosol-infected with wild-type M. tuberculosis, M. tuberculosis:ΔRD1, and M. kansasii, as previously described (Hansen et al. 2014). Five mice from each group were sacrificed 1 day postinfection and their lung homogenates were plated out for the enumeration of bacteria. At days 21 and 42 postinfection, the lungs and spleens of infected mice were homogenized for bacterial quantification by serial dilution. All animal experiments were in accordance with the regulations of the Canadian Council of Animal Care and approved by the McGill University Animal Committee.
Results
Mycobacterium kansasii Is among the Closest Species to M. tuberculosis
We identified the proteins conserved across 40 available mycobacterial genomes and two outgroups (R. equi and N. farcinica). We then used this information to build a rooted phylogenetic tree of the Mycobacterium genus. This tree can be divided into three groups: The outgroup actinobacteria (R. equi and N. farcinica), the rapid-growing mycobacteria (such as M. smegmatis), and emerging from this group, the slow-growing mycobacteria. In this slow-growing sublineage, members of the M. tuberculosis complex (M. tuberculosis, M. bovis, and M. africanum) clustered together, next to the early branching tubercle bacilli of the M. canettii clade (Fabre et al. 2004; Supply et al. 2013). The next most closely related organisms include M. kansasii and M. marinum (fig. 1), the latter a pathogenic species that can cause skin disease and lymphangitis in humans, and grows optimally at 30 °C.
Fig. 1.—
Phylogenetic relationships among Mycobacterium genus. The phylogenetic tree was generated using RAxML. The scale bar represents amino acid changes per site. Rapid growing species are shadowed in orange, slow growing species are shadowed in yellow, and outgroup species are shadowed in green. Mycobacterium tuberculosis, M. canettii, and M. kansasii are boxed in red.
The M. kansasii Genome
The M. kansasii ATCC 12478 genome consists of a chromosome of 6,432,277 bp (NCBI Reference Sequence NC_022663.1) plus a plasmid, pMK12478, which is 144,951 bp in size (NCBI Reference Sequence: NC_022654.1). The circular representation of the genome was generated using DNAplotter (Carver et al. 2009) and is shown in figure 2A. The annotated genome indicated the existence of 5,712 CDS in the chromosome, 3 rRNAs (5S, 16S, and 23S), 46 tRNAs, and 2 ncRNAs. Additionally, there were 10 disrupted pseudogenes, previously encoding 2 membrane proteins, 1 dihydrodipicolinate reductase, 1 serine/threonine protein kinase, 1 imidazole glycerol phosphate synthase, 1 transposase, 1 integrase, and 3 hypothetical proteins.
Fig. 2.—
(A) Circular representation and annotation features of M. kansasii ATCC 12478 chromosome. The two outermost circles represent forward and reverse-strand CDS (red and blue, respectively). tRNA is shown in green, rRNA in orange, and ncRNA in black. The two innermost blue/gray and olive/purple circles represent G + C content and GC skew, respectively. (B) Circular representation of the pMK12478 plasmid. Forward and reverse CDS and G + C content and GC skew are labeled in the same color scheme as the chromosome. Genes coding for putative sigma factors are coded in black; MCE in dark purple; replication protein in green; toxin–antitoxin in yellow, putative T4SS in pink; putative T7SS in orange (NCBI annotation; Ummels et al. 2014).
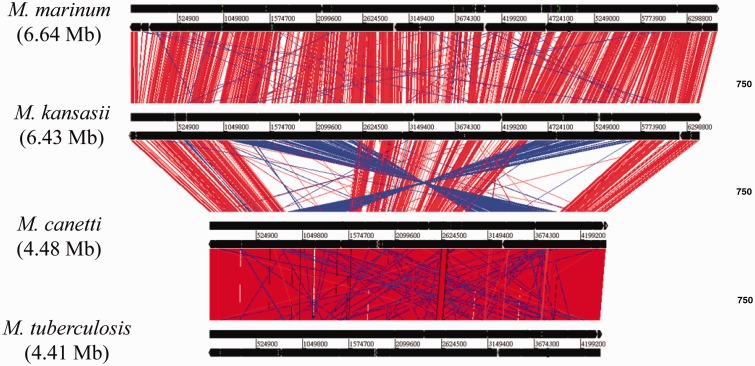
The comparison of the genome organization of M. tuberculosis, M. canettii CIPT 140010059, M. kansasii, and M. marinum was visualized by Artemis Comparison Tool (Carver et al. 2005). As shown in figure 3, two large chromosomal inversions separate the group of M. canettii and M. tuberculosis from that of M. marinum and M. kansasii. Mycobacterium kansasii and M. marinum genomes display large regions of similarity whereas M. canettii and M. tuberculosis exhibit high synteny conservation, consistent with their phylogenetic relationships. Individual genome comparisons between M. marinum, M. kansasii, and M. tuberculosis/M. canettii are presented in supplementary figure S1, Supplementary Material online.
Fig. 3.—
Comparisons of the genomes of M. tuberculosis, M. canettii, M. kansasii, and M. marinum. Red lines indicate local colinear blocks of DNA–DNA similarity and blue lines indicate rearranged regions.
The M. kansasii plasmid, pMK12478, is predicted to encode 154 proteins, including 4 mammalian cell entry (MCE) proteins, 1 PE family protein, several secretion-associated proteins, toxin–antitoxin systems, as well as transcriptional regulators and sigma factors (fig. 2B). Mycobacterium kansasii ATCC 12478 belongs to the subgroup 1, a subgroup strongly associated with diseases (Taillard et al. 2003). To assess whether the plasmid is associated with pathogenicity, we tested four recent clinical isolates from the McGill University Health Centre mycobacterial laboratory. Although all isolates were from subgroup 1, in accordance with previous report (Taillard et al. 2003), two of these isolates did not contain the plasmid, suggesting that the plasmid is dispensable for clinical disease in humans.
Changes in Genetic Content during the Step-Wise Emergence of M. tuberculosis
To compare the proportion of genes in different functional metabolic categories, RAST was used (Aziz et al. 2008), and visualized by SEED Viewer (Overbeek et al. 2014). RAST estimated 5,888 CDS in the genome. Of these, 1,953 were assigned into 27 subsystems, a collection of functional roles that constitute a certain biological pathway or structural complex, providing predictions on just over one-third of the genes (1,953/5,888 = 34%) (Overbeek et al. 2005; Meyer et al. 2009). Applying the same methodology to the smaller genome of M. tuberculosis H37Rv (Cole et al. 1998), there was a lower number, but higher proportion, of predicted CDS assigned to subsystems (1,619/4,317 = 38%). Differences in proportions of genes in each subsystem were assessed using the two-sample Z-test (two-tailed). A Bonferroni correction was used to account for multiple testing; therefore, a result with P < 0.05/27, or P < 0.0019 was considered significant at an α of 0.05. Compared with M. kansasii, M. tuberculosis has a greater proportion of its genome devoted to regulation and cell signaling. A closer inspection of genes in this category revealed that this difference is mostly due to the large number of genes involved in toxin–antitoxin systems (P < 0.0001) (fig. 4 and supplementary table S1, Supplementary Material online). To evaluate whether these differences were specific to the comparison between the two genomes, or representative of a larger dichotomy between M. tuberculosis and nontuberculous mycobacteria (NTM), we examined the genomes of the slow-growing M. marinum, M. avium subsp. hominissuis, M. intracellulare, M. avium subsp. paratuberculosis, and the fast-growing M. abscessus, M. vanbaalenii, M. gilvum, and M. smegmatis. Compared with these genomes, M. tuberculosis still shows a higher proportion of genes in the regulation and cell signaling subsystem, again driven by the number of toxin–antitoxin genes, whereas all other mycobacteria devote much more of their genomes to metabolism of various compounds. We also noted that the proportion of genes in all subsystems examined was the same in M. tuberculosis and M. canettii (P > 0.1) (supplementary table S1 and fig. S2, Supplementary Material online).
Fig. 4.—
The proportion of genes in each subsystem in M. tuberculosis, M. canettii, and M. kansasii, Asterisks indicate the results of the statistical tests comparing M. tuberculosis and M. kansasii. Sample proportions were compared using a two-sample Z-test. ****P < 0.0001.
To assess which M. kansasii genes absent from M. tuberculosis were most likely deleted during the emergence of M. tuberculosis, we identified 349 genes present in M. kansasii and 13 other nontuberculous mycobacteria that are exclusively absent from M. tuberculosis (346 out of these 349 genes are also lost in M. canettii CIPT 140010059) (supplementary table S2, Supplementary Material online). Based on their COG functional categories, many of these genes are predicted to be involved in energy production and conversion (n = 34), lipid metabolism (n = 46), and biosynthesis, transport, and catabolism of secondary metabolites (n = 37), consistent with the previously published observation of a greater metabolic capacity of M. kansasii (Tsukamura et al. 1969; Tsukamura 1983).
Comparison of Virulence Determinants of M. tuberculosis
A growing number of M. tuberculosis genes have been experimentally shown to be necessary for full virulence of this organism, so we investigated whether any of these was absent from the genome of M. kansasii. We found that the two-component signal transduction system PhoPR and dormancy regulon DosR/S/T are conserved in the low-virulence M. kansasii (summarized in table 1) (Perez et al. 2001; Ryndak et al. 2008; Converse et al. 2009; Leistikow et al. 2010). Based on RAST annotation, the M. tuberculosis genome has 87 PE and 66 PPE proteins, implicated in pathogen–host interaction (Sampson 2011); in comparison, the M. kansasii genome is predicted to encode 72 PE and 141 PPE proteins. Mycobacterium tuberculosis has 36 mce-associated genes (Zhang and Xie 2011) and we found a similar number (36 + 1 on the plasmid) in M. kansasii. In addition, more than 65% of the antigens encoded by M. tuberculosis are also present in M. kansasii including MPT70, MPT60, TB8.4, antigen 85, ESAT-6, and CFP-10, with greater than 75% amino acid homology and 80% coverage (Comas et al. 2010). Finally, in M. tuberculosis, the ESX-1 locus has been shown to be crucial for full virulence (Brodin et al. 2006; MacGurn and Cox 2007), with a variety of phenotypes shown to depend upon the presence of an intact ESX-1 locus (Abdallah et al. 2007). At the genomic level M. kansasii has an intact ESX-1 locus encoding building blocks of the Type VII secretion system (T7SS) (MKAN_14470–MKAN_14850), as well as the coregulated espA–espC–espD gene cluster (MKAN_12680, 12685, 12690), with protein similarities ranging from 58% to 95% (fig. 5B). Therefore, M. kansasii encodes orthologs for all these recognized virulence determinants of M. tuberculosis. Notably, it was noted that the putative ortholog of the espG1 (Rv3866) gene of M. tuberculosis, MKAN_07615 is encoded outside the ESX-1 locus. As the espG-gene products were suggested to represent potential chaperon proteins that interact with ESX-system-specific PE/PPE proteins and guide specific T7SS effectors to their “correct” ESX secretion system, this observation is of potential interest (Daleke et al. 2012).
Table 1.
Summary of Shared and Discordant Genomic and Phenotypic Features of the Mycobacterium kansasii and Mycobacterium tuberculosis
| M. tuberculosis | M. kansasii | |
|---|---|---|
| Disease | Host-adapted | Opportunistic |
| Transmission | Human–human | Water–human |
| Reservoir | Human | Environment |
| Genome size | 4.4 Mb | 6.4 Mb |
| Protein genes | 4,018 | 5,866 |
| % GC content | 66 | 66 |
| Presence of PhoPR 2-component system | Yes (Rv0757/8) | Yes (Mkan_11335/30) |
| Presence of DosR/S/T regulon | Yes (Rv3133c/3132c/2027c) | Yes (Mkan_22405/22410/22410) |
| # PE/PPE proteins | 153 | 213 |
| # mce-associated genes | 36 | 36 |
| Proportion of metabolism genes | 0.002316423 | 0.0084918 |
| Proportion of toxin/antitoxin genes | 0.013899 | 0.001019 |
| Presence of ESX-1 | Yes | Yes |
| T-cell antigens | 77 | 53 |
| Phagosomal disruption | Yes | Yes |
| Replication in mouse lungs | 4.5 log | 0.5 log |
Note.—The number of PE/PPE proteins and mce-associated genes are based on RAST annotation.
Fig. 5.—
(A) Comparative growth of M. kansasii and M. tuberculosis in 7H9 at 37 °C. (B) Morphological characteristics of M. kansasii (top panel) and M. tuberculosis (bottom panel) on 7H10 agar.
Changes in the In Vitro Phenotype during the Step-Wise Emergence of M. tuberculosis
The growth patterns and morphologies of M. kansasii and M. tuberculosis are shown in figure 5. In 7H9 broth with rolling, M. kansasii and M. tuberculosis grew at similar rates with doubling times of 19.6 and 20.9 h, respectively (fig. 5A). After growth on 7H10 agar for 8–10 weeks at 37 °C, M. kansasii colonies appeared slightly raised, smooth, with a light yellow color whereas M. tuberculosis colonies were more textured, wrinkled with no pigmentation (fig. 5B). Although the chemical basis for the rough morphology has not been elucidated in M. tuberculosis (Lemassu et al. 1992), it has been reported that M. kansasii strains, such as ATCC 12478 (also designated as TMC 1204) that contain acyltrehalose-containing lipooligosaccharides, exhibit a smooth morphology (Belisle and Brennan 1989).
To verify that the ESX-1 system is functionally intact in M. kansasii ATCC 12478, we conducted immunoblotting for the 6-kDa early secreted antigenic target ESAT-6 protein, one of the most abundant T7SS-associated proteins (Sorensen et al. 1995; Harboe et al. 1996). As expected, we detected ESAT-6 in the culture filtrate in both species, but not M. tuberculosis:ΔRD1 (region of difference containing the esat-6 gene as well as several other indispensable components of the T7SS [Pym et al. 2002; Guinn et al. 2004]), or M. tuberculosis:Δesat-6 (fig. 6A and B).
Fig. 6.—
(A) Top panel: SDS-PAGE analysis of mycobacterial culture filtrate proteins; lane 1: M. tuberculosis; lane 2: M. tuberculosis:ΔRD1; lane 3: M. tuberculosis:Δesat-6 ; lane 4: M. kansasii. Middle panel: Western blot showing the presence of ESAT-6 in M. tuberculosis and M. kansasii culture filtrates but not M. tuberculosis:ΔRD1 or M. tuberculosis:Δesat-6. Bottom panel: Band intensities measured by ImageJ and represented in histogram. (B) Schematic illustration of the organization of ESX-1 and orthologous genes in M. kansasii. Each gene is color-coded according to the degree of protein similarity when aligning M. kansasii amino acid sequence to that of M. tuberculosis. Mycobacterium kansasii genes that are orthologous to M. tuberculosis ESX genes but not organized in a gene cluster are boxed. Regions deleted in M. tuberculosis:ΔRD1 and M. tuberculosis:Δesat-6 are also indicated.
ESAT-6 is required for M. tuberculosis to access the cytosol after phagocytosis (van der Wel et al. 2007; Houben et al. 2012; Simeone et al. 2012). The production and secretion of ESAT-6 in M. kansasii led us to test whether this species is also capable of compromising the phagosomal membrane inside the macrophage using a previously established FRET reporter assay (Ray et al. 2010; Simeone et al. 2012). Indeed, as shown in figure 7, over the course of a 7-day infection, macrophages containing the FRET reporter infected with M. tuberculosis and M. kansasii began to exhibit a decrease in FRET signal in the cytosol from 535 to 450 nm with time plotted as fluorescent ratios of the two measured channels, suggesting that the integrity of the phagosomal membrane was compromised. This phenotype was not observed in macrophages infected with M. tuberculosis:ΔRD1, as has been previously published (van der Wel et al. 2007), indicating that the behavior of M. kansasii within a macrophage parallels that of fully virulent M. tuberculosis, extending previous observations with M. kansasii type I strains (Houben et al. 2012).
Fig. 7.—
Mycobacterium tuberculosis and M. kansasii induced a change in fluorescence ratio starting on day 4 postinfection whereas M. tuberculosis:1 did not show such activity.
Changes in the In Vivo Phenotype during the Step-Wise Emergence of M. tuberculosis
In preliminary experiments with M. kansasii, infection of C57BL/6 mice with the low dose of bacteria used for M. tuberculosis (∼50–100 colony forming units) did not result in a consistently detectable infection at 3 weeks after aerosolization, suggesting either a failure to deliver the organism to the airways or the rapid elimination of M. kansasii upon infection. However, assessment at 1 and 24 h postinfection with a higher inoculum showed comparable bacterial loads in mouse lungs, arguing against rapid elimination after exposure (supplementary table S3, Supplementary Material online). Therefore, to monitor and quantify M. kansasii burden over time, we used a higher infectious dose than we use for M. tuberculosis. As shown in figure 8, following aerosol infection with M. kansasii, there was less than 10-fold difference in bacterial burden from day 1 through day 42 of infection. In contrast, for M. tuberculosis we observed over 10,000-fold increase in bacterial numbers by day 21. To contrast the phenotype of ESX-1-containing M. kansasii with an ESX-1 mutant of M. tuberculosis, we infected a third group of mice with an intermediate dose of M. tuberculosis:ΔRD1. In the first 21 days, the M. tuberculosis mutant had increased in burden by 100-fold, and by day 42, it had reached the same bacterial burden as wild-type M. tuberculosis, as has been previously described (Lewis et al. 2003). Thus, M. kansasii, with an intact ESX-1 locus manifest orders of magnitude reduced in vivo fitness compared with both wild-type M. tuberculosis and the ESX-1 mutant of M. tuberculosis following experimental aerosol infection in mice.
Fig. 8.—
Bacterial burden in the lungs of mice infected through an aerosol route with M. tuberculosis, M. tuberculosis:ΔRD1, and M. kansasii over the course of 42 days.
Discussion
In this study, we analyzed the complete genome sequence of M. kansasii, an opportunistic pathogen that causes a sporadic, nontransmissible TB-like disease in humans. In agreement with a previous report, M. kansasii is one of the phylogenetically closest species to M. tuberculosis, an observation supported by the two genomes sharing extended regions of collinearity and synteny. The similarity between these genomes affords an opportunity to infer the common ancestor of these organisms and thereby suggest a sequence of events that resulted in the emergence from an environmental mycobacterium of a host-adapted professional pathogen.
Using the M. kansasii genome as a representative of a slow-growing environmental mycobacterium, the major genetic events that characterize the emergence of M. tuberculosis comparison are gene loss, gene acquisition, and gene amplification. These same processes are inferred when comparing M. tuberculosis to other nontuberculous mycobacteria, such as M. marinum, suggesting that much of the excess genetic material found in environmental mycobacteria codes for functions not required to survive in, and transmit between, mammalian hosts. Although it is possible that gene loss was selected because of the deletion of unnecessary functions, it also remains possible that certain antivirulence systems were present in environmental mycobacteria, such that their deletion actively contributed to enhanced virulence. As one putative example, the loss of the cobF gene during the evolution toward the MTBC should be mentioned. The gene cobF, which is absent from all MTBC members, is part of a gene cluster that is conserved in many environmental mycobacteria, including M. kansasii (MKAN_09045), and was also found present in all tested M. canettii strains (Supply et al. 2013). It remains to be determined whether the loss of this gene, whose gene product is predicted to be involved in cobalamin synthesis and endogenous vitamin B12 production (Gopinath et al. 2013), might have resulted in MTBC strains that depended upon the host for host-supplied substrates (Boritsch et al. 2014).
A complementary possibility to explain the enhanced virulence of M. tuberculosis involves that acquisition, through horizontal gene transfer, of novel M. tuberculosis-specific genes. The observation that many of these genes, including Rv3377-8c and Rv0987/88, are also found in M. canettii, which is considerably more virulent than M. kansasii in experimental infection models, supports the potential role of these acquired genes in the adaptation to a pathogenic lifestyle. A final possibility relates to the expansion of existing gene families. A previously published evolutionary analysis indicated that the considerable number of PE/PPE genes in M. tuberculosis is likely the result of gene expansion, rather than gene acquisition (Gey van Pittius et al. 2006). Our comparative genomic analysis detected a comparable amount of PE/PPE genes are seen between M. kansasii and M. tuberculosis, suggesting that this particular gene expansion did not coincide with the emergence of M. tuberculosis. Instead, we found that between these organisms there has been a markedly increased number of toxin–antitoxin genes in M. tuberculosis, as has been previously noted (Ramage et al. 2009). The apparent expansion of this gene family since their common ancestor argues for further investigation of the role toxin–antitoxin systems in the ability of M. tuberculosis to infect and cause disease.
Our genomic comparisons were also notable for the conservation of genes that have been experimentally implicated in TB pathogenesis. A nonexhaustive list of putative virulence factors of M. tuberculosis that were detected in M. kansasii is provided in table 1. Among these, one notes in the M. kansasii genome the presence of all five known M. tuberculosis ESX loci encoding the T7SS (fig. 6 and supplementary fig. S3, Supplementary Material online) (Bitter et al. 2009). Remarkably, the pMK12478 plasmid also contains a locus that putatively encodes a T7SS that is the most homologous to ESX-5 (Ummels et al. 2014) (supplementary fig. S3, Supplementary Material online). The presence of this plasmid carrying elements of T7SS and T4SS in M. kansasii adds new elements to the discussion on the origin and function of T7SS in mycobacteria. Beyond evaluating genomic conservation, we conducted functional studies of one of these ESX loci, named ESX-1, which has been implicated in a wide variety of M. tuberculosis virulence phenotypes. Previous studies have reported the presence of the ESX-1 secreted protein ESAT-6 in M. kansasii bacterial pellets as well as 3- to 5-week-old culture filtrates (Sorensen et al. 1995; Arend et al. 2005). Here, we showed that M. kansasii secreted similar amounts of ESAT-6 as M. tuberculosis after 4 days of growth in Sauton’s medium, a growth medium known to support ESAT-6 secretion (Champion 2013). As ESAT-6 has been shown to be necessary for M. tuberculosis to cause phagosomal perturbation, leading to bacterial relocalization to the cytosol (van der Wel et al. 2007; Simeone et al. 2012), we hypothesized that M. kansasii would likewise perturb the phagosomal membrane during macrophage infection. Like many other mycobacteria, M. kansasii produces endogenous beta-lactamase (Kwon et al. 1995), permitting us to use a FRET-based assay to detect whether M. kansasii could disrupt the phagosomal membrane, allowing the beta-lactamase to cleave the cytosolic FRET substrate and thereby change fluorescence emission upon excitation. Both M. kansasii and M. tuberculosis were able to disrupt the phagosomal membrane, which is likely a prerequisite for cytosol translocation as demonstrated by a M. kansasii clinical isolate (Houben et al. 2012). Thus, both genomic comparisons and intracellular infection studies point to key similarities between these organisms, begging the question of what additional virulence determinants have been acquired by M. tuberculosis since their common ancestor.
In experimental infections, M. kansasii showed considerably less growth in vivo, resulting in a persistent infection without clinical disease. These findings, along with the absence of a number of M. tuberculosis-specific genes, suggest that M. kansasii may lend itself to mechanistic studies through gain-of-function investigations involving the addition of M. tuberculosis-specific genes. Although M. marinum has been well utilized as a surrogate bacteria for a number of TB pathogenesis studies (Ramakrishnan 2012; Cambier et al. 2014), there are a number of differences between M. marinum and M. tuberculosis, including the faster growth rate of the former at a lower temperature, and the use of nonmammalian hosts. In contradistinction, M. kansasii grows in the same conditions, and at the same rate as M. tuberculosis, offering the possibility of a number of in vitro investigations, including transcriptomic, proteomic, and lipidomic comparisons. Additionally, M. kansasii can be used to generate productive pulmonary infection of standard mice used for experimental infection studies, albeit with lesser bacterial growth following infection, providing an in vivo phenotyping possibility to formally test the importance of in silico-derived differences between the organisms.
In conclusion, our comparative phenotypic and genomic study of M. kansasii and M. tuberculosis revealed that although these two species share numerous similarities, including the conservation of many M. tuberculosis virulence factors, they differ markedly in terms of in vivo fitness. These data support the search for novel M. tuberculosis-specific virulence factors not found in other mycobacteria, both for their fundamental value in understanding the pathogenesis of TB, but also for their potential translational value in species-specific diagnostic assays and novel immunization approaches.
Supplementary Material
Supplementary tables S1–S3 and figures S1–S3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
J.W. was supported by a PhD studentship award from the Fonds de la Recherche en Sante du Quebec (FRSQ) where M.B. is a Chercheur National. M.B. acknowledges the support of an operating grant from the Canadian Institutes for Health Research, MOP-97813. R.B. acknowledges the support by the Fondation pour la Recherche Médicale FRM no. DEQ20130326471. J.E. was supported by an ERC starting grant (Rupteffects, number 261166), and both J.E. and R.B are members of the LabEx consortium IBEID at the Institut Pasteur. The authors thank members of the Behr, Reed and Schurr labs for assistance and suggestions.
Literature Cited
- Abdallah AM, et al. Type VII secretion—mycobacteria show the way. Nat Rev Microbiol. 2007;5:883–891. doi: 10.1038/nrmicro1773. [DOI] [PubMed] [Google Scholar]
- Alcaide F, et al. Heterogeneity and clonality among isolates of Mycobacterium kansasii: implications for epidemiological and pathogenicity studies. J Clin Microbiol. 1997;35:1959–1964. doi: 10.1128/jcm.35.8.1959-1964.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arend SM, et al. ESAT-6 and CFP-10 in clinical versus environmental isolates of Mycobacterium kansasii. J Infect Dis. 2005;191:1301–1310. doi: 10.1086/428950. [DOI] [PubMed] [Google Scholar]
- Aziz RK, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belisle JT, Brennan PJ. Chemical basis of rough and smooth variation in mycobacteria. J Bacteriol. 1989;171:3465–3470. doi: 10.1128/jb.171.6.3465-3470.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besemer J, Lomsadze A, Borodovsky M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29:2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitter W, et al. Systematic genetic nomenclature for type VII secretion systems. PLoS Pathog. 2009;5:e1000507. doi: 10.1371/journal.ppat.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boritsch EC, et al. A glimpse into the past and predictions for the future: the molecular evolution of the tuberculosis agent. Mol Microbiol. 2014;93:835–852. doi: 10.1111/mmi.12720. [DOI] [PubMed] [Google Scholar]
- Brodin P, et al. Dissection of ESAT-6 system 1 of Mycobacterium tuberculosis and impact on immunogenicity and virulence. Infect Immun. 2006;74:88–98. doi: 10.1128/IAI.74.1.88-98.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambier CJ, Falkow S, Ramakrishnan L. Host evasion and exploitation schemes of Mycobacterium tuberculosis. Cell. 2014;159:1497–1509. doi: 10.1016/j.cell.2014.11.024. [DOI] [PubMed] [Google Scholar]
- Canueto-Quintero J, et al. Epidemiological, clinical, and prognostic differences between the diseases caused by Mycobacterium kansasii and Mycobacterium tuberculosis in patients infected with human immunodeficiency virus: a multicenter study. Clin Infect Dis. 2003;37:584–590. doi: 10.1086/376987. [DOI] [PubMed] [Google Scholar]
- Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics (Oxford, England) 2009;25:119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver TJ, et al. ACT: the Artemis Comparison Tool. Bioinformatics (Oxford, England) 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- Cassidy PM, Hedberg K, Saulson A, McNelly E, Winthrop KL. Nontuberculous mycobacterial disease prevalence and risk factors: a changing epidemiology. Clin Infect Dis. 2009;49:e124–e129. doi: 10.1086/648443. [DOI] [PubMed] [Google Scholar]
- Champion PA. Disconnecting in vitro ESX-1 secretion from mycobacterial virulence. J Bacteriol. 2013;195:5418–5420. doi: 10.1128/JB.01145-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JM, et al. EspD is critical for the virulence-mediating ESX-1 secretion system in Mycobacterium tuberculosis. J Bacteriol. 2012;194:884–893. doi: 10.1128/JB.06417-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole ST, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- Comas I, et al. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet. 2010;42:498–503. doi: 10.1038/ng.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comas I, et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat Genet. 2013;45:1176–1182. doi: 10.1038/ng.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Converse PJ, et al. Role of the dosR-dosS two-component regulatory system in Mycobacterium tuberculosis virulence in three animal models. Infect Immun. 2009;77:1230–1237. doi: 10.1128/IAI.01117-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett EL, et al. Risk factors for pulmonary mycobacterial disease in South African gold miners. A case-control study. Am J Respir Crit Care Med. 1999;159:94–99. doi: 10.1164/ajrccm.159.1.9803048. [DOI] [PubMed] [Google Scholar]
- Criscuolo A, Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 2010;10:210. doi: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daleke MH, et al. Specific chaperones for the type VII protein secretion pathway. J Biol Chem. 2012;287:31939–31947. doi: 10.1074/jbc.M112.397596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies PD. Infection with Mycobacterium kansasii. Thorax. 1994;49:435–436. doi: 10.1136/thx.49.5.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis SM. The spectrum of tuberculosis and non-tuberculous mycobacterial infection. Eur Radiol. 2004;14(Suppl. 3):E34–E42. doi: 10.1007/s00330-003-2042-1. [DOI] [PubMed] [Google Scholar]
- Evans AJ, Crisp AJ, et al. Pulmonary Mycobacterium kansasii infection: comparison of radiological appearances with pulmonary tuberculosis. Thorax. 1996;51:1243–1247. doi: 10.1136/thx.51.12.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SA, Colville A, Evans AJ, Crisp AJ, Johnston ID. Pulmonary Mycobacterium kansasii infection: comparison of the clinical features, treatment and outcome with pulmonary tuberculosis. Thorax. 1996;51:1248–1252. doi: 10.1136/thx.51.12.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre M, et al. High genetic diversity revealed by variable-number tandem repeat genotyping and analysis of hsp65 gene polymorphism in a large collection of “Mycobacterium canettii” strains indicates that the M. tuberculosis complex is a recently emerged clone of “M. canettii.”. J Clin Microbiol. 2004;42:3248–3255. doi: 10.1128/JCM.42.7.3248-3255.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre M, et al. Molecular characteristics of “Mycobacterium canettii” the smooth Mycobacterium tuberculosis bacilli. Infect Genet Evol. 2010;10:1165–1173. doi: 10.1016/j.meegid.2010.07.016. [DOI] [PubMed] [Google Scholar]
- Gagneux S. Host-pathogen coevolution in human tuberculosis. Philos Trans R Soc Lond B Biol Sci. 2012;367:850–859. doi: 10.1098/rstb.2011.0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gey van Pittius NC, et al. Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions. BMC Evol Biol. 2006;6:95. doi: 10.1186/1471-2148-6-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good RC, Snider DE., Jr Isolation of nontuberculous mycobacteria in the United States, 1980. J Infect Dis. 1982;146:829–833. doi: 10.1093/infdis/146.6.829. [DOI] [PubMed] [Google Scholar]
- Gopinath K, Moosa A, Mizrahi V, Warner DF. Vitamin B(12) metabolism in Mycobacterium tuberculosis. Future Microbiol. 2013;8:1405–1418. doi: 10.2217/fmb.13.113. [DOI] [PubMed] [Google Scholar]
- Griffith DE, et al. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med. 2007;175:367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- Guinn KM, et al. Individual RD1-region genes are required for export of ESAT-6/CFP-10 and for virulence of Mycobacterium tuberculosis. Mol Microbiol. 2004;51:359–370. doi: 10.1046/j.1365-2958.2003.03844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez MC, et al. Ancient origin and gene mosaicism of the progenitor of Mycobacterium tuberculosis. PLoS Pathog. 2005;1:e5. doi: 10.1371/journal.ppat.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JM, et al. N-glycolylated peptidoglycan contributes to the immunogenicity but not pathogenicity of Mycobacterium tuberculosis. J Infect Dis. 2014;209:1045–1054. doi: 10.1093/infdis/jit622. [DOI] [PubMed] [Google Scholar]
- Harboe M, Oettinger T, Wiker HG, Rosenkrands I, Andersen P. Evidence for occurrence of the ESAT-6 protein in Mycobacterium tuberculosis and virulent Mycobacterium bovis and for its absence in Mycobacterium bovis BCG. Infect Immun. 1996;64:16–22. doi: 10.1128/iai.64.1.16-22.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben D, et al. ESX-1-mediated translocation to the cytosol controls virulence of mycobacteria. Cell Microbiol. 2012;14:1287–1298. doi: 10.1111/j.1462-5822.2012.01799.x. [DOI] [PubMed] [Google Scholar]
- Joynson DH. Water: the natural habitat of Mycobacterium kansasii? Tubercle. 1979;60:77–81. doi: 10.1016/0041-3879(79)90039-4. [DOI] [PubMed] [Google Scholar]
- Kaustova J, et al. Endemic occurrence of Mycobacterium kansasii in water-supply systems. J Hyg Epidemiol Microbiol Immunol. 1981;25:24–30. [PubMed] [Google Scholar]
- Kwon HH, Tomioka H, Saito H. Distribution and characterization of beta-lactamases of mycobacteria and related organisms. Tuber Lung Dis. 1995;76:141–148. doi: 10.1016/0962-8479(95)90557-x. [DOI] [PubMed] [Google Scholar]
- Labrie SJ, El Haddad L, Tremblay DM, Plante PL, Wasserscheid J, Dumaresq J, Dewar K, Corbeil J, Moineau S. First complete genome sequence of Staphylococcus xylosus, a meat starter culture and a host to propagate Staphylococcus aureus phages. Genome Announc. 2014;2(4) doi: 10.1128/genomeA.00671-14. : e00671–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leistikow RL, et al. The Mycobacterium tuberculosis DosR regulon assists in metabolic homeostasis and enables rapid recovery from nonrespiring dormancy. J Bacteriol. 2010;192:1662–1670. doi: 10.1128/JB.00926-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemassu A, Levy-Frebault VV, Laneelle MA, Daffe M. Lack of correlation between colony morphology and lipooligosaccharide content in the Mycobacterium tuberculosis complex. J Gen Microbiol. 1992;138:1535–1541. doi: 10.1099/00221287-138-7-1535. [DOI] [PubMed] [Google Scholar]
- Lewis KN, et al. Deletion of RD1 from Mycobacterium tuberculosis mimics bacille Calmette-Guerin attenuation. J Infect Dis. 2003;187:117–123. doi: 10.1086/345862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillo M, Orengo S, Cernoch P, Harris RL. Pulmonary and disseminated infection due to Mycobacterium kansasii: a decade of experience. Rev Infect Dis. 1990;12:760–767. doi: 10.1093/clinids/12.5.760. [DOI] [PubMed] [Google Scholar]
- MacGurn JA, Cox JS. A genetic screen for Mycobacterium tuberculosis mutants defective for phagosome maturation arrest identifies components of the ESX-1 secretion system. Infect Immun. 2007;75:2668–2678. doi: 10.1128/IAI.01872-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marras TK, Chedore P, Ying AM, Jamieson F. Isolation prevalence of pulmonary non-tuberculous mycobacteria in Ontario, 1997 2003. Thorax. 2007;62:661–666. doi: 10.1136/thx.2006.070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSwiggan DA, Collins CH. The isolation of M. kansasii and M. xenopi from water systems. Tubercle. 1974;55:291–297. doi: 10.1016/0041-3879(74)90038-5. [DOI] [PubMed] [Google Scholar]
- Meyer F, Overbeek R, Rodriguez A. FIGfams: yet another set of protein families. Nucleic Acids Res. 2009;37:6643–6654. doi: 10.1093/nar/gkp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) Nucleic Acids Res. 2014;42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez E, et al. An essential role for phoP in Mycobacterium tuberculosis virulence. Mol Microbiol. 2001;41:179–187. doi: 10.1046/j.1365-2958.2001.02500.x. [DOI] [PubMed] [Google Scholar]
- Pollak A, Buhler VB. The cultural characteristics and the animal pathogenicity of an atypical acid-fast organism that causes human disease. Trans Annu Meet Natl Tuberc Assoc. 1953;49:86–88. [PubMed] [Google Scholar]
- Pym AS, Brodin P, Brosch R, Huerre M, Cole ST. Loss of RD1 contributed to the attenuation of the live tuberculosis vaccines Mycobacterium bovis BCG and Mycobacterium microti. Mol Microbiol. 2002;46:709–717. doi: 10.1046/j.1365-2958.2002.03237.x. [DOI] [PubMed] [Google Scholar]
- Ramage HR, Connolly LE, Cox JS. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet. 2009;5:e1000767. doi: 10.1371/journal.pgen.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan L. Revisiting the role of the granuloma in tuberculosis. Nat Rev Immunol. 2012;12:352–366. doi: 10.1038/nri3211. [DOI] [PubMed] [Google Scholar]
- Ray K, et al. Tracking the dynamic interplay between bacterial and host factors during pathogen-induced vacuole rupture in real time. Cell Microbiol. 2010;12:545–556. doi: 10.1111/j.1462-5822.2010.01428.x. [DOI] [PubMed] [Google Scholar]
- Richter E, Niemann S, Rusch-Gerdes S, Hoffner S. Identification of Mycobacterium kansasii by using a DNA probe (AccuProbe) and molecular techniques. J Clin Microbiol. 1999;37:964–970. doi: 10.1128/jcm.37.4.964-970.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricketts WM, O’Shaughnessy TC, van Ingen J. Human-to-human transmission of Mycobacterium kansasii or victims of a shared source? Eur Respir J. 2014;44:1085–1087. doi: 10.1183/09031936.00066614. [DOI] [PubMed] [Google Scholar]
- Roberts GD. Mycobacteria and Nocardia. In: Washington JA, editor. Laboratory procedures in clinical microbiology; Section of Clinical Microbiology. New York: Springer-Verlag; 1981. p. 856. [Google Scholar]
- Robledo JA, Murillo AM, Rouzaud F. Physiological role and potential clinical interest of mycobacterial pigments. IUBMB Life. 2011;63:71–78. doi: 10.1002/iub.424. [DOI] [PubMed] [Google Scholar]
- Ryndak M, Wang S, Smith I. PhoP, a key player in Mycobacterium tuberculosis virulence. Trends Microbiol. 2008;16:528–534. doi: 10.1016/j.tim.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Sampson SL. Mycobacterial PE/PPE proteins at the host-pathogen interface. Clin Dev Immunol. 2011;2011:497203. doi: 10.1155/2011/497203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Buso L, Comas I, Jorques G, Gonzalez-Candelas F. Recombination drives genome evolution in outbreak-related Legionella pneumophila isolates. Nat Genet. 2014;46:1205–1211. doi: 10.1038/ng.3114. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone R, et al. Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathog. 2012;8:e1002507. doi: 10.1371/journal.ppat.1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokurenko EV, Gomulkiewicz R, Dykhuizen DE. Source-sink dynamics of virulence evolution. Nat Rev Microbiol. 2006;4:548–555. doi: 10.1038/nrmicro1446. [DOI] [PubMed] [Google Scholar]
- Sorensen AL, Nagai S, Houen G, Andersen P, Andersen AB. Purification and characterization of a low-molecular-mass T-cell antigen secreted by Mycobacterium tuberculosis. Infect Immun. 1995;63:1710–1717. doi: 10.1128/iai.63.5.1710-1717.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supply P, et al. Genomic analysis of smooth tubercle bacilli provides insights into ancestry and pathoadaptation of Mycobacterium tuberculosis. Nat Genet. 2013;45:172–179. doi: 10.1038/ng.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taillard C, et al. Clinical implications of Mycobacterium kansasii species heterogeneity: Swiss National Survey. J Clin Microbiol. 2003;41:1240–1244. doi: 10.1128/JCM.41.3.1240-1244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson RM, et al. Factors associated with the isolation of Nontuberculous mycobacteria (NTM) from a large municipal water system in Brisbane, Australia. BMC Microbiol. 2013;13:89. doi: 10.1186/1471-2180-13-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamura M. Numerical classification of 280 strains of slowly growing mycobacteria. Proposal of Mycobacterium tuberculosis series, Mycobacterium avium series, and Mycobacterium nonchromogenicum series. Microbiol Immunol. 1983;27:315–334. doi: 10.1111/j.1348-0421.1983.tb00591.x. [DOI] [PubMed] [Google Scholar]
- Tsukamura M. Numerical identification of slowly growing mycobacteria. Microbiol Immunol. 1985;29:1039–1050. doi: 10.1111/j.1348-0421.1985.tb00894.x. [DOI] [PubMed] [Google Scholar]
- Tsukamura M, Mizuno S, Tsukamura S. Numerical classification of slowly growing mycobacteria. Am Rev Respir Dis. 1969;99:299–303. doi: 10.1164/arrd.1969.99.2.299. [DOI] [PubMed] [Google Scholar]
- Tsukamura S. Biological significance of pigments of mycobacteria. (Iii). Resistance to ultraviolet irradiation of various unclassified mycobacteria. Jpn J Tuberc Chest Dis. 1964;12:7–9. [PubMed] [Google Scholar]
- Ummels R, et al. Identification of a novel conjugative plasmid in mycobacteria that requires both type IV and type VII secretion. mBio. 2014;5:e01744–01714. doi: 10.1128/mBio.01744-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wel N, et al. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell. 2007;129:1287–1298. doi: 10.1016/j.cell.2007.05.059. [DOI] [PubMed] [Google Scholar]
- Veyrier F, Pletzer D, Turenne C, Behr MA. Phylogenetic detection of horizontal gene transfer during the step-wise genesis of Mycobacterium tuberculosis. BMC Evol Biol. 2009;9:196. doi: 10.1186/1471-2148-9-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira-Silva S, Touchon M, Abby SS, Rocha EP. Investment in rapid growth shapes the evolutionary rates of essential proteins. Proc Natl Acad Sci U S A. 2011;108:20030–20035. doi: 10.1073/pnas.1110972108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth T, et al. Origin, spread and demography of the Mycobacterium tuberculosis complex. PLoS Pathog. 2008;4:e1000160. doi: 10.1371/journal.ppat.1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Global tuberculosis report. Geneva (Switzerland): World Health Organization; 2013. [Google Scholar]
- Zhang F, Xie JP. Mammalian cell entry gene family of Mycobacterium tuberculosis. Mol Cell Biochem. 2011;352:1–10. doi: 10.1007/s11010-011-0733-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.