Abstract
Sequence capture and restriction site associated DNA sequencing (RADseq) are popular methods for obtaining large numbers of loci for phylogenetic analysis. These methods are typically used to collect data at different evolutionary timescales; sequence capture is primarily used for obtaining conserved loci, whereas RADseq is designed for discovering single nucleotide polymorphisms (SNPs) suitable for population genetic or phylogeographic analyses. Phylogenetic questions that span both “recent” and “deep” timescales could benefit from either type of data, but studies that directly compare the two approaches are lacking. We compared phylogenies estimated from sequence capture and double digest RADseq (ddRADseq) data for North American phrynosomatid lizards, a species-rich and diverse group containing nine genera that began diversifying approximately 55 Ma. Sequence capture resulted in 584 loci that provided a consistent and strong phylogeny using concatenation and species tree inference. However, the phylogeny estimated from the ddRADseq data was sensitive to the bioinformatics steps used for determining homology, detecting paralogs, and filtering missing data. The topological conflicts among the SNP trees were not restricted to any particular timescale, but instead were associated with short internal branches. Species tree analysis of the largest SNP assembly, which also included the most missing data, supported a topology that matched the sequence capture tree. This preferred phylogeny provides strong support for the paraphyly of the earless lizard genera Holbrookia and Cophosaurus, suggesting that the earless morphology either evolved twice or evolved once and was subsequently lost in Callisaurus.
Keywords: coalescence, ddRADseq, incomplete lineage sorting, RADseq, species tree, single nucleotide polymorphism, ultraconserved elements
Introduction
New methods for obtaining comparative genomics data are transforming phylogenetic studies of nonmodel organisms. Sequence capture and restriction site associated DNA sequencing (RADseq) are emerging as two of the most useful reduced-representation genome sequencing methods for phylogenetic and population-level studies. Sequence capture methods use short probes (60–120 nt) to hybridize to specific genomic regions that are subsequently sequenced, and therefore these methods require some advanced level of knowledge of the genomes under investigation (Gnirke et al. 2009; Mamanova et al. 2010). Sequence capture has been applied to a variety of studies aiming to resolve phylogenetic relationships at relatively “deep” evolutionary timescales, including mammals (McCormack et al. 2012), birds (McCormack et al. 2013), turtles and archosaurs (Crawford et al. 2012), fishes (Li et al. 2013), and squamates (Leaché et al. 2014; Pyron et al. 2014). RADseq methods (Baird et al. 2008) rely on restriction enzyme digestion of genomic DNA followed by the subsequent size-selection and sequencing of fragments that are of a certain size range (Miller et al. 2007; Puritz et al. 2014). The approach requires limited to no previous knowledge of the genome, which has made it a popular choice for studying recent speciation in organisms that lack existing genomic resources, including mosquitos (Emerson et al. 2010), plants (Eaton and Ree 2013), cichlids (Wagner et al. 2013), and beetles (Cruaud et al. 2014).
Sequence capture and RADseq data have great utility for phylogenetic investigations at different evolutionary timescales, yet the boundary separating the utility of each approach is unclear. Sequence capture using ultraconserved elements (UCEs) was originally described as an approach for resolving deep phylogenies (Faircloth et al. 2012); however, recently it has been shown to be useful for phylogeographic studies (Smith et al. 2014). Likewise, the application of RADseq methods has been extended from shallow timescales to divergences dating back to 50–60 Ma (Rubin et al. 2012; Cariou et al. 2013). Whether the two approaches provide similar results (i.e., congruent phylogenetic trees) for relationships across any particular timescale is unknown, because both data types have not been collected for the same study system (but see Harvey et al. 2013). The properties of the DNA sequence data alignments provided by the methods are quite different, which could result in different biases during phylogenetic analysis. For example, sequence capture provides relatively long loci (hundreds to thousands of nucleotides) with little missing data, whereas RADseq has the potential to recover thousands of short loci (50–150 nt, depending on sequencing effort), with large amounts of missing data resulting from allelic dropout (Arnold et al. 2013). Resolving difficult phylogenetic problems such as rapid speciation events requires sampling hundreds or thousands of loci (Liu and Edwards 2009), but whether the increased number of loci offered by RADseq methods is offset by the short length of the loci and missing data have not been explored.
The iguanian lizard family Phrynosomatidae is composed of 9 genera and 148 species and is therefore the most diverse and species-rich family of lizards in North America (Uetz 2014). This family is distributed broadly across North and Central America from southern Canada to Panama, and most diversity is centered in arid regions of the American Southwest and Mexico. The broad distribution and high species diversity of phrynosomatid lizards have made them an important focal group for comparative studies in ecology and evolutionary biology (e.g., Sinervo and Lively 1996; Lambert and Wiens 2013; Wiens et al. 2013). However, despite numerous phylogenetic studies, the relationships among the nine genera have been difficult to resolve. The relationships among the sand lizard genera Cophosaurus, Callisaurus, Holbrookia, and Uma are unclear, and previous studies based on morphology (de Queiroz 1989), allozymes (de Queiroz 1992), and mitochondrial DNA (mtDNA; Reeder 1995; Reeder and Wiens 1996; Wilgenbusch and de Queiroz 2000; Leaché and McGuire 2006; Wiens et al. 2010) have produced conflicting results. Identifying the order of divergence events within the sand lizards, and whether or not the two “earless” genera with concealed tympanic membranes (Cophosaurus and Holbrookia) form a clade are the two main questions that remain unanswered. Recent phylogenetic studies utilizing mitochondrial and nuclear genes converge on a common topology for these genera and support both Uma as sister to the other sand lizards and monophyly of the earless lizards (Wiens et al. 2010, 2013). The relationships among the sceloporines (Petrosaurus, Sceloporus, Urosaurus, and Uta) have been difficult to resolve due to rapid and successive speciation. These studies support a clade containing Urosaurus and Sceloporus (Wiens et al. 2010, 2013). However, determining whether Petrosaurus or Uta is the sister group to other sceloporines has remained uncertain (Wiens et al. 2010). Analyses based on concatenating independent loci differ from coalescent-based species trees, which indicates that gene tree conflict from incomplete lineage sorting could be affecting this part of the phrynosomatid tree.
In this study, we use new molecular data collected using sequence capture and double digest RADseq (ddRADseq; Peterson et al. 2012) to estimate the phylogenetic relationships among phrynosomatid lizard genera. We estimate phylogenetic trees for the sequence capture data using concatenation and coalescent-based species tree inference techniques, and we examine the genome-wide support for competing phylogenetic hypotheses for phrynosomatid lizards. The ddRADseq data are assembled using a variety of thresholds that govern the homology, paralogy, and levels of missing data. The phylogenetic trees estimated from the ddRADseq data assemblies are compared against each other and to the sequence capture data.
Materials and Methods
Sampling
We sampled one species from each of the nine genera of the Phrynosomatidae (table 1), including Callisaurus draconoides, Cophosaurus texanus, Holbrookia maculata, Petrosaurus thalassinus, Phrynosoma sherbrookei, Sceloporus occidentalis, Uma notata, Urosaurus ornatus, and Uta stansburiana. Two additional species, Gambelia wislizenii and Liolaemus darwinii, were included as outgroups for the sequence capture experiment, and G. wislizenii was included in the ddRADseq protocol for the same purpose. DNA was extracted from tissues using a NaCl extraction method (MacManes 2013) or a Qiagen DNeasy kit.
Table 1.
Species Included in the Analysis and an Overview of the Sequence Capture Data
| Species | Voucher | Raw Reads | Clean Reads | Nuclear Loci Captureda | Nuclear Loci k-mer Depthb | mtDNA (bp)c | mtDNA k-mer Depthd |
|---|---|---|---|---|---|---|---|
| Phrynosomatidae | |||||||
| Callisaurus draconoides | MVZ 265543 | 9,622,116 | 9,035,068 | 575 | 23,280 | 13,106 | 1,502,772 |
| Cophosaurus texanus | UWBM 7347 | 9,176,180 | 8,625,204 | 573 | 24,401 | 15,609 | 2,482,706 |
| Holbrookia maculata | UWBM 7362 | 12,314,136 | 11,604,340 | 573 | 31,000 | 12,865 | 1,307,531 |
| Petrosaurus thalassinus | MVZ 161183 | 4,500,868 | 3,959,796 | 523 | 8,281 | 7,898 | 248,342 |
| Phrynosoma sherbrookei | MZFC 28101 | 7,634,142 | 6,971,920 | 579 | 14,107 | 12,967 | 47,287 |
| Sceloporus occidentalis | UWBM 6281 | 13,531,214 | 12,733,646 | 540 | 30,235 | 7,422 | 113,757 |
| Uma notata | SDSNH 76166 | 2,332,400 | 2,099,068 | 577 | 4,232 | 7,296 | 20,763 |
| Urosaurus ornatus | UWBM 7587 | 3,427,288 | 3,042,766 | 577 | 6,673 | 6,286 | 28,028 |
| Uta stansburiana | UWBM 7605 | 12,927,696 | 12,085,734 | 538 | 25,034 | 16,703 | 1,144,368 |
| Outgroups | |||||||
| Gambelia wislizenii | UWBM 7353 | 9,874,902 | 7,824,714 | 549 | 5,180 | 15,790 | 581,925 |
| Liolaemus darwinii | LJAMM-CNP 14634 | 3,253,800 | 2,935,874 | 581 | 8,715 | 11,751 | 41,572 |
aTotal loci targeted = 585.
bAverage number of 90-bp k-mers across all captured loci.
cTotal base pairs; aligned length = 17,187 bp.
dNumber of 90-bp k-mers.
Sequence Capture Data Collection
To obtain a large collection of homologous loci from throughout the genome, we designed a set of RNA probes specific for iguanian lizards. The probes are a subset of the 5,472 UCE probes published by Faircloth et al. (2012) with ≥99% sequence similarity to published genomes for Anolis carolinensis (Alföldi et al. 2011) and S. occidentalis (Genomic Resources Development Consortium et al. 2015). We excluded loci that were within 100 kb of one another to reduce any chance of linkage. We identified 541 UCE loci that matched both published genomes, and we tiled two 120-bp probes for each locus that overlapped by 60 bp. We included probes for 44 additional genes used in the squamate Tree of Life project (Wiens et al. 2012). The loci were included to increase the overlap between our new data with existing genetic resources for squamate reptiles. In total, we synthesized 1,170 custom probes (targeting 585 loci) using the MYbaits target enrichment kit (MYcroarray Inc., Ann Arbor, MI).
Genomic DNA (400 ng) was sonicated to a target peak of 400 bp using a Bioruptor Pico (Diagenode Inc.). Genomic libraries were prepared using an Illumina Truseq Nano library preparation kit. The samples were hybridized to the RNA-probes in the presence of a blocking mixture composed of forward and reverse compliments of the Illumina Truseq Nano Adapters, with inosines in place of the indices, as well as chicken blocking mix (Chicken Hybloc, Applied Genetics Lab Inc.) to reduce repetitive DNA binding to beads. Libraries were incubated with the RNA probes for 24 h at 65 °C. Post-hybridized libraries were enriched using Truseq adapter primers with Phusion Taq polymerase (New England Biolabs Inc.) for 20 cycles. Enriched libraries were cleaned with AMPure XP beads. We quantified enriched libraries using quantitative polymerase chain reaction (qPCR) (Applied Biosystems Inc.) with primers targeting five loci mapping to different chromosomes in the Anolis genome. Library quality was verified using an Agilent TapeStation 2200 (Agilent Tech.). These samples were pooled in equimolar ratios and sequenced using an Illumina HiSeq2000 (100-bp, paired-end reads) at the QB3 facility at UC Berkeley.
Sequence Capture Bioinformatics
The raw DNA sequences were processed using Casava (Illumina), which demultiplexes the sequencing run based on sequence tags. The program Trimmomatic (Bolger et al. 2014) was used to remove low-quality reads, trim low-quality ends, and remove adapter sequences. The cleaned paired-reads were organized by individual and then assembled with the de novo assembler IDBA (Peng et al. 2010). We ran IDBA iteratively over k-mer values from 50 to 90 with a step length of 10. We used phyluce (Faircloth et al. 2012) to assemble loci across species. We started by aligning species-specific assemblies to the probe sequences using the program LASTZ (available from http://www.bx.psu.edu/miller_lab/ last accessed February 20, 2015). After creating an SQL relational database of assembly-to-probe matches for each species, we queried the database for loci that were shared for a minimum of three species across all samples, and for those that were present across all species. We performed multiple sequence alignments for each locus using MAFFT (Katoh and Standley 2013), and long ragged-ends were trimmed to reduce missing or incomplete data.
We authenticated the identity of each sample by aligning our new data for one of the protein-coding nuclear genes (PRLR) with data published by Wiens et al. (2010). This is an important step when using exemplar sampling to verify the identity of each sample. We conducted a multiple sequence alignment with MAFFT, and performed a maximum likelihood (ML) analysis using RAxML v8.0.2 (Stamatakis 2014) with 100 bootstrap replicates under the GTRGAMMA model. As expected, the phrynosomatid lizards in our study each formed a clade with their proper genus (results not shown).
Sequence Capture Phylogenetic Analysis
ML phylogenetic analyses were conducted using RAxML v8.0.2 (Stamatakis 2014) with the GTRGAMMA model. We estimated gene trees for each locus separately, and also conducted an analysis of the concatenated data. Branch support was estimated using the automatic bootstrap function, which calculates a stopping rule to determine when sufficient replicates have been generated (Pattengale et al. 2010). The individual sequence capture ML trees were filtered in PAUP* v.4b10 (Swofford 2003) to calculate the number of loci that supported particular topological arrangements for phrynosomatid lizards found by previous studies using morphology, allozymes, mtDNA, or nuclear loci. The concatenated data were also analyzed using Bayesian inference (BI) with MrBayes v3.2 (Ronquist et al. 2012). The MrBayes analysis was run for 2 million generations with two independent runs (each with four chains), sampling every 1,000 generations. Summaries of the posterior distribution excluded the first 25% of samples as burn-in. We also conducted phylogenetic analyses of mtDNA genome data using ML and BI (as described above). The mtDNA genomes are present in high copy number during library preparation, and fragments of this locus are sequenced as “by-catch” along with the nuclear loci. All trees were rooted with G. wislizenii.
We estimated divergence times for the concatenated sequence capture data using BEAST v1.8.1 (Drummond et al. 2012). We repeated the analysis for the mtDNA data to obtain a time-calibrated gene tree for this locus. We used marginal likelihood estimation (Baele et al. 2013) to compare a strict clock to the uncorrelated lognormal relaxed clock. Marginal likelihoods were estimated using path sampling and stepping-stone analyses (Baele et al. 2012), both with 100 sampling steps with 100,000 generations for each step. The strict clock was rejected for the sequence capture data (2 × loge Bayes Factor = 872) and for the mtDNA data (2 × loge Bayes Factor = 34). All analyses used an uncorrelated lognormal relaxed clock, Yule tree prior, and an HKY (Hasegawa–Kishino–Yano)+Γ model of nucleotide substitution. We applied one calibration point to obtain divergence times across the tree using the molecular dating results of previous studies that included up to four fossil calibrations (Wiens et al. 2013). We assumed that the crown group age for phrynosomatid lizards was on average 55 Ma (normal distribution, mean = 55, SD = 4), resulting in a 95% highest probability density ranging from 48.4 to 61.6 Ma. Two replicate analyses of 40 million generations each were run (2 million for the mtDNA), sampling every 4,000 steps (1,000 for the mtDNA), and discarding the first 25% prior to combining the results using LogCombiner v1.8. We calculated a maximum clade credibility tree using TreeAnnotator v1.8.
We estimated a species tree using MPEST v1.4 (Liu et al. 2010). This method estimates a coalescent species tree using the gene tree topology for each locus as the starting input. Using gene tree topologies instead of DNA sequences decreases the computation time of estimating a species tree and makes the approach advantageous for large phylogenomic data sets. However, the method does not account for gene tree estimation error, and this can reduce the accuracy of the species tree. We used the best ML gene tree estimated for each locus as the input for MPEST. To obtain support measures on the species tree, we ran MPEST 100 times using each of the 100 ML bootstrap trees obtained for each locus. The support measures were obtained by calculating an extended majority-rule consensus tree for the 100 species trees estimated by MPEST. The resulting taxon bipartitions measure the percentage of times that each bipartition occurred across the 100 species trees.
We also estimated a species tree for the sequence capture data using BP&P v3 (Rannala and Yang 2003; Yang and Rannala 2014). This method estimates a species tree using the multispecies coalescent model directly from the DNA sequence alignments while accounting for incomplete lineage sorting due to ancestral polymorphism. This full-Bayesian procedure accommodates uncertainty in gene tree estimation during species tree estimation and provides posterior probability values for species relationships. The method assumes the Jukes–Cantor model for the substitution process, with no rate variation across sites within a locus. Prior distributions are required for the population sizes and the age of the root of the tree in units of expected substitutions. A gamma prior G(2, 1,000), with mean 2/2,000 = 0.001, was used for the population size parameters. The age of the root in the species tree was assigned the gamma prior G(2, 100). After an initial burn-in of 1,000 steps we ran the analysis for 1 million generations, sampling every 100 steps. The analysis was repeated four times with random starting seeds to confirm adequate mixing and consistent results.
We also estimated a species tree using SVDquartets (Chifman and Kubatko 2014). This method infers the topology among randomly sampled quartets of species using a coalescent model, and then a quartet method is used to assemble the randomly sampled quartets into a species tree. We randomly sampled 10,000 quartets from the data matrix, and used the program Quartet MaxCut v.2.1.0 (Snir and Rao 2012) to infer a species tree from the sampled quartets. We measured uncertainty in relationships using nonparametric bootstrapping with 100 replicates. The bootstrap values were mapped to the species tree estimated from the original data matrix using SumTrees v.3.3.1 (Sukumaran and Holder 2010).
ddRADseq Data Collection
We collected ddRADseq data following the protocol described by Peterson et al. (2012). We double-digested 500 ng of genomic DNA for each sample with 20 units each of a rare cutter SbfI (restriction site 5′-CCTGCAGG-3′) and a common cutter MspI (restriction site 5′-CCGG-3′) in a single reaction with the manufacturer recommended buffer (New England Biolabs) for 4 h at 37 °C. Fragments were purified with Agencourt AMPure beads before ligation of barcoded Illumina adaptors onto the fragments. The oligonucleotide sequences used for barcoding and adding Illumina indexes during library preparation are provided in Peterson et al. (2012). The libraries were size-selected (between 415 and 515 bp after accounting for adapter length) on a Pippin Prep size fractionator (Sage Science). Precise size selection is critical with ddRADseq, because it minimizes variation in fragment size-based locus selection among libraries and increases the likelihood of obtaining homologous loci across samples (Puritz et al. 2014). The final library amplification used proofreading Taq and Illumina’s indexed primers. The fragment size distribution and concentration of each pool were determined on an Agilent 2200 TapeStation or 2100 Bioanalyzer, and qPCR was performed to determine sequenceable library concentrations before multiplexing equimolar amounts of each pool for sequencing on a single Illumina HiSeq 2500 lane (50-bp, single-end reads; pooled with 60 other samples) at the QB3 facility at UC Berkeley.
ddRADseq Bioinformatics
We processed raw Illumina reads using the program pyRAD v.2.17 (Eaton 2014). An advantage of pyRAD over other RADseq data set assembly tools such as Stacks (Catchen et al. 2013) is that it is designed to assemble data for phylogenetic studies containing divergent species using global alignment clustering, which may include indel variation. We demultiplexed samples using their unique barcode and adapter sequences, and sites with Phred quality scores under 99% (Phred score = 20) were changed into “N” characters, and reads with ≥10% N’s were discarded. Each locus was reduced from 50 to 39 bp after the removal of the 6-bp restriction site overhang and the 5-bp barcode. The filtered reads for each sample were clustered using the program USEARCH v.6.0.307 (Edgar 2010), and then aligned with MUSCLE (Edgar 2004). This clustering step establishes homology among reads within a species. We assembled the ddRADseq data using three different clustering thresholds (clustering = 80%, 90%, and 95%) to determine the impact of this parameter on phylogeny inference. As an additional filtering step, consensus sequences were discarded that had low coverage (<6 reads), excessive undetermined or heterozygous sites (>3), or too many haplotypes (>2 for diploids). The consensus sequences were clustered across samples using the same three thresholds used to cluster data within species (80%, 90%, and 95%). This step establishes locus homology among species. Each locus was aligned with MUSCLE, and a filter was used to exclude potential paralogs. The paralog filter removes loci with excessive shared heterozygosity among samples. The justification for this filtering method is that shared heterozygous single nucleotide polymorphisms (SNPs) across species are more likely to represent a fixed difference among paralogs than shared heterozygosity within orthologs among species. We applied two paralog filter levels to determine the potential impact of paralog detection on phylogeny inference, including a strict filter that allowed no shared heterozygosity (paralog = 1), and a more relaxed filter that allowed a maximum of three species to be heterozygous at a given site (paralog = 3).
The final ddRADseq loci were assembled by adjusting a minimum individual (min. ind.) value, which specifies the minimum number of individuals that are required to have data present at a locus in order for that locus to be included in the final matrix. Our ddRADseq data set contains ten species (nine phrynosomatid lizard genera and one outgroup), and setting min. ind. = 10 retains loci with data present for all ten species ( = 100% complete matrix). In contrast, setting min. ind. = 3 retains any locus with data present for three or more species. We compiled data matrices with min. ind. values ranging from 3 to 10 to study the sensitivity of missing data on phylogenetic analysis.
ddRADseq Phylogenetic Analysis
We estimated phylogenetic trees for the concatenated ddRADseq data using RAxML with the GTRGAMMA model. We did not attempt to estimate gene trees for the individual RAD loci, because each locus was only 39 bp after removing the 5-bp barcode and 6-bp restriction enzyme recognition sequences. The data were concatenated and branch support was estimated with the automatic bootstrap function. We estimated phylogenetic trees using 36 combinations of assembly parameters, including 1) six different min. ind. values that modulated the amount of missing data tolerated at any given locus (min. ind. values ranged from 3 to 8; higher values produce too few loci for meaningful comparisons), 2) two paralog filter values (paralogs = 1, paralogs = 3), and 3) three locus clustering thresholds (80%, 90%, and 95%).
Species trees were estimated from the ddRADseq data using SVDquartets. An advantage of this approach for analyses of ddRADseq data is that it seems to be able to handle large amounts of missing data. We randomly sampled 10,000 quartets from the data matrix, and used Quartet MaxCut to infer a species tree from the sampled quartets. We used nonparametric bootstrapping with 100 replicates to measure uncertainty in the tree. The bootstrap values were mapped to the species tree estimated from the original data matrix using SumTrees.
Results
Sequence Capture
Of the 585 loci targeted by the probes, the sequence capture protocol resulted in 584 loci shared among a minimum of three species. A total of 471 loci were shared among all phrynosomatid and outgroup species included in the study. These 584 loci provided a total of 358,363 bp for phylogenetic analysis, and they varied in length from 284 to 1,054 bp (mean = 615 bp). On average, the loci contained 11.2% variation (parsimony informative and uninformative sites; min = 0.8%; max = 31.2%; table 2). The number of parsimony informative sites ranged from 0 to 70 (mean = 20). The mtDNA data alignment was 17,187 bp in length, and these data contained 3,773 parsimony informative characters (19.4% variation; table 2).
Table 2.
Characteristics of the Sequence Capture Loci
| Data | Length (bp) | Variation (%) | PI |
|---|---|---|---|
| Nuclear locia | 615 (284–1,054)b | 11.2 (0.8–31.2) | 20 (0–70) |
| Combined nuclear loci | 358,363 | 11.2% | 11,850 |
| Mitochondrial DNA | 17,187 | 19.4% | 3,773 |
Note.—PI, parsimony-informative characters.
aLoci captured for ≥3 species = 584.
BMean (min–max).
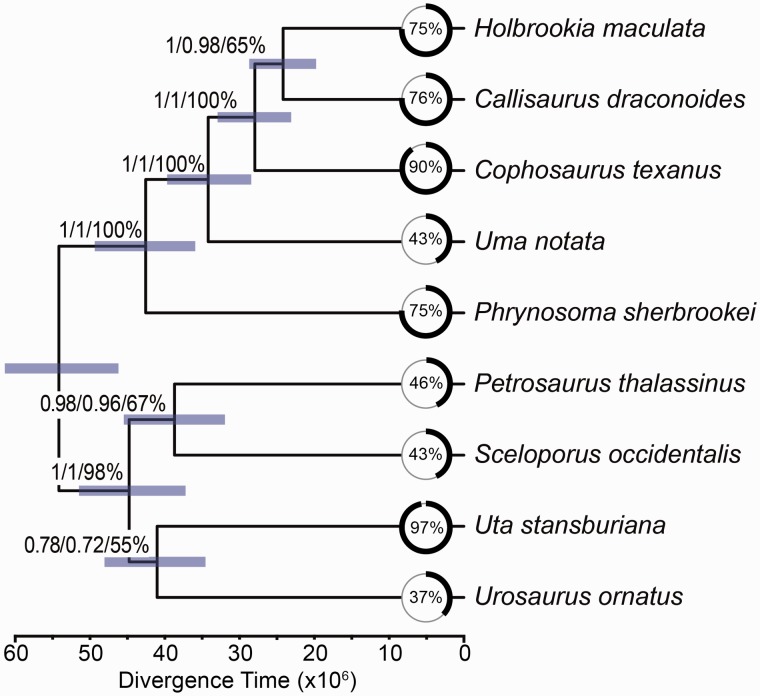
Phylogenetic analyses of the concatenated sequence capture loci using ML and BI (MrBayes and BEAST) provided strong support (ML bootstraps = 100%; posterior probabilities = 1.0) for a fully resolved phylogeny (fig. 1). Within the sceloporines, Sceloporus and Urosaurus are sister taxa, and Uta is sister to this clade, followed by Petrosaurus (fig. 1). The divergence time for the sceloporine crown group is 40.1 Ma (95% highest posterior density [HPD] = 33.2–46.9), and the subsequent times between speciation events leading to Uta and the Sceloporus + Urosaurus clade are short (1.7 and 3.7 Ma, respectively; fig. 1). These short divergence times are likely responsible for the difficulties that previous studies faced when trying to resolve this phylogeny with fewer loci. Within the Phrynosomatinae, Phrynosoma is the sister taxon to the remaining genera that form the sand lizards (i.e., Uma, Callisaurus, Cophosaurus, and Holbrookia) with a divergence time estimated at 38.2 Ma (95% HPD = 31.9–45.0 Ma). Within the sand lizards, Uma is sister to the remaining genera, followed by Cophosaurus. The clade containing Callisaurus and Holbrookia results in the paraphyly of the earless genera Holbrookia and Cophosaurus (fig. 1). The internal branch separating these three genera is short (2.7 Ma).
Fig. 1.—
Phylogenomic relationships among phrynosomatid lizards estimated with sequence capture data using BEAST. Bars on nodes indicate the 95% HPD for divergence times. Analyses using concatenation (RAxML, MrBayes, BEAST; 584 or 471 loci) and coalescent methods (SVDquartets, MPEST, BP&P; 471 loci) support the same topology. Concatenation provides absolute support on each node (bootstrap = 100%; posterior probability = 1.0), whereas the coalescent methods provide lower support for three short internal branches. Numbers on nodes are support values from SVDquartets (top), MPEST (middle), and BP&P posterior probabilities (bottom). Photographs by C.W.L., J.A.G., and A.D.G.
The coalescent-based species tree analyses supported the same topology as the concatenated data analyses, although the support was not as decisive for the shorter internal branches of the tree. Only three branches were not supported by 100% of the replicate MPEST or SVDquartet analyses. First, the clade containing Sceloporus and Urosaurus was only recovered 89% of time using MPEST. Second, the placement of Uta sister to the Sceloporus + Urosaurus clade received 99% bootstrap support from MPEST and 91% from SVDquartets. Third, the sister group relationship between Holbrookia and Callisaurus received 92% from MPEST and 99% from SVDquartets. The species tree analyses conducted with the Bayesian method BP&P provided posterior probabilities for relationships, and all relationships received a posterior probability of 1.0 with the exception of the clade containing Uta, Sceloporus, and Urosaurus (posterior probability = 0.54).
We quantified the number of gene trees that supported the estimated and alternative phylogenetic relationships to gauge the level of gene tree discordance among the sequence capture data (table 3). The relationship of Callisaurus + Holbrookia was represented by 137 loci (37.2%), the highest proportion of the possible relationships. The primary alternative relationship that we tested was the monophyly of the earless lizard genera, Holbrookia + Cophosaurus. A total of 103 of the sequence capture loci (21.9% of all loci examined) supported this alternative topology (table 3). An alternative that was even more common among the gene trees was a clade containing Cophosaurus + Callisaurus (120 loci), an untraditional grouping that also renders the earless lizards paraphyletic. We also quantified the number of nuclear loci that supported the alternative groupings recovered by the mtDNA gene tree (fig. 2). For example, the mtDNA clade containing Sceloporus + Petrosaurus is supported by 55 nuclear loci, and the Urosaurus + Uta clade is supported by 74 loci. The phylogenetic signal in the mtDNA gene tree is present in some of the sequence capture loci, but at very low frequency (<20% of all loci examined).
Table 3.
The Number of Nuclear Gene Trees Supporting Alternative Phrynosomatid Lizard Topologies
| Clade | Number of Loci | Frequency (%)a |
|---|---|---|
| Holbrookia + Callisaurusb | 175 | 37.2 |
| Holbrookia + Callisaurus + Cophosaurusb | 340 | 72.2 |
| Sand lizardsb | 210 | 44.6 |
| Sand lizards + Phrynosomab | 319 | 67.7 |
| Sceloporinesb | 226 | 48.0 |
| Sceloporus + Urosaurus + Utab | 91 | 19.3 |
| Sceloporus + Urosaurusb | 130 | 27.6 |
| Cophosaurus + Callisaurus | 120 | 25.5 |
| Holbrookia + Cophosaurusc | 103 | 21.9 |
| Urosaurus + Utad | 74 | 15.7 |
| Sceloporus + Uta | 63 | 13.4 |
| Sceloporus + Petrosaurusd | 55 | 11.7 |
| Uma + Cophosaurus | 19 | 4.0 |
aCalculated from complete loci only (471 total).
bClade supported by the sequence capture data in figure 1.
cEarless lizard clade.
dMitochondrial gene tree relationship.
Fig. 2.—
Gene tree estimated from mtDNA data fragments. Bars on nodes indicate the 95% HPD for divergence times. Support values are shown on branches (BEAST/MrBayes/RAxML), and the overall completeness for the mtDNA genomes is shown on the tips.
Double Digest RADseq
The number of loci assembled for each species with the ddRADseq data scales with the sequence similarity threshold used to determine homology while clustering reads (table 4). Conservative clustering (e.g., 95% clustering vs. 80% clustering) produces more loci per species, but as a consequence the mean sequencing depth per locus is reduced (table 4). The characteristics of the ddRADseq data matrices assembled using different thresholds for among-sample clustering, paralog filtering, and sequence coverage are provided in table 5. Although we recovered thousands of ddRADseq loci for each sample (table 4), there are no shared loci recovered across all ten species (i.e., min. ind. = 10) using conservative clustering. Allowing one individual to have missing data at a locus (i.e., min. ind. = 9) only increases the total number of loci to 3, which demonstrates the difficulty in obtaining homologous loci using the ddRADseq approach for distantly related species (table 5). Setting min. ind. = 3 and relaxing the clustering threshold to 80% produce over 2,600 loci containing 16,002 or 15,725 SNPs depending on the paralog filter (table 5). Increasing the stringency on the min. ind. parameter provides fewer loci and reduces the amount of missing data in the final data matrix. The coverage values for the ddRADseq assemblies are high (table 4), indicating that sequencing effort is probably not the main contributor to the high levels of missing data that we observed. It seems more likely that allelic dropout due to mutations at restriction sites (or mutations causing changes in the size of loci) is responsible for the patterns of missing data that we observed.
Table 4.
Summary of ddRADseq Data within Sample Clustering
| Species | Clusteringa = 80% |
Clustering = 90% |
Clustering = 95% |
||||
|---|---|---|---|---|---|---|---|
| Readsb | Locic | Depthd | Loci | Depth | Loci | Depth | |
| Callisaurus draconoides | 1,883,604 | 10,723 | 43.4 | 12,449 | 36.9 | 13,100 | 17.8 |
| Cophosaurus texanus | 1,452,471 | 8,686 | 41.8 | 10,048 | 35.9 | 10,553 | 18.4 |
| Holbrookia maculata | 699,921 | 4,657 | 27.5 | 7,880 | 24.2 | 11,156 | 14.2 |
| Petrosaurus thalassinus | 2,590,961 | 11,929 | 51.9 | 14,168 | 46.2 | 14,868 | 20.3 |
| Phrynosoma sherbrookei | 814,375 | 6,043 | 31.6 | 7,257 | 26.9 | 7,692 | 14.9 |
| Sceloporus occidentalis | 1,404,985 | 6,852 | 52.8 | 5,368 | 45.0 | 5,561 | 20.0 |
| Uma notata | 806,846 | 3,751 | 40.3 | 4,698 | 35.9 | 5,298 | 25.3 |
| Urosaurus ornatus | 3,465,996 | 7,695 | 122.5 | 9,512 | 102.6 | 8,305 | 28.9 |
| Uta stansburiana | 4,818,547 | 9,177 | 119.5 | 11,878 | 96.5 | 14,058 | 29.7 |
| Gambelia wislizenii | 5,406,187 | 14,306 | 88.4 | 19,823 | 66.9 | 23,088 | 23.6 |
aThreshold for clustering of reads within a species.
bRaw read counts after sample demultiplexing.
cLoci passing quality filters.
dMean sequencing depth.
Table 5.
The Number of Loci (and SNPs) Obtained from Different Assemblies of the ddRADseq Data
| Minimum Individualsa |
||||||||
|---|---|---|---|---|---|---|---|---|
| 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| 95% clusteringb, paralog = 1c | 1,079 (2,228) | 375 (841) | 173 (404) | 72 (182) | 27 (73) | 9 (26) | 3 (7) | 0 (0) |
| 95% clustering, paralog = 3 | 1,100 (2,282) | 384 (860) | 177 (413) | 74 (186) | 28 (76) | 10 (29) | 3 (7) | 0 (0) |
| 90% clustering, paralog = 1 | 1,826 (6,506) | 674 (2,637) | 306 (1,212) | 154 (632) | 68 (306) | 28 (128) | 7 (27) | 1 (3) |
| 90% clustering, paralog = 3 | 1,856 (6,655) | 693 (2,733) | 312 (1,244) | 158 (655) | 69 (313) | 29 (135) | 8 (34) | 1 (3) |
| 80% clustering, paralog = 1 | 2,629 (15,725) | 1,057 (6,893) | 478 (3,037) | 227 (1,409) | 109 (722) | 50 (348) | 13 (75) | 2 (13) |
| 80% clustering, paralog = 3 | 2,670 (16,002) | 1,083 (7,079) | 493 (3,155) | 234 (1,458) | 113 (752) | 53 (371) | 13 (75) | 2 (13) |
aMinimum number of individuals (min. ind.) required to retain a locus in the final alignment (out of ten sequences total).
bThreshold for both within-sample and across-sample clustering.
cMaximum number of shared polymorphic bases.
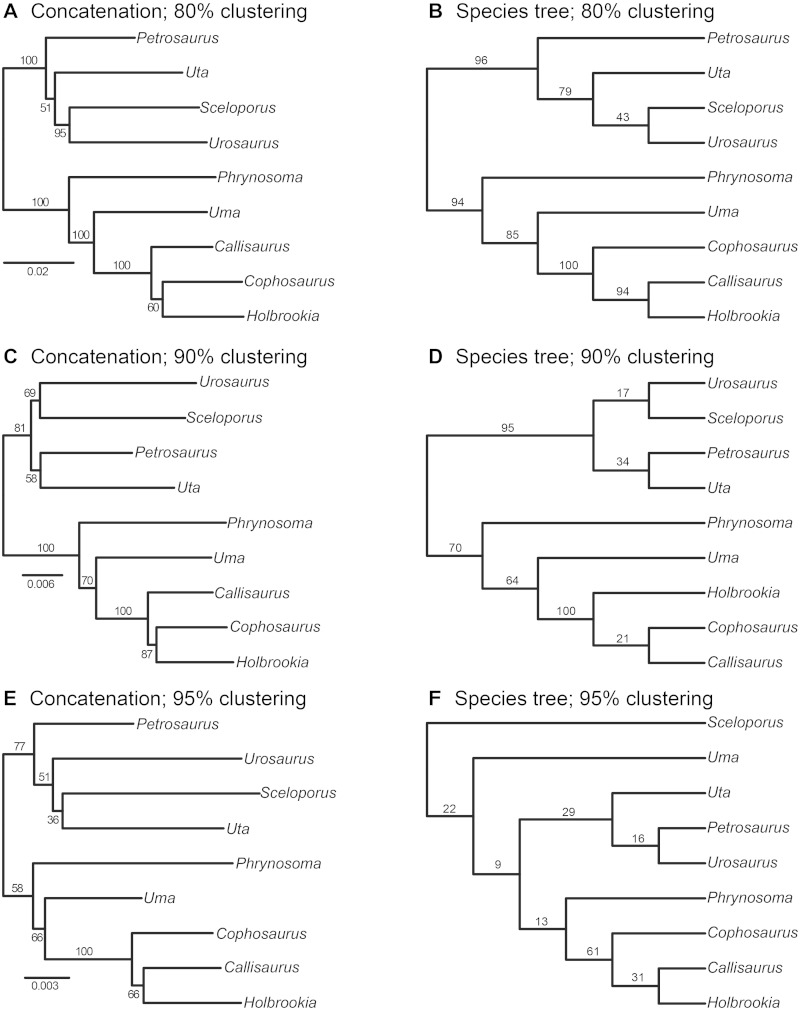
We estimated phylogenetic trees for the ddRADseq data using concatenation and a coalescent-based species tree approach (fig. 3). We present a comparison of phylogenies estimated using three different clustering threshold (i.e., 80%, 90%, and 95%) in figure 3. The phylogenetic trees estimated for SNP alignments assembled using different clustering thresholds, and with different methods, are in conflict. For example, the earless lizard genera, Cophosaurus and Holbrookia, form a clade with 80% and 90% clustering when using concatenation, but the species tree analysis supports a clade containing Holbrookia and Callisaurus (similar to the sequence capture and mtDNA results; figs. 1 and 2). Concatenation also supports a Holbrookia + Callisaurus clade, but only with a 95% clustering threshold (fig. 3E). The phylogenetic relationships for the sceloporine lizards are consistent and congruent with the sequence capture data when using 80% clustering (fig. 3A and B), but more conservative clustering thresholds (i.e., 90% and 95%) result in conflicting topologies, none of which are strongly supported.
Fig. 3.—
Phylogenetic trees estimated from the ddRADseq data using concatenation and coalescent-based species tree inference. For each clustering threshold (80%, A and B; 90%, C and D; 95%, E and F), results are shown for concatenation with RAxML (A, C, and E) and species tree inference with SVDquartets (B, D, and F). All results are from assemblies with min. ind. = 4 (minimum needed to form a quartet) and paralog filtering assuming no shared heterozygous sites (paralog = 1). Numbers on nodes are bootstrap values.
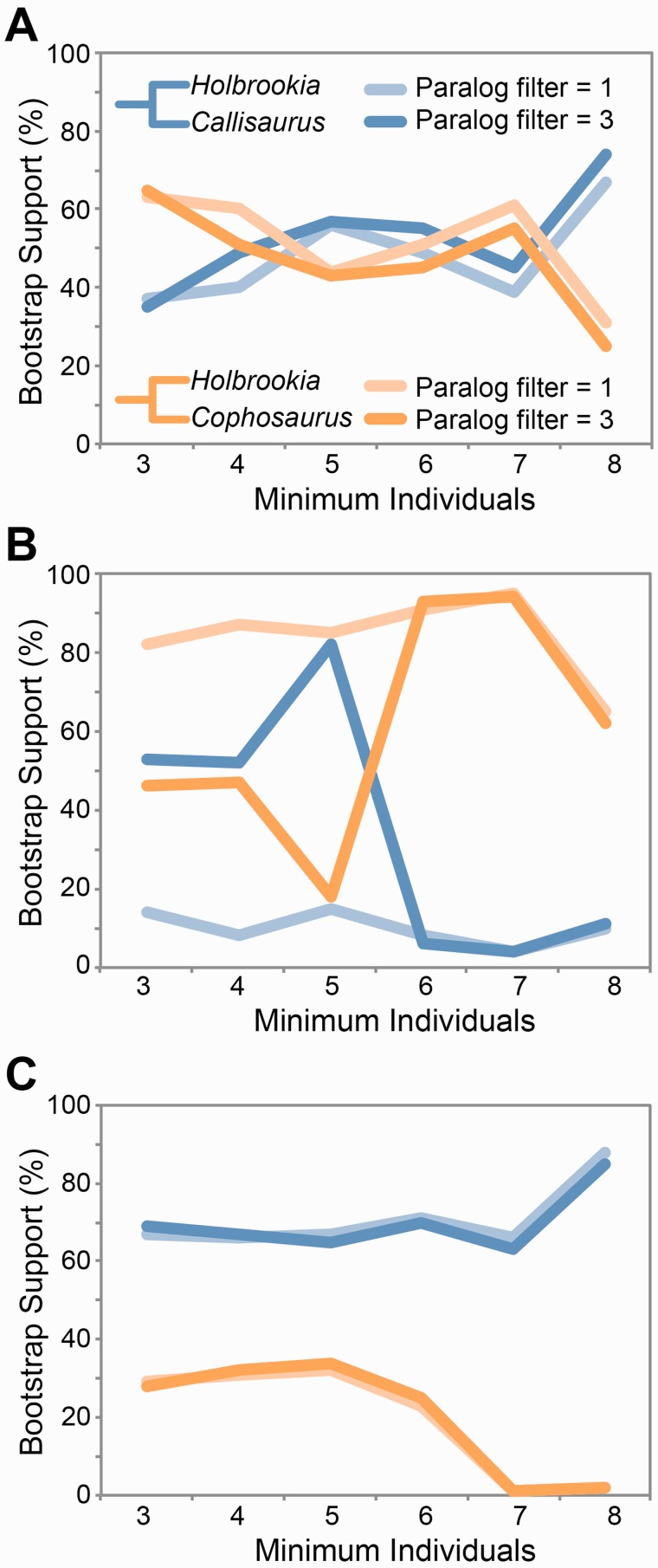
We compared the variation in bootstrap support from the concatenation analyses for the clade containing Callisaurus and Holbrookia with that of the earless lizard clade (i.e., Cophosaurus and Holbrookia) across different pyRAD assembly parameters (fig. 4). Data assembly parameters have an influence on the topology and bootstrap support for these alternative clades. The results are most consistent when the clustering threshold is high (fig. 4C), and as expected, there is still some variation across data assemblies containing different amounts of data. The paralog filter did not play a significant role in changing the bootstrap support values when using a clustering threshold of 80% or 95% (fig. 4). However, for the intermediate clustering threshold of 90% (fig. 4B), the paralog filter introduces large differences in the support for the alternative topologies. The most stringent clustering threshold (i.e., 95%) favors the Holbrookia + Callisaurus clade over the earless clade over all parameter settings that we explored.
Fig. 4.—
Variability in ddRADseq data support for monophyly of the earless lizards (Cophosaurus + Holbrookia) as a function of clustering threshold (A, 80%; B, 90%, C, 95%), minimum individuals (x axis), and paralog filtering. Results are from ML analyses of the concatenated data.
Discussion
Comparison of Approaches
Sequence capture and RADseq are two reduced-representation genome sequencing approaches for obtaining large numbers of homologous loci for phylogenetic inference. The utilities of the methods for phylogenetic inference are well established at opposite timescales, with sequence capture showing great promise for resolving relationships among distantly related species (Faircloth et al. 2012), and RADseq for phylogeographic and population-level investigations (Davey and Blaxter 2010). The methods have also been shown to work at largely overlapping timescales, but they have not been studied in a comparative manner, with the exception of a phylogeographic comparison by Harvey et al. (2013). For example, in silico studies of RADseq data have been applied to divergences dating back to 55–60 Ma in mammals, Drosophila, and fungi (Rubin et al. 2012; Cariou et al. 2013), and sequence capture has shown to be useful for phylogeographic studies of Pleistocene divergence in birds (Smith et al. 2014). We have conducted a comparison of these approaches using phrynosomatid lizards as a model system.
We found that the sequence capture data collected here were sufficient for resolving the relationships among phrynosomatid genera with strong support whether the loci were concatenated and assumed to share the same underlying genealogical history, or whether they were allowed to have independent histories and analyzed within a coalescent framework (fig. 1). The coalescent-based analyses provided lower support for the short internal branches of the tree, but there were no biases in terms of the support at particular timescales that might be expected if these data were insufficient for resolving recent divergences. However, as a consequence of sampling only one species per genus we excluded recent divergences within genera that occurred within the last 10 million years. Therefore, the phylogeny that we investigated was skewed toward containing relatively deeper divergences. The ddRADseq also showed no bias at different timescales. These data were able to resolve the deepest divergence in the phylogeny, but the short internal branches caused problems for the ddRADseq data; different data assemblies and different types of analyses of the same data assembly (concatenation vs. species tree inference) resulted in different topologies (figs. 3 and 4).
Incomplete lineage sorting is an important factor that can cause gene trees to conflict with the species tree. The time intervals between speciation events together with ancestral population sizes modulate the amount of incomplete lineage sorting that is expected; therefore, more data are required to resolve some speciation histories than others (Leaché and Rannala 2011). There is a substantial amount of gene tree discordance in the sequence capture loci presented here, and nearly 250 loci (approximately 50% of all loci sampled) support a topology for the sand lizards that conflicts with the estimated species tree (table 3). Gene tree discordance can cause phylogenetic inference error (Degnan and Rosenberg 2009), and the majority of gene trees could support an incorrect species tree if the phylogeny is in the anomaly zone (Degnan and Rosenberg 2006). Incidentally, the most common topology for sand lizards found across the sequence capture data support a clade containing Holbrookia and Callisaurus (table 3). The phrynosomatid genera do not appear to be in the anomaly zone, because if they were we would expect concatenation and coalescent inference to support different topologies (Kubatko and Degnan 2007; Liu and Edwards 2009).
The large amount of loci generated through RADseq approaches is particularly valuable for phylogeography, migration assessment, and phylogenetic inference among closely related species (e.g., Rheindt et al. 2014). In terms of their applications to nonmodel organisms, RADseq methods are more amenable to a broader set of evolutionary systems (Cruaud et al. 2014), since genomic resources are not needed to design probes as is the case with sequence capture. For phylogenetic investigations, ddRADseq data are most useful for studies of relatively closely related taxa, because the number of homologous loci obtained decreases in relation to time since divergence (Wagner et al. 2013). Furthermore, the pattern of missing data may be nonrandom, as the rate of allelic dropout is positively correlated with sequence divergence (Arnold et al. 2013).
A large assumption of RADseq approaches is that homologous loci are those that share a restriction site and high sequence similarity near the conserved restriction site. However, a reasonable possibility of clustering with nonhomologous genomic regions exists with this approach, particularly with short sequence reads (e.g., 50-bp single-end sequence reads, as used here). Bioinformatic postprocessing of ddRADseq data is the critical step that determines sequence homology (Ilut etal. 2014); as seen here, the thresholds selected for assembly parameters can have a strong influence on the size of the resulting data set and inferred phylogenetic relationships (table 5; fig. 4). Assembling sequence capture is more straightforward, because we know the number of loci, and a reference sequence is available for each locus (the 180-bp probe sites).
Phylogenetic inference with RADseq is feasible at the relatively deep evolutionary timescales studied here, and these branches did not seem particularly difficult for the SNP data to resolve. However, different assemblies of the ddRADseq data provided conflicting topologies for the short internal branches of the phylogeny. This suggests that the limitations of ddRADseq data are not focused on a particular timescale in the phylogeny, but are instead related to the length of the internal branches of the phylogeny. Even for studies focusing on recent population-level divergences, current RADseq protocols (reviewed by Puritz et al. 2014; Andrews et al. 2014) are highly susceptible to allelic dropout resulting from mutations at restriction sites (Arnold et al. 2013). The problem is exacerbated when attempting to assemble ddRADseq data for distantly related species (Rubin et al. 2012). Simulation work has shown that the loci with the highest mutation rates are those that have the most missing data (Huang and Knowles 2014), but those same loci may be the least valuable for resolving relationships among distantly related species. Only two loci were recovered for all ten species included in our ddRADseq experiment; these loci were obtained when the clustering threshold was reduced to 80% similarity (table 5). Different enzymes are expected to yield substantially different numbers of loci (Davey et al. 2011), and the enzyme combination selected here does not represent the optimum potential at which any RAD method will perform. Based on the phrynosomatid lizard data presented here, and the specific enzyme combination that we used (SbfI and MspI), there seems to be a low probability of obtaining large numbers of shared loci among distantly related species using ddRADseq.
At least for phrynosomatid lizards, phylogenetic relationships are sensitive to the parameter settings used during RADseq data assembly (fig. 4), especially for the short internal branches on the tree. We found conflicting topologies and variable levels of bootstrap support when changing the clustering threshold, paralogy filter, and the minimum number of individuals needed to retain a locus in the final alignment (fig. 3). The most consistent phylogenetic signal that we recovered for the short internal branch located within the Cophosaurus, Callisaurus, and Holbrookia clade was obtained when the sequence similarity threshold was high (95%); the phylogenetic relationships and bootstrap values stabilized across the various parameter settings (fig. 4C). Using lower sequence similarity thresholds doubled the number of loci, and this may seem beneficial, but this increase comes at the cost of introducing “RAD noise” that at worst produces conflicting topologies (fig. 3), and at the best only changes the support for the topology (fig. 4). Of course, we do not necessarily know the correct phylogeny, and this is why simulation studies are needed to quantify the errors and understand the consequences resulting from RADseq data misassembly on phylogeny inference.
Overall, RADseq data can be collected faster and are less expensive than sequence capture data, and RADseq has the potential to provide an order of magnitude more SNPs for evolutionary inference. There is no limit on the number of loci that can be targeted for sequence capture experiments, and in some model systems (e.g., humans) the method is used for sequencing the entire exome (Ng et al. 2009). However, for phylogeographic studies, it is possible that the sequence capture protocols that target highly conserved genomic regions (Lemmon et al. 2012) and/or UCEs (Faircloth et al. 2012) will provide relatively few SNPs. For example, a phylogeography study of Neotropical rainforest birds using sequence capture data recovered approximately 4,500 SNPs (1,500 UCE loci containing 2–3 variable sites per locus; Smith et al. 2014). In contrast, a phylogeographic study of Zimmerius flycatchers using RADseq recovered over 37,000 SNPs (Rheindt et al. 2014). If the goal of a study is to discern fine-scale phylogeographic patterns, then RADseq methods have the potential to provide more data at lower cost and effort. Although the number of loci that we targeted using sequence capture is lower than what we obtained using ddRADseq, the loci are longer and were more straightforward to analyze under a variety of inference techniques, including coalescent-based models that benefit from complete sampling at each locus. In the case of higher-level relationships among phrynosomatid lizard genera, we found sequence capture data to provide a more consistent phylogenetic signal compared with ddRADseq data.
Phylogenomics of Phrynosomatids
The phylogenomic signal from the sequence capture data and the mtDNA data provides strong support for the paraphyly of the earless lizard genera Holbrookia and Cophosaurus (fig. 1). Determining whether these two “earless” genera with concealed tympanic membranes form a clade has been difficult to resolve. Previous studies using mtDNA have provided contradictory, ambiguous, or spurious support for the resolution of these taxa (Reeder 1995; Wilgenbusch and de Queiroz 2000; Leaché and McGuire 2006; Wiens et al. 2010). The spurious relationships for sand lizards supported by the Leaché and McGuire (2006) study were the result of sample mislabeling errors that occurred during specimen collection (the tissues for Uma and Callisaurus were swapped during specimen collection), and those data were removed from GenBank in 2008. These new sequence capture data and partial mtDNA genomes presented here, all collected from authenticated samples, recover a clade containing Holbrookia and Callisaurus to the exclusion of Cophosaurus. Some of the SNP assemblies also support this relationship, including the coalescent-based analysis of the largest SNP matrix. The largest ddRADseq assembly also supports this relationship when analyzed using a species tree approach (fig. 3B). The preferred topology suggests that the earless morphology either evolved twice independently in Holbrookia and Cophosaurus or that evolved once in the common ancestor of Holbrookia, Callisaurus, and Cophosaurus, and was subsequently lost in Callisaurus. Either reconstruction requires the same number of character state transitions, and in the context of parsimony they are equivalent explanations for the evolution of the earless morphology.
The divergence times separating the sceloporine genera Sceloporus, Petrosaurus, Urosaurus, and Uta are on the order of 1.7–3.7 Myr (fig. 1), and these short time intervals have resulted in a difficult phylogenetic problem. Previous studies attempting to resolve these relationships with either a single locus (mtDNA) or a handful of nuclear loci have not been able to obtain strong support for the relationships among these groups (Wiens et al. 2010). Simulation studies have shown that rapid speciation events are difficult to resolve without hundreds or thousands of loci (Liu and Edwards 2009), and the new sequence capture data collected here provide strong support for the relationships among these genera using concatenation and coalescent-based analyses. The new mtDNA data (fig. 2) continue to struggle with resolving these relationships, and although these data are still fragmentary, it is unlikely that this single locus will be sufficient for resolving this part of the tree with strong support even after being sequenced to completion. The largest SNP assembly that we analyzed supported the same topology as the sequence capture and mtDNA data. These three new data sets provide compelling evidence for a new phyrnosomatid lizard phylogeny that contains a novel relationship among the sand lizards.
Acknowledgments
Scientific specimens were collected with permission from the Arizona Game and Fish Department (SP568189) and the California Department of Fish and Wildlife (SC-9768). The authors thank the following for assistance with tissue sample loans: S. Birks, M. Morando, A. Nieto-Montes de Oca, C. Spencer, and J. McGuire. They thank members of the Leaché lab for useful comments and discussion. The manuscript benefitted greatly from comments by C. Ané, J. Wiens, and one anonymous reviewer. This work was supported by grants from the National Science Foundation (DEB-1144630 awarded to A.D.L. and BIO-1202754 awarded to C.W.L.).
Literature Cited
- Alföldi J, et al. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature. 2011;477:587–591. doi: 10.1038/nature10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews KR, et al. Trade-offs and utility of alternative RADseq methods. Mol Ecol. 2014;23:5943–5946. doi: 10.1111/mec.12964. [DOI] [PubMed] [Google Scholar]
- Arnold B, Corbett-Detig RB, Hartl D, Bomblies K. RADseq underestimates diversity and introduces genealogical biases due to nonrandom haplotype sampling. Mol Ecol. 2013;22:3179–3190. doi: 10.1111/mec.12276. [DOI] [PubMed] [Google Scholar]
- Baele G, et al. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol Biol Evol. 2012;29:2157–2167. doi: 10.1093/molbev/mss084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baele G, Li WLS, Drummond AJ, Suchard MA, Lemey P. Accurate model selection of relaxed molecular clocks in Bayesian phylogenetics. Mol Biol Evol. 2013;30:239–243. doi: 10.1093/molbev/mss243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NA, et al. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One. 2008;3:e3376. doi: 10.1371/journal.pone.0003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cariou M, Duret L, Charlat S. Is RAD-seq suitable for phylogenetic inference? An in silico assessment and optimization. Ecol Evol. 2013;3:846–852. doi: 10.1002/ece3.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA. Stacks: an analysis tool set for population genomics. Mol Ecol. 2013;22:3124–3140. doi: 10.1111/mec.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chifman J, Kubatko L. Quartet inference from SNP data under the coalescent model. Bioinformatics. 2014;30:3317–3324. doi: 10.1093/bioinformatics/btu530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NG, et al. More than 1000 ultraconserved elements provide evidence that turtles are the sister group of archosaurs. Biol Lett. 2012;8:783–786. doi: 10.1098/rsbl.2012.0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruaud A, et al. Empirical assessment of RAD sequencing for interspecific phylogeny. Mol Biol Evol. 2014;31:1272–1274. doi: 10.1093/molbev/msu063. [DOI] [PubMed] [Google Scholar]
- Davey JW, Blaxter ML. RADSeq: next-generation population genetics. Brief Funct Genomics. 2010;9:416–423. doi: 10.1093/bfgp/elq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey JW, et al. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet. 2011;12:499–510. doi: 10.1038/nrg3012. [DOI] [PubMed] [Google Scholar]
- de Queiroz K. 1989. Morphological and biochemical evolution in the sand lizards. [PhD dissertation]. [Berkeley (CA)]: University of California, Berkeley. [Google Scholar]
- de Queiroz K. Phylogenetic relationships and rates of allozyme evolution among the lineages of sceloporine sand lizards. Biol J Linn Soc. 1992;45:333–362. [Google Scholar]
- Degnan JH, Rosenberg NA. Discordance of species trees with their most likely gene trees. PLoS Genet. 2006;2:e68. doi: 10.1371/journal.pgen.0020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan JH, Rosenberg NA. Gene tree discordance, phylogenetic inference, and the multispecies coalescent. Trends Ecol Evol. 2009;24:332–340. doi: 10.1016/j.tree.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton DAR. PyRAD: assembly of de novo RADseq loci for phylogenetic analyses. Bioinformatics. 2014;30:1844–1849. doi: 10.1093/bioinformatics/btu121. [DOI] [PubMed] [Google Scholar]
- Eaton DAR, Ree RH. Inferring phylogeny and introgression using RADseq data: an example from flowering plants (Pedicularis: Orobanchaceae) Syst Biol. 2013;62:689–706. doi: 10.1093/sysbio/syt032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Emerson KJ, Merz CR, Catchen JM, Hohenlohe PA, Cresko WA, Bradshaw WE, Holzapfel CM. Resolving postglacial phylogeography using high-throughput sequencing. Proc Natl Acad Sci U S A. 2010;107:16196–16200. doi: 10.1073/pnas.1006538107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faircloth BC, McCormack JE, Crawford NG, Harvey MG, Brumfield RT, Glenn TC. Ultraconserved elements anchor thousands of genetic markers spanning multiple evolutionary timescales. Syst Biol. 2012;61:717–726. doi: 10.1093/sysbio/sys004. [DOI] [PubMed] [Google Scholar]
- Gnirke A, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomic Resources Development Consortium, et al. Genomic resources notes accepted 1 August 2014–30 September 2014. Mol Ecol Resour. 2015;15:228–229. doi: 10.1111/1755-0998.12340. [DOI] [PubMed] [Google Scholar]
- Harvey MG, Smith BT, Glenn TC, Faircloth BC, Brumfield RT. Sequence capture versus restriction site associated DNA sequencing for phylogeography. arXiv preprint arXiv:1312.6439. 2013 doi: 10.1093/sysbio/syw036. [DOI] [PubMed] [Google Scholar]
- Huang H, Knowles LL. Unforeseen consequences of excluding missing data from next-generation sequences: simulation study of RAD sequences. Syst Biol. 2014 doi: 10.1093/sysbio/syu046. Advance Access published July 4, 2014, doi: 10.1093/sysbio/syu046. [DOI] [PubMed] [Google Scholar]
- Ilut DC, Nydam ML, Hare MP. Defining loci in restriction-based reduced representation genomic data from nonmodel species: sources of bias and diagnostics for optimal clustering. BioMed Res Int. 2014;2014:675158. doi: 10.1155/2014/675158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubatko LS, Degnan JH. Inconsistency of phylogenetic estimates from concatenated data under coalescence. Syst Biol. 2007;56:17–24. doi: 10.1080/10635150601146041. [DOI] [PubMed] [Google Scholar]
- Lambert SM, Wiens JJ. Evolution of viviparity: a phylogenetic test of the cold-climate hypothesis in phrynosomatid lizards. Evolution. 2013;67:2614–2630. doi: 10.1111/evo.12130. [DOI] [PubMed] [Google Scholar]
- Leaché AD, et al. A hybrid phylogenetic-phylogenomic approach for species tree estimation in African Agama lizards with applications to biogeography, character evolution, and diversification. Mol Phylogenet Evol. 2014;79:215–230. doi: 10.1016/j.ympev.2014.06.013. [DOI] [PubMed] [Google Scholar]
- Leaché AD, McGuire JA. Phylogenetic relationships of horned lizards (Phrynosoma) based on nuclear and mitochondrial data: evidence for a misleading mitochondrial gene tree. Mol Phylogenet Evol. 2006;39:628–644. doi: 10.1016/j.ympev.2005.12.016. [DOI] [PubMed] [Google Scholar]
- Leaché AD, Rannala B. The accuracy of species tree estimation under simulation: a comparison of methods. Syst Biol. 2011;60:126–137. doi: 10.1093/sysbio/syq073. [DOI] [PubMed] [Google Scholar]
- Lemmon AR, Emme S, Lemmon EM. Anchored hybrid enrichment for massively high-throughput phylogenomics. Syst Biol. 2012;61:727–744. doi: 10.1093/sysbio/sys049. [DOI] [PubMed] [Google Scholar]
- Li C, Hofreiter M, Straube N, Corrigan S, Naylor GJP. Capturing protein-coding genes across highly divergent species. Biotechniques. 2013;54:321–326. doi: 10.2144/000114039. [DOI] [PubMed] [Google Scholar]
- Liu L, Edwards SV. Phylogenetic analysis in the anomaly zone. Syst Biol. 2009;58:452–460. doi: 10.1093/sysbio/syp034. [DOI] [PubMed] [Google Scholar]
- Liu L, Yu L, Edwards SV. A maximum pseudo-likelihood approach for estimating species trees under the coalescent model. BMC Evol Biol. 2010;10:302. doi: 10.1186/1471-2148-10-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacManes M. 2013. MacManes salt extraction protocol. Figshare. Available from: http://dx.doi.org/10.6084/m9.figshare.658946. [Google Scholar]
- Mamanova L, et al. Target-enrichment strategies for next-generation sequencing. Nat Methods. 2010;7:111–118. doi: 10.1038/nmeth.1419. [DOI] [PubMed] [Google Scholar]
- McCormack JE, et al. Ultraconserved elements are novel phylogenomic markers that resolve placental mammal phylogeny when combined with species-tree analysis. Genome Res. 2012;22:746–754. doi: 10.1101/gr.125864.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack JE, et al. A phylogeny of birds based on over 1,500 loci collected by target enrichment and high-throughput sequencing. PLoS One. 2013;8:e54848. doi: 10.1371/journal.pone.0054848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MR, Dunham JP, Amores A, Cresko WA, Johnson EA. Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Res. 2007;17:240–248. doi: 10.1101/gr.5681207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattengale ND, Alipour M, Bininda-Emonds ORP, Moret BME, Stamatakis A. How many bootstrap replicates are necessary? J Comput Biol. 2010;17:337–354. doi: 10.1089/cmb.2009.0179. [DOI] [PubMed] [Google Scholar]
- Peng Y, Leung HCM, Yiu SM, Chin FYL. IDBA—a practical iterative de Bruijn graph de novo assembler. Res Comput Mol Biol. 2010;6044:426–440. [Google Scholar]
- Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE. Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS One. 2012;7:e37135. doi: 10.1371/journal.pone.0037135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puritz JB, et al. Demystifying the RAD fad. Mol Ecol. 2014;23:5937–5942. doi: 10.1111/mec.12965. [DOI] [PubMed] [Google Scholar]
- Pyron RA, et al. Effectiveness of phylogenomic data and coalescent species-tree methods for resolving difficult nodes in the phylogeny of advanced snakes (Serpentes: Caenophidia) Mol Phylogenet Evol. 2014;81:221–231. doi: 10.1016/j.ympev.2014.08.023. [DOI] [PubMed] [Google Scholar]
- Rannala B, Yang Z. Bayes estimation of species divergence times and ancestral population sizes using DNA sequences from multiple loci. Genetics. 2003;164:1645–1656. doi: 10.1093/genetics/164.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder TW. Phylogenetic relationships among phyrynosomatid lizards as inferred from mitochondrial ribosomal DNA sequences: substitutional bias and information content of transitions relative to transversions. Mol Phylogenet Evol. 1995;4:203–222. doi: 10.1006/mpev.1995.1020. [DOI] [PubMed] [Google Scholar]
- Reeder TW, Wiens JJ. Evolution of the lizard family Phrynosomatidae as inferred from diverse types of data. Herpetol Monogr. 1996;10:43–84. [Google Scholar]
- Rheindt FE, Fujita MK, Wilton PR, Edwards SV. Introgression and phenotypic assimilation in Zimmerius flycatchers (Tyrannidae): population genetic and phylogenetic inferences from genome-wide SNPs. Syst Biol. 2014;63:134–152. doi: 10.1093/sysbio/syt070. [DOI] [PubMed] [Google Scholar]
- Ronquist F, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin BER, Ree RH, Moreau CS. Inferring phylogenies from RAD sequence data. PLoS One. 2012;7:e33394. doi: 10.1371/journal.pone.0033394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinervo B, Lively CM. The rock-paper-scissors game and the evolution of alternative male strategies. Nature. 1996;380:240–243. [Google Scholar]
- Smith BT, Harvey MG, Faircloth BC, Glenn TC, Brumfield RT. Target capture and massively parallel sequencing of ultraconserved elements for comparative studies at shallow evolutionary time scales. Syst Biol. 2014;63:83–95. doi: 10.1093/sysbio/syt061. [DOI] [PubMed] [Google Scholar]
- Snir S, Rao S. Quartet MaxCut: a fast algorithm for amalgamating quartet trees. Mol Phylogenet Evol. 2012;62:1–8. doi: 10.1016/j.ympev.2011.06.021. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran J, Holder MT. Dendropy: a Python library for phylogenetic computing. Bioinformatics. 2010;26:1569–1571. doi: 10.1093/bioinformatics/btq228. [DOI] [PubMed] [Google Scholar]
- Swofford DL. Sunderland (MA): Sinauer Associates; 2003. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. [Google Scholar]
- Uetz P. 2014. The Reptile Database. Available from: http://www.reptile-database.org. [Google Scholar]
- Wagner CE, et al. Genome-wide RAD sequence data provide unprecedented resolution of species boundaries and relationships in the Lake Victoria cichlid adaptive radiation. Mol Ecol. 2013;22:787–798. doi: 10.1111/mec.12023. [DOI] [PubMed] [Google Scholar]
- Wiens JJ, et al. Resolving the phylogeny of lizards and snakes (Squamata) with extensive sampling of genes and species. Biol Lett. 2012;8:1043–1046. doi: 10.1098/rsbl.2012.0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiens JJ, Kozak KH, Silva N. Diversity and niche evolution along aridity gradients in North American lizards (Phrynosomatidae) Evolution. 2013;67:1715–1728. doi: 10.1111/evo.12053. [DOI] [PubMed] [Google Scholar]
- Wiens JJ, Kuczynski CA, Arif S, Reeder TW. Phylogenetic relationships of phrynosomatid lizards based on nuclear and mitochondrial data, and a revised phylogeny for Sceloporus. Mol Phylogenet Evol. 2010;54:150–161. doi: 10.1016/j.ympev.2009.09.008. [DOI] [PubMed] [Google Scholar]
- Wilgenbusch J, de Queiroz K. Phylogenetic relationships among the phrynosomatid sand lizards inferred from mitochondrial DNA sequences generated by heterogeneous evolutionary processes. Syst Biol. 2000;49:592–612. doi: 10.1080/10635159950127411. [DOI] [PubMed] [Google Scholar]
- Yang Z, Rannala B. Unguided species delimitation using DNA sequence data from multiple loci. Mol Biol Evol. 2014;31:3125–3135. doi: 10.1093/molbev/msu279. [DOI] [PMC free article] [PubMed] [Google Scholar]